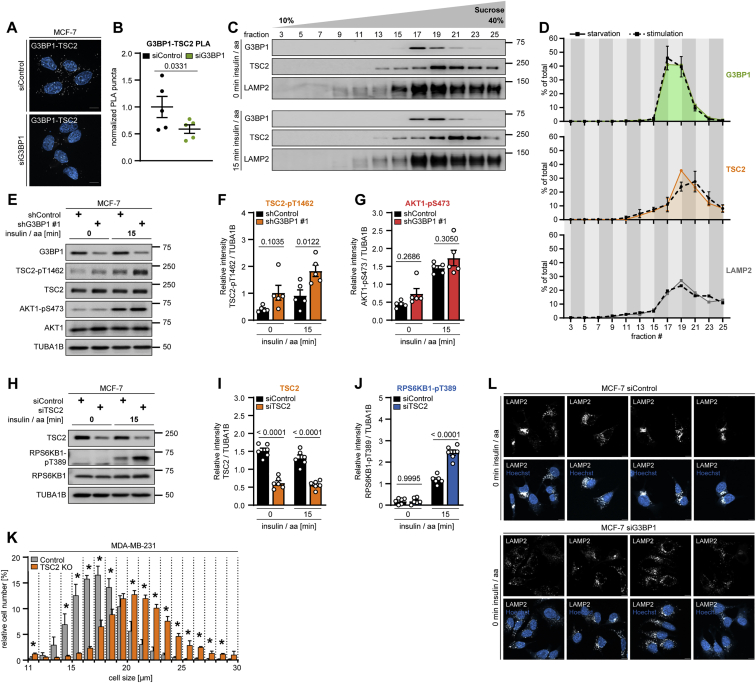
Figure S4.
G3BP1 phenocopies lysosomal TSC functions, related to Figures 3 and 4
(A) PLA of G3BP1-TSC2 in serum/aa-starved siG3BP1 cells. PLA puncta, white dots; nuclei, blue (DAPI). Scale bar, 10 μm. n = 3.
(B) Quantitation of data in (A). Shown are data points and mean ± SEM. n = 5 technical replicates.
(C) Sucrose density gradient of serum/aa-starved or insulin/aa-stimulated MCF-7 cells. n = 3.
(D) Quantitation of data in (C). Area under the curve highlighted in green (G3BP1), orange (TSC2), and gray (LAMP2), starved condition; dashed lines, stimulated condition. Mean ± SEM.
(E) Insulin/aa-stimulated shG3BP1 #1 cells. n = 5.
(F) Quantitation of TSC2-pT1462 in (E). Shown are data points and mean ± SEM.
(G) Quantitation of AKT1-pS473 in (E). Data shown as in (F).
(H) Insulin/aa-stimulated siTSC2 cells. n = 6.
(I) Quantitation of TSC2 in (H). Shown are data points and mean ± SEM.
(J) Quantitation of RPS6KB1-pT389 in (H). Data shown as in (I).
(K) Cell size of TSC2 KO cells. Mean ± SEM. *p < 0.05. n = 3.
(L) IF of LAMP2 positioning in serum/aa-starved siG3BP1 cells. White, LAMP2; blue (Hoechst), nuclei. Scale bar, 10 μm. n = 3.