Abstract
miR-19a/b belong to the miR-17-92 family. We have demonstrated previously that miR-19a/b are overexpressed in glioma and glioma cell lines. However, the role of miR-19a/b in glioma remains unclear. In the present study, we aim to identify the biological function and molecular mechanism of miR-19a/b in glioma cell proliferation and epithelial-mesenchymal transition (EMT). Knocking down miR-19a/b in LN308 glioblastoma (GBM) cells with higher expression of miR-19a/b inhibits cell proliferation and invasion, induces apoptosis, and suppresses EMT by downregulating the expression of Akt, phosphorylated p-Akt, nuclear factor κB (NF-κB), Snail, N-cadherin, and Vimentin and upregulating E-cadherin in vitro and in vivo. Enhanced proliferation and EMT are also observed when miR-19a/b are transfected into SNB19 GBM cells, with lowered expression of miR-19a/b. miR-19a is more effective than miR-19b in the regulation of biological behavior of glioma cells. miR-19a/b modulate molecular events for the promotion of EMT via the Akt-NF-κB pathway. SEPT7 has been confirmed as the target gene of miR-19a/b. The effect of miR-19a/b on proliferation and EMT of glioma cells and the Akt-NF-κB pathway could be reversed by transfection with SEPT7. Our study strongly suggests that miR-19a/b play a significant role in glioma progression and EMT through regulating target gene-SEPT7 and the SEPT7-Akt-NF-κB pathway.
Keywords: miR-19a/b, glioma, SEPT7-AKT-NF-κB pathway
Graphical Abstract
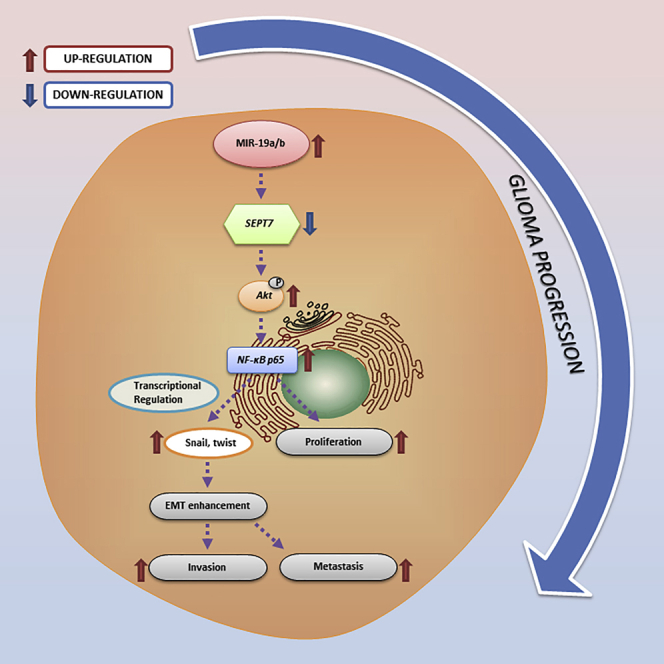
miR-19a/b promote proliferation, invasion, and EMT of glioma cells via upregulating the AKT-NF-κB pathway by targeting SEPT7.
Introduction
MicroRNAs (miRNAs) are a class of small (18–24 nt) non-coding RNAs that act as a novel class of gene expression regulators. These RNA molecules regulate numerous biological processes. There are about 2,599 unique mature human miRNAs that have been identified.1 The miRNA genome is often located at fragile genomic sites and genomic regions involved in cancers. Their expression is dysregulated due to genomic instabilities.2,3 The role of miRNAs in cancer was first speculated when it was observed in C. elegans and Drosophila that miRNAs control aspects of cell proliferation and apoptosis.4,5
Expression of various miRNAs has been reported to be differentially altered across a variety of tumor types, suggesting their direct involvement in oncogenesis.6, 7, 8
miR-19a/b have been regarded as members of a pan-cancer oncogenic miRNA superfamily-miR-17-92 cluster, which was the first miRNA cluster that was considered to have oncogenic properties, and the primary transcript generated by this family encodes six mature miRNAs, including miR-19a/b, which are the most important oncogenic miRNAs.9 miR-19a/b are upregulated in bladder cancer, gastric cancer, throat squamous cell carcinoma, and breast cancer.10, 11, 12, 13, 14 There are a few reports about the role of miR-19a and miR-19b in glioma. miR-19a/b have been identified to be upregulated in pediatric low-grade glioma.15 Upregulation of serum miR-19a in astrocytoma is associated with poor patient survival and may serve as a diagnostic and prognostic biomarker for human astrocytoma.16 Expression of miR-19a has been demonstrated to be upregulated with glioma progression in patients with primary World Health Organization (WHO) grade II gliomas that have spontaneously progressed to secondary glioblastomas (GBMs), i.e., WHO grade IV.17
We have confirmed that miR-19a/b expression is upregulated in glioma tissue samples and cell lines, and we identified PTEN and RUNX3 as the target genes of miR-19a/b.18,19
In addition, based on the bioinformatics analyses with HuMiTar, a sequence-based method for prediction of human miRNA targets,20 SEPT7 has been predicted to be the target gene of miR-19a/b. SEPT7 is a member of the septin family, a filament-forming cytoskeletal GTPase involved in the formation of intracellular microfilaments, and it plays a key role in a variety of cellular processes. Our previous study has identified that the expression of SEPT7 is significantly decreased in gliomas relative to normal brain tissues and its expression is negatively correlated with the ascending order of glioma grades. Moreover, cell proliferation and invasion are inhibited and apoptosis is induced either in vitro or in vivo when glioma cells are transfected with SEPT7.21, 22, 23, 24
This evidence strongly supports that SEPT7 plays a tumor suppressor role in gliomagenesis, while its downstream signaling pathway and effectors should be further explored. Additionally, epithelial-mesenchymal transition (EMT) is also an important mechanism to promote the migration and invasion of tumor cells. The relationship of miR-19a/b to SEPT7 and EMT that has not been elaborated before was emphasized in this study.
Additionally, Akt and nuclear factor κB (NF-κB) have been demonstrated to be constitutively activated and act as critical factors in the development or progression of various cancers and gliomas as well.25, 26, 27, 28, 29 Several studies have also validated the Akt/NF-κB signaling pathway in promoting EMT.30 Thus far, there was no report on whether Akt/NF-κB signaling is involved in the miR-19a/b/SEPT7 pathway in glioma cells.
In the current study, we sought to further study the effect of miR-19a/b on glioma cell proliferation and invasion in vitro and in vivo, and to identify whether miR19a/b exert their oncogenic role on glioma cells via SEPT7 and the Akt/NF-κB pathway.
Results
miR-19a plays a more crucial role in prompting glioma progression than does miR-19b
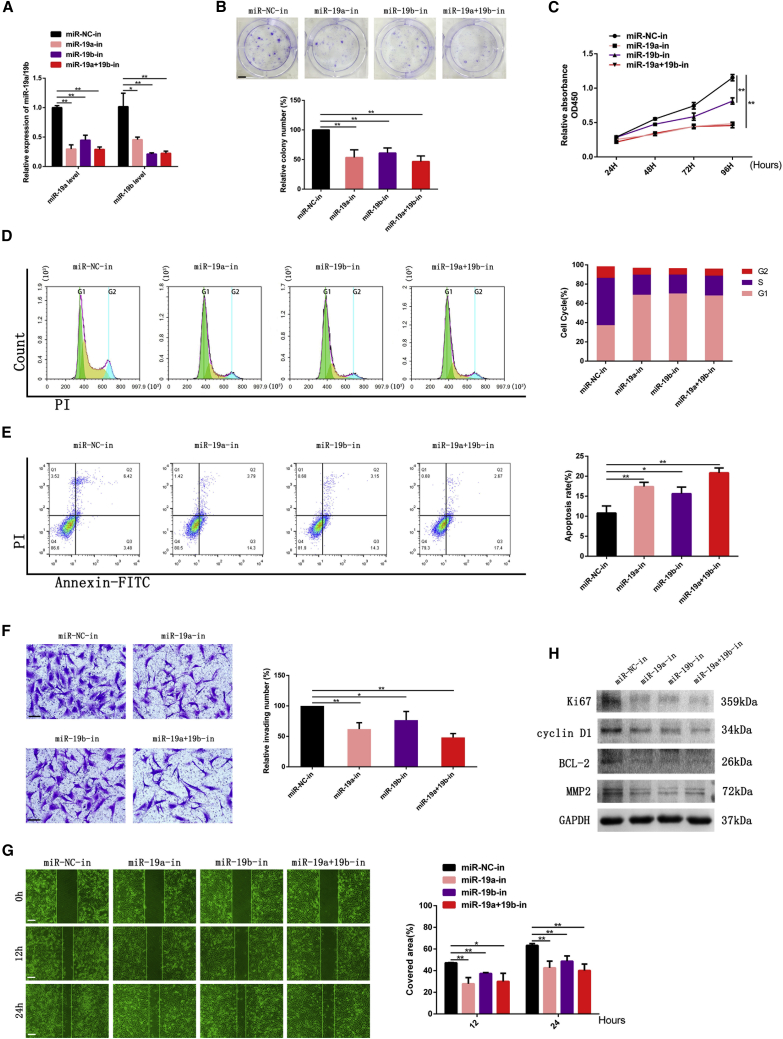
Inhibitors of miR-19a/b were transfected to LN308 glioma cells, which have been identified with higher expression of miR-19a/b, while mimics of miR-19a/b were transfected to SNB19 glioma cells with lower expression of miR-19a/b examined in our previous work.18 The results determined by real-time polymerase chain reaction (RT-PCR) showed that the expression level of miR-19a/b in LN308 cells transfected with inhibitors of miR-19a/b were significantly lowered, whereas SNB19 cells transfected with mimics of miR-19a/b were upregulated as compared to their negative control cells (Figures 1A and 2A).
Figure 1.
Knocking down miR-19a/b suppressed growth, invasion, and migration and promoted apoptosis of LN308 cells
(A) LN308 cells were transfected with miR-NC and miR-19a/19b inhibitors followed by RT-PCR analysis to detect miR-19a/b levels. U6 served as the internal control. (B) Colony formation assay of LN308 cells transfected with miR-NC and miR-19a/b inhibitors. Scale bar, 5 mm. (C) Cell proliferation detected by CCK-8 assay. (D) Cell-cycle assay of LN308 GBM cells transfected with miR-NC and miR-19a/b inhibitors. (E) Apoptosis of LN308 cells transfected with miR-19a/b inhibitors. (F) Effect of miR-19a/b inhibitors on GBM cell invasion examined by Transwell assay. Scale bars, 200 μm. (G) Migration of GBM cells of miR-NC and miR-19a/b inhibitor groups detected by wound-healing assay. Scale bars, 200 μm. (H) Expression of Ki67, cyclin D1, BCL-2, and MMP2 in LN308 cells transfected with miR-NC and miR-19a/b inhibitors detected by western blotting. GAPDH was used as the loading control. ∗p < 0.05, ∗∗p < 0.01.
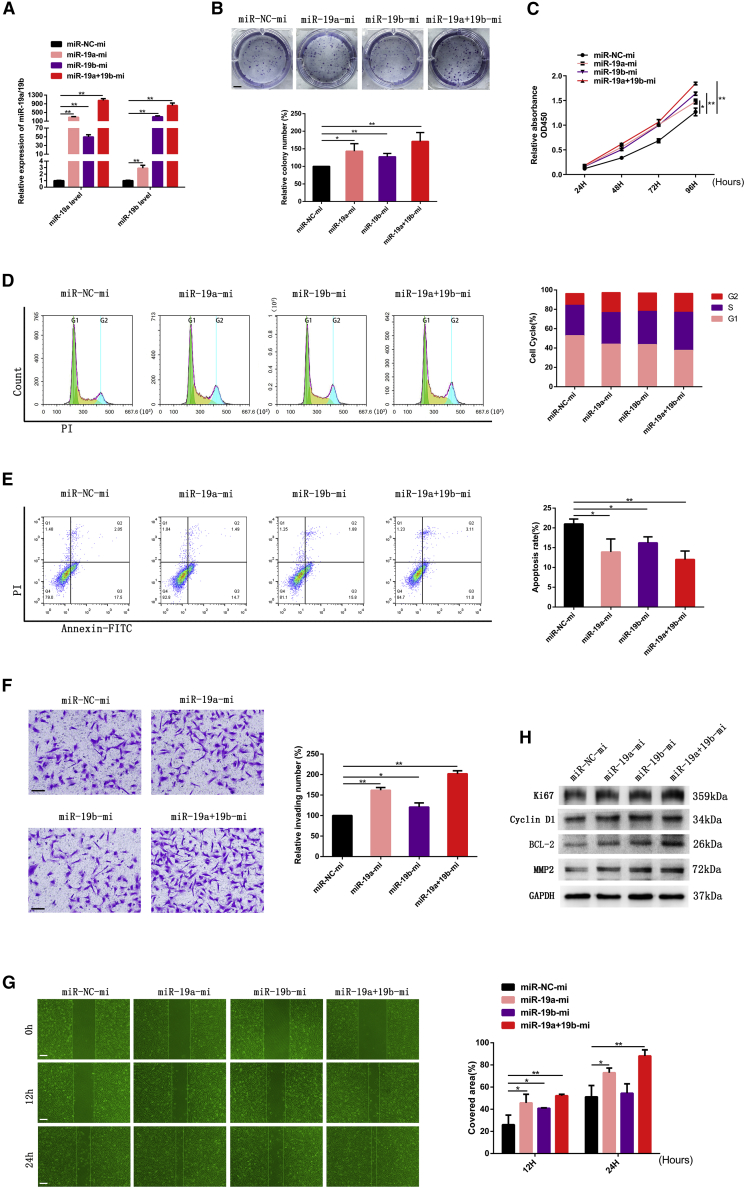
Figure 2.
Overexpression of miR-19a/b enhanced growth, invasion, and migration and inhibited apoptosis of GBM cells
(A) SNB19 cells were transfected with miR-NC mimics and miR-19a/b mimics followed by RT-PCR analysis to detect miR-19a/b expression levels. U6 served as the internal control. (B) Colony formation assay of SNB19 cells transfected with miR-NC mimics and miR-19a/b mimics. Scale bar, 5 mm. (C) Cell proliferation detected by CCK-8 assay. (D) Cell-cycle assay of SNB19 GBM cells transfected with miR-NC mimics and miR-19a/b mimics, respectively. (E) Apoptosis of SNB19 cells transfected with miR-19a/b mimics. (F) Effect of miR-19a/b overexpression on SNB19 cell invasion examined by Transwell assay. Scale bars, 200 μm. (G) Migration of SNB19 cells transfected with miR-19a/b mimics was monitored by wound-healing assay. Scale bars, 200 μm. (H) Protein levels of Ki67, cyclin D1, BCL-2, and MMP2 in SNB19 cells transfected with miR-NC mimics and miR-19a/b mimics detected by western blotting. GAPDH was used as the loading control. ∗p < 0.05, ∗∗p < 0.01.
The LN308 cell group transfected with miR-19a/b inhibitors showed a significant decrease in viability as compared to the number of cells and colonies formed in control group (Figures 1B and 1C) as detected by a Cell Counting Kit-8 (CCK-8) assay and colony formation assay, respectively. We also found that the effect of miR-19a inhibitor was stronger than that of the miR-19b inhibitor, and a combination of both miR-19a and miR-19b inhibitors was the most effective.
Flow cytometry examination demonstrated that the cell cycle was arrested in the G0/G1 phase, and the cell population in the S phase decreased when miR-19a/b expression levels were knocked down in LN308 cells, as shown in Figure 1D.
We also analyzed the effect of miR-19a/b inhibitors on cell apoptosis conducted by annexin V and propidium iodide (PI) double staining. Compared with the negative control (NC) group, miR-19a/b inhibitor groups showed a significant increase of apoptotic cell death, suggesting that the downregulation of miR-19a/b induced cell apoptosis (Figure 1E).
We assessed the glioma cell invasion by a Transwell assay and wound-healing assay. The numbers of invading cells in the miR-19a/b inhibitor groups were drastically reduced compared with that in the control group, as shown in the Transwell assay (Figure 1F). Meanwhile, the mobility of LN308 cells was also decreased when miR-19a/b were knocked down as detected by a wound-healing assay (Figure 1G).
miR-19a/b inhibitors downregulated the expression of molecular markers Ki67, matrix metallopeptidase 2 (MMP2), cyclin D1, and BCL-2 in LN308 cells (Figure 1H), coincided with the results that miR-19a/b inhibitors suppressed cell proliferation and invasion, arrested cell cycle progression, and induced apoptosis as described above.
For further elucidating the role of miR-19a/b in gliomagenesis, we examined the proliferation, cell cycle progression, apoptosis, and invasion of SNB19 glioma cells being treated with miR-19a/b mimics.
On the contrary, miR-19a/b mimics were identified to promote proliferation and suppress apoptosis of SNB19 glioma cells significantly (Figures 2B, 2C, and 2E).
It was also demonstrated that the S phase fraction was increased while the G0/G1 fraction decreased (Figure 2D) in SNB19 cells treated with miR-19a/b mimics, as determined by cell cycle analysis. The migratory and invasive abilities of SNB19 cells transfected with miR-19a/b mimics were increased, as shown in Figures 2F and 2G.
Meanwhile, SNB19 cells transfected with miR-19a/b mimics simultaneously increased the expression of Ki67, cyclin D1, BCL-2, and MMP2 (Figure 2H).
In short, glioma cell lines transfected with miR-19a/b mimics demonstrated enhanced cell viability and invasion, driving the cell cycle and suppressing apoptosis in vitro. Therefore, the oncogenic role of miR-19a/b was demonstrated from both negative and positive sides.
Moreover, we found that miR-19a was more effective than miR-19b in promoting glioma progression. Perhaps this result indicated that miR-19a played a more crucial role in gliomagenesis, while the effect of miR-19a combined with miR-19b was more potent than that of either miR-19a or miR-19b alone.
SEPT7 is identified as the target gene of miR-19a/b
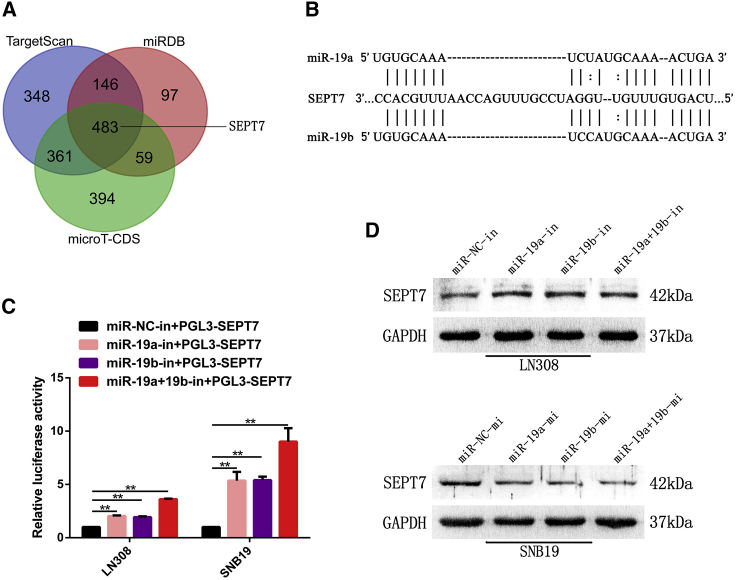
SEPT7 has been predicted to be a target gene of miR-19a/b by bioinformatics analyses with HuMiTar, a sequence-based method for the prediction of human miRNA targets.20 This finding was corroborated by the predicted results of TargetScan, miRDB, and microT-CDS (Figure 3A). There was complementarity between miR-19a/b and the SEPT7 3′ UTR (Figure 3B). A fragment of the SEPT7 3′ UTR containing the miR-19a/b putative binding site was cloned to construct pGL3-SEPT7 and conducted luciferase reporter assays. As shown in Figure 3C, a luciferase reporter assay revealed a remarkable increase of luciferase activity in cells co-transfected with pGL3-SEPT7 and miR-19a/b inhibitors. Moreover, western blot analysis showed that SEPT7 expression was upregulated in LN308 cells treated with miR-19a/b inhibitors, while SEPT7 was downregulated in SNB19 cells transfected with miR-19a/b mimics (Figure 3D). This evidence confirmed that miR-19a/b modulated SEPT7 expression by directly binding to the 3′ UTR of SEPT7.
Figure 3.
miR-19a/b directly targeted SEPT7 in GBM cells
(A) Venn diagram displaying miR-19a/b computationally predicted to target SEPT7 by three different prediction algorithms: TargetScan, miRDB, and microT-CDS. (B) Predicted miR-19a/b binding sequence in 3′ UTR of SEPT7 as shown by HuMiTar. (C) Luciferase assay of cells transfected with SEPT7 3′ UTR luciferase reporter plasmid together with miR-19a/b inhibitors or miR-NC inhibitor. (D) SEPT7 expression in glioma cells transfected with miR-19a/b inhibitors and miR-19a/b mimics was detected by western blotting . GAPDH was used as the loading control. ∗∗p < 0.01.
SEPT7 reverses the effect of miR-19a/b on glioma progression
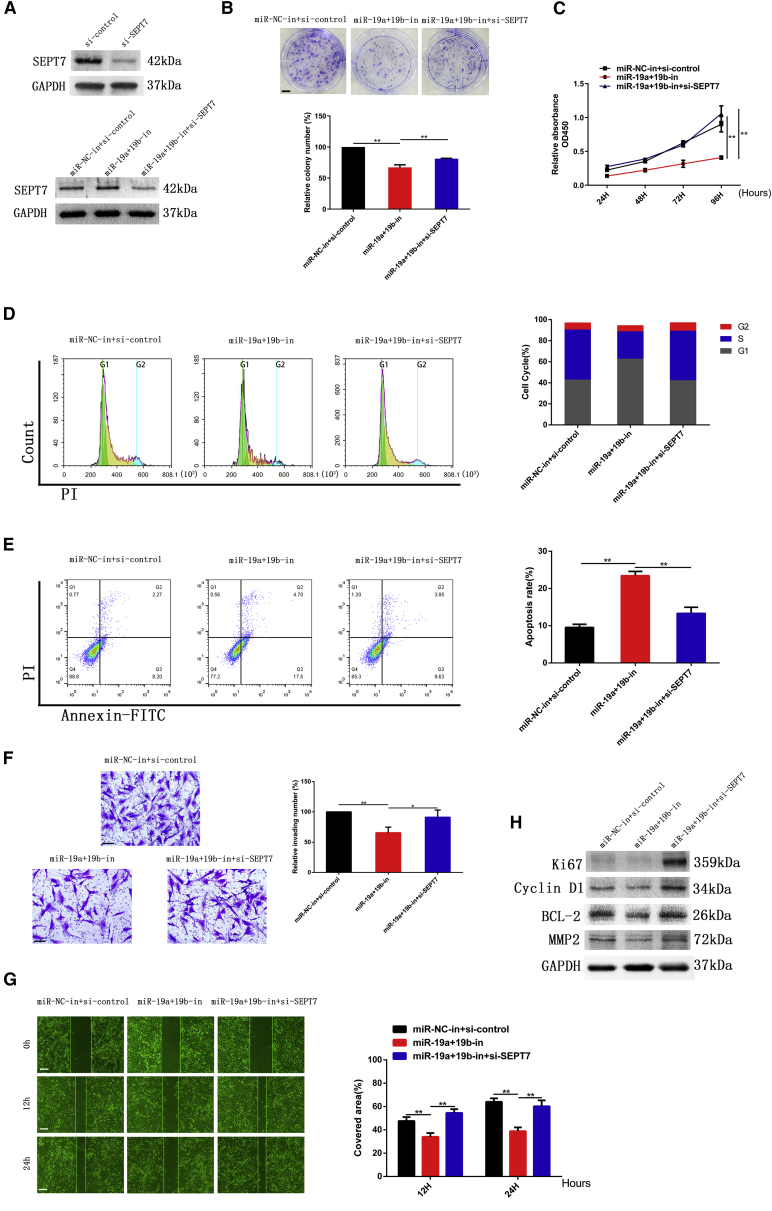
The expression of SEPT7 was increased when miR-19a/b were knocked down (Figure 3D). In order to explore the reverse effect of SEPT7 on miR-19a/b, SEPT7 small interfering RNA (siRNA) and inhibitors of miR-19a/b were co-transfected to LN308 cells. The effects of miR-19a/b inhibitors on proliferation, invasion, cell cycle kinetics, and apoptosis of glioma cells were partially abrogated by downregulation of SEPT7 (Figures 4A–4G). Suppression of the expression of Ki67, cyclin D1, BCL-2, and MMP2 induced by miR-19a/b inhibitors was also reversed by SEPT7 siRNA (Figure 4H).
Figure 4.
Knocking down miR-19a/b suppressed proliferation and invasion of glioma cells by targeting SEPT7
(A) Expression of SEPT7 in glioma cells transfected with SEPT7 siRNA detected by western blotting. (B) Colony formation assay of LN308 cells co-transfected with miR-19a/b inhibitors and SEPT7 siRNA. Scale bar, 5 mm. (C) Cell proliferation of LN308 cells co-transfected with miR-19a/b inhibitors and SEPT7 siRNA detected by CCK-8 assay. (D) Cell-cycle analysis of LN308 GBM cells transfected with miR-19a/b inhibitors and miR-19a/b inhibitors + SEPT7 siRNA. (E) Cell apoptosis of LN308 cells transfected with miR-19a/b inhibitors and miR-19a/b inhibitors + SEPT7 siRNA. (F) Effect of miR-19a/b inhibitors and miR-19a/b inhibitors + SEPT7 siRNA on invasion of LN308 cell was examined by Transwell assay. Scale bars, 200 μm. (G) Migration of LN308 cells transfected with miR-19a/b inhibitors and miR-19a/b inhibitors + SEPT7 siRNA was monitored by wound-healing assay. Scale bars, 200 μm. (H) Protein levels of Ki67, cyclin D1, BCL-2, and MMP2 in LN308 cells transfected with miR-19a/b inhibitors or miR-19a/b inhibitors + SEPT7 siRNA examined by western blot analysis. GAPDH was used as the loading control. ∗p < 0.05, ∗∗p < 0.01.
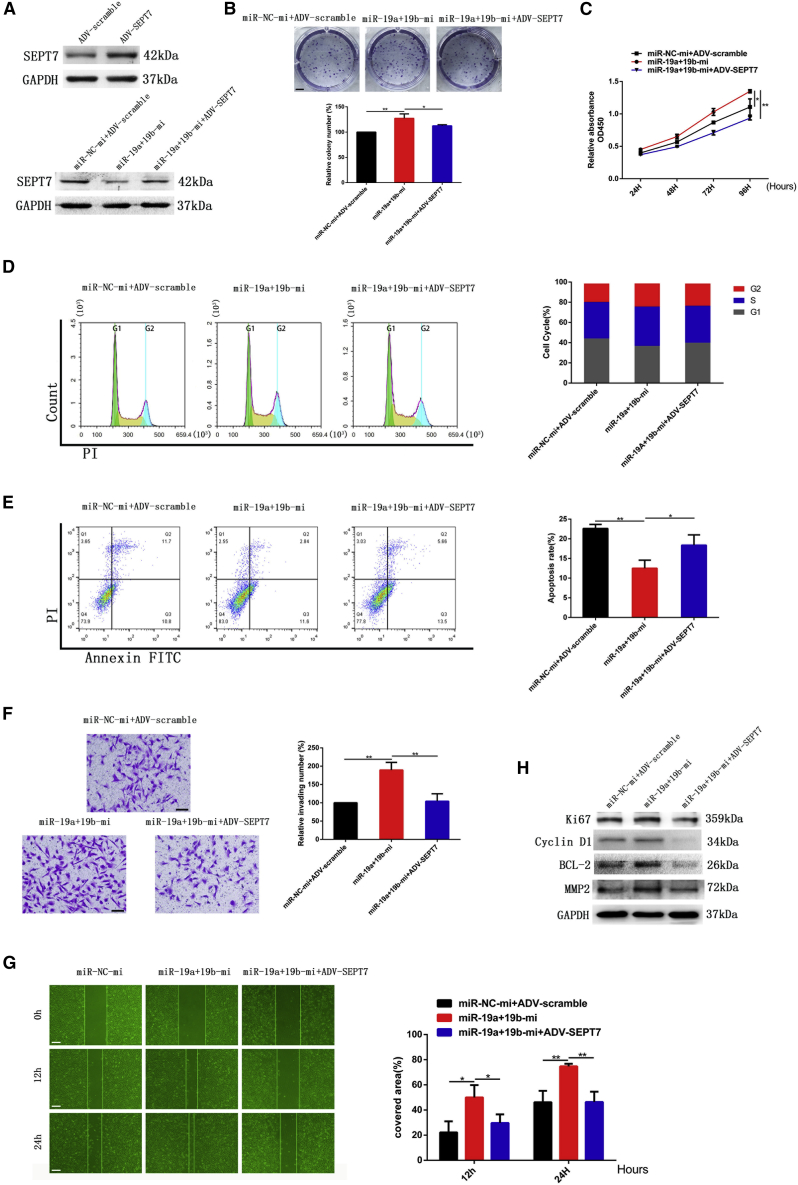
As shown in Figure 3D, SEPT7 was downregulated when miR-19a/b mimics were transfected into SNB19 cells. The reverse effect of SEPT7 on miR-19 mimics was observed by upregulation of SEPT7 in SNB19 cells transfected with SEPT7 recombinant adenovirus (ADV-SEPT7) (Figure 5A).
Figure 5.
miR-19a/b promoted cell proliferation, invasion, and migration and reduced apoptosis of GBM cells by targeting SEPT7
(A) Expression of SEPT7 was detected by western blotting when ADV-SEPT7 was transfected to SNB19 cells. GAPDH served as the loading control. (B) Colony formation assay of SNB19 cells transfected with miR-19a/b mimics and miR-19a/b mimics + ADV-SEPT7. Scale bar, 5 mm. (C) Cell proliferation of SNB19 cells detected by CCK-8 assay. (D) Cell-cycle kinetic analysis of SNB19 cells after transfection with miR-19a/b mimics and miR-19a/b mimics + ADV-SEPT7. (E) Apoptosis of miR-19a/b mimics group and miR-19a/b mimics + ADV-SEPT7 group. (F) Effect of miR-19a/b mimics and miR-19a/b mimics + ADV-SEPT7 on invasion of SNB19 cells was examined by Transwell assay. Scale bars, 200 μm. (G) Migration of SNB19 cells transfected with miR-19a/b mimics and miR-19a/b mimics + ADV-SEPT7 monitored by wound healing assay. Scale bars, 200 μm. (H) Protein levels of Ki67, cyclin D1, BCL-2, and MMP2 in LN308 cells of miR-19a/b mimics group and miR-19a/b mimics + ADV-SEPT7 group examined with western blot analysis. GAPDH was used as the loading control. ∗p < 0.05, ∗∗p < 0.01.
As above mentioned, glioma cells transfected with miR-19a/b mimics alone demonstrated increased cell viability and invasion, accelerated cell cycle progression, and decreased apoptosis in vitro. Conversely, glioma cells co-transfected with miR-19a/b mimics and ADV-SEPT7 showed inhibition of cell proliferation, invasion, and cell cycle progression and induction of cell apoptosis as compared with the miR-19 mimics-treated group. Additionally, the expression of their relevant protein markers, including Ki67, cyclin D1, BCL-2, and MMP2, was altered accordingly (Figures 5B–5H). The results were similar to the inhibitory effect of SEPT7 on glioma progression.24 This evidence indicates that the effect of miR-19a/b mimics on glioma progression can be reversed to a considerable degree by upregulation of SEPT7.
Taken together, both the results from transfection of miR-19a/b inhibitors plus SEPT7 siRNA as well as miR-19a/b mimics co-transfected with ADV-SEPT7 in vitro indicated that miR-19a/b affected the biological behavior of glioma cells through, at least partially, the negative regulation of SEPT7.
miR-19a/b activate the Akt /NF-κB pathway and promote EMT by targeting SEPT7
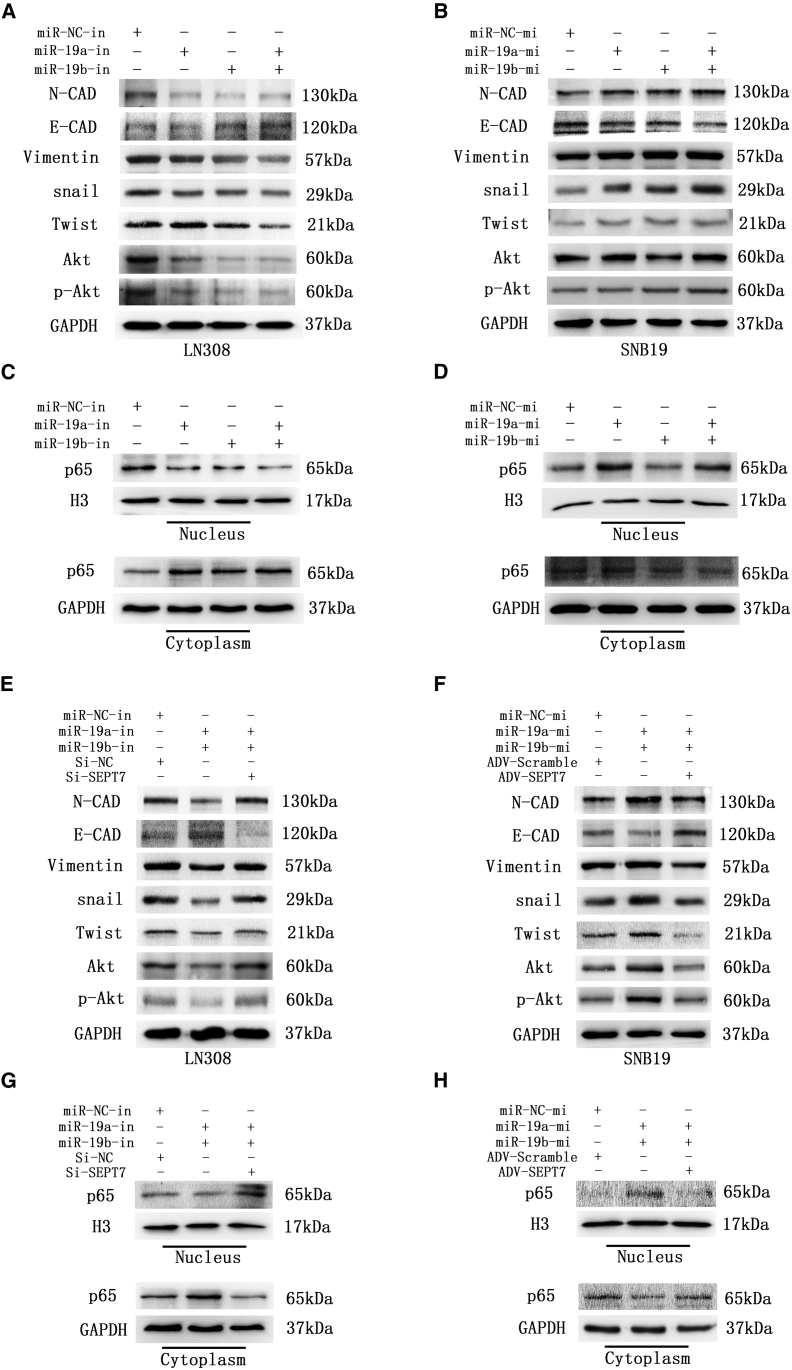
We further investigated the effect of miR-19a/b on EMT, a process that contributes to tumor cell invasion and migration. The classic EMT markers N-cadherin and Vimentin were upregulated while E-cadherin was downregulated when miR-19a/b expression strengthened. On the contrary, N-cadherin and Vimentin were downregulated and E-cadherin was upregulated when miR-19a/b expression was suppressed (Figures 6A and 6B).
Figure 6.
miR-19a/b were promoted EMT by activation of the Akt /NF-κB pathway, and SEPT7 reversed the effect of miR-19a/b on the Akt /NF-κB pathway and EMT
(A) Akt, p-Akt, and EMT-associated proteins in LN308 cells transfected with inhibitors of miR-19a/b were determined by western blotting. GAPDH was used as the loading control. (B) Aktp-Akt, and EMT-associated proteins in SNB19 cells transfected with mimics of miR-19a/b were determined by western blotting. GAPDH was used as the loading control. (C) Expression of NF-κB (p65) in cytoplasm and nucleus in LN308 cells transfected with inhibitors of miR-19a/b as detected by western blot analysis. GAPDH and histone 3 (H3) were used as loading controls. (D) Expression of NF-κB (p65) in cytoplasm and nucleus in SNB19 cells transfected with mimics of miR-19a/b as detected by western blot analysis. GAPDH and H3 were used as loading controls. (E) Expression of Akt, p-Akt, and EMT-associated proteins in LN308 cells transfected with miR-19a/b inhibitors and miR-19a/b inhibitors + SEPT7 siRNA as detected by western blot analysis. GAPDH was used as the loading control. (F) Expression of Akt, p-Akt, and EMT-associated proteins in SNB19 cells transfected with miR-19a/b mimics and miR-19a/b mimics + ADV-SEPT7 as detected by western blot analysis. GAPDH was used as the loading control. (G) Expression of NF-κB (p65) in cytoplasm and nucleus of LN308 cells transfected with miR-19a/b inhibitors and miR-19a/b inhibitors + SEPT7 siRNA as detected by western blot analysis. GAPDH and H3 were used as the loading controls, respectively. (H) Expression of NF-κB (p65) in cytoplasm and nucleus of SNB19 cells transfected with miR-19a/b mimics and miR-19a/b mimics + ADV-SEPT7 as detected by western blot analysis. GAPDH and H3 were used as the loading controls, respectively.
Expression levels of the EMT-activating transcription factors Snail and Twist were simultaneously enhanced by miR-19a/b (Figures 6A and 6B). We further co-transfected SEPT7 siRNA and miR-19a/b inhibitors to LN308 cells, and ADV-SEPT7 and miR-19a/b mimics to SNB19 cells as well, and found that SEPT7 reversed the effect of miR-19a/b on the expression of Snail, Twist, and the biomarkers of EMT (Figures 6E and 6F). These results indicated that miR-19a/b activated the EMT process to promote glioma invasion and migration by targeting SEPT7.
The Akt/NF-κB (p65) pathway was regarded as a positive regulator of EMT. To explore the underlying mechanism of miR-19a/b promoting EMT, the impact of miR-19a/b on the Akt/NF-κB (p65) signaling pathway was investigated. miR-19a/b were demonstrated to upregulate the expression of Akt and phosphorylated p-Akt and promote the nuclear translocation of NF-κB (p65) (Figures 6A–6D). SEPT7 has been identified to reverse the effect of miR-19a/b on Akt, p-Akt, and nuclear translocation of NF-κB (p65) (Figures 6E–6H).
miR-19a-targeted therapy in an orthotopic xenograft model
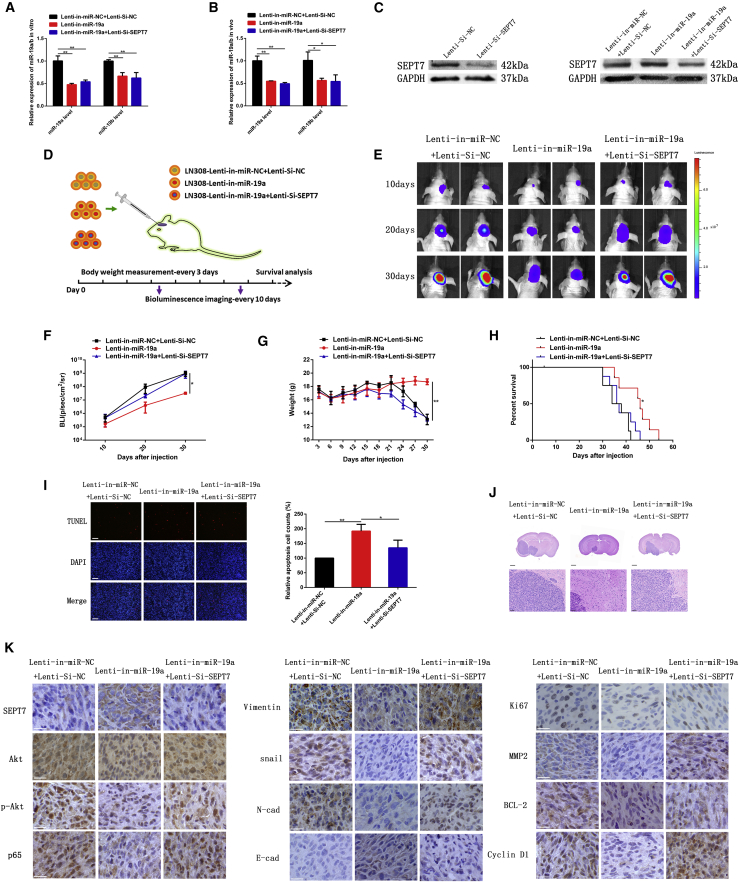
miR-19a has been shown to be more effective in glioma progression compared with miR-19b, further identifying miR-19a as a potential target for the treatment of malignant gliomas. Additionally, the oncogenic effect of miR-19a is mediated mostly through the negative regulation of SEPT7, and Lipofectamine-mediated targeted therapies for orthotopic xenograft glioma models were performed.
The expression levels of miR-19a and SEPT7 were knocked down as detected by Real-time PCR (RT-PCR) and western blotting in LN308 cells after either being transfected with miR-19a inhibitor alone or co-transfected with SEPT7 siRNA (Figures 7A and 7C). Likewise, the expression of miR-19a was downregulated in xenograft tumor samples of the Lenti-in-miR-19a group (the mice were implanted with LN308 cells transfected with lentivirus bearing the inhibitor of miR-19) (Figure 7B), and the expression levels of miR-19a and SEPT7 were downregulated in the Lenti-in-miR-19a + Lenti-Si-SEPT7 group (the mice were treated with lentivirus carrying both the inhibitor of miR-19a and siRNA of SEPT7) (Figures 7B and 7K). The procedure of targeted therapies for xenograft glioma models is demonstrated in Figure 7D. During the observation period, the signals of bioluminescence imaging (BLI) were weaker in the Lenti-in-miR-19a group than the control group (Figures 7E and 7F, p < 0.05, compared with the lenti-in-miR-NC control group), while the signals of the Lenti-in-miR-19a + Lenti-Si-SEPT7 group were much stronger than those in the Lenti-in-miR-19a group (Figures 7E and 7F, p < 0.05, compared with the Lenti-in-miR-19a group), which indicated that knocking down miR-19a decelerated tumour growth, and downregulation of SEPT7 reversed the inhibitory effect of miR-19a inhibitor on glioma proliferation. At the end of the observation period, the Lenti-in-miR-19a group inhibited tumor growth significantly. However, there was no difference in tumor growth between the Lenti-in-miR-19a + Lenti-Si-SEPT7 group and the control group. The body weights of mice in three groups were quantified, which showed that mice of the Lenti-in-miR-19a group gained weight, while the mice of the control group and the Lenti-in-miR-19a + Lenti-Si-SEPT7 group lost weight at the end of observation (Figure 7G, p < 0.01, compared with the Lenti-in-miR-19a group).
Figure 7.
miR-19a downregulation inhibited xenograft tumor growth and induced cell apoptosis in vivo by targeting SEPT7
(A) Expression of miR-19a in LN308 cells transfected with Lenti-in-miR-NC, Lenti-Si-NC, Lenti-in-miR-19a, and Lenti-Si-SEPT7. (B) Expression of miR-19a in tumor samples of the Lenti-miR-NC + Lenti-Si-NC group, the Lenti-in-miR-19a group, and the Lenti-in-miR-19a + Lenti-Si-SEPT7 group detected by RT-PCR analysis. (C) Expression of SEPT7 in LN308 cells transfected with Lenti-Si-SEPT7 or Lenti-in-miR-19a. (D) Schematic representation of miR-19a-targeted therapy in vivo. The mice were divided randomly into three groups, i.e., the Lenti-miR-NC + Lenti-Si-NC group, the Lenti-in-miR-19a group, and the Lenti-in-miR-19a + Lenti-Si-SEPT7 group. (E) Representative bioluminescence images of orthotopic xenografts on the days indicated. (F) Bioluminescence signals were quantified in tumors from three groups. (G) Mouse weight of three groups was recorded as an indicator of tumor progression. (H) Kaplan-Meier survival curve of mice from three groups. (I) Apoptosis of glioma samples was detected using a TUNEL assay. Scale bars, 200 μm. (J) Representative HE staining of tumor gross dissection from three groups. Scale bars, 1 mm (upper) and 50 μm (lower). (K) Immunohistochemistry analysis of the expression of SEPT7, Akt, p-Akt, p65, E-cadherin, N-cadherin, Vimentin, Snail, Ki67, MMP2, BCL-2, and cyclin D1 in tumor samples from three groups. Scale bars, 50 μm. ∗p < 0.05, ∗∗p < 0.01.
The median survival of the control group was 36 days, while mice bearing orthotopic xenografts transfected with miR-19a inhibitor had a median survival of 46 days (Figure 7H, p < 0.01, compared with the control group), and mice bearing orthotopic xenografts co-transfected with Lenti-in-miR-19a and Lenti-Si-SEPT7 had a median survival of 36.5 days (Figure 7H, p < 0.05, compared with the Lenti-in-miR-19a group).
Hematoxylin and eosin (HE) staining of tumor tissues confirmed that tumor volumes of the Lenti-in-miR-19a group were reduced as compared with those of the control group. Nevertheless, mice in the miR-19a inhibitor + SEPT7 siRNA group regained tumor growth, and their tumor volumes were enlarged compared with those in the Lenti-in-miR-19a group (Figure 7J).
Cell apoptosis was detected by the TUNEL (terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling) method in xenograft tumors. Tumors in the Lenti-in-miR-19a group exhibited an increased apoptotic rate as compared with that in control tumor group, while the apoptotic rate of tumors in the miR-19a inhibitor + SEPT7 siRNA group was lowered as compared with that in tumors of the Lenti-in-miR-19a group (Figure 7I). This result also suggested that the miR-19a inhibitor promoted cell apoptosis at least partially through negative regulation on SEPT7.
Immunohistochemical analysis revealed that the expression of SEPT7 was upregulated in xenograft tumor samples derived from the Lenti-in-miR-19a group. The expression levels of Akt, p-Akt, Snail, N-cadherin, Vimentin, Ki67, BCL-2, cyclin D1, MMP2, and NF-κB (p65) in tumor samples were significantly suppressed by miR-19a inhibitor and partially restored by downregulation of SEPT7 (Figure 7K). The findings were coincident with the effect of co-transfected miR-19a inhibitor and SEPT7 siRNA on the expression of these biomarkers in vitro. Accordingly, all results from in vivo experiments suggested that downregulation of miR-19a inhibited tumor growth and malignant progression through the SEPT7-AKT-NF-κB (p65) pathway.
Discussion
miRNAs play important roles in gene modulation. It is estimated that potentially up to one-third of all protein-coding genes are regulated by miRNAs according to computational predictions.31, 32, 33 Dysregulation of miRNAs has also been associated with glioma formation and progression.34, 35, 36 Several studies have revealed that miR-19a/b directly affect target genes such as PPARα, RUNX3, PTEN, and RhoB to regulate the proliferation and invasion of glioma cells.18,19,37,38 Additionally, it has been reported that miR-19a/b themselves can be modulated by long non-coding RNA MEG3 and linc-NeD125.39,40
In our previous work, we determined the expression of miR-19a and miR-19b in eight glioma cell lines, 43 freshly resected glioma samples, and 75 archival paraffin-embedded glioma specimens with different grades of malignancy by both real-time PCR and in situ hybridization, and we found that the expression of miR-19a/b is upregulated in glioma tissues and cell lines. LN308 glioma cells are identified to have the highest expression of miR19a/b, while their expression in SNB19 glioma cells is the lowest. Furthermore, miR-19a/b expression is positively correlated with glioma grades, that is, the more malignant the glioma, the higher is the expression of miR-19a/b, suggesting that miR-19a/b might act as oncomiRs in glioma.18
In the current study, cell proliferation and invasion were suppressed by knocking down miR-19a/b in LN308 glioma cells, whereas cell apoptosis was induced. Conversely, cell proliferation and invasion are enhanced, and cell apoptosis is suppressed, when miR-19a/b are overexpressed in SNB19 cells. Knockdown or overexpression of miR-19a exerts a more remarkable effect on the biological behaviors of GBM cells than miR-19b, while the effect of combined miR-19a and miR-19b is stronger than either one of them alone. These results suggest that miR-19a is likely to play a more critical role in glioma progression, but miR-19a combined with miR-19b can exert a synergistic effect on glioma progression. The expression of the biomarkers involved in cell proliferation, cell cycle progression, invasion, and apoptosis, including Ki67, cyclin D1, MMP2, and BCL-2, are downregulated when miR-19a/b are knocked down in vitro and in vivo. All evidence suggests that miR-19a/b play an oncogenic role in gliomagenesis.
For further exploring the underlying mechanism of the oncogenic effects of miR-19a/b on glioma cells, the major target genes that miR-19a/b negatively regulated in gliomagenesis were predicted on the basis of bioinformatics analyses with HuMiTar,20 and SEPT7 has been found to be one of the predicted target genes of miR-19. In the present study, SEPT7 was validated to be directly regulated by miR-19a/b using a luciferase reporter assay. Meanwhile, when knocking down miR-19a/b with corresponding inhibitors, the expression of SEPT7 was significantly upregulated, as shown in western blot analysis. These results indicate that SEPT7 is a target gene of miR-19a/b. Additionally, our previous study also demonstrated that SEPT7 is downregulated and plays a tumor suppressor role in glioma.24
On the basis of previous work, we further identified whether SEPT7 is one of the major effectors of miR-19a/b for the regulation of biological behavior of glioma cells. We found that proliferation and invasion of glioma cells co-transfected with miR-19a/b inhibitors and SEPT7 siRNA are suppressed, but to a significantly less extent compared with transfected miR-19a/b inhibitors alone. Meanwhile, proliferation and invasion of the cells co-transfected with miR-19a/b mimics and SEPT7 are enhanced, but much less effectively than when transfected only with the miR-19a/b mimics group. These results suggest that the effect of miR-19a/b on biological behaviors of glioma cells can be reversed to a considerable degree by SEPT7. miR-19a inhibitor can inhibit growth of the LN308 glioma xenograft, and the effect of miR-19a inhibitor on suppressing tumor growth and inducing cell apoptosis also can be reversed by SEPT7 siRNA in vivo. All evidence has strongly indicated that SEPT7 might play an important role in miR-19a/b by affecting biological behaviors of glioma cells.
The EMT enables tumor cells to acquire the characteristics of mesenchymal cells and lose the features of epithelial cells, such as adherens junctions and apical-basal polarity. The EMT is identified as a key step in the progression of glioma and plays a major role in promoting the migration and invasion of tumor cells.41 The overexpression of mesenchymal cell traits such as N-cadherin and Vimentin coincided with loss of the epithelial cell biomarker E-cadherin, which is the classic pattern of EMT activation.42 Our data have shown that knockdown of miR-19a/b results in downregulation of N-cadherin and Vimentin, as well as upregulation of E-cadherin expression, in glioma cells. On the contrary, overexpression of miR-19a/b upregulates N-cadherin and Vimentin expression, and it downregulates E-cadherin expression. Thus, miR-19a/b are identified to promote EMT in glioma cells. SEPT7 has been demonstrated to reverse the effect of miR-19a/b on EMT by antagonizing the regulation of miR-19a/b on N-cadherin, Vimentin, and E-cadherin. These results support that miR-19a/b enhances EMT by suppression of SEPT7.
To further elucidate the underlying mechanism of miR-19a/b’s regulation on EMT, we also studied the effects of miR-19a/b and SEPT7 on the Akt/NF-κB signaling pathway.
The phosphatidylinositol 3-kinase (PI3K)/Akt/NF-κB signaling pathway is activated and plays an important role in EMT and the progression of gliomas.43 Akt has been reported to promote EMT by enhancing the expression of Twist, one of the major transcription factors to induce EMT. Twist also can exert a significant effect on the Akt signaling pathway, and Aktactivity was inhibited when Twist was knocked down in glioma cells in vivo.44,45 As the downstream factor of Akt, NF-κB also plays a crucial role in glioma progression.46 NF-κB is activated by PI3K/AktT pathway, and Akt phosphorylates IκB kinase (IKK)α and IκBα successively, leading to IκBα degradation and followed by nuclear translocation of one of the NF-κB family members, p65 (RelA), and transcription of its target genes.47 p65 has been identified to promote EMT of glioma cells by inducing the transcription of Snail and Twist, two important EMT transcription factors.48,49
Akt/NF-κB activate EMT of glioma cells by decreasing E-cadherin expression and enhancing vimentin and N-cadherin expression so as to enhance their mesenchymal phenotype. Our results demonstrate that Akt and its activating phenotype, p-Akt,50 are upregulated when miR-19a/b mimics are transfected to glioma cells and downregulated when miR-19a/b is knocked down. Akt is identified to be activated by miR-19a/b and, in turn, promoted NF-κB (p65) translocation into the cell nucleus. When miR-19a/b mimics are transfected into glioma cells, it has been found that they upregulate Akt and p-Akt, promote nuclear translocation of NF-κB (p65), and enhance expression of major EMT transcription factors, including Snail and Twist, and then the important EMT markers N-cadherin and Vimentin are upregulated, while E-cadherin is inhibited, thereby accelerating the EMT process. When miR-19a/b are knocked down, the opposite results are found. These findings fully illustrate that miR-19a/b activate the Akt/NF-κB (p65) signaling pathway to promote the EMT process.
NF-κB (p65) also promotes invasion of glioma cells by activating MMPs, a group of enzymes necessary for extracellular matrix (ECM) degradation and facilitating tumor cell migration.46 The expression of MMP2 is positively correlated with that of NF-κB (p65) when miR-19a/b are overexpressed or knocked down, which indicates that miR-19a/b might regulate MMP2 via modulating NF-κB (p65). Cyclin D1 is also regulated by NF-κB (p65).46 Accordingly, it can be speculated that miR-19a/b enhance cyclin D1 through NF-κB (p65) to promote cell cycle progression.
PTEN has been identified as the principal negative regulator of the PI3K-AKT signaling pathway in glioma, and it was also the target gene of miR-19a/b in our previous study,19 so PTEN is naturally thought to play an important role in miR-19a/b regulating the Akt/NF-κB (p65) signaling pathway. However, in the present study we also observed that the effect of knocking down miR-19a/b on the expression of Akt, NF-κB (p65), Twist, Snail, E-cadherin, N-cadherin, Vimentin, MMP2, and cyclin D1 can be reversed to a certain extent by knocking down SEPT7, while the effect of miR-19a/b mimics can be reduced to a considerable degree by overexpression of SEPT7 (Figure 8). These results indicate that, in addition to PTEN, miR-19a/b also activate AKT/NF-κB (p65) EMT through suppressing SEPT7.
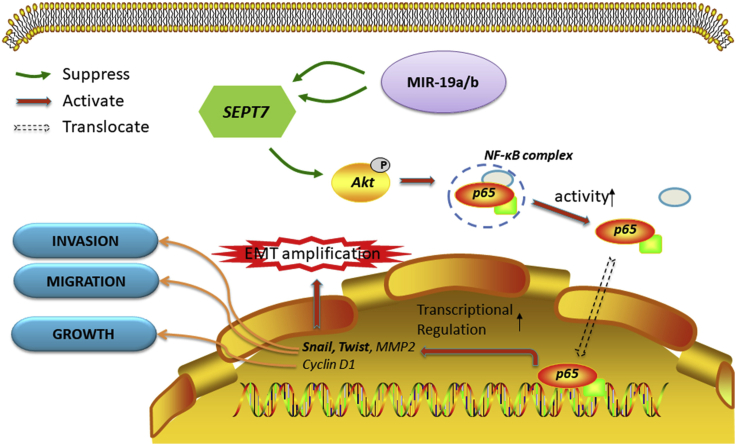
Figure 8.
Schematic model of miR-19a/b/SEPT7 axis-induced glioma cell malignant progression
In glioma cells, upregulation of miR-19a/b promoted glioma malignant progression via Akt-NF-κB pathway by targeting SEPT7.
In summary, upregulation of miR-19a/b plays a vital role in the malignant progression of glioma. miR-19a exerts a more prominent effect than does miR-19b on biological behaviors of glioma cells. Combined miR-19a with miR-19b have a synergistic effect on glioma growth and malignant progression. SEPT7 is identified as one of the major target genes of miR-19a/b, and miR-19a/b activates Akt/NF-κB to promote EMT partially by suppressing SEPT7. Understanding the dysregulation of the miR-19a/b/SEPT7/Akt/NF-κB pathway in glioma is important to improve our knowledge of the molecular pathophysiology of GBM development and progression, and it may offer potential novel targets for the treatment of GBM.
Materials and methods
Cell culture
The human SNB19 and LN308 GBM cell lines were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Science. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Invitrogen) and 2 mM glutamine (Sigma-Aldrich, St. Louis, MO, USA), and incubated at 37°C with 5% CO2 and subcultured every 2–3 days.
Cell transfection with oligonucleotides and ADV-SEPT7
The specific inhibitors of miR-19a/b, mimics of miR-19a/b and SEPT7 siRNA, and scramble constructs were chemically synthesized by GenePharma (Shanghai, P.R. China). The RNA interference sequences are listed in Table S2.
Oligonucleotides were transfected into SNB19 and LN308 cells (75 nM, 48 h) at 70% confluency using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. ADV-SEPT7 and a scrambled sequence adenovirus (ADV-scramble) were purchased from GenePharma (P.R. China). For cell transfection, cells were incubated with diluted viral constructs in serum-free medium at 37°C for 1 h, the necessary amount of complete medium was added, and cells were incubated for a further 48–72 h. For the in vivo study, the lentivirus with inhibitor of mir-19a and siRNA of SEPT7 (GeneChem, Shanghai, P.R. China) were designed to knock down the expression of miR-19a and SEPT7 in GBM cells. The constructed lentiviral vectors were transfected into LN308 cells according to the manufacturer’s instructions.
miR-19 expression determined by quantitative real-time PCR
Total RNA was extracted from parental GBM cells and cells transfected with inhibitors of miR-19a/b or miR19a/b mimics using TRIzol reagent (Invitrogen). Then RNA was reverse transcribed into strand cDNA using a GoTaq reverse transcription kit (Promega, Beijing, P.R. China). miRNA-specific reverse transcription primers and quantitative PCR primers were obtained from GenePharma. The primer sequences are listed in Table S1.
To quantify the expression level of miR-19a/b, quantitative real-time PCR was conducted on a CFX96 PCR cycler (Bio-Rad, Hercules, CA, USA) using GoTaq qPCR master mix (Promega), and fold changes were calculated by relative quantification. Each sample was analyzed in triplicate. Data are shown as fold change (2−ΔΔCT method).
LN308 and SNB19 cells were divided into miR-19a/b inhibitors and miR-19 a/b mimics sets. The miR-19a/b inhibitors set was further divided into five groups as follows: (1) NC group: LN308 cells transfected with scramble sequence of miR-19a/b inhibitors (miR-NC-in) and LN308 cells transfected with scramble sequence of SEPT7 siRNA (si-control). (2) miR-19a inhibitor group (miR-19a-in): LN308 cells transfected with miR-19a inhibitor. (3) miR-19b inhibitor group (miR-19b-in): LN308 cells transfected with miR-19b inhibitor. (4) miR-19a/b inhibitors group (miR-19a + 19b-in): LN308 cells transfected with both miR-19a inhibitor and miR-19b inhibitor. (5) miR-19a/b-in + SEPT7 siRNA group (miR-19a + 19b-in + si-SEPT7): LN308 cells co-transfected with inhibitors of miR-19a/b and SEPT7 siRNA simultaneously.
The miR-19a/b mimics set was also divided into five groups as follows: (1) NC group: SNB19 cells transfected with scramble sequence of miR-19a/b mimics (miR-NC-mi) and SNB19 transfected with scramble sequence of recombinant adenovirus (ADV-scramble). (2) miR-19a mimics group (miR-19a-mi): SNB19 cells transfected with miR-19a mimics. (3) miR-19b mimics group (miR-19b-mi): SNB19 cells transfected with miR-19b mimics. (4) miR-19a/b mimics group (miR-19a + 19b-mi): SNB19 cells transfected with miR-19a/b mimics. (5) miR-19a/b mimics + ADV-SEPT7 group (miR-19a + 19b-mi + ADV-SEPT7): SNB19 cells co-transfected with miR-19a/b mimics and SEPT7 recombinant adenovirus.
Colony formation assays and cell proliferation analysis
Parental cells and transfected cells were seeded into six-well plates (500 cells for each well) and continuously cultured for 10 days. After washing with PBS, cells were fixed with ice-cold methanol and stained with crystal violet solution (Sigma-Aldrich, St. Louis, MO, USA). Images were taken and colony dot numbers were counted, and results were expressed as percentage of control. For the CCK-8 assay, cells were seeded in 96-well plates (3 × 103 for each well). Optical density of the cells was measured in triplicate wells at 450 nm for different suitable time points using the CCK-8 kit (Dojindo, Kumamoto, Japan) following the manufacturer’s protocol.
Cell cycle kinetic analysis
Parental or transfected cells in the log phase of growth were harvested, and the cells were fixed with ice-cold 70% ethanol and incubated at 4°C overnight. Before staining, the cells were spun down in a cooled centrifuge and resuspended in cold PBS. RNAase (Becton Dickinson, Franklin Lakes, NJ, USA) was added, followed by incubation of cells in PI (Becton Dickinson). Samples were detected using a FACScan flow cytometer (Becton Dickinson), and the data were analyzed with FlowJo software (FlowJo, USA).
Detection of apoptotic cell death
Parental and transfected cells in the log phase of growth were harvested, centrifuged, and resuspended in binding buffer contains annexin-Vfluorescein isothiocyanate (FITC) (KeyGEN BioTECH, Nanjing, P.R. China) and PI (KeyGEN BioTECH).
Flow cytometric assays were performed using a FACScan flow cytometer (Becton Dickinson). The experiments were repeated three times for each group of cells, and the data were analyzed by FlowJo software (FlowJo, USA).
The apoptotic cell death for the tumor specimens from the in vivo study was examined by the TUNEL method using an in situ cell death kit (KeyGEN BioTECH). The tumor tissue slices were incubated with Proteinase K for 15 min at room temperature, then washed with PBS. Endogenous peroxidase was blocked by 3% H2O2 in methanol for 30 min at room temperature. Sections were then incubated with TUNEL reaction mixture for 60 min at 37°C in a humidified atmosphere in the dark. Positive cells were visualized under fluorescence microscopy. Nuclei were counterstained with a DAPI karyotyping kit (Genmed, Shanghai, P.R. China) and visualized using an FV-1200 laser scanning confocal biological microscope (Olympus, Tokyo, Japan).
Transwell assay and wound-healing assay
Transwell chambers were precoated with Matrigel on the upper surface of the polycarbonate membrane (24-well insert, 8-μm pore size, Becton Dickinson). Transfected and control cells suspending in serum-free DMEM were added to the upper chamber, and DMEM containing 10% fetal bovine serum was placed into the lower chamber as a chemoattractant. After 24 h of incubation, the cells that had invaded the lower surface of the filter were fixed with 4% paraformaldehyde and stained with crystal violet. The average number of migrated cells was determined in three randomly chosen visual fields under an inverted light microscope (Olympus, Tokyo, Japan).
Transfected and control cells seeded in six-well plates were allowed to proliferate freely until they reached 80%–90% confluency. The scratch wounds were created in the monolayer of confluent glioma cells with a 200-μL pipette tip. Then, these cells were incubated as usual. A wound healing assay was measured and imaged using an inverted light microscope (Olympus) at 0, 12, and 24 h. The data were obtained from three repeated experiments.
Western blot analysis
Total proteins were extracted from the parental and transfected cells lysed in radioimmunoprecipitation assay (RIPA) buffer (Solarbio, Beijing, P.R. China). Protein lysates from each sample were subjected to SDS-PAGE separation and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA). The membrane was incubated with relevant primary antibodies, including Ki67, MMP2, cyclin D1, BCL-2, Akt, p-Akt, Vimentin, Snail, E-cadherin, N-cadherin, and Twist (Abcam, Shanghai, P.R. China), SEPT7 and p65 (Santa Cruz Biotechnology, Dallas, TX, USA), and GAPDH and histone 3 (H3) (ABclonal, Wuhan, P.R. China), followed by incubation with a horseradish peroxidase (HRP)-conjugated secondary antibody (Abcam). Proteins were detected using a SuperSignal protein detection kit (Pierce, Rockford, IL, USA).
Plasmid constructs and luciferase reporter assay
A fragment of the SEPT7 3′ UTR containing a putative miR-19a and miR-19b binding site was cloned into XbaI-treated pGL3-control luciferase reporter vector (pGL3) and located immediately downstream of the stop codon of luciferase (pGL3-SEPT7).
For luciferase reporter experiments, LN308 and SNB19 cells were transfected with pGL3-SEPT7 and inhibitors of miR-19a and miR-19b, and the pGL3-SEPT7 was separately transfected as a control. Luciferase activity was determined by using a Dual-Luciferase reporter assay system (Promega) according to the manufacturer’s instructions.
Orthotopic GBM mouse model and miR-19a targeted therapy in vivo
Four-week-old male immune-deficient nude mice (BALB/c-nu) were purchased from the animal center of the Cancer Institute of Chinese Academy of Medical Science (Beijing, P.R. China), and they were bred at the Experimental Animal Center of the Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences. The experimental protocols were approved by the Ethics Committee for Animal Experimentation of Tianjin Medical University General Hospital.
The LN308 cell line was selected to establish a nude mouse orthotopic GBM model. The method was the same as previously described,51 with LN308 cells transfected with luciferase-encoding lentivirus (GeneChem) at first.
Then, the mice were divided randomly into three groups as follows, and each group consisted of six mice: (1) Lenti-NC group: LN308 cells transfected with lentivirus carrying scramble sequence of miRNA inhibitor (Lenti-in-miR-NC) and scramble sequence of SEPT7 siRNA (Lenti-Si-NC) were implanted. (2) Lenti-in-miR-19a group: the mice were implanted with LN308 cells transfected with lentivirus bearing the inhibitor of miR-19a. (3) Lenti-in-miR-19a + Lenti-Si-SEPT7 group: the mice were treated with lentivirus carrying both inhibitor of miR-19a and siRNA of SEPT7.
Quantitative RT-PCR (qRT-PCR) and western blotting were performed to confirm transfection efficiency.
The survival of mice in each group was observed. Intracranial tumor growth was monitored by the BLI method on days 10, 20, and 30, respectively. The mice were injected intraperitoneally with d-luciferin (Promega) and imaged with the in vivo imaging system (IVIS) Spectrum live imaging system (PerkinElmer, Waltham, MA, USA) to measure the tumor volume. Survival period and weight of mice were recorded through the whole experiment. At the end of the observation period, tumor tissues were removed to extract total RNA for detecting miR-19a expression and for preparation of paraffin-embedded sections.
Immunohistochemistry (IHC) and HE staining
An IHC assay was performed as previously described21 for detection of SEPT7 and p65 (NF-κB) (Santa Cruz Biotechnology, Dallas, TX, USA) and Akt, p-Akt, Snail, E-cadherin, N-cadherin, Vimentin, Ki67, BCL2, MMP2, and Cyclin D1 (Abcam, Shanghai, P.R. China) expression.
HE staining was also routinely performed. The processed slides were observed under an optical microscope (Olympus, Japan).
Statistical analysis
Data are expressed as the means ± SD of three independent experiments. Statistical analyses were performed using SPSS version 13.0 (SPSS, Chicago, IL, USA). Differences between experimental groups and controls were assessed by a Student’s t test or ANOVA analysis, and the survival analysis was performed using log-rank tests. p-value <0.05 was considered statistically significant (single asterisks in the figures) and p <0.01 was considered strongly significant (double asterisks in the figures).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant nos. 30872985 and 81101915).
Author contributions
W. Wang and Z.J. designed research and drafted the paper. W. Wang, Y.H., and A.Z. performed experiments and collected data. W. Wang and Z.J. provided advise on the design of research and analyzed data. W.Y., W. Wei, G.W., and Z.J. reviewed and revised the paper. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2021.01.005.
Supplemental information
References
- 1.Fridrichova I., Zmetakova I. MicroRNAs contribute to breast cancer invasiveness. Cells. 2019;8:1361. doi: 10.3390/cells8111361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C.M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J.J., Yang J., Lin J., Yao N., Zhu Y., Zheng J., Xu J., Cheng J.Q., Lin J.Y., Ma X. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv. Syst. 2009;25:13–20. doi: 10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
- 4.Hunter S.E., Finnegan E.F., Zisoulis D.G., Lovci M.T., Melnik-Martinez K.V., Yeo G.W., Pasquinelli A.E. Functional genomic analysis of the let-7 regulatory network in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003353. doi: 10.1371/journal.pgen.1003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 6.Macharia L.W., Wanjiru C.M., Mureithi M.W., Pereira C.M., Ferrer V.P., Moura-Neto V. MicroRNAs, hypoxia and the stem-like state as contributors to cancer aggressiveness. Front. Genet. 2019;10:125. doi: 10.3389/fgene.2019.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dharmawardana N., Ooi E.H., Woods C., Hussey D. Circulating microRNAs in head and neck cancer: a scoping review of methods. Clin. Exp. Metastasis. 2019;36:291–302. doi: 10.1007/s10585-019-09961-6. [DOI] [PubMed] [Google Scholar]
- 8.Schwarzenbach H., Nishida N., Calin G.A., Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X., Li Y., Qi P., Ma Z. Biology of miR-17-92 cluster and its progress in lung cancer. Int. J. Med. Sci. 2018;15:1443–1448. doi: 10.7150/ijms.27341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y., Liu J., Kang Y., He Y., Liang B., Yang P., Yu Z. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J. Exp. Clin. Cancer Res. 2014;33:67. doi: 10.1186/s13046-014-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q., Liu M., Ma F., Luo Y., Cai R., Wang L., Xu N., Xu B. Circulating miR-19a and miR-205 in serum may predict the sensitivity of luminal A subtype of breast cancer patients to neoadjuvant chemotherapy with epirubicin plus paclitaxel. PLoS ONE. 2014;9:e104870. doi: 10.1371/journal.pone.0104870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan Y., Yin H., Zhang H., Fang J., Zheng W., Li D., Li Y., Cao W., Sun C., Liang Y. Sp1-driven up-regulation of miR-19a decreases RHOB and promotes pancreatic cancer. Oncotarget. 2015;6:17391–17403. doi: 10.18632/oncotarget.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Q., Yang Z., An Y., Hu H., Yin J., Zhang P., Nie Y., Wu K., Shi Y., Fan D. miR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014;5:e1144. doi: 10.1038/cddis.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu T.Y., Zhang T.H., Qu L.M., Feng J.P., Tian L.L., Zhang B.H., Li D.D., Sun Y.N., Liu M. miR-19a is correlated with prognosis and apoptosis of laryngeal squamous cell carcinoma by regulating TIMP-2 expression. Int. J. Clin. Exp. Pathol. 2013;7:56–63. [PMC free article] [PubMed] [Google Scholar]
- 15.Tantawy M., Elzayat M.G., Yehia D., Taha H. Identification of microRNA signature in different pediatric brain tumors. Genet. Mol. Biol. 2018;41:27–34. doi: 10.1590/1678-4685-GMB-2016-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhi F., Shao N., Wang R., Deng D., Xue L., Wang Q., Zhang Y., Shi Y., Xia X., Wang S. Identification of 9 serum microRNAs as potential noninvasive biomarkers of human astrocytoma. Neuro-oncol. 2015;17:383–391. doi: 10.1093/neuonc/nou169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malzkorn B., Wolter M., Liesenberg F., Grzendowski M., Stühler K., Meyer H.E., Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia Z., Wang K., Zhang A., Wang G., Kang C., Han L., Pu P. miR-19a and miR-19b overexpression in gliomas. Pathol. Oncol. Res. 2013;19:847–853. doi: 10.1007/s12253-013-9653-x. [DOI] [PubMed] [Google Scholar]
- 19.Sun J., Jia Z., Li B., Zhang A., Wang G., Pu P., Chen Z., Wang Z., Yang W. miR-19 regulates the proliferation and invasion of glioma by RUNX3 via β-catenin/Tcf-4 signaling. Oncotarget. 2017;8:110785–110796. doi: 10.18632/oncotarget.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan J., Chen H., Kurgan L., Chen K., Kang C., Pu P. HuMiTar: a sequence-based method for prediction of human microRNA targets. Algorithms Mol. Biol. 2008;3:16. doi: 10.1186/1748-7188-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu S., Jia Z.F., Kang C., Huang Q., Wang G., Liu X., Zhou X., Xu P., Pu P. Upregulation of SEPT7 gene inhibits invasion of human glioma cells. Cancer Invest. 2010;28:248–258. doi: 10.3109/07357900903179609. [DOI] [PubMed] [Google Scholar]
- 22.Jiang R.C., Pu P.Y., Jiao B.H. The expression of CDC10 in gliomas. Chin. J. Clin. Neurosci. 2002;10:227–229. [Google Scholar]
- 23.Jiang R.C., Pu P.Y., Shen C.H. Preliminary study on cancer-related gene expression profiles in 63 cases of gliomas by cDNA array. Chin. J. Neurosurg. 2004;20:18–21. [Google Scholar]
- 24.Jia Z.F., Huang Q., Kang C.S., Yang W.D., Wang G.X., Yu S.Z., Jiang H., Pu P.Y. Overexpression of septin 7 suppresses glioma cell growth. J. Neurooncol. 2010;98:329–340. doi: 10.1007/s11060-009-0092-1. [DOI] [PubMed] [Google Scholar]
- 25.Fresno Vara J.A., Casado E., de Castro J., Cejas P., Belda-Iniesta C., González-Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004;30:193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Karin M., Cao Y., Greten F.R., Li Z.W. NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 27.Orlowski R.Z., Baldwin A.S., Jr. NF-κB as a therapeutic target in cancer. Trends Mol. Med. 2002;8:385–389. doi: 10.1016/s1471-4914(02)02375-4. [DOI] [PubMed] [Google Scholar]
- 28.Tilborghs S., Corthouts J., Verhoeven Y., Arias D., Rolfo C., Trinh X.B., van Dam P.A. The role of nuclear factor-kappa B signaling in human cervical cancer. Crit. Rev. Oncol. Hematol. 2017;120:141–150. doi: 10.1016/j.critrevonc.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Wu J.T., Kral J.G. The NF-κB/IκB signaling system: a molecular target in breast cancer therapy. J. Surg. Res. 2005;123:158–169. doi: 10.1016/j.jss.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Yu M., Qi B., Xiaoxiang W., Xu J., Liu X. Baicalein increases cisplatin sensitivity of A549 lung adenocarcinoma cells via PI3K/Akt/NF-κB pathway. Biomed. Pharmacother. 2017;90:677–685. doi: 10.1016/j.biopha.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 32.Berezikov E., van Tetering G., Verheul M., van de Belt J., van Laake L., Vos J., Verloop R., van de Wetering M., Guryev V., Takada S. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shirdel E.A., Xie W., Mak T.W., Jurisica I. NAViGaTing the micronome—using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS ONE. 2011;6:e17429. doi: 10.1371/journal.pone.0017429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner J.D., Williamson R., Almefty K.K., Nakaji P., Porter R., Tse V., Kalani M.Y. The many roles of microRNAs in brain tumor biology. Neurosurg. Focus. 2010;28:E3. doi: 10.3171/2009.10.FOCUS09207. [DOI] [PubMed] [Google Scholar]
- 35.Hummel R., Maurer J., Haier J. MicroRNAs in brain tumors : a new diagnostic and therapeutic perspective? Mol. Neurobiol. 2011;44:223–234. doi: 10.1007/s12035-011-8197-x. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y.S., Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y., Tao T., Liu N., Luan W., Qian J., Li R., Hu Q., Wei Y., Zhang J., You Y. PPARα, a predictor of patient survival in glioma, inhibits cell growth through the E2F1/miR-19a feedback loop. Oncotarget. 2016;7:84623–84633. doi: 10.18632/oncotarget.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q., Guo W., Zhang Y., Wu Y., Xiang J. miR-19a promotes cell proliferation and invasion by targeting RhoB in human glioma cells. Neurosci. Lett. 2016;628:161–166. doi: 10.1016/j.neulet.2016.06.031. [DOI] [PubMed] [Google Scholar]
- 39.Qin N., Tong G.F., Sun L.W., Xu X.L. Long noncoding RNA MEG3 suppresses glioma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of miR-19a. Oncol. Res. 2017;25:1471–1478. doi: 10.3727/096504017X14886689179993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laneve P., Po A., Favia A., Legnini I., Alfano V., Rea J., Di Carlo V., Bevilacqua V., Miele E., Mastronuzzi A. The long noncoding RNA linc-NeD125 controls the expression of medulloblastoma driver genes by microRNA sponge activity. Oncotarget. 2017;8:31003–31015. doi: 10.18632/oncotarget.16049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahlert U.D., Nikkhah G., Maciaczyk J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 2013;331:131–138. doi: 10.1016/j.canlet.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 42.Moustakas A., de Herreros A.G. Epithelial-mesenchymal transition in cancer. Mol. Oncol. 2017;11:715–717. doi: 10.1002/1878-0261.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu L., Chen G., Yang J., Ma Z., Yang Y., Hu Y., Lu Y., Cao Z., Wang Y., Wang X. Bone marrow mesenchymal stem cells suppress growth and promote the apoptosis of glioma U251 cells through downregulation of the PI3K/AKT signaling pathway. Biomed. Pharmacother. 2019;112:108625. doi: 10.1016/j.biopha.2019.108625. [DOI] [PubMed] [Google Scholar]
- 44.Xu W., Yang Z., Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adhes. Migr. 2015;9:317–324. doi: 10.1080/19336918.2015.1016686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikheev A.M., Mikheeva S.A., Severs L.J., Funk C.C., Huang L., McFaline-Figueroa J.L., Schwensen J., Trapnell C., Price N.D., Wong S., Rostomily R.C. Targeting TWIST1 through loss of function inhibits tumorigenicity of human glioblastoma. Mol. Oncol. 2018;12:1188–1202. doi: 10.1002/1878-0261.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., Liu Y., Ren Y., Kang L., Zhang L. Transmembrane protein with unknown function 16A overexpression promotes glioma formation through the nuclear factor-κB signaling pathway. Mol. Med. Rep. 2014;9:1068–1074. doi: 10.3892/mmr.2014.1888. [DOI] [PubMed] [Google Scholar]
- 47.Viatour P., Merville M.P., Bours V., Chariot A. Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 2005;30:43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y., Mayo M.W., Xiao A., Hall E.H., Amin E.B., Kadota K., Adusumilli P.S., Jones D.R. Loss of BRMS1 promotes a mesenchymal phenotype through NF-κB-dependent regulation of Twist1. Mol. Cell. Biol. 2015;35:303–317. doi: 10.1128/MCB.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prasad P., Vasas A., Hohmann J., Bishayee A., Sinha D. Cirsiliol suppressed epithelial to mesenchymal transition in B16F10 malignant melanoma cells through alteration of the PI3K/Akt/NF-κB signaling pathway. Int. J. Mol. Sci. 2019;20:608. doi: 10.3390/ijms20030608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antonelli M., Massimino M., Morra I., Garrè M.L., Gardiman M.P., Buttarelli F.R., Arcella A., Giangaspero F. Expression of pERK and pAKT in pediatric high grade astrocytomas: correlation with YKL40 and prognostic significance. Neuropathology. 2012;32:133–138. doi: 10.1111/j.1440-1789.2011.01252.x. [DOI] [PubMed] [Google Scholar]
- 51.Emdad L., Janjic A., Alzubi M.A., Hu B., Santhekadur P.K., Menezes M.E., Shen X.N., Das S.K., Sarkar D., Fisher P.B. Suppression of miR-184 in malignant gliomas upregulates SND1 and promotes tumor aggressiveness. Neuro-oncol. 2015;17:419–429. doi: 10.1093/neuonc/nou220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.