Abstract
Circulating microRNAs (miRNAs) have been proposed as biomarkers for type 2 diabetes (T2D). Adipose tissue (AT), for which dysfunction is widely associated with T2D development, has been reported as a major source of circulating miRNAs. However, the role of dysfunctional AT in the altered pattern of circulating miRNAs associated with T2D onset remains unexplored. Herein, we investigated the relationship between T2D-associated circulating miRNAs and AT function, as well as the role of preadipocytes and adipocytes as secreting cells of candidate circulating miRNAs. Among the plasma miRNAs related to T2D onset in the CORonary Diet Intervention with Olive oil and cardiovascular PREVention (CORDIOPREV) cohort, baseline miR-223-3p levels (diminished in patients who next developed T2D [incident-T2D]) were significantly related to AT insulin resistance (IR). Baseline serum from incident-T2D participants induced inflammation and IR in 3T3-L1 adipocytes. We demonstrated that tumor necrosis factor (TNF)-α inhibited miR-223-3p secretion while enhancing miR-223-3p intracellular accumulation in 3T3-L1 (pre)adipocytes. Overexpression studies showed that an intracellular increase of miR-223-3p impaired glucose and lipid metabolism in these cells. Our findings provide mechanistic insights into the alteration of circulating miRNAs preceding T2D, unveiling both preadipocytes and adipocytes as miR-223-3p-secreting cells and suggesting that inflammation promotes miR-223-3p intracellular accumulation, which might contribute to (pre)adipocyte dysfunction and body metabolic dysregulation.
Keywords: adipose tissue, adipocyte, circulating microRNAs, inflammation, miR-223-3p, preadipocytes, type 2 diabetes
Graphical Abstract
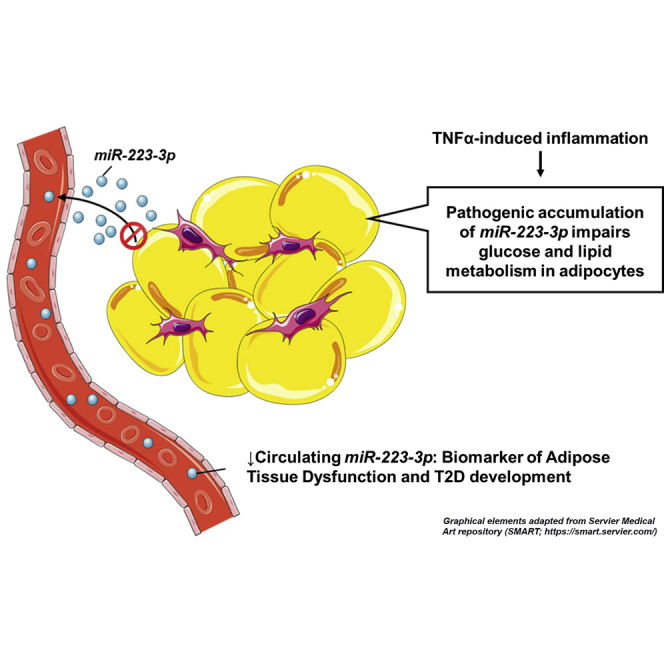
The relationship between adipose dysfunction and circulating miRNAs associated with T2D onset was investigated. Low circulating miR-223-3p related to adipose tissue insulin resistance index (ATIRI) in incident-T2D subjects. Both preadipocytes and adipocytes secrete miR-223-3p, and inflammation-induced intracellular miR-223-3p accumulation leads to cell dysfunction, which might contribute to body metabolic dysregulation.
Introduction
Type 2 diabetes (T2D) mellitus prevalence has increased during the last years, and it has been estimated that in the next decade the number of cases will double if no action is taken.1 Therefore, it is critical to identify the early mechanisms that precede T2D onset in order to design targeted preventive therapeutic strategies.
MicroRNAs (miRNAs), a class of small non-coding RNAs that post-transcriptionally regulate gene expression, have emerged as key molecules for cell function.2 miRNAs recognize specific target sites in mRNA sequences and mediate gene-expression silencing.2 Anomalous miRNA levels and alterations in miRNA biogenesis machinery have been related to a number of metabolic diseases, including T2D, obesity, and dyslipidemia.2, 3, 4, 5
miRNAs are also actively secreted into the circulation and have been proposed to act as messengers for intercellular communication.3,6 This characteristic has pointed out cell-free circulating miRNAs as potential biomarkers for disease, and altered circulating levels of numerous miRNAs have been associated with metabolic disorders.4,7, 8, 9
Recently, we assessed the predictive value of a number of miRNAs for the diagnosis of T2D incidence in the CORonary Diet Intervention with Olive oil and cardiovascular PREVention (CORDIOPREV) study (ClinicalTrials.gov: NCT00924937) and demonstrated that when combined with glycosylated hemoglobin (HbA1c), a group of nine miRNAs (miR-9, miR-28-3p, miR-29a, miR-30a-5p, miR-103, miR-126, miR-150, miR-223-3p, and miR-375) provided a higher predictive value in the T2D diagnosis than clinical parameters (fasting glucose, 2h glucose of oral glucose tolerance test [OGTT], and HbA1c).10 The relationship between baseline levels of these miRNAs with markers of beta cell function and systemic and peripheral insulin resistance (IR) was also investigated. However, their potential association with adipose tissue (AT) deregulation was not analyzed. This is of interest, as AT dysfunction has been proposed as a major contributing factor for the development of T2D, and the AT miRNA expression profile is altered in obesity and T2D.5 Indeed, miRNAs are involved in the regulation of essential AT processes, such as adipogenesis.11 Previous studies have also demonstrated that AT is a major physiological source of circulating miRNAs.6 In this line, several cross-sectional human studies have shown that the obesity-related expression pattern of specific circulating miRNAs reflects their miRNA AT expression profiles, supporting a role for circulating miRNAs as AT biomarkers.12,13 However, the relationship between circulating miRNAs and AT functional state is not fully understood, yet it might be useful to identify AT-related metabolic complications.
In this scenario, we aimed at analyzing the relationship between previously established predictive miRNAs for T2D onset in the CORDIOPREV cohort10 and the loss of AT insulin sensitivity. Once established that circulating miR-223-3p was dysregulated in relation to AT function, we performed functional analysis to elucidate both the potential of pre- and adipocytes as miR-223-3p-secreting cells and the consequences of miR-223-3p dysregulation on adipocyte biology. Our results indicate that miR-223-3p secretion by preadipocytes and adipocytes is prevented under inflammatory conditions, which may underlie the role for this miRNA in adipocyte physiology and as a potential predictor of adipose dysfunction related to T2D development.
Results
Circulating miRNA levels and AT IR
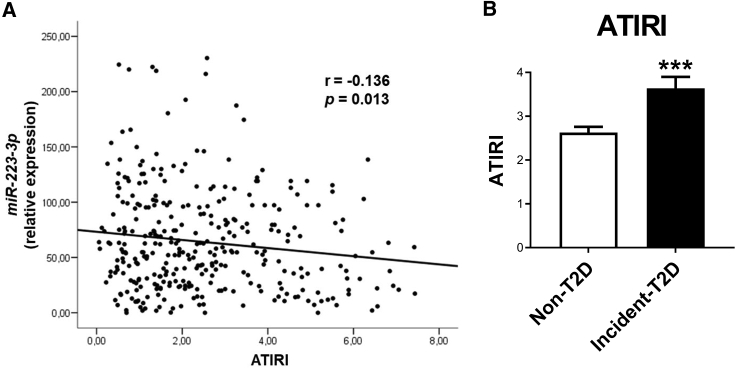
We first explored the correlation between plasma levels of the 9 miRNAs previously associated with the incidence of T2D in the CORDIOPREV study (miR-9, miR-28-3p, miR-29a, miR-30a-5p, miR-103, miR-126, miR-150, miR-223-3p, and miR-375),10 and AT IR index (ATIRI). None of the miRNAs, except baseline plasma miR-223-3p levels, which were 28.2% lower in incident-T2D subjects than in non-T2D subjects (p = 0.016),10 exhibited a significant and negative correlation with ATIRI (Figure 1A; Table S1). The relationship between ATIRI and miRNA plasma levels and clinical and anthropometric data was further examined by the Akaike Information Criterion (AIC) multivariate regression analysis with the stepAIC method.14,15 This analysis indicated that the best model (i.e., with the lowest AIC value) to explain ATIRI included miR-223-3p plasma levels together with gender, body mass index (BMI), glucose, and triglycerides (Table S2), supporting the role of miR-223-3p. ATIRI was also significantly higher in incident-T2D subjects (Figure 1B). These results suggested that miR-223-3p might be related to AT dysfunction.
Figure 1.
Analysis of miR-223-3p plasma levels and ATIRI in the CORDIOPREV-DIAB cohort
Correlation analysis between baseline circulating miR-223-3p expression levels and adipose tissue insulin resistance index (ATIRI) determined in CORDIOPREV-DIAB study patients (n = 462). (A) r, Pearson’s correlation coefficientBaseline ATIRI in patients from the CORDIOPREV-DIAB study who did not develop T2D (non-T2D; n = 355) and patients who developed T2D (incident-T2D; n = 107) during the median follow-up of 60 months. (B) Values are mean ± SEM, ∗∗∗p < 0.001 versus non-T2D. See also Tables S1 and S2.
In silico analysis of miR-223-3p target genes and pathways
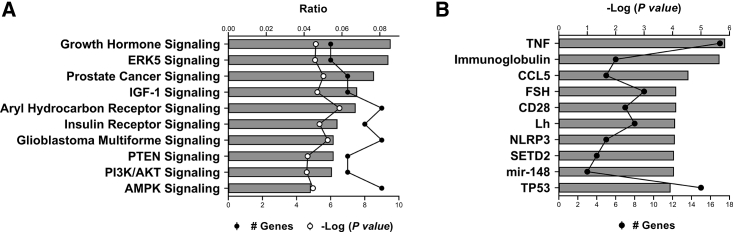
Given the relationship between plasma miR-223-3p and the marker of AT insulin responsiveness, ATIRI, we next examined its potential target genes to elucidate its putative involvement in pathways related to AT function. In silico analysis of the top 50 genes in four different databases (miRDB, microRNA, TargetScan, and miRTarBase) enabled the identification of 157 target genes of miR-223-3p, 50 of which have been experimentally validated, including GLUT4 and FOXO, and 107 were predicted in silico (Table S3), such as IRS-1. In this line, among the 166 significantly over-represented pathways revealed by Ingenuity Pathway Analysis (IPA) software analysis (Table S3), insulin receptor signaling and phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathways were ranked in the top 10 most significant over-represented pathways (Figure 2A).
Figure 2.
In silico analysis of miR-223-3p target genes
(A and B) The top 10 most significantly enriched canonical pathways (A) and upstream regulators (B) of the miR-223-3p target genes set according to Ingenuity Pathway Analysis (IPA). Bars indicate ratio (A) and −log (p value of overlap) (B), respectively. Black lines indicate number of proteins, and white line indicates −log (p value). See also Table S3.
IPA analysis also revealed tumor necrosis factor (TNF)-α as the second most significant upstream regulator of miR-223-3p target genes (Figure 2B; Table S3). Indeed, multiple over-represented pathways in the miR-223-3p regulatory network were related to inflammation (Table S3).
Exposure of adipocytes to baseline serum from non- and incident-T2D patients
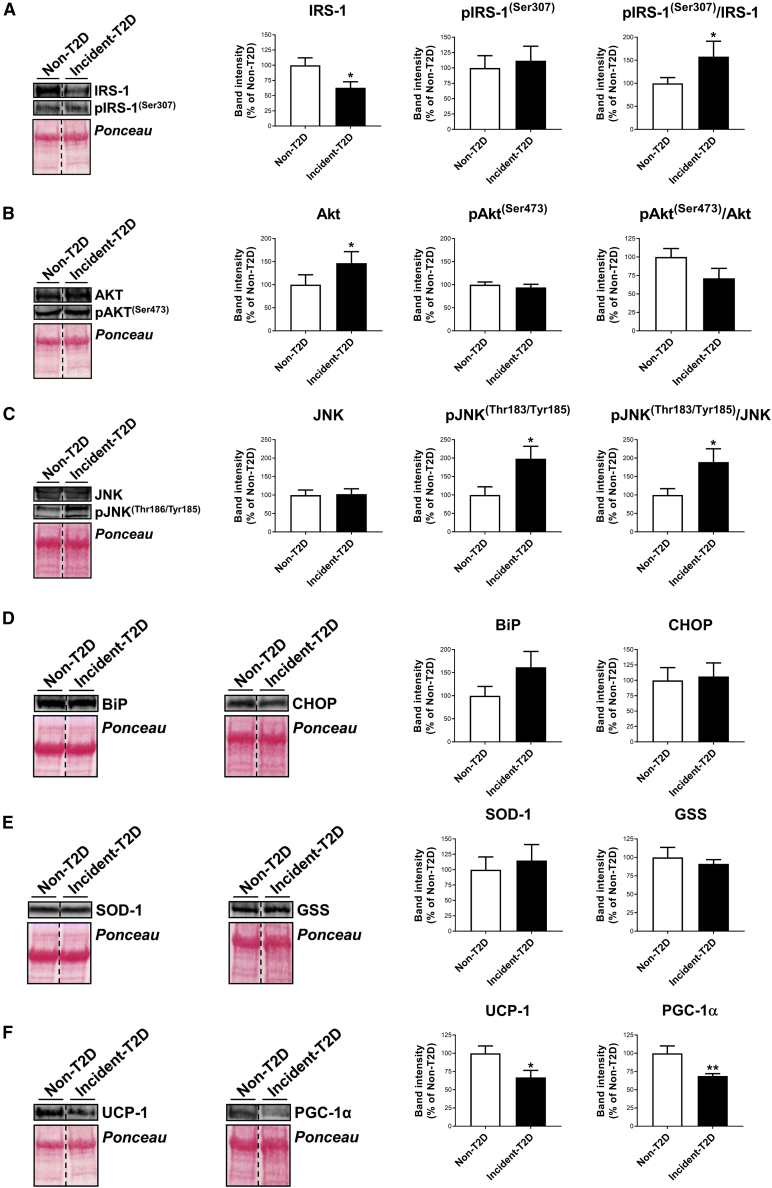
In order to investigate the influence of circulating factors on AT function previous to T2D onset, we next assessed the response of adipocytes upon exposure to sera obtained from a subgroup of non-T2D and incident-T2D subjects from the CORDIOPREV-Diabetes (DIAB) cohort (n = 32 for each group) at baseline, when differences in the biochemical and anthropometric profiles have not yet become apparent (Table S4). These experiments showed significant reduced protein content of the insulin signaling mediator, IRS-1, in 3T3-L1 adipocytes treated with incident-T2D sera as compared to non-T2D sera (Figure 3A). Akt content was also altered in the former group (Figure 3B). Notably, cells exposed to incident-T2D sera exhibited significantly higher levels of the active (i.e., phosphorylated [p]) form of the inflammatory marker, JNK, which resulted in a higher pJNK/JNK ratio (Figure 3C). No effects were observed in relation to markers of endoplasmic reticulum (ER) stress (BIP, CHOP) or oxidative stress (SOD-1, GSS) (Figures 3D and 3E), yet a decrease in mitochondrial markers (UCP-1, PGC-1α) was observed after treatment with incident-T2D versus non-T2D serum (Figure 3F).
Figure 3.
Adipocyte response to in vitro treatment with baseline serum from non- and incident-T2D patients
(A–F) Representative blots and quantification of IRS-1 and pIRS-1(Ser307), Akt and pAkt(Ser473), JNK and pJNK(Thr183/Tyr185), and their corresponding ratios (A−C); BiP and CHOP (D); SOD-1 and GSS (E); and UCP-1 and PGC-1α (F) in 3T3-L1 cells at day 6 of differentiation upon 24 h of exposure to 10% baseline serum from non-T2D patients (n = 32) and incident-T2D (n = 32) during the median follow-up of 60 months. Values are mean ± SEM, ∗p < 0.05 and ∗∗p < 0.01 versus non-T2D. See also Table S4.
Regulation of miR-223-3p expression and secretion in adipocytes
Given the association between AT IR and low circulating miR-223-3p levels and the previous observations from Thomou et al.,6 demonstrating that AT is a major source of circulating miRNAs, we explored the ability of adipocytes to store and release miR-223-3p at different stages of differentiation. Likewise, based on the relationship observed in silico between miR-223-3p and inflammatory pathways and the proven link between AT dysfunction and inflammation,16 we also examined miR-223-3p secretory dynamics in adipocytes challenged with TNF-α.
miR-223-3p expression and secretory profiles during adipogenesis
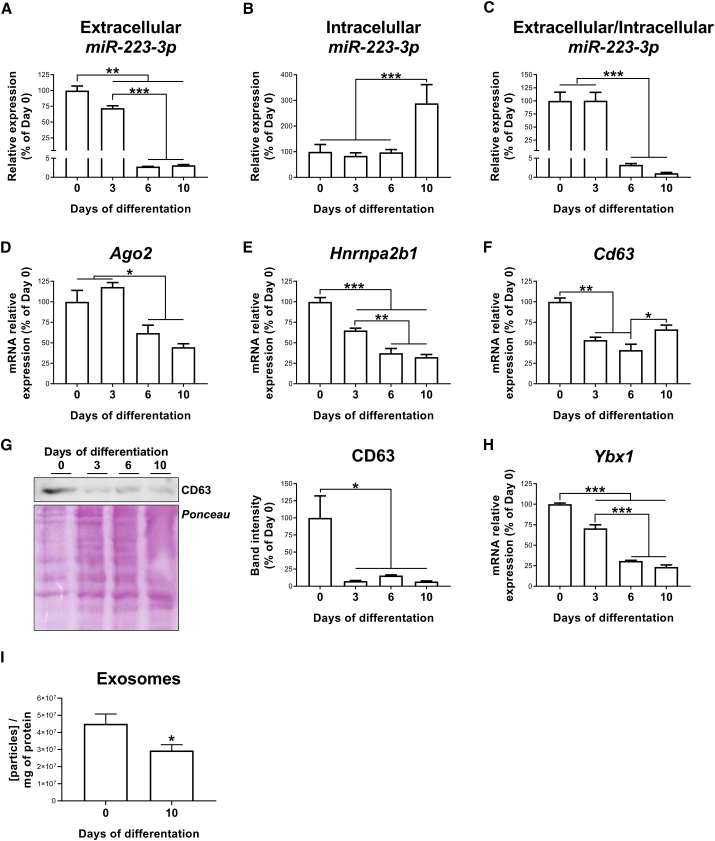
miR-223-3p expression, as a measure of intracellular miR-223-3p content, was investigated in 3T3-L1 cells at different days (D) of differentiation (D0, D3, D6, and D10). The amount of miR-223-3p released by these cells into the culture media in the previous 24-h period was also evaluated. As shown in Figure 4A, miR-223-3p released into the culture media was significantly higher for preadipocytes (D0−D3) than for adipocytes at later stages of differentiation (D6−D10). Intracellular miR-223-3p levels were 2.9-, 3.4-, and 2.9-fold higher in fully differentiated adipocytes than in adipose cells at D0, D3, and D6, respectively (Figure 4B). These results indicated that preadipocytes primarily secrete miR-223-3p rather than keeping it intracellularly, whereas adipocytes preferentially retain this miRNA intracellularly, as illustrated by the miR-223-3p extracellular/intracellular ratio (Figure 4C).
Figure 4.
Regulation of miR-223-3p, miRNA-exosome machinery, and exosome production during 3T3-L1 adipocyte differentiation
(A–C) Extracellular (A) and intracellular (B) levels of miR-223-3p were measured in the culture media and in the cell extracts, respectively, and the miR-223-3p extracellular/intracellular ratio was calculated during 3T3-L1 differentiation (C). (D–F) mRNA expression of Ago2 (D), Hnrnpa2b1 (E), and Cd63 (F) during 3T3-L1 differentiation. (G) Representative immunoblotting and quantification of CD63 during 3T3-L1 differentiation. (H) mRNA expression of Ybx1 during 3T3-L1 differentiation. (I) Exosome concentration in cell culture supernatant from 3T3-L1 cells at day 0 or 10 of differentiation (I). Values are given as mean ± SEM (n = 6), ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 versus previous days of differentiation. See also Figure S1.
In keeping with these results, we observed a decline in the mRNA and/or protein content of Ago-2 (involved in miRNA processing), Hnrnpa2b1 (involved in miRNA sorting into exosomes), and the exosome marker, CD63, during 3T3-L1 cell differentiation (Figures 4D−4G). Furthermore, the mRNA levels of Ybx1, which specifically participates in miR-223-3p sorting into exosomes,2,17,18 also decreased during adipogenesis (Figure 4H).
Extracellular vesicles (EVs), secreted by 3T3-L1 cells at D0 or D10 of differentiation, were isolated from cell culture medium by serial ultracentrifugation. The purified particles were examined by transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and immunoblotting (Figure S1). Consistent with other reports on exosomes,19,20 these analyses revealed that EVs released by 3T3-L1 cells displayed a typical exosome morphology (i.e., cup-shaped and rounded vesicles) and size (∼100 nm) (Figures S1A and S1B). A higher abundance of CD63 was found in the exosomal preparation in comparison with the total cell lysate (Figure S1C), thus validating the procedure employed for exosome isolation. In accordance with our expression data, exosome production was significantly lower at D10 compared to D0 (Figure 4I).
miR-223-3p expression and secretory profiles in response to inflammation
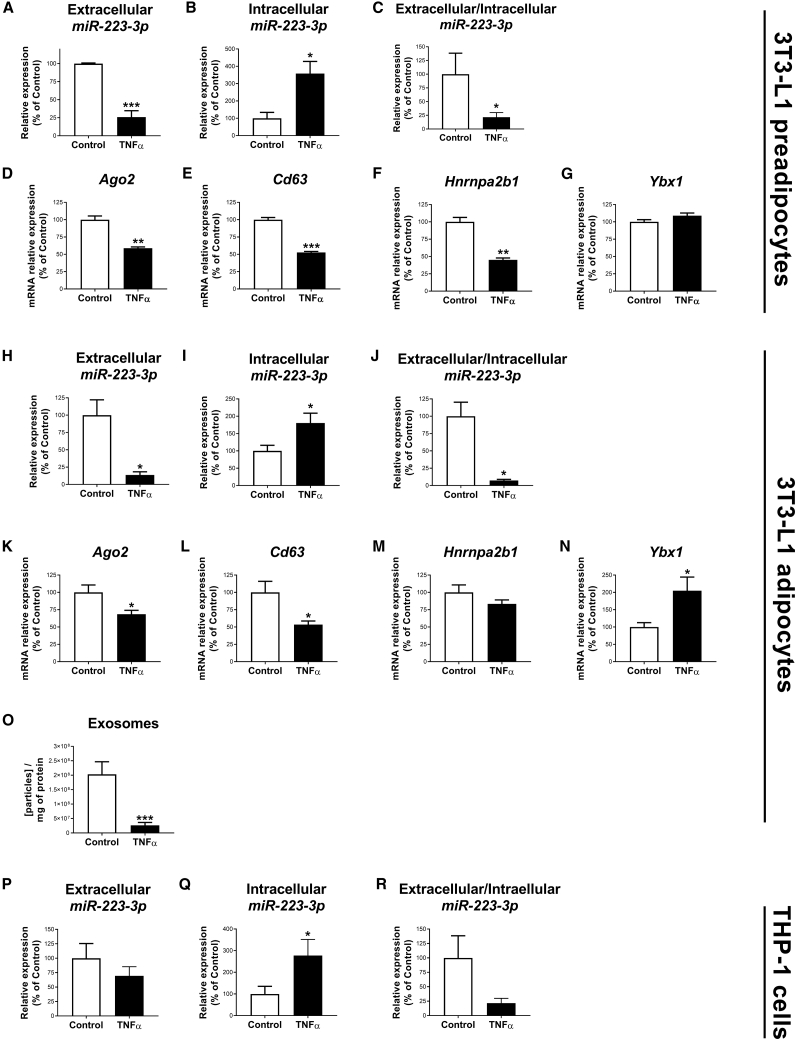
We next explored the influence of inflammation on miR-223-3p secretion by exposing 3T3-L1 adipocytes to TNF-α. The effects of TNF-α on inflammation and insulin signaling were validated by the increase in the pJNK/JNK ratio (Figure S2A) and the decrease in insulin-induced stimulation of Akt activation (i.e., pAkt/Akt ratio), respectively (Figure S2B). TNF-α significantly reduced miR-223-3p secretion while increasing miR-223-3p intracellular content, as compared to control condition when exposed to either preadipocytes or adipocytes (Figures 5A−5C and 5H−5J, respectively). Furthermore, Ago2, Cd63, and Hnrnpa2b1 mRNA expression levels were reduced by TNF-α (Figures 5D−5F for preadipocytes; and Figures 5K−5M for adipocytes), whereas Ybx1 mRNA levels were not changed (preadipocytes) or significantly increased (adipocytes) compared to the control condition (Figures 5G and 5N). Accordingly, exosome concentration was significantly lower in the culture supernatant from 3T3-L1 adipocytes exposed to TNF-α than controls (Figure 5O). Due to the relevant role of macrophages on AT inflammatory homeostasis and the proposed anti-inflammatory role of miR-223-3p,21, 22, 23, 24, 25 we also analyzed the miR-223-3p response to TNF-α in THP-1 macrophages. However, exposure of these cells to TNF-α, which promotes the expression of classic proinflammatory markers (Figure S3), did increase miR-223-3p intracellular content but not its secretion (Figures 5P−5R).
Figure 5.
Regulation of miR-223-3p upon TNF-α-induced insulin resistance in adipocytes
(A–C and H–J) Extracellular (A and H) and intracellular (B and I) levels of miR-223-3p measured in the culture media and in the cell extracts, respectively, and miR-223-3p extracellular/intracellular ratio in control or TNF-α-treated 3T3-L1 preadipocytes and adipocytes, respectively (C and J). (D–N) mRNA expression of Ago2 (D and K), Cd63 (E and L), Hnrnpa2b1 (F and M), and Ybx1 (G and N) in control or TNF-α-treated 3T3-L1 preadipocytes (D−G) or adipocytes (K−N). (O) Concentration of exosomes in cell culture supernatant from 3T3-L1 cells upon 24 h of treatment with 5 nM TNF-α or vehicle (control). (P–R) Extracellular (P) and intracellular (Q) levels of miR-223-3p measured in the culture media and in the cell extracts, respectively, and miR-223-3p extracellular/intracellular ratio (R) in control or TNF-α-treated THP-1 cells. Values are given as mean ± SEM (n = 6), ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 versus control. See also Figures S1−S3.
Effects of miR-223-3p overexpression on glucose metabolism in 3T3-L1 adipocytes
To address whether intracellular miR-223-3p accumulation might have consequences on adipose cell function, we overexpressed miR-223-3p in 3T3-L1 adipocytes at different stages of differentiation using a miR-223-3p mimic (Figure 6, Figure 7, Figure 8). Transfection efficiency was validated as shown in Figure S4, confirming that these cells accumulate miR-223-3p intracellularly.
Figure 6.
Effect of miR-223-3p overexpression on glucose metabolism in adipocytes at an early stage of differentiation
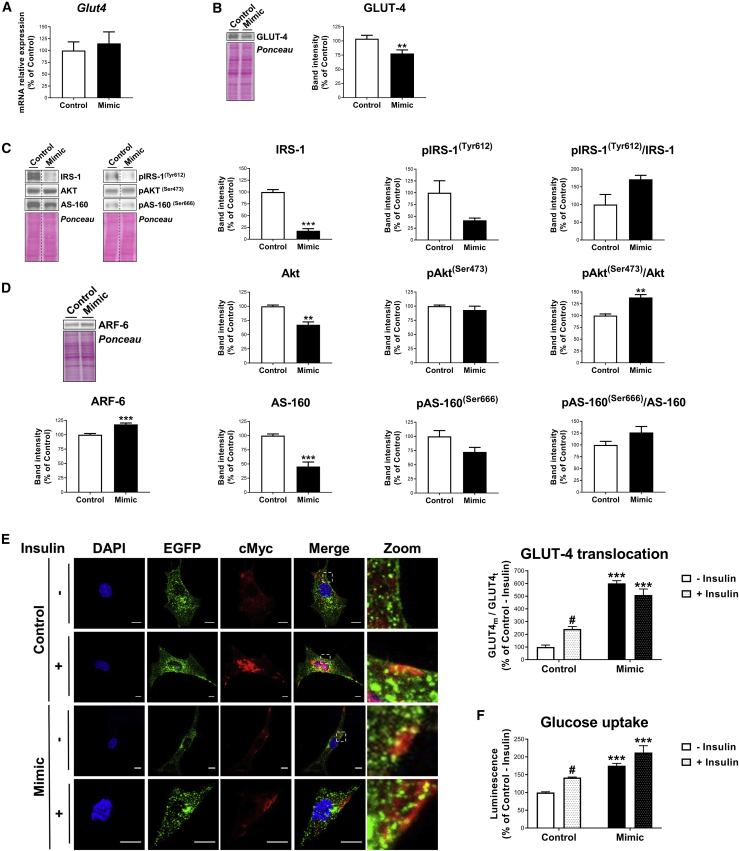
(A–D) mRNA expression of Glut4 (A); representative blot and quantification of GLUT4 (B); representative blots and quantification of IRS-1, pIRS-1(Tyr608), Akt, pAkt(Ser473), AS-160, pAS-160(Ser666), and their respective ratios (C); and representative blot and quantification of ARF6 (D) in 3T3-L1 cells transfected with negative miRNA-control (control) or miR-223-3p mimic (mimic) at day 3 of differentiation. (E and F) Representative confocal micrographs and quantification of GLUT4 translocation (E) and glucose uptake (F) of cells transfected with negative miRNA-control (control) or miR-223-3p mimic (mimic) at day 3 of differentiation, stimulated or not with insulin (100 nM, 20 min). Cells were co-transfected with cMyc-GLUT4-EGFP expression plasmid and immunostained with anti-cMyc antibody, showing total GLUT4 (GLUT4t) in green and membrane-associated GLUT4 (GLUT4m) in red. Nuclei were stained with DAPI (blue). Scale bars, 10 μm. Values are given as mean ± SEM (n = 6), ∗∗p < 0.01 and ∗∗∗p < 0.001 versus negative miRNA control; #p < 0.05 versus noninsulin-stimulated cells. See also Figures S4A−S4C.
Figure 7.
Effect of miR-223-3p overexpression on glucose metabolism in adipocytes at a late stage of differentiation
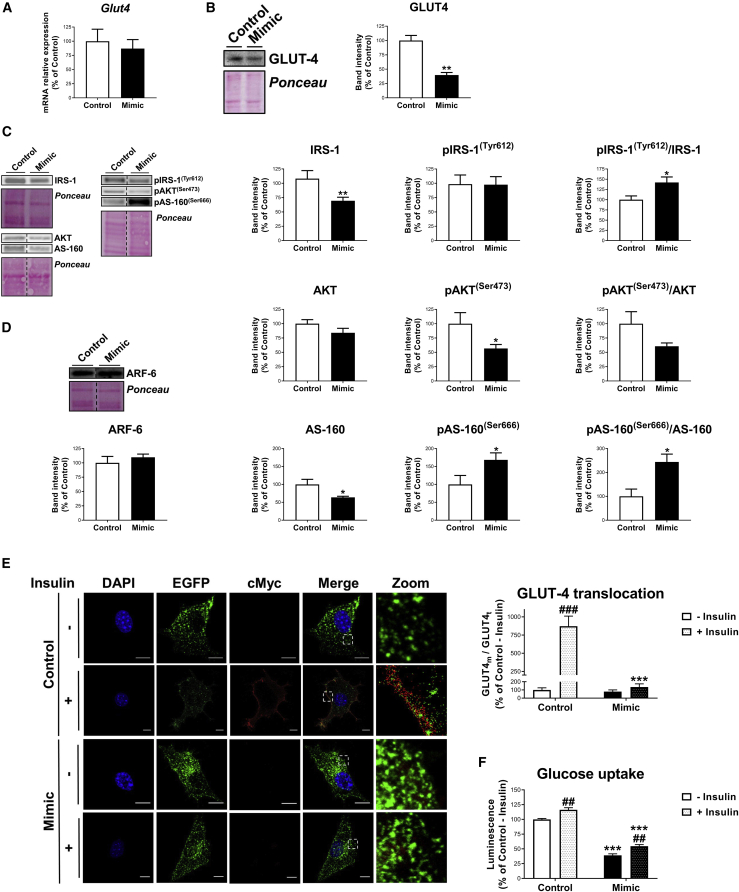
(A–D) mRNA expression of Glut4 (A); representative blot and quantification of GLUT4 (B); representative blots and quantification of IRS-1, pIRS-1(Tyr608), Akt, pAkt(Ser473), AS-160, pAS-160(Ser666), and their respective ratios (C); and representative blot and quantification of ARF6 (D) in 3T3-L1 cells transfected with negative miRNA-control (control) or miR-223-3p mimic (mimic) at day 6 of differentiation. (E and F) Representative confocal micrographs and quantification of GLUT4 translocation (E) and glucose uptake (F) of cells transfected with negative miRNA-control (control) or miR-223-3p mimic (mimic) at day 6 of differentiation, stimulated or not with insulin (100 nM, 20 min). Cells were co-transfected with cMyc-GLUT4-EGFP expression plasmid and immunostained with anti-cMyc antibody, showing GLUT4t in green and GLUT4m in red. Nuclei were stained with DAPI (blue). Scale bars, 10 μm. Values are given as mean ± SEM (n = 6), ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 versus negative miRNA control; ##p < 0.01 and ###p < 0.001 versus noninsulin-stimulated cells. See also Figures S4D and S4E.
Figure 8.
Effects of miR-223-3p overexpression on lipid metabolism in adipocytes at an early and a late stage of differentiation
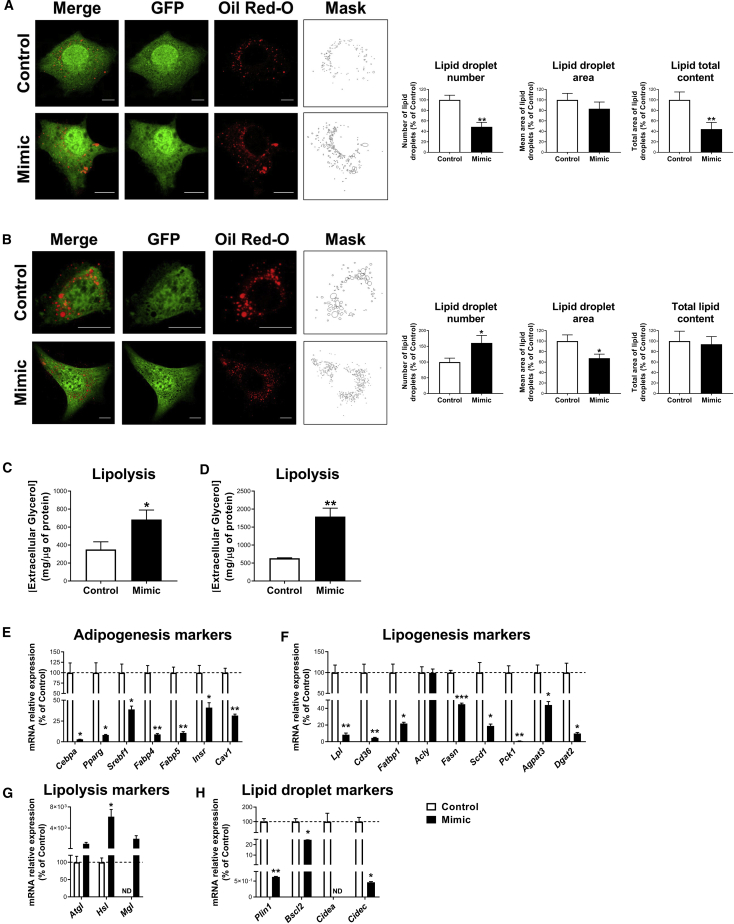
(A and B) Representative confocal micrographs and lipid droplet morphological analysis in 3T3-L1 cells transfected with negative miRNA-control (control) or miR-223-3p mimic (mimic) at day 3 (A) and 6 (B) of differentiation. Cells were co-transfected with phrGFP-N1 encoding GFP alone (green) to validate transfection and lipid droplets stained with Oil Red O. Scale bars, 10 μm. (C and D) Lipolysis rate, determined by extracellular glycerol content, of 3T3-L1 cells transfected with negative miRNA-control (control) or miR-223-3p mimic (mimic) at day 3 (C) and 6 (D) of differentiation. (E–H) mRNA levels of genes related to adipogenesis (E), lipogenesis (F), lipolysis (G), and lipid droplet biogenesis and growth (H) in 3T3-L1 cells transfected with negative miRNA-control (control) or miR-223-3p mimic (mimic) at day 3 of differentiation. Values are given as mean ± SEM (n = 6), ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 versus control. See also Figure S4.
3T3-L1 cells transfected with miR-223-3p mimic at D3 of differentiation exhibited similar mRNA levels but lower protein content of the miR-223-3p target, GLUT4, than control cells (Figures 6A and 6B). miR-223-3p mimic-overexpressing cells also exhibited significant decreases of several insulin-signaling mediators, including IRS-1, Akt, and AS-160 protein content, as compared to their control counterparts (Figure 6C). In contrast, a significant increase in the protein content of ARF6, which has been involved in GLUT4 incorporation into intracellular GLUT4 storage vesicles,26 was observed in cells overexpressing miR-223-3p (Figure 6D).
To evaluate whether miR-223-3p may also affect GLUT4 translocation to the plasma membrane, we employed a cMyc-GLUT4-EGFP expression vector, which was co-transfected with the miR-223-3p mimic or negative miRNA control. Quantification of intracellular and plasma membrane-associated GLUT4 in confocal micrographs revealed that 3T3-L1 cells overexpressing miR-223-3p exhibited more GLUT4 located in the plasma membrane than control adipocytes under both basal and insulin-stimulated conditions. In contrast to that observed in control cells, plasma membrane-associated GLUT4 levels in miR-223-3p-overexpressing cells were similar in the absence or presence of insulin (Figure 6E). In all groups, glucose uptake values changed in parallel to GLUT4 translocation to the plasma membrane (Figure 6F).
The effects of miR-223-3p overexpression were also examined in 3T3-L1 adipocytes at later stages of differentiation (D6) (Figure 7). 3T3-L1 cells at D6 overexpressing miR-223-3p also exhibited decreased GLUT4 protein levels but similar GLUT4 mRNA levels (Figures 7A and 7B). There were also significant decreases in IRS-1 and AS-160 protein content, as well as in Akt phosphorylation levels (Figure 7C), whereas ARF6 levels remained unchanged (Figure 7D). Regarding GLUT4 translocation, no differences were observed in basal values between 3T3-L1 adipocytes overexpressing miR-223-3p mimic and control cells (Figure 7E). However, insulin-induced GLUT4 translocation was reduced by 6.4-fold in adipocytes overexpressing miR-223-3p (Figure 7E). Functional studies showed that both basal and insulin-stimulated glucose uptake was lower in adipocytes expressing miR-223-3p mimic than in control cells (Figure 7F).
Effects of miR-223-3p overexpression on lipid metabolism in 3T3-L1 adipocytes
Lipid accumulation assessed by quantification of Oil Red O staining in confocal micrographs revealed lower lipid content in cells transfected at D3 with miR-223-3p mimic than control cells, which was attributable to a reduction in lipid droplet (LD) number (Figure 8A). When transfected at D6, 3T3-L1 adipocytes, expressing miR-223-3p mimic, contained more but smaller LDs than control cells, so that no changes in total lipid content were observed as a consequence of miR-223-3p overexpression (Figure 8B). Finally, 3T3-L1 cells, overexpressing miR-223-3p at either early or late differentiation stages, exhibited enhanced lipolytic rates (measured as glycerol release into the medium) compared to their respective control cells (Figures 8C and 8D). These observations were in agreement with a significantly reduced mRNA content of several markers of adipogenesis, lipogenesis, and LD biogenesis and growth, and the increase of lipolysis-related genes in these cells (Figures 8E−8H).
Discussion
Herein, we provide experimental evidence supporting that AT dysfunction precedes T2D onset and that decreased circulating miR-223-3p may represent a potential biomarker for this event. More specifically, low circulating miR-223-3p levels have been associated with compromised AT insulin sensitivity previous to T2D onset. Furthermore, decreased miR-223-3p secretion concomitant with its intracellular accumulation, as occurs under inflammatory conditions, has pathological effects on both preadipocyte and adipocyte physiology, which may have further consequences on systemic metabolic homeostasis.
We10 and others7,27,28 have previously reported an association between plasma levels of various miRNA and T2D incidence. However, despite the increasing number of studies reporting circulating miRNAs as metabolic biomarkers, the cause and physiological consequences of the alterations in these circulating miRNAs have been poorly explored. This knowledge might be of relevance to elucidate the underlying mechanisms of T2D development and to establish novel therapeutic strategies. In this scenario, AT is especially relevant due to its role as a major contributor of circulating miRNAs, as stated by foregoing reports,6,12,13 and as a regulator of whole-body metabolic homeostasis.5,29 In this line, we specifically found that low circulating miR-223-3p levels (previously associated with T2D incidence10) are related to impaired AT insulin sensitivity, placing circulating miR-223-3p as a potential biomarker for AT functionality.
Concordant with this hypothesis, our in vitro analyses revealed that adipocytes secrete miR-223-3p, especially at early stages of differentiation, whereas an inflammatory stimulus hampering insulin signaling, i.e., TNF-α, prevented adipocyte miR-223-3p secretion, likely due to the dysregulation of general miRNA processing and exosome production. This is noteworthy since inflammation is a hallmark of dysfunctional AT that has been widely proposed as a central mechanism leading to metabolic disturbances.30,31 Notably, inflammation was triggered and insulin signaling cascade altered in adipocytes upon exposure to baseline sera collected from the patients who were going to develop T2D in the next 5 years. In view of these results, it is tempting to speculate that, unlike patients who did not develope T2D, a proinflammatory environment present before the establishment of T2D would favor AT IR and would impair miR-223-3p secretion. This is in accordance to previous reports supporting that AT dysregulation may precede the development of metabolic complications.32 Therefore, low circulating miR-223-3p levels may not only represent a marker for T2D onset, as previously reported,10 but also for AT IR. In addition, our results raised the question as to whether an inflamed AT with diminished miR-223-3p secretion capacity might contribute to the decreased levels of circulating miR-223-3p. Taking into consideration the ubiquitous expression of this miRNA, the possible contribution of metabolic organs, other than the AT, to the circulating miR-223-3p pool should also be assessed in future studies. Likewise, other miRNAs, among other factors produced by the AT, might be involved in the metabolic dysregulation preceding T2D onset. Previous studies showed a correspondence between the AT expression of specific miRNAs and their circulating levels,12,13 but the relationship of these miRNAs and the functional state of AT were not explored. Apart from the pathogenic alteration of miR-223-3p secretion, the differences found in miR-223-3p dynamics in relation to adipocyte differentiation stage suggest that this miRNA may play distinct roles in preadipocytes and adipocytes. These differences might have physiological relevance and concur with the previously described distinct secretory profile, including EVs, of preadipocytes and adipocytes,33 as it has been confirmed in the present study.
Interestingly, intracellular miR-223-3p accumulation in adipocytes, rather than diminished miR-223-3p expression, accounted for the impaired miR-223-3p secretion by these cells during adipogenesis and in response to inflammation-induced IR. Although adipocyte miR-223-3p secretion has not been previously analyzed, several studies have explored intracellular miR-223-3p expression in adipocytes and AT. In agreement with our results, an increase in miR-223-3p expression during adipogenesis34,35 and after TNF-α treatment in adipocytes36 has been reported. Furthermore, higher AT miR-223-3p expression in IR obese patients compared to either lean or non-IR obese subjects21,36 and a decrease after bariatric surgery37 were described, supporting the notion of miR-223-3p retention in adipose cells during metabolic dysregulation.
Previous studies found that stromal vascular fraction (SVF), rather than mature adipocytes, accounted for most miR-223-3p expression in obese visceral AT.21 Taking this and the anti-inflammatory function attributed to this miRNA into consideration, a role for miR-223-3p in macrophages and AT inflammation has been proposed.3,21,24,25 In line with previous studies, herein we observed that, similar to that found in (pre)adipocytes, THP-1 macrophages release miR-223-3p, suggesting that adipose macrophages might also contribute to the circulating miR-223-3p pool. Nevertheless, TNF-α exposure did not affect miR-223-3p secretion but increased its accumulation inside these cells, further supporting the previously proposed role for this miRNA in inflammation-related processes.21, 22, 23, 24, 25 By contrast, the consequences of miR-223-3p dysregulation on adipocyte differentiation and function have been scarcely explored.34, 35, 36 However, previous findings34, 35, 36 and our results suggest a relevant regulatory role of this miRNA on AT physiology beyond the already-reported role in modulating AT immunity.3,21,24,25 Therefore, we also aimed at carrying out a comprehensive analysis of the effect of intracellular miR-223-3p accumulation on preadipocyte and adipocyte function by overexpressing miR-223-3p.
We experimentally demonstrated that miR-223-3p overexpression impaired the expression and activation of insulin signaling mediators at both early and late stages of adipocyte differentiation. Nonetheless, the consequences of insulin signaling disruption, determined by glucose uptake and GLUT4 translocation, differed depending on the differentiation stage. Early adipocytes exhibited constitutive insulin-independent glucose uptake and GLUT4 translocation to the plasma membrane. This could be construed as a compensatory mechanism mediated by the upregulation of the GLUT4 recycling marker, ARF6, to fulfil the concomitant-diminished GLUT4 protein content. In keeping with this, increased insulin-independent glucose uptake has been previously associated with decreased insulin responsiveness in adipose cells.38,39 In accordance to previous reports,36 this contrasts the reduced glucose uptake, either basal or insulin stimulated, by late adipocytes upon miR-223-3p overexpression. Our results indicate that this phenomenon is likely due to impaired insulin signaling and reduced presence of GLUT4 at the plasma membrane, which is in line with the reduced glucose uptake seen in AT from diabetic humans.40 The apparent contradictory findings relative to adipocyte differentiation stage could be accounted for by the demonstrated increase in insulin-independent glucose uptake at earlier stages of adipogenesis that declines during adipogenic differentiation.41 This suggests that despite the fact that GLUT4 is a validated miR-223-3p target gene, cell type-specific miR-223-3p regulatory networks can be triggered with disparate effects on GLUT4 expression and activity.
Importantly, proper adipocyte lipid storage into LDs is crucial for maintaining metabolic homeostasis, and defects on AT lipogenesis have been related to metabolic disorders.30,42 Thus, we also examined the effect of miR-223-3p on lipid metabolism, finding a reduction in both the expression of markers of adipogenesis and lipogenesis and LD biogenesis and growth in early adipocytes, which was consistent with their reduced number of LDs. This suggests that early miR-223-3p overexpression during adipogenesis may prevent adipocyte differentiation due to an impaired capacity of these cells for accumulating lipids, which represents a hallmark of AT-associated metabolic disorders.43 LD size was reduced by miR-223-3p overexpression in late differentiated adipocytes. These changes were concomitant to an increased lipolytic rate regardless of the differentiation stage. Both AT inability to store lipids and increased lipolysis are associated with ectopic lipid storage and lipotoxic effects on other metabolic tissues and are considered risk factors for IR and T2D.44 Whether the effects of miR-223-3p on lipid turnover are directly mediated or secondary to the effects on GLUT4 translocation and glucose uptake, required for de novo lipogenesis, remains to be clarified.
To sum up, our in vitro analyses have shown, for the first time, that miR-223-3p has an effect not only on GLUT4 protein expression in differentiated adipocytes36 but also globally affects early and late adipocyte physiology by altering both glucose and lipid metabolism. Our results suggest that pathogenic intracellular miR-223-3p accumulation impairs adipogenesis and energy storage in differentiated adipocytes. This could have detrimental consequences for metabolic health, as it has been proposed that it is the AT expandability, rather than AT size, which led to AT dysfunction.30,45 This hypothesis explains the existence of lean diabetic patients and so-called metabolically healthy obese subjects. Within the context of our cohort study, this could explain why some individuals developed T2D, whereas others remained healthy after a median follow-up of 60 months.
In conclusion, we have undertaken a comprehensive study of the relationship between circulating and AT miR-223-3p in the context of T2D predisposition. Our results indicate that AT IR may precede T2D development. Within this context, we identified that adipocyte miR-223-3p release was prevented under pathological conditions (i.e., inflammation), which might contribute to the lower circulating miR-223-3p levels observed in patients who are going to develop T2D. These results are in line with the previously proposed notion of AT as an important source of circulating miRNAs and pave the way to future studies on miRNA release from this organ in the field of metabolic diseases. We also reported that the resulting pathological accumulation of miR-223-3p in preadipocytes and adipocytes under inflammatory conditions compromises cellular function. miR-223-3p overexpression not only inhibited GLUT4 and insulin-stimulated glucose uptake in these cells, but also led to impaired insulin signaling, GLUT4 trafficking, and lipid metabolism. With the consideration of the major role of adipocyte dysfunction in the development of multiple metabolic diseases, these results unveil adipose miR-223-3p as a relevant player in tissue dysfunction and consequently, in the associated establishment of T2D. Altogether, our results provide novel molecular and cellular cues underlying the development of metabolic disturbances that may be helpful to design novel therapeutic strategies to prevent T2D.
Materials and methods
Patients
Participants of this study belonged to the CORDIOPREV study (ClinicalTrials.gov: NCT00924937). Detailed study design and biochemical and anthropometric characteristics of participants have been published elsewhere.46 Briefly, the CORDIOPREV study is a prospective study carried out in 1,002 patients with coronary heart disease and high cardiovascular risk. Patients who had their last coronary event more than 6 months previously to the inclusion, aged between 20 and 75 years old, and without other serious diseases or a life expectancy of less than 5 years were included. Subjects were randomized into two different dietary models (Mediterranean and low-fat diets). For the present study, those patients without diagnosed T2D at baseline (n = 462) were included (CORDIOPREV-DIAB).10,47,48 Patients were classified according to T2D incidence (American Diabetes Association [ADA] diagnosis criteria)49 during the median follow-up of 60 months in the non-T2D group (participants who did not develop T2D during the follow-up; n = 355) and the incident-T2D group (participants who developed T2D during the follow-up; n = 107). As reported previously, no statistical differences were found in the demographic and metabolic characteristics between dietary interventions in the CORDIOPREV-DIAB cohort (chi-square test = 1.948; p = 0.163).
For in vitro experiments, serum samples from a subcohort (n = 64) of the CORDIOPREV-DIAB study were used as detailed below, and biochemical and anthropometric characteristics from this subcohort are shown in Table S4.
Written consent was obtained from all of the subjects prior to recruitment, and the study protocol and all amendments were approved by the Ethics Committee of Reina Sofia University Hospital, all of which follow the Helsinki Declaration and good clinical practices.
Biochemical and anthropometric measurements
Venous blood from the participants was collected in tubes containing EDTA after a 12-h overnight fast. Anthropometric (BMI, waist circumference) and biochemical variables (total cholesterol, high-density lipoprotein [HDL] cholesterol, low-density lipoprotein [LDL] cholesterol, triglyceride, nonesterified fatty acid [NEFA], apoA1, apoB, glucose, HbA1c, and insulin levels), as well as homeostatic model assessment for IR (HOMA-IR), were assessed, as previously described.10 The high-sensitivity C-reactive protein (hs-CRP) was determined by high-sensitivity ELISA (BioCheck, San Francisco, CA, USA). AT IR index (ATIRI) was determined according to the following formula: ATIRI = fasting plasma NEFA (mM) × fasting plasma insulin (pmol/L), which has been proposed as a suitable and useful method in clinical practice to estimate AT insulin sensitivity.50
Quantification of circulating miRNAs by quantitative real-time PCR
miRNAs were isolated using the miRNeasy Mini Kit (QIAGEN, Hilden, Germany) from plasma collected at baseline following the manufacturer’s guidelines. RNA retrotranscription using the TaqMan MicroRNA Reverse Transcription Kit (Life Technologies, Carlsbad, CA, USA), preamplification using the TaqMan PreAmp Master Mix (Life Technologies, Carlsbad, CA, USA), and quantitative real-time PCR using the OpenArray Platform (Life Technologies, Carlsbad, CA, USA) were performed as previously detailed.10 Specifically, we investigated the nine miRNAs (miR-9, miR-28-3p, miR-29a, miR-30a-5p, miR-103, miR-126, miR-150, miR-223-3p, and miR-375), which were included in the predictive model for T2D previously established by us.10 miR-143 and miR-144 were selected as housekeeping genes according to the NormFinder Bioinformatic tool and BestKeeper method.10,51, 52, 53 Relative expression was analyzed with OpenArray Real-Time qPCR Analysis Software (Life Technologies, Carlsbad, CA, USA).
Among the nine miRNAs analyzed, miR-223-3p was the only miRNA exhibiting significant association with ATIRI, and therefore, this miRNA was selected for further analyses.
In silico analysis of miR-223-3p target genes
miRDB (www.mirdb.org),54 microRNA (https://bigd.big.ac.cn/databasecommons/database/id/1426 ),55 and TargetScan (www.targetscan.org)56 database searches were performed in order to identify predicted miR-223-3p target genes. In addition, miRTarBase database (http://mirtarbase.cuhk.edu.cn/php/index.php)57 was employed for identification of previously validated miR-223-3p target genes. To increase the predictive power, the search of each database was limited to the 50 genes with the highest corresponding score value. Canonical pathway and upstream regulator prediction analyses were performed using IPA (software version [v.]43605602 by Ingenuity Systems [QIAGEN, Hilden, Germany]).58 The significance of the association between the dataset and each canonical pathway was measured in two ways: (1) a ratio of the number of target genes that mapped to the pathway divided by the total number of genes that mapped to the canonical pathway displayed, and (2) Fisher’s exact test was used to calculate a p value. Pathways with a false discovery rate (FDR) < 0.05 by the Benjamini-Hochberg method58 were considered statistically significant.
Upstream regulator analysis was also carried out based on the Ingenuity Knowledge Base. The significance of the prediction between the dataset and upstream regulators was measured in two ways: (1) the number of genes from our dataset that mapped to the upstream regulator target genes, and (2) p value computed based on significant overlap between dataset genes and known targets regulated by each upstream regulator.
3T3-L1 cell differentiation and experimental treatments
3T3-L1 cells were purchased from ATCC (Manassas, VA, USA). Cells were differentiated into adipocytes according to our previously established protocols.59 Cells were harvested at D0, D3, D6, and D10 of differentiation for mRNA, miRNA, and protein analyses, and culture supernatants collected for miR-223-3p quantification.
Exposure of 3T3-L1 adipocytes to baseline serum from non-T2D and incident-T2D patients
To evaluate whether circulating factors from those patients who are going to develop T2D may affect adipocyte function, 3T3-L1 adipocytes were exposed to serum collected at baseline from incident-T2D (n = 32) or from non-T2D (n = 32) patients of the CORDIOPREV-DIAB cohort, matched by BMI, waist circumference, and biochemical variables. Briefly, differentiated 3T3-L1 adipocytes cultured in DMEM containing 10% fetal bovine serum (FBS) were incubated for 2 h in medium alone and then treated for 24 h with medium containing 10% inactivated patients’ serum (incubated at 56°C, 30 min), as previously described.60 Subsequently, cells were collected for protein analyses. The clinical characteristics of patients are summarized in Table S4.
Inflammation-induced IR model in 3T3-L1 adipocytes
In order to evaluate the influence of inflammation on the regulation of miR-223-3p, 3T3-L1 adipocytes at D6 were subjected to a previously validated model of inflammation-induced IR based on a 24-h exposure to TNF-α (5 nM).61 Controls exposed to vehicle alone were included. To test insulin sensitivity, cells were exposed to 100 nM insulin or medium alone during 5 min.61 At the end of the experiments, cells were harvested for miRNA, mRNA, and protein analyses. Cell culture supernatants were collected for miR-223-3p measurement.
miR-223-3p overexpression studies in 3T3-L1 cells
Overexpression studies were carried out to assess the intracellular functions of miR-223-3p. To this end, 3T3-L1 cells at D3 were exposed to 30 nM miR-223-3p mimic (mirVana miRNA mimic, assay identification [ID] MC12301; Invitrogen, Carlsbad, CA, USA) or negative control (Pre-miR miRNA Precursor Negative Control #1; Ambion, Waltham, MA, USA) in the presence of Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s guidelines. Overexpression studies at later stages of differentiation (D6) were carried out by electroporation using 100 nM miR-223-3p mimic or negative control, according to our established protocols.59 miR-223-3p mimic overexpression was validated by quantitative real-time PCR, as detailed below, and by confocal imaging upon cotransfection with Cy3 Dye-labeled Pre-miR Negative Control #1 (Ambion, Waltham, MA, USA) under the same conditions (Figure S1). After treatments (48 h), cells were harvested for protein isolation, functional analyses (glucose uptake), and Oil Red O staining for confocal imaging and LD quantification. Cell culture supernatants were collected for lipolysis assessment.
For analysis of the effect of miR-223-3p on GLUT4 traffic, a cMyc-GLUT4-EGFP expression plasmid, kindly donated by Dr. J. A. Pessin (Albert Einstein College of Medicine, NY, USA), was employed. This plasmid encodes GLUT4 recombinant protein containing cMyc epitope tag in the first exofacial loop, as well as EGFP fused in-frame at the carboxy terminus.62 A mixture containing the cMyc-GLUT4-EGFP expression plasmid and miR-223-3p mimic or negative control was employed for co-expression studies in 3T3-L1 cells at D3 or D6 using the same procedures of those employed for single overexpression experiments. 48 h after transfection/electroporation, cells were exposed to 100 nM insulin or medium alone during 20 min and processed for evaluation of GLUT4 translocation by confocal microscopy as detailed below.
THP-1 cell culture
THP-1 cells were purchased from ATCC (Manassas, VA, USA) and grown in RPMI-1640 culture medium (Biowest, France) supplemented with FBS (10%), L-glutamine (2 mM), glucose (4,500 mg/L), and penicillin/streptomycin (1%) (Sigma-Aldrich, Spain), following the supplier’s recommendation. To induce macrophage differentiation, THP-1 cells were transferred to RPMI-1640 medium supplemented with FBS (3%), L-glutamine (2 mM), glucose (4,500 mg/L), and penicillin/streptomycin (1%); seeded in 12-well plates at 2 × 105 cells/mL; and treated with phorbol 12-myristate 13-acetate (PMA, 10 ng/mL) (Sigma-Aldrich) for 48 h. Differentiated macrophages were then exposed to TNF-α (5 nM) or vehicle for 24 h. At the end of the experiments, cells were harvested and cell culture supernatants collected for miR-223-3p determination.
Quantification of miRNA and mRNA in adipocyte cultures by quantitative real-time PCR
TRIzol Reagent (Sigma-Aldrich, St. Louis, MO, USA) was used for total RNA isolation, as detailed elsewhere.63 RNA was retrotranscribed into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) and quantitative real-time PCR performed using GoTaq qPCR Master Mix Kit (Promega, Madrid, Spain) on a LightCycler 480 System (Roche, Basilea, Switzerland), according to previous studies.60 Expression of genes of interest was normalized by the geometrical average of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), β-actin (ACTB), and hypoxanthine-guanine phosphoribosyltransferase (HPRT) using the BestKeeper tool.53 Primer details are depicted in Table S5.
miRNA isolation from 3T3-L1 adipocytes (intracellular) and culture supernatants (extracellular) was performed using miRNeasy kit (QIAGEN, Hilden, Germany). cDNA synthesis and preamplification were carried out, as described above, and miRNA levels were assessed by quantitative real-time PCR using TaqMan Universal PCR Master Mix II and the iQ5 Multicolor Real Time PCR Detection System (Bio-Rad, Hercules, CA, USA), according to the manufacturer’s guidelines. Commercially available and prevalidated TaqMan primer/probe sets for retrotranscription and quantitative real-time PCR were used (miRBase: mmu-miR-223-3p, assay 002295; Applied Biosystems, Foster City, CA, USA). miR-17-3p (miRBase: mmu-miR-17-3p, assay 002543; Applied Biosystems, Foster City, CA, USA) was used as a housekeeping control to normalize miR-223-3p relative expression based on previous studies.64 Fold changes compared with the housekeeping controls were determined by calculating 2−ΔCt, according to the manufacturer’s guidelines. Replicates and negative controls were included in each reaction.
Immunoblotting
Total protein from 3T3-L1 adipocytes was extracted using SDS-DTT buffer (62.5 mM Tris-HCl , 100 mM DTT , 2% SDS [Sigma-Aldrich, St. Louis, MO, USA], 20% glycerol [VWR International, Radnor, PA, USA], pH 7.6) at 65°C. Immunoblotting was performed as previously reported.61,65 Briefly, 30−50 μg of protein per sample was loaded into 4%–20% precasted SDS-PAGE gels and transferred to nitrocellulose membranes using the Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, CA, USA). Membranes were stained with Ponceau to ensure equal sample loading. Blots were incubated overnight at 4°C with specific antibodies (Table S6), and immunoreactive bands were visualized using the Clarity Enhanced Chemiluminescence (ECL) Western Blotting Substrate (Bio-Rad, Hercules, CA, USA). The intensity of the bands was quantified by densitometry using ImageJ v.1.50 software (NIH) and normalized by Ponceau staining.61
Functional assays
Glucose uptake upon insulin stimulation (100 nM, 1 h) by 3T3-L1 adipocytes was estimated by luminescence using the Glucose Uptake-Glo Assay (Promega, Madrid, Spain). To determine nonspecific glucose uptake, a subset of cells was exposed to 25 mM cytochalasin B (Sigma-Aldrich, St. Louis, MO, USA) for 5 min prior to insulin stimulation.
For lipolysis assessment, cell culture supernatants were collected at the end of the experiments, and free glycerol was measured using the Free Glycerol Determination Kit (Sigma-Aldrich, St. Louis, MO, USA). The fluorescent reagent Amplex UltraRed (Invitrogen, Carlsbad, CA, USA) was used to increase the sensitivity of lipolysis assessments. Flex Station 3 (BioNova Scientific, Madrid, Spain) was employed to measure both luminescence and fluorescence.
EV isolation
EVs were isolated from cell culture media of 3T3-L1 cells at D0 or D10 of differentiation and at D6 of differentiation upon exposure to TNF-α (5 nM) or control medium (24 h), as previously described.66 Serum-free culture media were employed to avoid contamination from FBS, and the culture media corresponding to a 24 h-period were collected and centrifuged at 300 × g for 10 min to remove cells in suspension. Thereafter, supernatants were centrifuged at 2,000 × g for 20 min and at 10,000 × g for 30 min to remove cell debris. Finally, supernatants were ultracentrifuged at 100,000 × g for 1 h to pellet EVs. All centrifugations were performed at 4°C. EV pellets were resuspended in 1 mL sterile PBS, stored at 4°C, and utilized within 1 week of isolation for subsequent analysis.
TEM
EVs were negatively stained as previously described.67 Isolated EVs were fixed in glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) (1%, v/v) for 30 min at room temperature. Then, 10 μL droplets of fixed, isolated EVs were placed onto 200 mesh copper grids with carbon film (Agar Scientific, Essex, UK) and allowed to air dry for 30 min. Grids were washed (2 × 2 min in Milli-Q water) before negative staining with 2% (v/v) uranyl acetate (Electron Microscopy Sciences) for 4 min. Surplus stain was washed (2 × 2 min in Milli-Q water), and grids were allowed to air dry before being examined in a Jeol JEM-1400 TEM (Jeol, Tokyo, Japan) with Gatan Microscope Suite software (Gatan, Pleasanton, CA, USA).
EV size and concentration analysis
Size and concentration distributions of EVs were determined using NTA, as described before.68 Briefly, NTA uses the light-scattering properties and Brownian motion of laser-illuminated particles in suspension to determine EV size and concentration. NTA was undertaken using the NanoSight NS300 configured with a 488-nm laser (Malvern Panalytical, Madrid, Spain) using NTA software v.3.3 in the facilities of the University Institute of Nanochemistry (IUNAN; University of Córdoba). Camera shutter speed was maintained at 30 ms. Samples were diluted in PBS to concentrations between 2 × 108 and 3 × 109 particles/mL. 60 s videos were recorded in replicates of 5 per sample with camera sensitivity and detection threshold set to 13 and 8, respectively. Temperature was monitored manually and ranged from 24°C to 26°C. EV concentration and distribution were normalized to the cell protein content as determined by the Bradford assay (Bio-Rad, Hercules, CA, USA) and expressed as [particles]/mg of protein.
Confocal microscopy
miR-223-3p mimic overexpression was validated by confocal imaging upon cotransfection with Cy3 Dye-labeled Pre-miR Negative Control #1 (Ambion, Waltham, MA, USA).
GLUT4 translocation was evaluated in 3T3-L1 cells overexpressing miR-223-3p mimic or control using the cMyc-GLUT4-EGFP expression plasmid as a reporter. Briefly, 3T3-L1 cells grown on glass coverslips were fixed in 4% paraformaldehyde (PFA), processed without using detergent to preserve membrane integrity, and labeled with primary anti-Myc antibody (1:500) (Bio-Rad, Hercules, CA, USA) and Alexa Fluor 594-conjugated secondary antibody (1:500) (Thermo Fisher Scientific, Waltham, MA, USA). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
For LD visualization, PFA 4% fixed cells were washed with 60% isopropanol and incubated with oil red O solution (Sigma-Aldrich, St. Louis, MO, USA) for 30 min.59 Expression of miR-223-3p mimic was detected by cotransfection with an expression vector encoding GFP (phrGFP-N1; Agilent Technologies, Santa Clara, CA, USA).
Samples were examined with a LSM 5 Exciter confocal microscope (Carl Zeiss, Oberkochen, Germany). After acquisition, images underwent a deconvolution process by the software Huygens Essential 2.4.4 (Scientific Volume Imaging, Hilversum, Netherlands). Image analysis was performed using ImageJ v.1.50 software (NIH, Bethesda, MD, USA) for GLUT4 distribution and LD morphometric analysis.65
Statistical analysis
Results are expressed as mean ± SEM. Kolmogorov-Smirnov or Shapiro-Wilk test of normality was carried out in order to determine the use of parametric or nonparametric tests for statistical assessment of the CORDIOPREV-DIAB or in vitro data, respectively. Comparisons of variables between the study groups were assessed by one-way ANOVA and between the different in vitro experimental conditions by unpaired two-tailed Student’s t test for those variables that showed a normal distribution or the Mann-Whitney U-test for those that did not follow a normal distribution. For multiple group comparison between the different experimental conditions, either one-way ANOVA followed by Tukey’s multiple comparisons post hoc test for parametric data or Kruskal-Wallis with Dunn’s multiple comparisons post hoc test for nonparametric data was performed. Pearson’s correlation tests were performed to analyze the associations between the study variables. The relationship of ATIRI with the nine miRNA plasma levels, clinical (glucose, total cholesterol, HDL cholesterol, triglycerides, LDL cholesterol, apoA, apoB, Lp(A), CRP, insulin, HbA1c, and HOMA-IR) and anthropometric (age, gender, BMI, and waist circumference) data, was further examined in the R packet Modern Applied Statistics with S (MASS) by the AIC multivariate regression analysis with the stepAIC method.14,15 For in vitro experiments, at least four replicates were obtained for each condition. Values were considered significant when the p value <0.05. Analyses were performed with SPSS 23.0 (SPSS Iberica, Madrid, Spain) or GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA).
Acknowledgments
The authors are grateful to the Andalusian Bioinformatics Platform (PAB) Centre located at the University of Málaga (Spain) for the IPA software. We also thank Dr. Gema García (Microscopy, Cytomics and Scientific Imaging Unit of the IMIBIC, Cordoba, Spain) for her help with confocal microscopy studies and Drs. Francisco J. Romero Salguero and Irene Humanes-Pérez and Dr. José A. González Reyes for their help with NTA and TEM studies, respectively. CIBEROBN is an initiative of ISCIII, Spain. This work was supported by Ministerio de Ciencia, Innovación y Universidades/FEDER (BFU2016-76711-R and BFU2017-90578-REDT to M.M.M., AGL2012/39615 to J.L.-M., and AGL2015-67896-P to J.L.-M. and A.C.); Consejería de Salud y Bienestar Social/Junta de Andalucía/FEDER (PI-0159-2016 to R.G.-R. and PI-0092-2017 to M.C.-P.); Plan Propio de Investigación de la Universidad de Córdoba 2019 (Mod 2.5 to R.G.-R.); and Instituto de Salud Carlos III (ISCIII)/FEDER (PIE14/00005 to J.L.-M. and M.M.M.). M.C.-P. was the recipient of a postdoctoral grant Juan de la Cierva Formación (FJCI-2017-32194) from the MICINN (Spain). This study was cofunded by European Regional Development Fund/European Social Fund “Investing in your future” and Consejería de Economía, Conocimiento, Empresas y Universidad/Junta de Andalucía/FEDER (BIO-0139).
Author contributions
M.M.M., J.L.-M., and R.G.-R. designed research. J.L.-M. coordinated the CORDIOPREV-DIAB study. J.S.-C., O.A.R., A.C., J.F.A.-D., and A.P.-H. performed research and analyzed data. M.M.M., J.S.-C., and M.C.-P. wrote the manuscript. R.G.-R., J.L.-M., O.A.R.-Z., A.C., and J.F.A.-D. reviewed/edited the manuscript. All of the authors discussed the results and the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2021.01.014.
Contributor Information
José López-Miranda, Email: md1lomij@uco.es.
María M. Malagón, Email: bc1mapom@uco.es.
Supplemental information
References
- 1.Ginter E., Simko V. Type 2 diabetes mellitus, pandemic in 21st century. Adv. Exp. Med. Biol. 2012;771:42–50. doi: 10.1007/978-1-4614-5441-0_6. [DOI] [PubMed] [Google Scholar]
- 2.Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mori M.A., Ludwig R.G., Garcia-Martin R., Brandão B.B., Kahn C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019;30:656–673. doi: 10.1016/j.cmet.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortega F.J., Mercader J.M., Moreno-Navarrete J.M., Rovira O., Guerra E., Esteve E., Xifra G., Martínez C., Ricart W., Rieusset J. Profiling of Circulating MicroRNAs Reveals Common MicroRNAs Linked to Type 2 Diabetes That Change With Insulin Sensitization. Diabetes Care. 2014;37:1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 5.Gentile A.M., Lhamyani S., Coín-Aragüez L., Clemente-Postigo M., Oliva Olivera W., Romero-Zerbo S.Y., García-Serrano S., García-Escobar E., Zayed H., Doblado E. miR-20b, miR-296, and Let-7f Expression in Human Adipose Tissue is Related to Obesity and Type 2 Diabetes. Obesity (Silver Spring) 2019;27:245–254. doi: 10.1002/oby.22363. [DOI] [PubMed] [Google Scholar]
- 6.Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C., Rao T.N., Winnay J.N., Garcia-Martin R., Grinspoon S.K. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zampetaki A., Kiechl S., Drozdov I., Willeit P., Mayr U., Prokopi M., Mayr A., Weger S., Oberhollenzer F., Bonora E. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 8.Jones A., Danielson K.M., Benton M.C., Ziegler O., Shah R., Stubbs R.S., Das S., Macartney-Coxson D. miRNA Signatures of Insulin Resistance in Obesity. Obesity (Silver Spring) 2017;25:1734–1744. doi: 10.1002/oby.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Rawaf H.A. Circulating microRNAs and adipokines as markers of metabolic syndrome in adolescents with obesity. Clin. Nutr. 2019;38:2231–2238. doi: 10.1016/j.clnu.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 10.Jiménez-Lucena R., Rangel-Zúñiga O.A., Alcalá-Díaz J.F., López-Moreno J., Roncero-Ramos I., Molina-Abril H., Yubero-Serrano E.M., Caballero-Villarraso J., Delgado-Lista J., Castaño J.P. Circulating miRNAs as Predictive Biomarkers of Type 2 Diabetes Mellitus Development in Coronary Heart Disease Patients from the CORDIOPREV Study. Mol. Ther. Nucleic Acids. 2018;12:146–157. doi: 10.1016/j.omtn.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arner P., Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol. 2015;11:276–288. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 12.Kim N.H., Ahn J., Choi Y.M., Son H.J., Choi W.H., Cho H.J., Yu J.H., Seo J.A., Jang Y.J., Jung C.H. Differential circulating and visceral fat microRNA expression of non-obese and obese subjects. Clin. Nutr. 2020;39:910–916. doi: 10.1016/j.clnu.2019.03.033. [DOI] [PubMed] [Google Scholar]
- 13.Heneghan H.M., Miller N., McAnena O.J., O’Brien T., Kerin M.J. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J. Clin. Endocrinol. Metab. 2011;96:E846–E850. doi: 10.1210/jc.2010-2701. [DOI] [PubMed] [Google Scholar]
- 14.Akaike H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974;19:716–723. [Google Scholar]
- 15.Heinze G., Wallisch C., Dunkler D. Variable selection - A review and recommendations for the practicing statistician. Biom. J. 2018;60:431–449. doi: 10.1002/bimj.201700067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longo M., Zatterale F., Naderi J., Parrillo L., Formisano P., Raciti G.A., Beguinot F., Miele C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019;20:2358. doi: 10.3390/ijms20092358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shurtleff M.J., Temoche-Diaz M.M., Karfilis K.V., Ri S., Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., Pérez-Hernández D., Vázquez J., Martin-Cofreces N., Martinez-Herrera D.J., Pascual-Montano A., Mittelbrunn M., Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colombo M., Raposo G., Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 20.Tkach M., Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 21.Deiuliis J.A., Syed R., Duggineni D., Rutsky J., Rengasamy P., Zhang J., Huang K., Needleman B., Mikami D., Perry K. Visceral Adipose MicroRNA 223 Is Upregulated in Human and Murine Obesity and Modulates the Inflammatory Phenotype of Macrophages. PLoS ONE. 2016;11:e0165962. doi: 10.1371/journal.pone.0165962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ismail N., Wang Y., Dakhlallah D., Moldovan L., Agarwal K., Batte K., Shah P., Wisler J., Eubank T.D., Tridandapani S. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121:984–995. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim G.D., Ng H.P., Patel N., Mahabeleshwar G.H. Kruppel-like factor 6 and miR-223 signaling axis regulates macrophage-mediated inflammation. FASEB J. 2019;33:10902–10915. doi: 10.1096/fj.201900867RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ying W., Tseng A., Chang R.C.-A., Morin A., Brehm T., Triff K., Nair V., Zhuang G., Song H., Kanameni S. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J. Clin. Invest. 2015;125:4149–4159. doi: 10.1172/JCI81656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang G., Meng C., Guo X., Cheruku P.S., Shi L., Xu H., Li H., Wang G., Evans A.R., Safe S. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125:2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 26.Leto D., Saltiel A.R. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat. Rev. Mol. Cell Biol. 2012;13:383–396. doi: 10.1038/nrm3351. [DOI] [PubMed] [Google Scholar]
- 27.Zhang T., Lv C., Li L., Chen S., Liu S., Wang C., Su B. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. BioMed Res. Int. 2013;2013:761617. doi: 10.1155/2013/761617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y., Gao G., Yang C., Zhou K., Shen B., Liang H., Jiang X. The role of circulating microRNA-126 (miR-126): a novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. Int. J. Mol. Sci. 2014;15:10567–10577. doi: 10.3390/ijms150610567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrier J.-F., Derghal A., Mounien L. MicroRNAs in Obesity and Related Metabolic Disorders. Cells. 2019;8:859. doi: 10.3390/cells8080859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghaben A.L., Scherer P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019;20:242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 31.Clemente-Postigo M., Oliva-Olivera W., Coin-Aragüez L., Ramos-Molina B., Giraldez-Perez R.M., Lhamyani S., Alcaide-Torres J., Perez-Martinez P., El Bekay R., Cardona F., Tinahones F.J. Metabolic endotoxemia promotes adipose dysfunction and inflammation in human obesity. Am. J. Physiol. Endocrinol. Metab. 2019;316:E319–E332. doi: 10.1152/ajpendo.00277.2018. [DOI] [PubMed] [Google Scholar]
- 32.Korf H., van der Merwe S. Adipose-derived exosomal MicroRNAs orchestrate gene regulation in the liver: Is this the missing link in nonalcoholic fatty liver disease? Hepatology. 2017;66:1689–1691. doi: 10.1002/hep.29343. [DOI] [PubMed] [Google Scholar]
- 33.Sarantopoulos C.N., Banyard D.A., Ziegler M.E., Sun B., Shaterian A., Widgerow A.D. Elucidating the Preadipocyte and Its Role in Adipocyte Formation: a Comprehensive Review. Stem Cell Rev. Rep. 2018;14:27–42. doi: 10.1007/s12015-017-9774-9. [DOI] [PubMed] [Google Scholar]
- 34.Qin L., Chen J., Tang L., Zuo T., Chen H., Gao R., Xu W. Significant Role of Dicer and miR-223 in Adipose Tissue of Polycystic Ovary Syndrome Patients. BioMed Res. Int. 2019;2019:9193236. doi: 10.1155/2019/9193236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan X., Gao Y., Zhou J., Wang J., Zheng F., Guo F., Chang A., Li X., Wang B. miR-223 Regulates Adipogenic and Osteogenic Differentiation of Mesenchymal Stem Cells Through a C/EBPs/miR-223/FGFR2 Regulatory Feedback Loop. Stem Cells. 2015;33:1589–1600. doi: 10.1002/stem.1947. [DOI] [PubMed] [Google Scholar]
- 36.Chuang T.-Y., Wu H.-L., Chen C.-C., Gamboa G.M., Layman L.C., Diamond M.P., Azziz R., Chen Y.-H. MicroRNA-223 Expression is Upregulated in Insulin Resistant Human Adipose Tissue. J. Diabetes Res. 2015;2015:943659. doi: 10.1155/2015/943659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macartney-Coxson D., Danielson K., Clapham J., Benton M.C., Johnston A., Jones A., Shaw O., Hagan R.D., Hoffman E.P., Hayes M. MicroRNA Profiling in Adipose Before and After Weight Loss Highlights the Role of miR-223-3p and the NLRP3 Inflammasome. Obesity (Silver Spring) 2020;28:570–580. doi: 10.1002/oby.22722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y.-H., Pratley R.E. The evolving role of inflammation in obesity and the metabolic syndrome. Curr. Diab. Rep. 2005;5:70–75. doi: 10.1007/s11892-005-0071-7. [DOI] [PubMed] [Google Scholar]
- 39.Talior I., Yarkoni M., Bashan N., Eldar-Finkelman H. Increased glucose uptake promotes oxidative stress and PKC-δ activation in adipocytes of obese, insulin-resistant mice. Am. J. Physiol. Endocrinol. Metab. 2003;285:E295–E302. doi: 10.1152/ajpendo.00044.2003. [DOI] [PubMed] [Google Scholar]
- 40.Ferrannini E., Iozzo P., Virtanen K.A., Honka M.-J., Bucci M., Nuutila P. Adipose tissue and skeletal muscle insulin-mediated glucose uptake in insulin resistance: role of blood flow and diabetes. Am. J. Clin. Nutr. 2018;108:749–758. doi: 10.1093/ajcn/nqy162. [DOI] [PubMed] [Google Scholar]
- 41.Hauner H., Röhrig K., Spelleken M., Liu L.S., Eckel J. Development of insulin-responsive glucose uptake and GLUT4 expression in differentiating human adipocyte precursor cells. Int. J. Obes. Relat. Metab. Disord. 1998;22:448–453. doi: 10.1038/sj.ijo.0800606. [DOI] [PubMed] [Google Scholar]
- 42.Ameer F., Scandiuzzi L., Hasnain S., Kalbacher H., Zaidi N. De novo lipogenesis in health and disease. Metabolism. 2014;63:895–902. doi: 10.1016/j.metabol.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Oliva-Olivera W., Coín-Aragüez L., Lhamyani S., Clemente-Postigo M., Torres J.A., Bernal-López M.R., El Bekay R., Tinahones F.J. Adipogenic Impairment of Adipose Tissue-Derived Mesenchymal Stem Cells in Subjects With Metabolic Syndrome: Possible Protective Role of FGF2. J. Clin. Endocrinol. Metab. 2017;102:478–487. doi: 10.1210/jc.2016-2256. [DOI] [PubMed] [Google Scholar]
- 44.Guilherme A., Virbasius J.V., Puri V., Czech M.P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virtue S., Vidal-Puig A. It’s not how fat you are, it’s what you do with it that counts. PLoS Biol. 2008;6:e237. doi: 10.1371/journal.pbio.0060237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delgado-Lista J., Perez-Martinez P., Garcia-Rios A., Alcala-Diaz J.F., Perez-Caballero A.I., Gomez-Delgado F., Fuentes F., Quintana-Navarro G., Lopez-Segura F., Ortiz-Morales A.M. CORonary Diet Intervention with Olive oil and cardiovascular PREVention study (the CORDIOPREV study): Rationale, methods, and baseline characteristics: A clinical trial comparing the efficacy of a Mediterranean diet rich in olive oil versus a low-fat diet on cardiovascular disease in coronary patients. Am. Heart J. 2016;177:42–50. doi: 10.1016/j.ahj.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camargo A., Jimenez-Lucena R., Alcala-Diaz J.F., Rangel-Zuñiga O.A., Garcia-Carpintero S., Lopez-Moreno J., Blanco-Rojo R., Delgado-Lista J., Perez-Martinez P., van Ommen B. Postprandial endotoxemia may influence the development of type 2 diabetes mellitus: From the CORDIOPREV study. Clin. Nutr. 2019;38:529–538. doi: 10.1016/j.clnu.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 48.Blanco-Rojo R., Alcala-Diaz J.F., Wopereis S., Perez-Martinez P., Quintana-Navarro G.M., Marin C., Ordovas J.M., van Ommen B., Perez-Jimenez F., Delgado-Lista J., Lopez-Miranda J. The insulin resistance phenotype (muscle or liver) interacts with the type of diet to determine changes in disposition index after 2 years of intervention: the CORDIOPREV-DIAB randomised clinical trial. Diabetologia. 2016;59:67–76. doi: 10.1007/s00125-015-3776-4. [DOI] [PubMed] [Google Scholar]
- 49.American Diabetes Association Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 2011;34(Suppl 1):S62–S69. doi: 10.2337/dc11-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ter Horst K.W., van Galen K.A., Gilijamse P.W., Hartstra A.V., de Groot P.F., van der Valk F.M., Ackermans M.T., Nieuwdorp M., Romijn J.A., Serlie M.J. Methods for quantifying adipose tissue insulin resistance in overweight/obese humans. Int. J. Obes. 2017;41:1288–1294. doi: 10.1038/ijo.2017.110. [DOI] [PubMed] [Google Scholar]
- 51.Danese E., Minicozzi A.M., Benati M., Paviati E., Lima-Oliveira G., Gusella M., Pasini F., Salvagno G.L., Montagnana M., Lippi G. Reference miRNAs for colorectal cancer: analysis and verification of current data. Sci. Rep. 2017;7:8413. doi: 10.1038/s41598-017-08784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Spiegelaere W., Dern-Wieloch J., Weigel R., Schumacher V., Schorle H., Nettersheim D., Bergmann M., Brehm R., Kliesch S., Vandekerckhove L., Fink C. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE. 2015;10:e0122515. doi: 10.1371/journal.pone.0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfaffl M.W., Tichopad A., Prgomet C., Neuvians T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 54.Liu W., Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20:18. doi: 10.1186/s13059-019-1629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:1–38. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pulido M.R., Diaz-Ruiz A., Jiménez-Gómez Y., Garcia-Navarro S., Gracia-Navarro F., Tinahones F., López-Miranda J., Frühbeck G., Vázquez-Martínez R., Malagón M.M. Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS ONE. 2011;6:e22931. doi: 10.1371/journal.pone.0022931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arias de la Rosa I., Escudero-Contreras A., Rodríguez-Cuenca S., Ruiz-Ponce M., Jiménez-Gómez Y., Ruiz-Limón P., Pérez-Sánchez C., Ábalos-Aguilera M.C., Cecchi I., Ortega R. Defective glucose and lipid metabolism in rheumatoid arthritis is determined by chronic inflammation in metabolic tissues. J. Intern. Med. 2018;284:61–77. doi: 10.1111/joim.12743. [DOI] [PubMed] [Google Scholar]
- 61.Díaz-Ruiz A., Guzmán-Ruiz R., Moreno N.R., García-Rios A., Delgado-Casado N., Membrives A., Túnez I., El Bekay R., Fernández-Real J.M., Tovar S. Proteasome Dysfunction Associated to Oxidative Stress and Proteotoxicity in Adipocytes Compromises Insulin Sensitivity in Human Obesity. Antioxid. Redox Signal. 2015;23:597–612. doi: 10.1089/ars.2014.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Procino G., Caces D.B., Valenti G., Pessin J.E. Adipocytes support cAMP-dependent translocation of aquaporin-2 from intracellular sites distinct from the insulin-responsive GLUT4 storage compartment. Am. J. Physiol. Renal Physiol. 2006;290:F985–F994. doi: 10.1152/ajprenal.00369.2005. [DOI] [PubMed] [Google Scholar]
- 63.Guzmán-Ruiz R., Ortega F., Rodríguez A., Vázquez-Martínez R., Díaz-Ruiz A., Garcia-Navarro S., Giralt M., Garcia-Rios A., Cobo-Padilla D., Tinahones F.J. Alarmin high-mobility group B1 (HMGB1) is regulated in human adipocytes in insulin resistance and influences insulin secretion in β-cells. Int. J. Obes. 2014;38:1545–1554. doi: 10.1038/ijo.2014.36. [DOI] [PubMed] [Google Scholar]
- 64.Andersen D.C., Jensen C.H., Schneider M., Nossent A.Y., Eskildsen T., Hansen J.L., Teisner B., Sheikh S.P. MicroRNA-15a fine-tunes the level of Delta-like 1 homolog (DLK1) in proliferating 3T3-L1 preadipocytes. Exp. Cell Res. 2010;316:1681–1691. doi: 10.1016/j.yexcr.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 65.Moreno-Castellanos N., Rodríguez A., Rabanal-Ruiz Y., Fernández-Vega A., López-Miranda J., Vázquez-Martínez R., Frühbeck G., Malagón M.M. The cytoskeletal protein septin 11 is associated with human obesity and is involved in adipocyte lipid storage and metabolism. Diabetologia. 2017;60:324–335. doi: 10.1007/s00125-016-4155-5. [DOI] [PubMed] [Google Scholar]
- 66.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006;Chapter 3 doi: 10.1002/0471143030.cb0322s30. Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 67.Cizmar P., Yuana Y. Detection and Characterization of Extracellular Vesicles by Transmission and Cryo-Transmission Electron Microscopy. Methods Mol. Biol. 2017;1660:221–232. doi: 10.1007/978-1-4939-7253-1_18. [DOI] [PubMed] [Google Scholar]
- 68.Mehdiani A., Maier A., Pinto A., Barth M., Akhyari P., Lichtenberg A. An innovative method for exosome quantification and size measurement. J. Vis. Exp. 2015:50974. doi: 10.3791/50974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.