Abstract
During virus infection in animals, the virus completes its life cycle in a host cell. A virus infection results in the metabolic deregulation of its host and leads to metabolic disorders, ultimately paving the way for cancer progression. Because metabolic disorders in virus infections occurring in animal are similar to metabolic disorders in human tumorigenesis, animal antiviral microRNAs (miRNAs), which maintain the metabolic homeostasis of animal cells, in essence, may have anti-tumor activity in humans. However, that issue has not been investigated. In this study, shrimp miR-34, a potential antiviral miRNA of shrimp against white spot syndrome virus (WSSV) infection, was identified. Overexpression of shrimp miR-34 in shrimp fed bacteria expressing miR-34 suppressed WSSV infection by targeting the viral wsv330 and wsv359 genes. Furthermore, the expression of shrimp miR-34 in mice fed miR-34-overexpressing shrimp suppressed breast cancer progression by targeting human CCND1, CDK6, CCNE2, E2F3, FOSL1, and MET genes. Therefore, our study suggests that the miRNAs in food could be an effective strategy for synchronously controlling viral diseases of economic animals and cancers in humans.
Keywords: shrimp miR-34, virus infection, breast cancer, food intake
Graphical Abstract

Food intake of shrimp miR-34, a small non-coding RNA, simultaneously suppresses virus infection in shrimp and human tumorigenesis in a cross-species manner, indicating that the intake of miRNAs in food might be an effective strategy for synchronously controlling viral diseases of economic animals and cancers in humans.
Introduction
It is well known that the life cycle of a virus ends in the host cell. Therefore, viral infections can cause metabolic disorders in host cells.1,2 As previously reported, virus replication is associated with enhanced cellular metabolism in its host because it depends on its host for the synthesis of viral genomes and viral proteins.3 In humans, hepatitis C virus (HCV) induces early interference of glycolysis, the pentose phosphate pathway, and the citric acid cycle, which is conducive to the biosynthetic activity of the host to support virus replication and reproduction.4 Drosophila C virus (DCV) infection disrupts cellular processes in Drosophila melanogaster, resulting in a decreased metabolic rate.5 In the aquaculture industry, shrimp is one of the main sources of revenue for many countries.6 During the past few decades, shrimp farming has rapidly grown on a global scale as a result of its high economic and nutritional values. However, since 1981, several new viral pathogens have emerged, increasing shrimp mortality.6 Among these viral pathogens, white spot syndrome virus (WSSV) threatens shrimp farming, and its outbreak severely limits the development of shrimp industry.7 WSSV contains a circular and double-stranded DNA genome that contains approximately 300-kb pairs and is one of the largest animal viruses fully sequenced.8 Because the WSSV genome is not homologous with any known virus, it is assigned to a new family (Nimaviridae) and genus (Whispovirus).9 Cancer is the main example of a common human disease with metabolic disorders.10,11 Changes in cell metabolism are a sign of cancer because this contributes to tumor initiation, growth, and maintenance.10 In this context, tumorigenesis and virus infection have some similarities in metabolic disorders.
Much evidence supports certain genes exerting regulatory effects on metabolism in normal cells. Once these genes are abnormally expressed, they lead to deregulated metabolic processes in cells, which cause metabolic disorders in the body.11,12 According to some reports, microRNAs (miRNAs) are endogenously expressed non-coding RNAs that have an important role in the regulation of gene expression. These miRNAs affect metabolic disorders associated with abnormal gene expression.13 Generally, an individual miRNA has multiple target genes,14,15 implying that a single miRNA may target various genes in different species. Shrimp miR-34, a homolog of human miR-34a, acts as a suppressor in breast cancer by targeting human genes in a cross-species manner.16 In essence, the antiviral activity of a molecule restores the metabolic disorder caused by the viral infection to a normal homeostasis. Therefore, antiviral miRNAs may have antitumor activity. Shrimp miR-S8 is a miRNA that only exists in shrimp and that can reduce the tumorigenic ability of melanoma stem cells by targeting the transcription factor YB-1.17 Shrimp, one of the most important human food sources of protein, are currently suffering diseases caused by viral infections. It is worth exploring whether miRNAs can bridge the control of shrimp diseases and human cancers.
In this study, overexpression of shrimp miR-34 through feeding promoted an antiviral immune response in shrimp. At the same time, the antiviral shrimp miR-34 exhibited anti-tumor activity in mice fed miR-34-overexpressing shrimp. Our findings showed that miRNA in food simultaneously controlled shrimp viral disease and human tumorigenesis.
Results
Control of shrimp viral disease by shrimp miR-34 in food
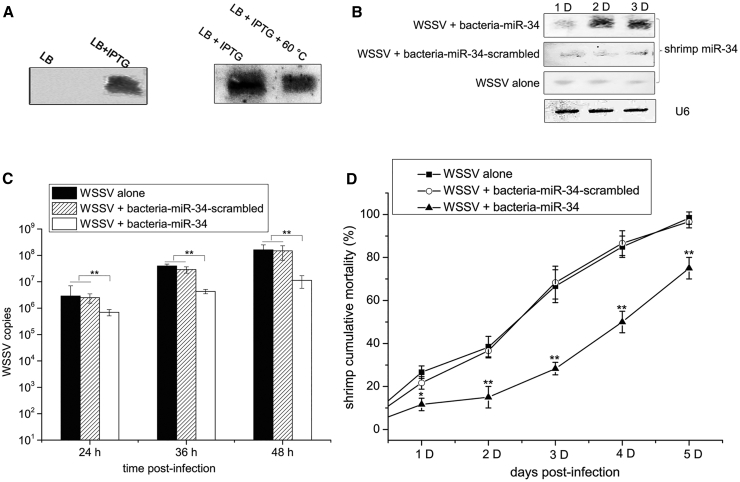
To control WSSV infection in shrimp by adding shrimp miR-34 to food, miR-34 was first expressed in bacteria, and then, shrimp were fed the recombinant bacteria expressing the miR-34 after an evaluation of the viral infection. Northern blot analysis indicated that mature shrimp miR-34 was expressed in isopropyl β-D-1-thiogalactopyranoside (IPTG)-induced recombinant bacteria (Figure 1A). To facilitate the preparation of the food, bacteria expressing mature miR-34 were heat-inactivated at 60°C. Northern blot data revealed that after heat inactivation, a sufficient amount of mature miR-34 was detected in the IPTG-induced recombinant bacteria (Figure 1B). The heat-inactivated bacteria expressing shrimp miR-34 (bacteria-miR-34) were mixed with the shrimp feed. Then, the shrimp were fed the mixed feed and were challenged with WSSV. The data indicated that mature miR-34 expression was significantly increased by feed containing bacteria-miR-34 compared with the controls (Figure 1C).
Figure 1.
Control of shrimp viral disease by miR-34 in food
(A) The expression of mature shrimp miR-34 in bacteria. The miR-34 construct was transformed into HT115 bacteria. The recombinant bacteria were cultured in Luria-Bertani (LB). Then, IPTG was used to induce the expression of miR-34 in bacteria. The expression level of miR-34 was detected by northern blot analysis. (B) The detection of mature shrimp miR-34 in heat-inactivated bacteria. The recombinant bacteria were cultured and induced by IPTG in LB medium and then heat inactivated at 60°C. Northern blot analysis was used to detect miR-34 expression levels. (C) The detection of mature shrimp miR-34 in shrimp fed shrimp feed containing miR-34. Shrimp were fed shrimp feed containing heat-inactivated bacteria expressing miR-34 (bacteria-miR-34), which was followed by challenge with WSSV. WSSV alone and WSSV + bacteria expressing the scrambled miR-34 (bacteria-miR-34-scrambled) were used as controls. At different days after feeding, shrimp muscle was subjected to northern blot analysis to detect miR-34 expression levels. (D) The influence of shrimp miR-34 on the virus copy number in shrimp. Shrimp were fed shrimp feed containing bacteria-miR-34 or bacteria-miR-34-scrambled and infected with WSSV (∗∗p < 0.01). (E) Shrimp mortality (∗∗p < 0.01).
The results showed that continuous high expression of miR-34 in shrimp led to significant decreases in WSSV copy number (Figure 1D) and virus-infected shrimp mortality (Figure 1E). However, WSSV copy number and the mortality of shrimp fed the feed containing bacteria-miR-34-scrambled were consistent with that of the control WSSV alone (Figures 1D and 1E). These data indicated that an elevated expression level of mature miR-34 in shrimp fed bacteria-expressing miR-34 promoted an antiviral immune response in shrimp in vivo.
Mechanism by which miR-34 defends shrimp against the WSSV infection
To explore the mechanisms behind miR-34 providing shrimp with antiviral immunity, targeted gene predictions were performed. The results indicated that two WSSV genes (wsv330 and wsv359) were targeted by shrimp miR-34 (Figure 2A). To examine the interaction between shrimp miR-34 and the WSSV genes, the 3′ UTRs of the wsv330 and wsv359 genes and their corresponding mutants were cloned as fused constructs with the EGFP gene (Figure 2B). Co-transfection of the synthesized miR-34 and EGFP-wsv330-3′ UTR construct or the EGFP-wsv359-3′ UTR construct led to a significant decrease in fluorescence intensity compared with the controls (Figures 2C and 2D). However, miR-34-scrambled did not affect EGFP expression (Figures 2C and 2D). These data indicated that shrimp miR-34 directly targeted the wsv330 and wsv359 genes.
Figure 2.
Mechanism of miR-34 in shrimp defending against WSSV infection
(A) The prediction of WSSV genes targeted by shrimp miR-34. The seed sequence of miR-34 was underlined. (B) Constructs of WSSV and EGFP genes. The underlined sequence represented that targeted by miR-34. (C) The direct interaction between shrimp miR-34 and virus genes in insect cells. Cells were co-transfected with miR-34 and the 3′ UTR of virus genes. At 48 h after co-transfection, fluorescence was examined. Scale bar, 100 μm. (D) The fluorescent intensities of cells. (E) The interaction between shrimp miR-34 and wsv330 and wsv359 genes in vivo. Shrimp were co-injected with WSSV and miR-34. At different post-infection times, the expression levels of wsv330 and wsv359 genes were evaluated by quantitative real-time PCR. As a control, miR-34-scrambled was included in the assays. (F) Western blot analysis of wsv330 /wsv359-encoded protein after shrimp were co-injected with WSSV and miR-34. (G) The evaluation of wsv330 and wsv359 silencing in vivo using northern blot analysis. Shrimp were injected with WSSV and wsv330-siRNA or wsv359-siRNA. WSSV alone and siRNA-scrambled were included in the injections. At different post-infection times, the expression of wsv330 and wsv359 was detected by northern blotting. Shrimp β-actin was used as a control. (H) The detection of wsv330 and wsv359 silencing in vivo by western blot analysis. (I) The effects of wsv330 and wsv359 silencing on virus infection in shrimp. The WSSV copy number in wsv330-silenced or wsv359-silenced shrimp was quantified by quantitative real-time PCR. WSSV alone and siRNA-scrambled were included in the injections as controls. (J) The influence of wsv330 and wsv359 silencing on WSSV-infected shrimp mortality. After silencing of wsv330 or wsv359, the cumulative mortality of WSSV-infected shrimp was monitored daily. (K) A model for miR-34-mediated antiviral activity in shrimp. Throughout, statistically significant differences among treatments are represented with asterisks: ∗p < 0.05, ∗∗p < 0.01.
To explore the interaction between shrimp miR-34 and its targets in vivo, miR-34 was overexpressed in shrimp, and the targeted gene expression was examined. Quantitative real-time PCR and western blots revealed that miR-34 overexpression resulted in a significant decrease in expression of wsv330 and wsv359 (Figures 2E and 2F). These findings suggested that viral wsv330 and wsv359 genes were targeted by shrimp miR-34.
To evaluate the roles of wsv330 and wsv359 in WSSV infection, expression of the two genes was repressed by gene-specific small interfering RNAs (siRNAs) (Figures 2G and 2H). The silencing of wsv330 or wsv359 led to significant decreases in WSSV copy number in shrimp (Figure 2I) and virus-infected shrimp mortality (Figure 2J) relative to the controls. These data demonstrated that wsv330 and wsv359 were required for WSSV infection in shrimp in vivo.
Taken together, these findings indicated that shrimp miR-34 directly targeted viral wsv330 and wsv359 genes to inhibit virus infection in shrimp, thereby leading to normal metabolism as represented by high survival rates and advantageous growth (Figure 2K).
Inhibitory effects on breast cancer progression by shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria
To evaluate the influence of shrimp miR-34 from shrimp fed bacteria expressing shrimp miR-34 (bacteria-miR-34) on cancer development, shrimp were cooked and the effects of shrimp miR-34 on the growth and metastasis of breast cancer cells were characterized. Northern blot analysis revealed that mature double-stranded miR-34 of cooked shrimp fed bacteria-miR-34 was detected at higher levels relative to the controls (shrimp injected with miR-34 and shrimp fed bacteria-miR-34) (Figure 3A), indicating that cooking did not destroy the mature miR-34 in shrimp because of the annealing of the double-stranded miR-34 at room temperature. Because shrimp were fed bacteria-miR-34 daily, shrimp miR-34 was continuously expressed in the shrimp. To explore the effects of residual miR-34 in shrimp on tumorigenesis, the miRNAs extracted from the muscles of shrimp fed the feed containing bacteria-miR-34 or bacteria-miR-34-scrambled, as well as shrimp fed only shrimp feed as a control, were transfected into MDA-MB-231 and MDA-MB-435 breast cancer cells. Northern blots revealed that shrimp miR-34 was expressed in cells transfected with the miRNAs from shrimp fed the feed containing bacteria-miR-34 compared with the controls (shrimp fed only shrimp feed and the negative control), whereas shrimp miR-34 levels in cells transfected with the miRNAs from shrimp fed feed containing bacteria-miR-34-scrambled was similar to group fed regular feed (Figure 3B). Mature miR-34 from the muscles of shrimp fed the feed containing bacteria-miR-34 significantly suppressed the growth and metastasis of breast cancer cells compared with the controls (Figures 3C and 3D), suggesting a suppressive effect on breast cancer progression from shrimp miR-34 in cooked shrimp fed miR-34-expressing bacteria.
Figure 3.
Inhibitory effects on breast cancer progression of shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria
(A) Northern blot analysis of mature shrimp miR-34 in shrimp. Shrimp were fed bacteria-miR-34. Two days later, shrimp were cooked in a microwave oven for 2 min. Shrimp injected with miR-34 were used as a control. Total RNA was extracted from shrimp muscle and subjected to northern blot analysis to detect miR-34 expression levels. U6 was used as a control. (B) Expression of shrimp miR-34 in breast cancer cells. Shrimp were fed shrimp feed alone or shrimp feed containing bacteria-miR-34 or bacteria-miR-34-scrambled for 2 days and then cooked. The miRNAs extracted from the cooked shrimp muscles were transfected into MDA-MB-231 or MDA-MB-435 cells. At 36 h after transfection, mature shrimp miR-34 level in cells was detected using northern blot. Cells without treatment served as a negative control. (C) Influence of shrimp miR-34 on breast cancer cell growth from cooked shrimp fed miR-34-expressing bacteria. A cell proliferation assay was performed with MDA-MB-231 or MDA-MB-435 cells transfected with miRNAs from muscle of cooked shrimp fed bacteria-miR-34. Shrimp fed shrimp feed alone and those fed shrimp feed containing bacteria-miR-34-scrambled were included in the assays. As a negative control, cells that did not receive treatment were also assayed. (D) Effects of shrimp miR-34 on breast cancer cell migration from cooked shrimp fed miR-34-expressing bacteria. A wound healing assay was performed to evaluate metastasis of MDA-MB-231 or MDA-MB-435 cells at 36 h after transfection with miRNAs from muscle of cooked shrimp fed bacteria-miR-34. Shrimp fed shrimp feed alone and those fed shrimp feed containing bacteria-miR-34-scrambled were used as controls. (E) The role of shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria in breast cancer cell cycle. MDA-MB-231/435 cells were treated with miRNAs extracted from cooked shrimp fed shrimp feed containing bacteria-miR-34 or bacteria-miR-34-scrambled; 36 h later, the cell cycle was examined. (F) The impact of shrimp miR-34 on breast cancer cell migration from cooked shrimp fed shrimp feed containing bacteria-miR-34 was determined with Boyden chamber assays. MDA-MB-231 or 435 cells were treated with miRNAs from muscles of cooked shrimp fed miR-34-expressing bacteria. At 36 h after transfection, a cell migration assay was performed. (G) Influence of miR-34 on breast cancer cell adhesion. At 36 h after MDA-MB-231 or 435 cells were transfected with miRNAs from cooked shrimp fed bacteria-miR-34, cell adhesive ability was assessed. (H) Role of miR-34 in breast cancer cell invasion. MiRNAs from cooked shrimp fed bacteria-miR-34 were transfected into MDA-MB-231 or 435 cells, and a cell invasion assay was performed 36 h after transfection. (I) Expression levels of miR-34 target genes in breast cancer cells. MDA-MB-231 or 435 cells were transfected with miRNAs from cooked shrimp fed bacteria-miR-34. At 36 h after transfection, expression levels of CCND1, CDK6, CCNE2, E2F3, MET, and FOSL1 were quantified by quantitative real-time PCR. (J) Protein levels of miR-34 target genes in breast cancer cells. Western blot analysis was conducted to detect protein levels. β-tubulin was used as a control. Cells without treatment served as negative control. Throughout, statistically significant differences among treatments are represented with asterisks: ∗p < 0.05, ∗∗p < 0.01. Scale bar, 100 μm.
To further investigate the anti-tumor activity of shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria, the effects of miR-34 on cell cycle and metastasis (migration, adhesion, and invasion) were examined. Flow cytometric analysis revealed that breast cancer cells treated with miRNAs from the muscles of shrimp fed shrimp feed containing bacteria-miR-34 were arrested in the G0/G1 phase, whereas cells transfected with miRNAs from the muscles of shrimp fed shrimp feed only or shrimp feed containing bacteria-miR-34-scrambled were similar to the negative control (Figure 3E), indicating that shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria influenced cell cycle arrest. Shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria inhibited the migration, adhesion, and invasion of breast cancer cells compared with the controls (Figures 3F–3H). These data indicated the inhibitory effects on the growth and metastasis of breast cancer cells from shrimp miR-34 in cooked shrimp fed miR-34-expressing bacteria.
To assess whether shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria targeted CCND1, CDK6, CCNE2, E2F3, MET, and FOSL1 genes, the expression of those six genes in breast cancer cells transfected with miRNAs from the muscle of shrimp fed food containing bacteria-miR-34 was investigated. Quantitative real-time PCR analyses showed that the six genes were downregulated in cells treated with miRNAs from the muscle of shrimp fed food containing the bacteria-miR-34 (Figure 3I), indicating miR-34-mediated regulation of targeted gene expression. Western blot analysis showed similar results (Figure 3J).
Altogether, these findings showed that shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria suppressed breast cancer progression by targeting the human CCND1, CDK6, CCNE2, E2F3, MET, and FOSL1 genes.
Inhibition of breast tumor growth and metastasis by shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria in vivo
To further evaluate the effects of shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria on tumor growth and metastasis in vivo, breast cancer cells (MDA-MB-231) were subcutaneously injected into non-obese diabetic/severe combined immunodeficiency (NOD/SCID) female mice, which were then fed cooked muscles from shrimp fed miR-34-expressing bacteria (shrimp miR-34 feed) and were examined for tumor growth and metastasis (Figure 4A). As a control, mice fed the cooked muscles of shrimp fed regular feed without miR-34 (shrimp feed) was included in the assays. The results demonstrated that the growth and size of solid tumors from mice fed shrimp miR-34 feed were significantly decreased compared with mice fed shrimp feed (Figures 4B and C). The miR-34 expression levels in the blood and solid tumors of the mice fed shrimp miR-34 feed were much higher than what was observed in mice fed shrimp feed (Figures 4D and 4E). These results indicated that exogenous shrimp miR-34 could be delivered into mice through food, leading to suppression of tumor growth in breast cancer in vivo. To examine the influence of miR-34 on expression of target genes in solid tumors, quantitative real-time PCR, western blot, and immunohistochemical analyses were performed. The results indicated that the expression levels of miR-34 target genes (CCND1, CDK6, CCNE2, E2F3, and MET) were significantly downregulated in the tumors of mice fed shrimp miR-34 feed compared with those of the controls (Figures 4F–4H). These results indicated that shrimp miR-34 inhibited the growth of breast cancer by downregulating the expression of target genes in vivo.
Figure 4.
Suppression of breast tumor growth and metastasis by shrimp miR-34 from cooked shrimp fed miR-34-expressing bacteria in vivo
(A) Model of the animal experiment. Breast cancer cells (MDA-MB-231) were injected into nude mice. Nude mice were fed cooked muscle from shrimp fed shrimp feed alone (shrimp feed) or those fed cooked muscle of shrimp fed miR-34-expressing bacteria (shrimp miR-34 feed) every 2 days. Subsequently, tumor growth and metastasis were examined. (B) Tumor growth curves measured weekly 2 weeks after cell injection. (C) Representative images of solid tumors harvested 6 weeks after cell injection. (D) Shrimp miR-34 expression levels in the blood of mice. Six weeks after injecting breast cancer cells (MDA-MB-231), blood was collected from mice with different treatments. The expression level of miR-34 in the blood was examined by quantitative real-time PCR. (E) Quantitative real-time PCR analysis of miR-34 expression level in solid tumors derived from mice with different treatments. (F) Quantitative real-time PCR analysis of miR-34 target genes in solid tumors of mice with different treatments. (G) The expression level of miR-34 target genes encoding proteins in solid tumors. Western blot was used to detect protein levels. β-tubulin was included as a control. (H) Immunohistochemical analysis of miR-34 target gene-encoding proteins in solid tumors. Brown and blue colors represented proteins and nuclei, respectively. Proteins that were detected were indicated on the left. Scale bar, 100 μm. (I) Representative images of mouse lungs harvested 4 weeks after cell injection. (J) Images displaying tumor nodules per lung for mice that received different treatments. (K) MiR-34 expression level in blood of nude mice. Four weeks after cell injection, RNA was extracted from the blood of mice and analyzed using quantitative real-time PCR. (L) Model expressing the role of shrimp miR-34 in tumorigenesis through food intake. Throughout, statistically significant differences among treatments are indicated with asterisks: ∗∗p < 0.01.
To evaluate the metastatic potential of breast cancer cells, cells with luciferase expressing MDA-MB-231 were intravenously injected through the tail vein of nude mice. Then, the mice were fed the cooked muscles of shrimp fed miR-34-expressing bacteria (shrimp miR-34 feed) or shrimp feed without miR-34 (shrimp feed). The luminescence flux data indicated that metastasis of breast cancer cells to the lungs of the mice treated with shrimp miR-34 feed was significantly suppressed compared with that of the control mice (regular feed) (Figure 4I). Furthermore, mice treated with shrimp feed produced more metastatic tumor nodules per lung compared with mice treated with shrimp miR-34 feed (Figure 4J), indicating that miR-34 significantly decreased tumor colonization in the lung. At the same time, higher expression levels of miR-34 were observed when mice were fed the miR-34 feed (Figure 4K).
In summary, these findings revealed that the consumption of shrimp miR-34 through food had the ability to suppress tumorigenesis in mice by targeting multiple genes (Figure 4L), showing that shrimp miR-34 acted as a cancer therapeutic molecule to inhibit breast cancer progression.
Discussion
During the past few decades, shrimp has grown to account for a major portion of aquaculture. The increased use of shrimp worldwide may be due to increased profits and scientific inputs provided to farmers.18 However, one of the most serious viral pathogen of farmed shrimp, known as WSSV, has led to sharp reductions in shrimp production and severe economic losses.19 Emerging evidence has shown that miRNAs, which are small non-coding RNAs, have key roles in defending against WSSV infection.20,21 As reported, a single miRNA can target multiple genes,14 implying that a single miRNA may exert biological functions in multiple animal species by targeting different genes. Our previous study showed that shrimp miR-34 possessed antiviral activity in shrimp and an anti-tumor capacity in humans.16 In that context, we decided to address whether shrimp miR-34 exerted anti-viral and anti-tumor function during food intake. The results of this study revealed that overexpression of miR-34 in shrimp by feeding them with bacteria expressing miR-34 inhibited virus infection and that food intake of shrimp miR-34 suppressed tumorigenesis in mice. In that context, miRNA in food simultaneously controlled shrimp viral disease and human tumorigenesis. Our study provided a novel strategy for the control of viral disease in aquatic animals and tumorigenesis in humans through food intake.
In almost all cell types, microvesicles (MVs), small vesicles in cells, mediate intercellular communication through cell surface molecules.22,23 During the delivery of extracellular miRNAs into target cells, miRNAs can be packaged into MVs, and subsequently, miRNA delivery is triggered.24 Therefore, in this study, shrimp miR-34 in animal feed may be delivered to the cells in shrimp and to human tumor cells in mice, leading to the prevention of virus infection in the shrimp and tumorigenesis in the mice. As previously reported, the oral delivery of the har-miR-2002b mimic, which is specifically expressed during the larval stages of Helicoverpa armigera, can significantly reduce the expression level of a trypsin-like serine protease in vivo.25 Feeding of Escherichia coli HT115 strain (DE3) expressing double-stranded RNAs (dsRNAs), which targets the gene encoding ADP/ATP protein, suppresses gene expression during Nosema bombycis infection in the silkworm.26 Ingestion of bacteria expressing dsRNA triggers specific RNA (shrimp Rab7 and STAT) silencing in shrimp.27 In Caenorhabditis elegans, extracellular dsRNAs enter cells through SID1 and SID2.28, 29, 30 SID1 may act as a key protein for transport into intestinal cells or other cell types, and SID2 is an intestinal lumen protein that is required for the initial uptake of dsRNA from the environment through endocytosis.28, 29, 30 SID5, an endosome-associated protein, facilitates dsRNAs that release into the body cavity or enhance RNAi.31 Recently, SID1 has been characterized in white shrimp.32 In this context, these processes may also be required for RNAi-mediated gene silencing through orally delivered dsRNA in shrimp. In this study, shrimp miR-34 in shrimp feed might have been absorbed by intestinal cells and delivered through MV, leading to inhibition of viral infection by targeting viral wsv330 and wsv359 genes. Recent studies have demonstrated that miR-29b and miR-200c are encapsulated in exosomes in cow milk, conferring protection against degradation, and then, miRNAs enter into cells through endocytosis to regulate human gene expression.33 The oral ingestion of miRNAs (miR-34a, miR-143, and miR-145), synthesized with a methyl group on the 2′ position of the ribose of the 3′-terminal nucleotide, suppresses tumor growth in a mouse model of colon cancer.34 In our study, shrimp miR-34 in animal feed might be delivered to target cells through the MV-mediated pathway, leading to suppression of tumorigenesis. In that context, our findings showed that an individual shrimp miRNA in food simultaneously exhibited antiviral activity in shrimp and anti-tumor capacity in mice by targeting various genes in a cross-species manner.
Materials and methods
Shrimp culture and WSSV infection
Marsupenaeus japonicus shrimp were cultured in 25°C air-pumped circulating seawater. All experiments included 20 shrimp per group. Virus-free shrimp were intramuscularly injected with WSSV virus (105 copies/mL). At different time points after infection, shrimp hemolymph and muscles were collected.
MiR-34 expression in bacteria
To express mature shrimp miR-34 in bacteria, shrimp miR-34 was cloned into the LITMUS 38i plasmid using annealed DNA oligonucleotides (5′-AGCTTTGGCAGT GTGGTTAGCTGGTTGTG-3′ and 5′-AATTCACAACCAGCTAACCACACTGCC AA-3′) corresponding to the miR-34 sequence (Takara Bio, Japan). As a control, miR-34-scrambled was used (miR-34-scrambled, 5′-AGCTTTTCTCCGAACGTG TCACGTTTG-3′ and 5′-AATTCAAACGTGACACGTTCGGAGAAA-3′) and cloned accordingly. The construct was then transformed into HT115 (DE3) cells, an RNase-III-deficient strain of E. coli. The recombinant bacteria were cultured in LB medium. IPTG was used to induce miR-34 expression in HT115 cells.
Overexpression of miR-34 in shrimp using bacteria expressing miR-34
HT115 (DE3) cells expressing miR-34 were inactivated by heating at 60°C for 90 min. The heat-inactivated bacteria were mixed with shrimp feed. The mixed feed, containing 3 × 1012 bacteria/g, was used to feed shrimp every day. During the first feeding, shrimp were simultaneously infected with WSSV. On the indicated days after feeding, shrimp muscles and hemolymph were collected for subsequent experiment.
Northern blot analysis
Total RNA was extracted from shrimp hemocytes, shrimp muscle, or human cells using a mirVanaPTMP miRNA isolation kit (Ambion, USA) and was separated on a denaturing 15% polyacrylamide gel. Then, RNAs were transferred to a nylon membrane (Amersham Biosciences, UK). After being cross-linked with ultraviolet light, the DNA probe labeled with digoxigenin was used for hybridization. Then DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Germany) was used for signal detection.
Quantification of WSSV copies
The viral genomic DNA was extracted using TIANmp Genomic DNA Kit (Tiangen Biotech, Beijing, People’s Republic of China). A linearized plasmid containing a 1,400-bp DNA fragment of the WSSV genome was diluted into 103, 104, 105, 106, 107, 108, and 109 copies/μL as an internal standard. The extracted viral genomic DNA was subjected to quantitative real-time PCR to detect WSSV copies using WSSV-specific primers (5′-CCACCAATTCTACTCATGTACCAAA-3′ and 5′-TCCTTGCAATGGG CAAAATC-3′) and a WSSV-specific TaqMan probe (5′-FAM-CTGGGTTACGAGTCT AA-TAMRA-3′). PCR conditions were as follows: 95°C for 1 min, followed by 45 cycles at 95°C for 15 s, 52°C for 30 s, and 72°C for 30 s.
Shrimp cumulative mortality assay
Shrimp mortality was monitored daily. These experiments were conducted three times.
Prediction of target genes
TargetScan 5.1 (http://www.targetscan.org) and miRanda (http://www.microrna.org/) were used to predict the WSSV genes targeted by shrimp miR-34. Based on the 3′ UTRs of WSSV genes, TargetScan was used to search for each miRNA seed sequence, and miRanda was used to match the entire miRNA sequence according to parameters. The genes obtained by both algorithms were the potential targets of shrimp miR-34.
Interaction between miR-34 and its target genes
The EGFP gene was cloned into a pIZ/V5-His vector (Invitrogen, USA) using EGFP-specific primers (5′-AAGAGCTC GGATCCCCGGGTAC-3′ and 5′-AATCTAGAGTCGCGGCCGCTTTA-3′). The EGFP-wsv330-3′ UTR plasmid was constructed by cloning wsv330 3′ UTR downstream of EGFP with sequence-specific primers (5′-GCGTCTAGAAGAATTA ACCTGCTTT-3′ and 5′-TATCCGCGGGAAGGGTCCTTTAG-3′). The EGFP-wsv359-3′ UTR plasmid was constructed using primers (5′-TATT CTAGAACGTGGGGTCCTTCTT-3′ and 5′-ATTCCGCGGCTGGATACCGTAGG-3′). The 3′ UTR mutations were generated by PCR with sequence- specific primers (wsv330 3′ UTR, 5′-TCTGGTCCTGGACAGAGTCATACTGTAC-3′ and 5′-CGACG CAGAGTACAGTATGACTCTGTCC-3′; wsv359 3′ UTR, 5′-GATTGTTGGGGTTG AAGACAATGTCTCC-3′ and 5′-AGAGTCGGCGGAGACAT TGTCTTCAACC-3′).
When insect High Five cells (Invitrogen) reached 70% confluence, they were co-transfected with 50 nM of synthesized shrimp miR-34 (5′-UGGCAGUGUGGUU AGCUGGUUGU-3′ and 5′-ACAACCAGCUAACCACACUGCCA-3′) and 200 ng of EGFP, EGFP-wsv330-3′ UTR, EGFP-wsv330-3′ UTR-mutation, EGFP-wsv359- 3′ UTR, or the EGFP-wsv359-3′ UTR-mutation construct. The synthesized miR-34-scrambled (5′-AUUUGACAGAUGCCUAGUACCAG-3′ and 5′-CUGGUACUAGG CAUCUGUCAAAU-3′) was included as a control. High Five cells were cultured at 27°C with Express Five SFM Medium (Invitrogen) supplemented with l-glutamine (Invitrogen); 48 h later, a Flex Station II microplate reader (Molecular Devices, USA) was used to detect the fluorescence at 480 nm and 520 nm. The experiment was biologically repeated three times.
To examine the interaction between miR-34 and wsv330 or wsv359 genes in vivo, shrimp were co-injected with WSSV and synthesized miR-34 or miR-34-scrambled. At different times after infection, the expression levels of wsv330 and wsv359 were detected using quantitative real-time PCR. Shrimp β-actin was used as a control. The wsv330 primers (5′-ACTCCCCTAACCCATTGACAT-3′ and 5′-GGAGA ACAGGAACTAGAGACGTACT-3′), wsv359 primers (5′-AG ATTTCCC TCCAGATCCTCC-3′ and 5′-TGACCAAGACAAATGCTACCCT-3′), or β-actin primers (5′-CGAGCACGGCATCGTTACTA-3′ and 5′-TTGTAGA AAGTG TGATGCCAGATCT-3′) were used to perform real-time PCR; 10 μL of PCR condition containing 5 μL SYBR Premix Ex Taq (Takara), 0.5 μL cDNA template, and 0.2 μL 10 mM primers was prepared. PCR was performed at 95°C for 5 min, followed by a 40 cycle at 95°C for 10 s and a 30-s cycle at 60°C.
RNAi assays in shrimp
siRNAs specifically targeting wsv330 (wsv330-siRNA, 5′-GGAUCAAGAACGCGGGUAA-3′ and 5′-UUACCCGCGUUCUUGAUCC-3′) or wsv359 (wsv359-siRNA, 5′-GCAGGUUGACUGUACAUUA-3′ and 5′-UAAUG UACAGUCAACCUGC-3′) were synthesized using an in vitro T7 transcription kit for siRNA synthesis (Takara) according to the manufacturer’s instructions. The siRNA sequence was randomly scrambled, generating siRNA-scrambled (5′-CUUCAGUGACCGGACCUACGACGAU-3′ and 5′-AUCGUCGUAGGUCCG GUCACUGAAG-3′). The synthesized siRNAs were assessed by agarose gel electrophoresis and quantified by spectrophotometry. Virus-free shrimp were co-injected with siRNA (30 μg) and WSSV (105 copies/mL). The injection of WSSV alone served as a positive control. At different time points after infection (0, 24, 36, and 48 h), shrimp muscles, gills, and hemolymph were collected. Shrimp mortality was monitored daily. All experiments were biologically replicated three times.
Cell culture
Human breast cancer cell lines (MDA-MB-231 and MDA-MB-435) were cultured at 37°C without CO2 using Leibovitz’s L-15 Medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS). MDA-MB-231 and MDA-MB-435, cells were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, People’s Republic of China).
Expression of shrimp miR-34 in cells
Shrimp were fed shrimp feed containing bacteria-miR-34 daily. Shrimp fed feed without bacteria-miR-34 were used as controls. Two days later, shrimp were collected and cooked by heating in a microwave for 2 min. The miRNAs extracted from the cooked shrimp muscles were transfected into tumor cells.
Cell proliferation assay
After cells (1 × 104/well) were plated onto a 96-well plate, miR-34 extracted from cooked shrimp muscles was transfected into cells using Lipofectamine RNAiMAX (Life Technologies, USA); 36 h later, cell proliferation was examined using a CellTiter 96 AQueous One Solution Cell Proliferation Assay Kit (Promega, USA) and monitored using an iMARKTM microplate reader (Bio-Rad, USA) at 490 nm.
Wound-healing assay
Cells were plated onto a 24-well plate. When the cells reached approximately 70% confluence, a 100 μL pipette tip was used to create a wound. Then, miR-34 extracted from cooked shrimp muscles was transfected into cells; 36 h later, the images were obtained with a Nikon Ti-S microscope (Nikon, USA).
Cell cycle assay
Fluorescence-activated cell sorting (FACS) analysis was performed to examine the cell cycle. Cells (4 × 105) were cultured in a 25-cm2 culture flask. Subsequently, miR-34 extracted from cooked shrimp muscles was transfected into cells using Lipofectamine RNAiMAX (Life Technologies, USA); 36 h later, the cells were collected and fixed with 70% ethanol at −20°C overnight, followed by incubation with RNaseA (200 μg/mL, Sangon Biotech, Shanghai, People’s Republic of China) for 30 min at 37°C and then stained with propidium iodide solution for 15 min at 37°C. Finally, the cells were examined with an FACSCalibur flow cytometer (BD Biosciences, USA).
Cell migration assay with a Boyden chamber
Migration of cancer cells was examined with a 24-well Boyden chamber (Corning, Corning, NY, USA) with 8-μm inserts (Corning). Cells were treated with miR-34 extracted from cooked shrimp muscles; 36 h later, the cells were collected and suspended in serum-free medium. The cells (5 × 104 for MDA-MB-231 and 8 × 104 for MDA-MB-435) were inoculated into the insert. Then, the insert was placed in a well containing 600 μL of medium with 10% serum. After incubation for 24 h, the insert was fixed with 4% paraformaldehyde for 15 min. The cells were stained with crystal violet (0.005%, Beyotime Biotechnology, Shanghai, People’s Republic of China) for 15 min. Images of migrating cells were obtained with a Nikon Ti-s microscope (Nikon, USA). The results showed the number of migrating cells per ×200 micrographs per sample.
Cell adhesion assay
The 24-well plate was pre-coated with fibrinogen (20 μg/mL; Sigma-Aldrich, USA) at 37°C for 2 h. Then 10% bovine serum albumin was added to the wells and incubated at 37°C for 0.5 h to block non-specific proteins. Meanwhile, miR-34 extracted from cooked shrimp was transfected into cells. After incubation for 36 h, the cells were collected and suspended in serum-free medium. The cells (5 × 104) were then transferred to the pre-coated plate. Non-adherent cells were removed after 30 min of culturing at 37°C. The cells were fixed with 4% paraformaldehyde and then stained with methylrosanilinium chloride solution (Beyotime Biotechnology, Shanghai, People’s Republic of China) for 15 min. The number of stained cells was obtained under a microscope.
Cell invasion assay
Cells were transfected with miR-34 extracted from cooked shrimp muscles. The invasion capacity was evaluated with 24-well matrigel invasion chambers with 8-μm inserts (Corning). The insert was pre-activated at 37°C for 2 h with Dulbecco’s modified eagle medium (DMEM, Gibco, USA). The suspended cells (5 × 104) were inoculated into the insert. The insert was placed into a well containing 600 μL of medium with 10% serum. After incubation for 24 h, the insert was fixed with 4% paraformaldehyde (Sigma-Aldrich, USA) for 15 min, followed by incubation for 15 min with crystal violet (0.005%, Beyotime Biotechnology, Shanghai, People’s Republic of China). A Nikon Ti-S microscope (Nikon, USA) was used to obtain images of invading cells. The results showed the number of invasions per ×200 micrographs per sample.
Animal experiment
To analyze tumor growth in vivo, 5 four-week-old NOD/SCID female mice were used to induce tumor growth after subcutaneously being injected with breast cancer cells (MDA-MB-231; 1 × 106). The mice were separated into two groups. After cell injection, the mice were fed cooked muscles of shrimp fed shrimp feed with or without miR-34-expressing bacteria every 2 days. Tumor growth was evaluated weekly 2 weeks after cell injection and was analyzed for tumor length (L) and width (W). The volume of the tumor was calculated according to the formula: volume = 0.52 × L × W2. Six weeks later, NOD/SCID nude mice were sacrificed. For tumor metastasis in vivo, lentiviral packaging was used to construct luciferase expressing MDA-MB-231 cells. Then, the cells (3 × 105) were intravenously injected into the tail vein of five nude mice. The mice were also separated into two groups and treated as described above. Metastatic tumors were observed weekly with an IVIS Spectrum CT Preclinical In Vivo Imaging System (PerkinElmer, USA).
Immunohistochemical analysis
Solid tumors were dissected and fixed with formalin and then embedded in paraffin. The paraffin sections were incubated with antibodies against human cyclin D1 (CCND1), cyclin-dependent kinase 6 (CDK6), cyclin E2 (CCNE2), transcription factor E2F3, or hepatocyte growth factor receptor (MET) (Proteintech Group, USA) at 4°C overnight. Goat anti-rabbit immunoglobulin G (IgG; Sigma-Aldrich, USA) conjugated with HRP (horseradish peroxidase) was incubated at room temperature for 2 h. Subsequently, 3,3′-diaminobenzidine (DAB) was used to test the sections. Nuclei were stained with hematoxylin solution. The sections were observed using a microscope (Zeiss).
Quantitative real-time PCR analysis of miR-34 target genes in solid tumors
Quantitative real-time PCR was conducted to assess the mRNA levels of miR-34 targets with gene-specific primers (CCND1, 5′-CCCTCGGTGTCCTACTTCAAAT G T-3′ and 5′-GGAAGCGGTCCAGGTAGTTCAT-3′; CDK6, 5′-AGAGCAAGATA ATAAAGGAGATGGG-3′ and 5′-CATGTGAGACTTTGAGTAGACCTGA-3′; CCNE2, 5′-GCATTATGACACCACCGAAGA-3′ and 5′-AGGGCAATCAATCACA GCAC-3′; E2F3, 5′-TGACTGCGTGAGCCTTAGAA-3′ and 5′-CAAGAGCCACAA CAAAGAACAG-3′; and MET, 5′-CTCTACTTTCATTGGGGAGCA-3′ and 5′-CCTCAT CATCAGCGTTATCTTC-3′). The human glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 5′-GGTATCGTGGAAGGACTCATGAC-3′ and 5′-AT GCCAGTGAGCTTCCCGTTCAG-3′) gene was used as a control. PCR was conducted by incubation at 95°C for 10 min, followed by 50 cycles at 95°C for 15 s and 60°C for 1 min.
Quantification of miR-34
Synthesized shrimp miR-34 was diluted at different concentrations (103, 104, 105, 106, 107, 108, and 109/mL). Subsequently, shrimp miR-34 was used as an internal standard to conduct quantitative real-time PCR with TaqMan Micro-RNA assay (Applied Biosystem, USA). The PCR conditions were incubation at 95°C for 10 min, 50 cycles at 95°C for 15 s, and 60°C for 1 min.
Western blot analysis
Proteins were separated by SDS-PAGE and then transferred to a nitrocellulose membrane (Bio-Rad Laboratories, USA). The membrane was first blocked with 5% milk for 1 h at room temperature and then incubated with a primary antibody for 2 h. Subsequently, HRP-conjugated goat anti-mouse or anti-rabbit IgG (Sigma-Aldrich, USA) was used to incubate the membrane for 2 h at room temperature. Proteins were detected using Western Lightning Plus-ECL Oxidizing Reagent Plus (PerkinElmer, USA).
Statistical analysis
The means and standard deviations of three replicates were calculated using one-way analysis of variance (ANOVA). Student’s t test was used to determine the statistical significance among different treatments.
Acknowledgments
This work was financially supported by the National Key Research and Development Program of China (2018YFD0900504) and the China Ocean Mineral Resources R & D Association (DY135-B-04).
Author contributions
X.Z. designed the experiments and wrote the manuscript. Y.C. and H.W. conducted the experiment and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Chen I.T., Aoki T., Huang Y.T., Hirono I., Chen T.C., Huang J.Y., Chang G.D., Lo C.F., Wang H.C. White spot syndrome virus induces metabolic changes resembling the warburg effect in shrimp hemocytes in the early stage of infection. J. Virol. 2011;85:12919–12928. doi: 10.1128/JVI.05385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong T., Ren X., Lin S., Li S., Gong Y. Elucidation of metabolic responses in mud crab Scylla paramamosain challenged to WSSV infection by integration of metabolomics and transcriptomics. Dev. Comp. Immunol. 2020;113:103799. doi: 10.1016/j.dci.2020.103799. [DOI] [PubMed] [Google Scholar]
- 3.Drakesmith H., Prentice A. Viral infection and iron metabolism. Nat. Rev. Microbiol. 2008;6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 4.Diamond D.L., Syder A.J., Jacobs J.M., Sorensen C.M., Walters K.A., Proll S.C., McDermott J.E., Gritsenko M.A., Zhang Q., Zhao R. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold P.A., Johnson K.N., White C.R. Physiological and metabolic consequences of viral infection in Drosophila melanogaster. J. Exp. Biol. 2013;216:3350–3357. doi: 10.1242/jeb.088138. [DOI] [PubMed] [Google Scholar]
- 6.Walker P.J., Winton J.R. Emerging viral diseases of fish and shrimp. Vet. Res. 2010;41:51. doi: 10.1051/vetres/2010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y., Zhang X. Comprehensive characterization of viral miRNAs involved in white spot syndrome virus (WSSV) infection. RNA Biol. 2012;9:1019–1029. doi: 10.4161/rna.20741. [DOI] [PubMed] [Google Scholar]
- 8.Yang F., He J., Lin X., Li Q., Pan D., Zhang X., Xu X. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 2001;75:11811–11820. doi: 10.1128/JVI.75.23.11811-11820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong Y., Ju C., Zhang X. Shrimp miR-1000 functions in antiviral immunity by simultaneously triggering the degradation of two viral mRNAs. Front. Immunol. 2018;9:2999. doi: 10.3389/fimmu.2018.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 11.DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Kawauchi K., Araki K., Tobiume K., Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 13.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Selcuklu S.D., Donoghue M.T., Rehmet K., de Souza Gomes M., Fort A., Kovvuru P., Muniyappa M.K., Kerin M.J., Enright A.J., Spillane C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J. Biol. Chem. 2012;287:29516–29528. doi: 10.1074/jbc.M111.335943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le M.T., Xie H., Zhou B., Chia P.H., Rizk P., Um M., Udolph G., Yang H., Lim B., Lodish H.F. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol. Cell. Biol. 2009;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y., Yang X., Zhang X. Shrimp miR-34 from shrimp stress response to virus infection suppresses tumorigenesis of breast cancer. Mol. Ther. Nucleic Acids. 2017;9:387–398. doi: 10.1016/j.omtn.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F., Wei J., Zhang S., Zhang X. Shrimp miR-S8 suppresses the stemness of human melanoma stem-like cells by targeting the transcription factor YB-1. Cancer Res. 2017;77:5543–5553. doi: 10.1158/0008-5472.CAN-17-1375. [DOI] [PubMed] [Google Scholar]
- 18.Pradeep B., Rai P., Mohan S.A., Shekhar M.S., Karunasagar I. Biology, host range, pathogenesis and diagnosis of white spot syndrome virus. Indian J. Virol. 2012;23:161–174. doi: 10.1007/s13337-012-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shekar M., Pradeep B., Karunasagar I. White spot syndrome virus: genotypes, epidemiology and evolutionary studies. Indian J. Virol. 2012;23:175–183. doi: 10.1007/s13337-012-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang T., Zhang X. Functional analysis of a crustacean microRNA in host-virus interactions. J. Virol. 2012;86:12997–13004. doi: 10.1128/JVI.01702-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L., Yang G., Zhang X. The miR-100-mediated pathway regulates apoptosis against virus infection in shrimp. Fish Shellfish Immunol. 2014;40:146–153. doi: 10.1016/j.fsi.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Cocucci E., Racchetti G., Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Théry C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., Lu J., Yin Y., Cai X. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Jayachandran B., Hussain M., Asgari S. An insect trypsin-like serine protease as a target of microRNA: utilization of microRNA mimics and inhibitors by oral feeding. Insect Biochem. Mol. Biol. 2013;43:398–406. doi: 10.1016/j.ibmb.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Pan Q., Wang L., Dang X., Ma Z., Zhang X., Chen S., Zhou Z., Xu J. Bacterium-expressed dsRNA downregulates microsporidia nosema bombycis gene expression. J. Eukaryot. Microbiol. 2017;64:278–281. doi: 10.1111/jeu.12346. [DOI] [PubMed] [Google Scholar]
- 27.Attasart P., Namramoon O., Kongphom U., Chimwai C., Panyim S. Ingestion of bacteria expressing dsRNA triggers specific RNA silencing in shrimp. Virus Res. 2013;171:252–256. doi: 10.1016/j.virusres.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Winston W.M., Molodowitch C., Hunter C.P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 29.Winston W.M., Sutherlin M., Wright A.J., Feinberg E.H., Hunter C.P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA. 2007;104:10565–10570. doi: 10.1073/pnas.0611282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McEwan D.L., Weisman A.S., Hunter C.P. Uptake of extracellular double-stranded RNA by SID-2. Mol. Cell. 2012;47:746–754. doi: 10.1016/j.molcel.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinas A., Wright A.J., Hunter C.P. SID-5 is an endosome-associated protein required for efficient systemic RNAi in C. elegans. Curr. Biol. 2012;22:1938–1943. doi: 10.1016/j.cub.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labreuche Y., Veloso A., de la Vega E., Gross P.S., Chapman R.W., Browdy C.L., Warr G.W. Non-specific activation of antiviral immunity and induction of RNA interference may engage the same pathway in the Pacific white leg shrimp Litopenaeus vannamei. Dev. Comp. Immunol. 2010;34:1209–1218. doi: 10.1016/j.dci.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 33.Baier S.R., Nguyen C., Xie F., Wood J.R., Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 2014;144:1495–1500. doi: 10.3945/jn.114.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mlotshwa S., Pruss G.J., MacArthur J.L., Endres M.W., Davis C., Hofseth L.J., Peña M.M., Vance V. A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res. 2015;25:521–524. doi: 10.1038/cr.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]