Summary
Almost all plants form trichomes, which protect them against insect herbivores by forming a physical barrier and releasing chemical repellents. Glandular trichomes produce a variety of specialized defensive metabolites, including volatile terpenes. Previous studies have shown that the defence hormone jasmonic acid (JA) affects trichome development and induces terpene synthases (TPSs) but the underlying molecular mechanisms remain unclear. Here, we characterized a loss‐of‐function allele of the HD‐ZIP IV transcription factor woolly (wo) and analysed its role in mediating JA signalling in tomato. We showed that knockout of wo led to extensive trichome defects, including structural and functional changes in type VI glandular trichomes, and a dramatic reduction in terpene levels. We further found that wo directly binds to TPS gene promoters to recruit SlMYC1, a JA signalling modulator, and that together these transcription factors promote terpene biosynthesis in tomato trichomes. The wo/SlMYC1 regulatory module is inhibited by SlJAZ2 through a competitive binding mechanism, resulting in a fine‐tuned JA response in tomato trichomes. Enhanced expression of SlMYC1 substantially increased terpene levels and improved tomato resistance to spider mites. Interestingly, we also found that SlMYC1 plays an additional role in glandular cell division and expansion in type VI trichomes, independent of JA. Together, our results reveal a novel, JA‐mediated regulatory mechanism that promotes insect resistance in tomato.
Keywords: woolly, SlMYC1, JA signalling, trichomes, pest resistance, tomato
Introduction
The sessile nature of plants forces them to develop various ways to adapt to the environment. In response to some stresses, plants produce a wide variety of secondary metabolites, which are often stored in specific cell lineages (Kang et al., 2010; Schweizer et al., 2013). Terpenoids constitute the most diverse and abundant class of plant metabolites and have important roles in the interaction between plants and their environments. In fruits and flowers, terpenoids serve as an important component of volatile aromas, signals that attract insects and mammals for pollination, fruit predation and seed dispersal (Aharoni et al., 2003; Chen et al., 2003). Terpenes are also produced by trichomes, cellular structures that project from the epidermal surface and provide a protective barrier against pathogens and pests. These structures synthesize a diverse array of specialized secondary metabolites that exert toxic or repellent effects on herbivores and microbes (Barba et al., 2019; Calo et al., 2006; Kang et al, 2010; Kang et al., 2014; Ma et al., 2016).
Tomato produces seven types of trichomes (named I–VII), of which I, IV, VI and VII are glandular and II, III and V are non‐glandular (Luckwill, 1943). Among the glandular trichomes, types I and IV are capitate trichomes with a small, single‐celled head, whereas types VI and VII have four and eight head cells, respectively. Glandular trichomes are the main factory of a variety of specialized metabolites, including acyl sugars, terpenoids, alkaloids and flavonoids (McDowell et al., 2011). In type I glandular trichomes, the principal chemical produced in the small glandular cells is acyl sugar (Schilmiller et al., 2012; Schilmiller et al., 2015). Most volatile chemicals, including terpenoids and flavonoids, are synthesized mainly in the glandular head of type VI trichomes (Balcke et al., 2017).
Terpenes can be divided into hemiterpenes (C5), monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20) and triterpenes (C30) based on their number of isoprene (C5) units (Bohlmann et al., 1998). Monoterpenes, sesquiterpenes and diterpenes are synthesized by terpene synthases (TPSs). There are 45 TPS genes, including 29 that are functional or potentially functional, in cultivated tomato (Solanum lycopersicum) (Falara et al., 2011; Vasiliki et al., 2011). TPS4 (MTS2) and TPS5 (MTS1) were reported to function as monoterpene synthases in tomato (Van et al., 2007). Other TPSs, including TPS9 and TPS12, were identified as sesquiterpene synthases (Schilmiller et al., 2010). Based on previous microarray data, many TPSs are expressed in a wide range of tissues but some, including SlTPS5 and SlTPS9, are mainly expressed in type VI trichomes and can be induced by JA (Matsuba et al., 2013; Spyropoulou et al., 2014; Van et al., 2007).
Jasmonate (JA) is a herbivory‐induced hormone that participates in terpene biosynthesis and trichome development (Hong et al., 2012). Methyl jasmonate (MeJA) treatment was found to significantly enhance the expression of a number of monoterpene and sesquiterpene synthases. In addition, treatment with multiple applications of MeJA was shown to increase trichome density in Arabidopsis and the density of type VI trichomes on newly formed leaves in tomato (Boughton et al., 2005; Yoshida et al., 2009). MeJA treatment leads to ubiquitin‐mediated degradation of the JA signalling repressor jasmonate‐ZIM domain (JAZ) protein (Chini et al., 2007; Fonseca et al., 2009; Seo et al., 2011; Song et al., 2011; Thines et al., 2007; Yan et al., 2007). A bHLH transcriptional factor, MYC2, has been reported as a direct target of JAZ repressors in Arabidopsis. AtMYC2 seemed to promote sesquiterpene biosynthesis by directly binding to the promoters of AtTPS21 and AtTPS11 (Hong et al., 2012).
Recently, several mechanisms regulating trichome development in tomato have emerged. A gain‐of‐function mutation in the Woolly (Wo) gene, which encodes an HD‐ZIP IV protein, was shown to substantially increase the number of type I trichomes in tomato (Yang et al., 2011). A C2H2 zinc finger transcription factor encoded by the Hair gene interacts with Wo and is also essential for type I trichome development (Chang et al., 2018). A search for downstream factors uncovered a CyclinB2 homolog in tomato that was induced by Wo and participated in Wo‐mediated type I trichome development (Gao et al., 2017; Yang et al., 2011). Recently, reduced SlMYC1 expression was observed to suppress type VI glandular trichome formation and terpene biosynthesis in tomato (Xu et al., 2018). Nevertheless, the mechanisms that regulate type VI trichome development and terpene biosynthesis are still unknown.
Despite the strong link between JA signalling and glandular trichome development, the detailed mechanism of this link remains a mystery. In this study, we identified a loss‐of‐function allele of woolly (wo) which affects the number of two major types of glandular trichomes (types VI and VII). In addition to influencing trichome development, wo directly regulates TPS gene expression via the JA signalling pathway by forming a JAZ‐regulated complex with SlMYC1.
Results
woolly is essential for trichome development
To investigate tomato trichome development, we screened M3 seeds from a mutagenized tomato population. We identified a mutant that had no long trichomes on its leaves or stems (termed a short‐hair phenotype) (Figure 1a–d). In 211 F2 progenies of a cross between the mutant and wild type (WT) tomato, we found 49 individuals that showed the mutant phenotype, indicating that the short‐hair phenotype was caused by a recessive mutation at a single locus. Through bulked‐segregant analysis sequencing (BSA‐Seq), we identified a signal peak in the end of chromosome 2. Further examination revealed there were three SNPs with a SNP index of 1 (Table S1). Two of these, SNP‐41695644 and SNP‐43067486, were located in the introns of Solyc02g072510 and Solyc02g078340, respectively. The third SNP, SNP‐44533258, was a T to C transversion at position 316 in the coding sequence of the wo gene (Figure 1e, f).
Figure 1.
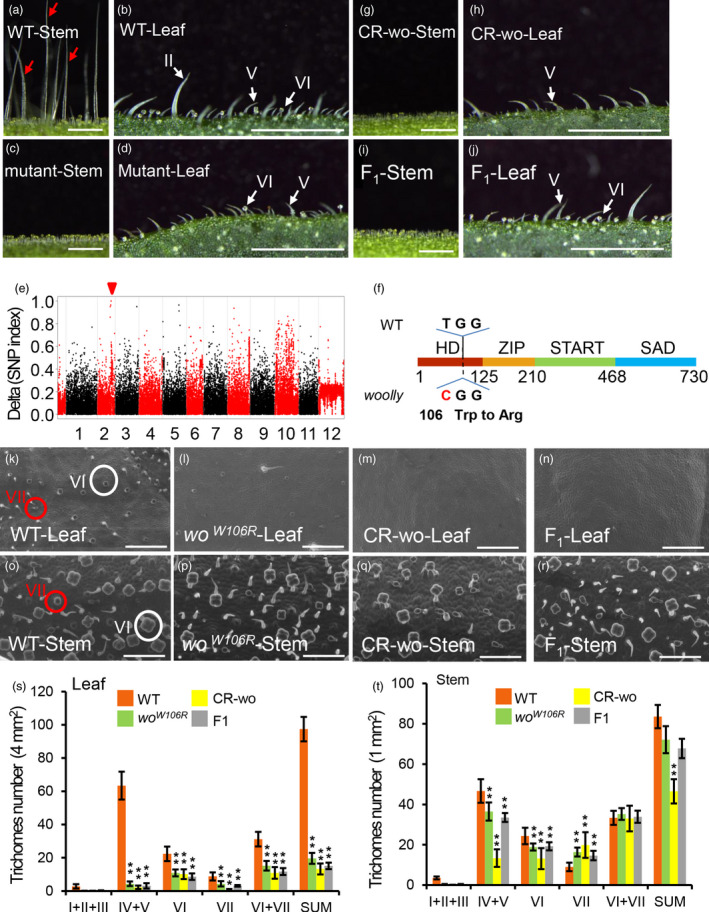
Identification of a new loss‐of‐function allele of woolly. (a–d) The phenotype of stems (a, c) and leaves (b, d) in wild‐type (WT) and short hair mutants. Red arrows in the Figure a point to the long trichomes. Scale bar: 1 mm. (e) Mapping of the mutation by BSA. The red triangle points to the causal mutation with very high frequency (ideally 100% having the variant allele). (f) The location of causal mutation of woolly. Filled colourful boxes represent four domains of woolly protein (HD, 1–125 aa; ZIP, 126–210 aa; START, 211‐468 aa and SAD, 469‐730 aa). A single‐base substitution (T 316 C) is shown in the HD domain, which results in the mis‐sense mutation (Trp 106 Arg). (g–h) wo mutants were generated by CRISPR/Cas9. Scale bar: 1mm. (i, j) The stem (i) and leaf (j) of the F1 plants (cross between CR‐wo plants and woW106R mutants). Scale bar: 1 mm. (k–n) Trichome phenotype on the leaves of WT (k), woW106R mutants (l), CR‐wo (m) and F1 plants (n) by SEM. White circles indicate type VI trichomes. Red circles show type VII trichomes. Scale bar: 125 μm. (o–r) Trichome phenotype on the stems of WT (o), woW106R mutants (p), CR‐wo (q) and F1 plants (r) by SEM. Scale bar: 100 μm. White circles show type VI trichomes. Red circles indicate type VII trichomes. Scale bar: 250 μm. (s, t) Statistic analysis of trichome abundance on the leaf (s) and stem (t) of WT, woW106R mutants, CR‐wo and F1 plants. Four sets of trichomes (I + II + III, IV + V, VI and VII) were quantified. ‘SUM’ on the X‐axis means the sum of all types of trichomes. Values are represented as means ± SD (n = 5). Asterisks indicate significant differences by t‐test: ** P < 0.01, * 0.01 < P < 0.05. [Colour figure can be viewed at wileyonlinelibrary.com]
We further verified the SNP‐44533258 mutation using PCR followed by sequencing of the F2 population mentioned above (Figure S1A). The SNP caused a W106R mutation in the HD domain of wo, which is responsible for protein‐DNA interaction and transcriptional activity (Ariel et al., 2007). To confirm the W106R mutation, we generated wo knockout lines by CRISPR‐Cas9 genome editing (Barrangou et al., 2007; Mali et al., 2013). We identified many edited lines including one with a single base deletion in the 48bp of the HD domain, leading to a frameshift mutation (Figure S1B). The CRISPR‐Cas9‐wo lines (called CR‐wo hereafter) and F1 progeny of a cross between CR‐wo and the woW106R mutant had no long trichomes on leaves or stems, resembling the homozygous woW106R mutant (Figure 1g–j).
The previously reported gain‐of‐function mutation of Wo led to a higher density of type I trichomes in tomato (Yang et al., 2011). To further characterize the role of wo in trichome development, we quantitated trichomes in four‐week‐old plants of woW106R mutants, CR‐wo lines, F1 crossed lines, and WT (Figure 1k–t). For this analysis, we grouped type I, II and III trichomes into one category (I + II+III) as all three types are substantially longer than the other types and could be easily recognized using a dissecting microscope. We also grouped type IV and V trichomes into one category (IV + V) because both types are morphologically similar, the only difference being the presence of glandular cells in type VI trichomes. In total, we quantified four sets of trichomes (I + II+III, IV + V, VI and VII). Compared with WT, the wo mutants (woW106R mutants, CR‐wo lines, and the crossed F1 plants) had almost no long trichomes (I + II+III) but still had short trichomes (IV, V, VI and VII) on their leaves, stems and fruits (Figure 1k–r; Figure S2). In addition, all wo mutants had substantially fewer type VI trichomes. The reduction was about 50% on leaves and about 25‐45% on stems, depending on the mutant background (Figure 1s–t). By contrast, the changes observed in the number of type VII trichomes in the wo mutants were more complex, with the density lower on the leaves but higher on the stems compared with WT (Figure 1s–t).
Next, we used qRT‐PCR to analyse the mRNA levels of wo in different tissues. The wo gene was highly expressed in type VI trichomes and young tissues, including meristems, young leaves and young flowers, but had a low level of expression in more mature tissues (Figure S3A). We also made a GUS reporter construct using the wo promoter (about 2500 bp upstream of wo gene. Similar to the qRT‐PCR result, GUS staining showed that wo was highly expressed in type VI trichomes, but it also showed expression in other types of trichomes as well as in leaves (Figure S3B). Together, our results provide forward genetic evidence supporting active participation of wo in trichome development in tomato.
woolly interacts with JA signalling to regulate trichome development and terpene biosynthesis
As type VI glandular trichomes are one of the main producers a variety of metabolites, including terpenes (Besser et al., 2009; Fridman et al., 2005; Sallaud et al., 2009; Schilmiller et al., 2009), we used Gas Chromatography‐Mass Spectrometry (GC‐MS) to quantify the volatile terpenes in leaves of WT and wo mutant plants. Our measurements revealed that the levels of monoterpenes (2‐carene, β‐phellandrene) and sesquiterpenes (α‐humulene) were significantly reduced in the woW106R mutant and in CR‐wo plants compared to WT (Figure 2a–d).
Figure 2.
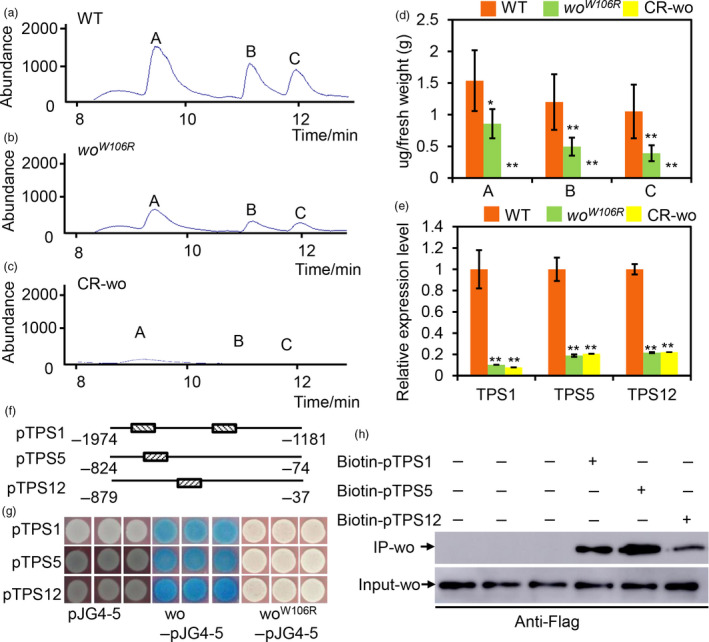
woolly directly regulates the terpene biosynthesis. (a–c) GC‐MS chromatograms of the leaves of WT (A), woW106R mutant (b) and CR‐wo plants (c). a: 2‐Carene; b: β‐Phellandrene; c: α‐humulene. (d) The quantification of terpenes base on the GC‐MS analysis. The Y‐axis represents the content of the terpenes (μg) per gram (g) of the fresh leaf. Values are represented as means ± SD (n = 5). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. (e) Relative expression level of TPS1, 5 and 12 in the gland of type VI trichomes of WT, woW106R mutants and CR‐wo mutants. Values are represented as means ± SD (n = 3). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. (f) The promoter region of TPSs bound by wo is represented by the narrow line. The box filled with the slash indicates the L1 box. The number shows the location of the promoter. (g) Y1H shows that wo binds to the promoter fragments of TPS genes. The full‐length coding sequence of wo and woW106R was cloned into the vector pJG4‐5 (marked as wo‐pJG4‐5 and woW106R‐pJG4‐5). The promoters of TPSs were cloned into the vector pLacZi. Empty vector pJG4‐5 was used as the negative control. The transformed yeast cells were incubated on SD/‐Trp‐Ura + X‐gal medium. The blue clones indicate the interaction between the proteins and the promoters. (h) The biotin‐labelled DNA IP assay. The negative control is in the left three lanes in which the beads were incubated with the total protein. The anti‐Flag antibody was used for the immunoblotting of Flag‐wo. [Colour figure can be viewed at wileyonlinelibrary.com]
To learn more about the regulatory mechanism of wo, we performed RNA‐seq on young leaves of the woW106R mutant, where trichomes were vigorously initiated. By pairwise comparisons of the RNA‐seq data, we identified 2517 up‐regulated genes and 2210 down‐regulated genes in the woW106R mutant (FDR adjusted P value < 0.05). Among the differentially expressed genes (DEGs), we identified four TPS genes (TPS1, TPS5, TPS12 and TPS41) that were both highly expressed in trichomes and down‐regulated in the woW106R mutant (Figure S4A; Table S2). TPS1/5 and TPS12 have previously been reported to regulate biosynthesis of monoterpenes and sesquiterpenes, respectively (Schie et al., 2007). We further examined the transcript levels of the three TPSs in leaves by qRT‐PCR and found that the expression of these genes was significantly reduced in both woW106R mutant and CR‐wo plants (Figure S4B). The reduced terpene levels in both woW106R mutant and CR‐wo plants could be caused by their reduced number of type VI trichomes, by lower production of terpenes by their remaining type VI trichomes, or both. To address these possibilities, we collected all glandular cells from type VI trichomes of woW106R , CR‐wo and WT plants for transcript analysis. Our qRT‐PCR results showed that expression of TPSs was indeed reduced in the glandular cells of type VI trichomes of woW106R mutants and CR‐wo plants (Figure 2e).
To determine whether this regulation of TPSs by wo is direct or indirect, we performed yeast one‐hybrid (Y1H) assays. We divided the about 2000‐bp promoter of each of the three TPS genes (TPS1, TPS5 and TPS12) into three fragments. To avoid disrupting cis‐elements in the promoters, we included a 100‐bp overlap between neighbouring fragments. The TPS promoter fragments were inserted into the pLacZi vector, and the wo cDNA was introduced into the pJG4‐5 vector. Transformants containing the pJG4‐5‐wo and pLacZi‐pTPS constructs became blue after 5 days of incubation on SD/‐Trp/‐Ura medium with X‐gal, indicating that wo can physically bind to the promoters of the three TPS genes (Figure 2f, g). It was previously reported that the HD‐ZIP IV transcription factor subfamily can interact with the L1 box motif (TAAATG(C/T)) (Nakamura et al., 2006). Such a motif was observed in the TPS promoter fragments bound by wo (Figure 2f). We next examined the effect of the W106R mutation on the binding ability of wo. Interestingly, the transformants containing pJG4‐5‐woW106R and the pLacZi‐pTPS constructs failed to turn blue after 5 days of incubation, suggesting that the W106R mutation disrupted the interaction between wo and the TPS promoters. To further verify the interaction, we performed a biotin‐IP assay and our results showed that the wo protein could be detected if the biotin‐promoter was added (Figure 2h).
It was previously reported that the number of type VI glandular trichomes decreased when JA signalling was disrupted (Bosch et al., 2014; Boughton et al., 2005; Li et al., 2004). In the RNA‐seq data, we identified 1610 co‐regulated genes in the overlap between the DEGs in the woW106R mutant and the previously reported JA‐responsive genes (Du et al., 2017) (Figure 3a). Among the co‐regulated genes, 42.5% (281 of 661) of the JA‐induced genes were down‐regulated in the woW106R mutant, while 34% (320 of 949) of the JA‐suppressed genes were up‐regulated in the woW106R mutant, indicating the high degree of overlap in the functions of wo and JA in trichome development (Figure 3b). Interestingly, the 262 JA‐induced and wo‐dependent genes were highly enriched in metabolic processes, suggesting that both JA and wo might regulate metabolism in tomato trichomes (Figure S5).
Figure 3.
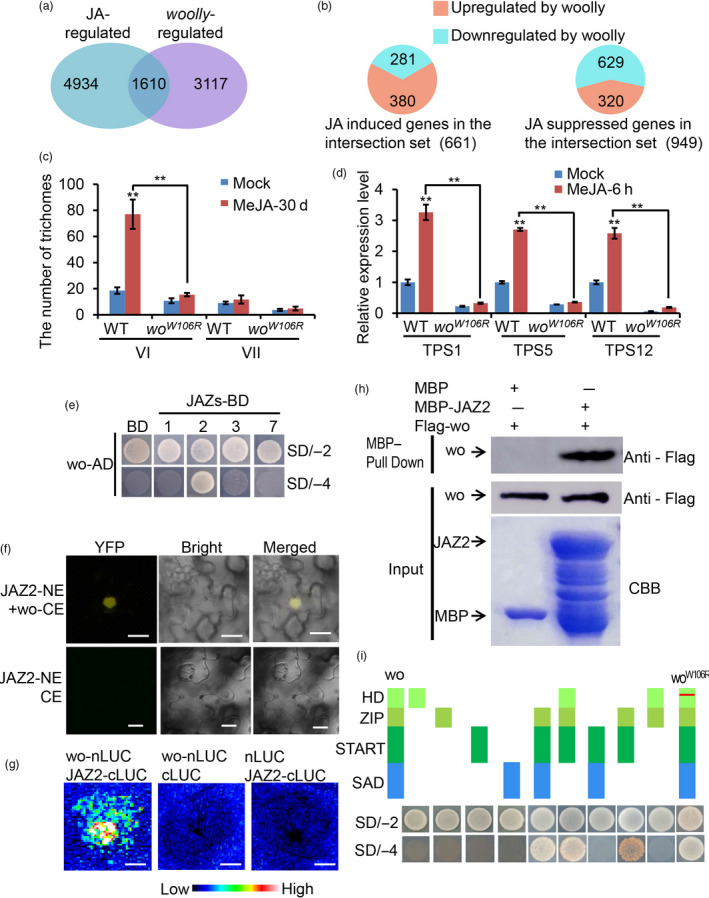
wo participates in JA signalling. (a) Venn diagram showing the overlap of wo‐regulated genes (differentially expressed genes between woW106R mutants and WT, FDR‐adjusted P value < 0.05) and JA‐induced genes (differentially expressed genes between MeJA‐treated and untreated WT). (b) Distribution of co‐regulated genes by JA and wo. (c) The number of type VI and VII trichomes on the leaf of WT and woW106R mutants with MeJA treatment. Values are represented as means ± SD (n = 10). Asterisks indicate significant differences by t‐test: ** P < 0.01, * 0.01 < P < 0.05. (d) Relative expression level of TPSs in the gland of type VI trichomes from WT and woW106R mutants with MeJA treatment. Values are represented as means ± SD (n = 3). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. (e) wo physically interacts with JAZ2 by Y2H. wo was cloned into pGADT7 vector (wo‐AD); the CDS of JAZs was inserted into pGBKT7 vector (JAZs‐BD). The transformants containing AD‐wo and empty pGBKT7 (BD) vector were used as negative control. All transformants were incubated on the SD/‐T‐L (SD/‐2) and SD/‐T‐L‐H‐A (SD/‐4) medium. The numbers (1, 2, 3 and 7) represent JAZ1, JAZ2, JAZ3 and JAZ7. (f) BiFC showing the interaction between wo and JAZ2. wo and JAZ2 were cloned into pUC‐SPYNE and pUC‐SPYCE respectively (wo‐CE and JAZ2‐NE). JAZ2‐NE and pUC‐SPYCE were used together as negative control. Scale bar = 10 μm. (g) The firefly luciferase complementation imaging (LCI) assay. JAZ2 and wo were cloned into the pCAMBIA1300‐cLUC (cLUC‐JAZ2) and pCAMBIA1300‐nLUC (nLUC‐wo) respectively. The combinations (nLUC‐wo + cLUC) and (cLUC‐JAZ2 + nLUC) were used as negative control. The colour bar below indicates the range of luminescence intensity in each image. Scale bar = 2.5 mm. (h) Pull‐down assay. Flag‐wo is pulled down by MBP‐JAZ2 (right lane), but not by MBP (left lane). The anti‐Flag antibody was used for the immunoblotting of Flag‐wo. (i) Y2H assay examining the interaction between the domains of wo or woW106R and JAZ2. The schematic diagram shows the four domains of wo and woW106R constructs (HD, ZIP, START and SAD). All domains were cloned into pGADT7 vector. The red line shows the W106R mutation in the HD domain of wo. [Colour figure can be viewed at wileyonlinelibrary.com]
To see if wo is essential for JA‐mediated regulation of trichome development and function, we quantified trichomes and TPS transcript levels in the leaves of WT and woW106R mutants after MeJA treatment (Figure 3c; Figure S4C). In the WT, MeJA treatment led to increased type VI trichomes while type VII trichomes seemed to be only marginally affected (Figure 3c; Figure S6). However, in the woW106R mutant, MeJA treatment had little effect on the number of type VI trichomes (Figure 3c). This result suggests that wo is indispensable for JA‐promoted development of type VI trichomes. Furthermore, qRT‐PCR revealed that the expression of three TPSs (TPS1, TPS5 and TPS12) was rapidly induced in WT leaves by MeJA, with a 3‐ to 30‐fold increase in expression after 6 h of MeJA treatment (Figure S4C). However, this induction of TPS1 and TPS5 was significantly inhibited in the woW106R mutant (Figure S4C). Compared to WT, the woW106R mutant showed only a slight up‐regulation of TPS12 expression after MeJA treatment (Figure S4C). Next, we extracted the glandular cells of the type VI trichomes for transcript analyses. Our results confirmed that the expression of the three TPS genes was induced in the glandular cells of the type VI trichomes by MeJA and that this induction was suppressed by the woW106R mutation (Figure 3d). Based on these observations, we inferred that functional wo is indispensable for JA‐induced trichome formation and TPS expression in tomato.
To further address the relationship between wo and JA signalling, we analysed the expression levels of eleven JAZs – the major repressors of the JA signalling pathway – in leaves and type VI trichomes (JAZ12 was not successfully amplified). JAZ1, JAZ2, JAZ3 and JAZ7 were highly expressed in type VI trichomes, while the remaining JAZs were mainly expressed in leaves (Figure S7). The overlapping expression patterns of the JAZs and wo prompted us to hypothesize that JAZs could serve as the integrators of JA signalling and wo function. To test this, we co‐transformed PGADT7‐wo and PGBKT7‐JAZ constructs into the yeast strain AH109 for a Y2H assay. Among the JAZs expressed in type VI trichomes (JAZs 1, 2, 3 and 7), only transformants carrying PGADT7‐wo and PGBKT7‐JAZ2 could grow on the SD/‐Ade/‐His/‐Leu/‐Trp medium, indicating that there was a specific interaction between wo and JAZ2 (Figure 3e). We further verified this specific binding in multiple systems including BiFC, LCI and pull‐down assays (Figure 3f–h). To pinpoint the interacting domain in wo, we performed Y2H using different domains of the wo protein. Our analysis showed that wo interacted with JAZ2 via a ZIP‐START domain and that a W106R mutation in the HD domain of wo had no effect on the interaction (Figure 3i). These results suggested that JA signalling interacts with wo via the repressor JAZ2 in tomato trichomes.
woolly and SlMYC1 form a regulatory module in tomato trichomes
Members of the MYC family of basic helix‐loop‐helix transcription factors were previously shown to act as vital components of JA signalling (Boter et al., 2004). In Arabidopsis, AtMYC2, AtMYC3 and AtMYC4 all interact with JAZ repressors in the JA signalling pathway (Fernández‐Calvo et al., 2011). Tomato has two homologous genes clustered in MYC2 subclade, SlMYC1 and SlMYC2 (Figure 4a). SlMYC2 has been demonstrated to regulate JA‐mediated plant immunity in tomato through a hierarchical transcriptional cascade (Du et al., 2017). It was recently reported that most of type VI trichomes were replaced by type VII‐Like (VII‐L) trichomes and terpene biosynthesis was impaired when SlMYC1 expression was reduced (Xu et al., 2018). Consistent with this observation, we found by qRT‐PCR that SlMYC1 was highly expressed in type VI trichomes (Figure S8A). To determine the spatiotemporal expression pattern of SlMYC1, we stably transformed tomato with the SlMYC1 promoter (about 2500 bp upstream of the SlMYC1 gene) fused with the GUS reporter (pSlMYC1:GUS). GUS staining clearly showed GUS expression in the type VI and VII trichomes (Figure S8C). In addition, we also detected a weak GUS signal in other tissues (Figure S8B), indicating widespread low level expression of SlMYC1 along with higher levels of expression in trichomes.
Figure 4.
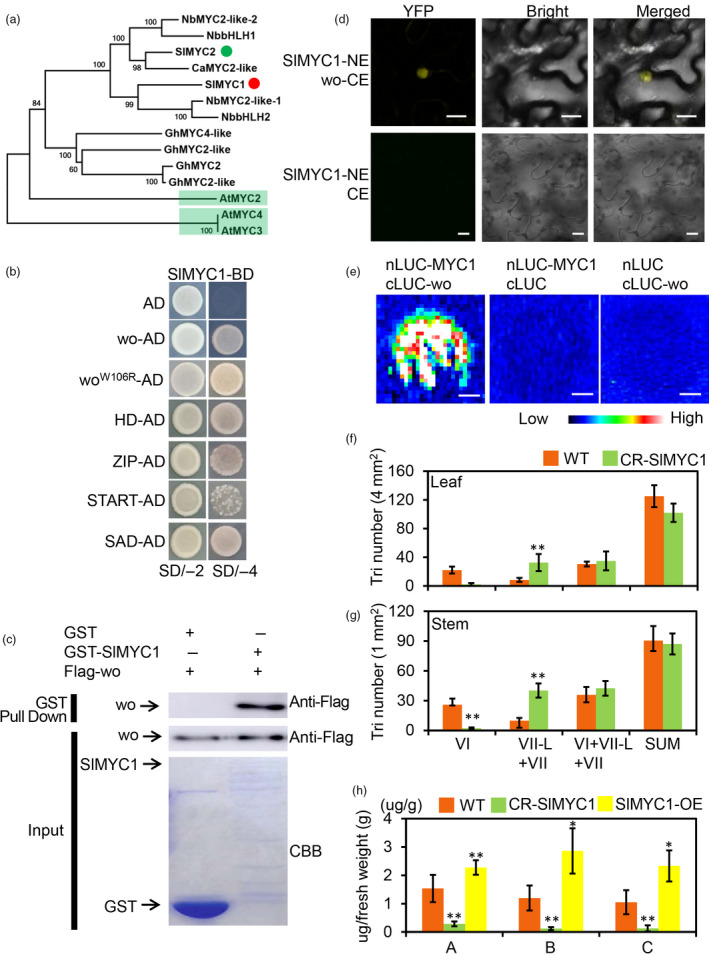
wo physically interacts with SlMYC1. (a) Cladogram of homologous genes of SlMYC1.The red and green dots represent SlMYC1 and SlMYC2 respectively. The light green highlights AtMYC2, AtMYC3 and AtMYC4. (b) wo, woW106R and the four domains of wo interact with SlMYC1 by Y2H. SlMYC1 was cloned into the pGBKT7 vector (SlMYC1‐BD). The coding sequence of wo, woW106R and four domains were inserted into pGADT7 (wo‐AD and woW106R‐AD). The transformant containing SlMYC1‐BD and pGADT7 (AD) was used as negative control. All transformants were incubated on the SD/‐T‐L (SD/‐2) and SD/‐T‐L‐H‐A (SD/‐4) medium. (c) Pull‐down assay. Flag‐wo is pulled down by GST‐SlMYC1 (right lane), but not by GST (left lane). The anti‐Flag antibody was used for the immunoblotting of Flag‐wo. The positions of various purified proteins separated by SDS‐PAGE are marked with asterisks on the CBB‐stained gel. (d) wo interact with SlMYC1 by BiFC. SlMYC1 and wo were cloned into pUC‐SPYNE and pUC‐SPYCE respectively (SlMYC1‐NE and wo‐CE). (pUC‐SPYCE (CE) + JAZ2‐NE) were used as negative control. Scale bar = 10 μm. (e) The firefly LCI assay. wo and SlMYC1 were cloned into the pCAMBIA1300‐cLUC and pCAMBIA1300‐nLUC respectively (cLUC‐wo and nLUC‐SlMYC1). The combinations (nLUC‐SlMYC1 + cLUC) and (cLUC‐wo + nLUC) were used as negative control. The colour bar below presents the range of luminescence intensity in each image. Scale bar: 2.5 mm. (f, g) The density of trichomes (tri) on the leaves (f) and stems (g) of WT and CR‐SlMYC1. ‘SUM’ on the X‐axis means the sum of all types of trichomes. VII‐L represents the type VII‐Like trichomes. The Y‐axis shows the density of the trichomes. Values are represented as means ± SD (n = 10). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. (h) GC‐MS analysis shows the terpene content in WT, CR‐SlMYC1 and SlMYC1 overexpression (SlMYC1‐OE) lines. a: 2‐Carene, b: β‐Phellandrene, c: α‐humulene. The Y‐axis shows the content of the terpenes (μg) per gram (g) of fresh leaf. Values are represented as means ± SD (n = 5). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. [Colour figure can be viewed at wileyonlinelibrary.com]
Because both wo and SlMYC1 were expressed in type VI trichomes and regulated the expression of TPS genes, we inferred that wo and SlMYC1 likely function in the same pathway in the regulation of terpene synthesis. To test this, we first analysed the potential interaction between these two transcriptional factors. We constructed a bait vector by cloning the full‐length CDS of SlMYC1 into the pGBKT7 vector and then co‐transformed this bait vector together with pGADT7 carrying the full‐length CDS of wo as well as four domains of wo into the AH109 yeast strain. The transformants carrying both wo‐pGADT7 and SlMYC1‐pGBKT7 could grow on SD/‐Trp/‐Leu and SD/‐Trp/‐Leu/‐His/‐Ade medium, indicating that there was interaction between wo and SlMYC1 (Figure 4b). We further corroborated this interaction by BiFC and LCI assays in 5‐week‐old N. benthamiana leaves and by an in vitro pull‐down assay (Figure 4c–e). In the Y2H assay, SlMYC1 seemed to interact with all four domains of wo, and a W106R mutation in wo failed to affect this interaction (Figure 4b).
To functionally characterize the interaction between wo and SlMYC1, we analysed the trichome phenotypes in SlMYC1 loss‐of‐function mutants. Using CRISPR‐Cas9, we obtained stable SlMYC1 knockout lines in tomato (named CR‐SlMYC1 lines) (Figure S9A, E). We found that the long trichomes on both stems and leaves were not considerably changed in CR‐SlMYC1 plants compared with WT (Figure S9F‐I). Under SEM, we rarely saw type VI trichomes but found many type VII‐like trichomes in CR‐SlMYC1 plants and fruits (Figure S9J‐O; Figure S10). Further quantification of mature adaxial leaves and stems showed that the density of type VI trichomes was significantly reduced in CR‐SlMYC1, while the number of small glandular trichomes (type VI, VII and VII‐like) was the same as in the WT (Figure 4f, g). Consistent with a previous report (Xu et al., 2018), GC‐MS showed that monoterpenes (2‐carene, β‐phellandrene) and sesquiterpenes (α‐humulene) were markedly reduced in CR‐SlMYC1 plants, indicating the vital role of SlMYC1 in terpene biosynthesis (Figure 4h).
The wo‐SlMYC1 module participates in JA‐mediated terpene biosynthesis
SlMYC1 has been reported to promote the expression of TPS3 and TPS5 (Spyropoulou et al., 2014). Here we examined the expression of three wo‐regulated TPSs (TPS1, TPS5 and TPS12) in WT and CR‐SlMYC1 plants. We found that the expression of these wo‐regulated TPSs was substantially down‐regulated in the small glandular trichomes (types VI, VII‐L, and VII) and leaves of the CR‐SlMYC1 plants (Figure 5a, Figure S11). We next performed Y1H to see if SlMYC1 directly regulates TPS genes. Unexpectedly, SlMYC1 failed to interact with TPS promoters in our system (Figure 5b). Even more surprisingly, our Dual‐LUC activation assay showed that SlMYC1 indeed activated the expression of TPS genes (Figure 5c). We also unexpectedly observed that wo was not able to activate the expression of TPS genes in the Dual‐LUC activation assay despite the fact that wo directly binds to TPS promoters (Figure 5d). Based on these observations, together with the fact that SlMYC1 physically associates with wo, we hypothesized that SlMYC1 may be recruited by wo to TPS promoters for the transcriptional activation. To test this hypothesis, we performed the Dual‐LUC activation assay using protoplasts of woW106R mutant. The result showed that neither wo nor SlMYC1 was sufficient to activate TPS1 and TPS5 expression in the woW106R mutant background, while TPS12 could be slightly induced by SlMYC1 (Figure 5e). However, the expression of the three TPS genes was substantially enhanced when both wo and SlMYC1 were added to the system, indicating that wo and SlMYC1 work together in TPS activation (Figure 5e).
Figure 5.
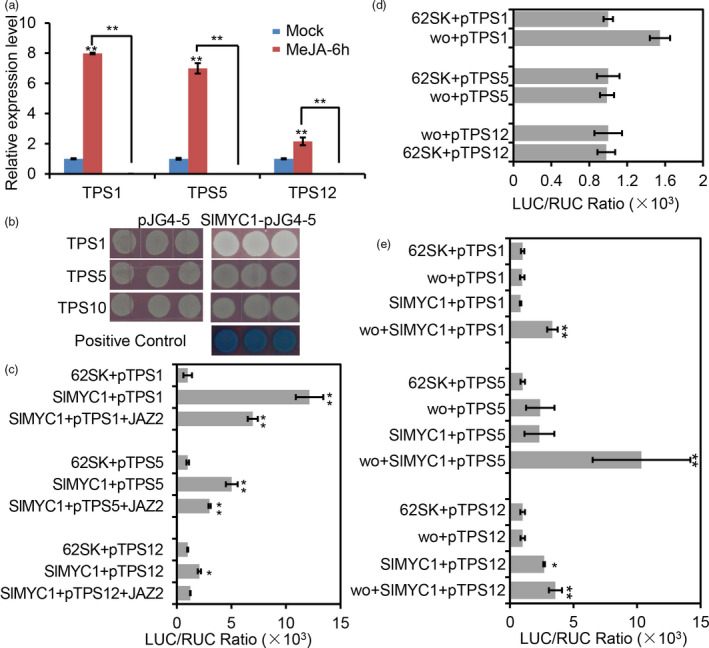
SlMYC1 participates in JA signalling. (a) Relative expression level of TPSs in the glandular cells of the type VI trichomes in WT and CR‐SlMYC1 plants with MeJA treatment. Values are represented as means ± SD (n = 3). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. (b) Y1H assay. The CDS of SlMYC1 was cloned into the vector pJG4‐5 (SlMYC1‐pJG4‐5). The promoters of TPSs were inserted into the vector pLacZi. Empty vector pJG4‐5 was used as negative control. The transformed yeast cells were grown on SD (‐Trp‐Ura + X‐gal). The blue clones show the positive interaction between wo and the promoters of TPSs. (c–e) The activation assay by dual‐LUC in tobacco leaf protoplasts (c, d) and woW106R mutants (e). The full‐length CDS of wo and SlMYC1 were cloned into the effector vector (pGreen II 62‐SK); the promoters of TPSs were introduced into the reporter vector (pGreen II 0800). The TPS promoter plus pGreen II 62‐SK (62SK) were used as control. LUC, firefly luciferase activity; RLU, Renilla luciferase activity. Values are represented as means ± SD (n = 5). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. [Colour figure can be viewed at wileyonlinelibrary.com]
Since wo participates in JA‐mediated trichome regulation, we further tested the potential involvement of SlMYC1 in JA signalling. We treated CR‐SlMYC1 plants with MeJA for 6 h and examined the TPS expression pattern. Knock out of SlMYC1 blocked the JA‐induced up‐regulation of TPS1, TPS5 and TPS12, indicating that SlMYC1 function is indispensable for JA‐induced TPS expression in tomato (Figure 5a, Figure S11). To functionally test the regulation of SlMYC1 by JAZ2, we performed a Dual‐LUC reporter assay. The expression of TPS genes induced by SlMYC1 was significantly repressed when JAZ2 was added in the Dual‐LUC assay, indicating that SlMYC1 activity can be repressed by JAZ2 (Figure 5c).
JAZs can disrupt the SlMYC1‐wo interaction
In other species and systems, a number of MYC members, including MYC2, MYC3 and MYC4, were reported to be repressed by JAZ JA‐signalling inhibitors. To understand how SlMYC1 is regulated in JA signalling, we first performed a Y2H screen to analyse the interaction between SlMYC1 and 10 JAZ proteins of tomato (JAZ11 and JAZ12 were not successfully cloned) (Du et al., 2017). Among the tomato JAZs, SlMYC1 could specifically interact with JAZ2, JAZ4, JAZ6, JAZ8 and JAZ10 (Figure 6a). Because wo specifically interacts with JAZ2, we focused on JAZ2 and further verified the interaction between JAZ2 and SlMYC1 in multiple systems. BiFC, LCI and pull‐down assays all supported the conclusion that JAZ2 could physically bind to SlMYC1 (Figure 6b–d).
Figure 6.
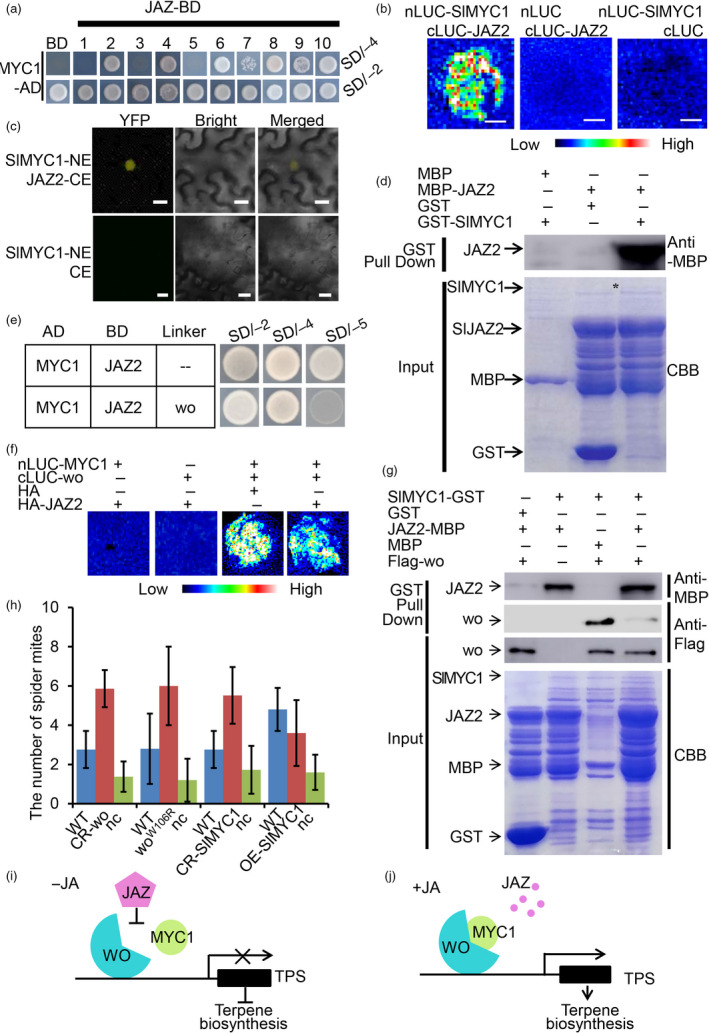
JAZ2 disrupts the interaction between wo and SlMYC1. (a) Y2H showing the interaction between SlMYC1 and ten JAZs. SlMYC1 and JAZs were cloned into pGADT7 and pGBKT7 vectors respectively. The transformants containing AD‐SlMYC1 and empty pGBKT7 (BD) vector were used as the negative control. All transformants were incubated on the SD/‐T‐L (SD/‐2) and SD/‐T‐L‐H‐A (SD/‐4) medium. (b) LCI assay. nLUC‐SlMYC1 represents SlMYC1 in the pCAMBIA1300‐nLUC vector. cLUC‐JAZ2 represents JAZ2 in the pCAMBIA1300‐cLUC vector. Combinations of (cLUC‐JAZ2 + nLUC) and (nLUC‐SlMYC1 + cLUC) were used as the negative control. The colour bar below presents the range of luminescence intensity in each image. (c) BiFC assay. SlMYC1‐NE represents SlMYC1 in pUC‐SPYNE vector. JAZ2‐CE represents JAZ2 in the pUC‐SPYCE vector. SlMYC1‐NE plus pUC‐SPYCE (CE) were used as the negative control. Scale bar = 10 μm. (d) Pull‐down assay. MBP‐JAZ2 is pulled down by GST‐SlMYC1 (right lane), not by GST (left lane). The anti‐MBP antibody was used for the immunoblotting of JAZ2. The positions of various purified proteins separated by SDS‐PAGE are marked with asterisks on the CBB‐stained gel. (e) Y3H assays showing that JAZ2 and wo compete to interact with SlMYC1. The AD‐SlMYC1 and pBridge wo‐JAZ2 were co‐transformed into the yeast strain AH109. The AD‐SlMYC1 and BD‐JAZ2 were used as the control. All transformants were incubated on the SD/‐T‐L (SD/‐2), SD/‐T‐L‐H‐A (SD/‐4) and SD/‐T‐L‐H‐A‐M (SD/‐5) medium. (f) LCI assay showing JAZ2 disrupts the SlMYC1‐wo interaction. cLUC‐wo represents wo in the pCAMBIA1300‐cLUC vector. nLUC‐SlMYC1 represents SlMYC1 in the pCAMBIA1300‐nLUC vector. HA‐JAZ2 represents JAZ2 in the pUC‐SPYCE vector. The colour bar below presents the range of luminescence intensity in each image. Scale bar: 2.5 mm. (g) In vitro pull‐down assay showing the competitive binding between JAZ2 and SlMYC1 with wo. The JAZ2‐MBP and Flag‐wo are pulled down by SlMYC1‐GST (the second lane and third lane). Fixed amounts of SlMYC1‐GST and Flag‐wo fusion proteins were incubated with JAZ2‐MBP fusion protein. The assay shows that the amount of Flag‐wo pulled down by SlMYC1‐GST is reduced (the right lane). The positions of various purified proteins separated by SDS‐PAGE are marked with asterisks on the CBB‐stained gel. (h) Two‐choice assays showing the differential spider resistance in WT, CR‐wo, woW106R mutants, CR‐SlMYC1 and OE‐SlMYC1 lines. nc represents the number of spiders that fail to make a choice. Values are represented as means ± SD (n = 5). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. (i, j) The model of the wo‐SlMYC1 module regulating terpene biosynthesis. Without JA, JAZ2 prevents the wo‐SlMYC1 interaction via protein–protein interaction, inhibiting the TPS activation and terpene biosynthesis (i). JA induces the degradation of JAZ2, so wo can interact with SlMYC1 to activate the expression of TPSs and terpene synthesis (j). [Colour figure can be viewed at wileyonlinelibrary.com]
Since both wo and SlMYC1 take part in JA signalling and interact with the suppressor JAZ2, we next investigated whether JAZ2 inhibits the interaction between wo and SlMYC1. To address this possibility, we conducted yeast three‐hybrid (Y3H) assays. In our experiments, SlMYC1 and JAZ2 interacted on SD/‐A‐H‐T‐L medium (SD/‐4) but induction of wo led to a dramatic reduction of the SlMYC1‐JAZ2 interaction on SD/‐A‐H‐T‐L‐M medium (SD/‐5) (Figure 6e). To corroborate this observation, we performed competitive pull‐down assays and an LCI experiment. Our results showed that the ability of SlMYC1‐GST to pull down Flag‐wo decreased as JAZ2‐MBP was added in the pull‐down system (Figure 6g). Consistently, LCI activity, which reflects wo‐SlMYC1 interaction, was weakened when JAZ2 was added in the LCI system (Figure 6f). Together, our results suggested that wo recruited SlMYC1 to TPS promoters for gene activation. Without JA, the repressor JAZ2 disrupts the interaction between SlMYC1 and wo (Figure 6i). In presence of JA, the repressor JAZ2 is degraded, the wo‐SlMYC1 module activates the expression of TPS genes, and terpene biosynthesis is promoted (Figure 6j).
Glandular trichome density and terpene levels are both thought to affect pest resistance in plants. To examine how the wo‐SlMYC1 module affects insect resistance, we conducted a two‐choice assay using spider mites feeding on tomato. Two hours after initiation of the feeding, more than 90% of the mites had moved to either WT or mutant leaves. Compared to WT, we found that the mites preferred to move to CR‐wo, woW106R mutant, and CR‐SlMYC1 leaves (Figure 6h). We also constructed SlMYC1 overexpression lines (OE‐SlMYC1) and quantified the number of type VI and VII trichomes. The results showed that overexpression of SlMYC1 did not increase the number of type VI and VII trichomes (Figure S9A&C). We also measured the levels of volatile terpenes in the overexpression lines using GC‐MS. Our results showed that the level of monoterpenes (2‐carene, β‐phellandrene) and sesquiterpenes (α‐humulene) was significantly increased in the OE‐SlMYC1 plants (Figure 4h, Figure S9D). The two‐choice assay also showed that spider mites preferred to move to leaves of the WT compared with those of the OE‐SlMYC1 plants (Figure 6h). To further verify this result, we performed a mite feeding assay. Nine‐leaf‐stage (~4‐week‐old) WT and OE‐SlMYC1 plants were infested with adult mites. After five days of feeding, all leaves of the WT plants, particularly the fifth leaf, became yellow and wilted and had visible spider webs, in sharp contrast to the mild symptoms of OE‐SlMYC1 plants (Figure S12A,B). Quantitation of the number of wounded leaves and the areas of the chlorotic lesions showed that WT leaves suffered significantly more damage than those of OE‐SlMYC1 plants (Figure S12C,D).
JA‐independent function of SlMYC1 in type VI glandular trichomes
The opposite changes in the number of type VI and type VII‐like trichomes observed in CR‐SlMYC1 plants raised the possibility that SlMYC1 could function to maintain the balance between these two types of trichomes. A previous report and our observations both showed that the knockout of SlMYC1 resulted in the disappearance of type VI trichomes and an increase of type VII or VII‐like trichomes (Xu et al., 2018). Xu et al. reported that SlMYC1‐RNAi resulted in a reduction in the diameter of the glandular cells and the length of the stalk cell in type VI trichomes. One possible explanation for this phenotype is that the additional type VII‐like trichomes were not true type VII trichomes. To address this, we carefully examined the type VII‐like trichomes in CR‐SlMYC1 plants. As shown in Figure 7e, f and Figure S13, almost all the type VII‐like trichomes exhibited typical features of type VI trichomes, including the arrangement of the glandular cells at the top and the presence of intermediate cells between the glandular cells and the stalk (Figure 7a–h; Figure S13). We also found that the glandular trichomes on WT fruit were mostly type VI trichomes but not type VII trichomes, while most of the glandular trichomes on the fruit of CR‐SlMYC1 were type VII‐like with type VI trichome features (Figure S13). However, the size of the glandular cells of the type VII‐like trichomes was significantly smaller, with an average diameter about 1/3 that of the glandular cells in the type VI trichomes in the WT (Figure 7i). In addition, about 55% of the smaller glandular trichomes (VII‐like plus VII) were type VII‐like trichomes in CR‐SlMYC1 leaves, similar to the percentage of typical type VI trichomes in the smaller glandular trichomes (VI plus VII) of WT (Figure 7j).
Figure 7.
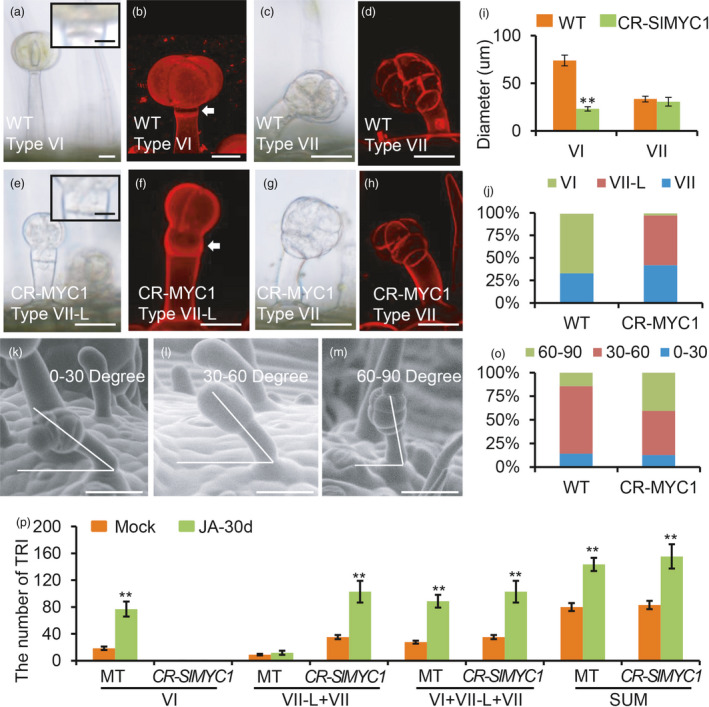
SlMYC1 regulates the morphology of type VI trichomes. (a‐h) The morphology of the type VI, VII and VII‐Like (VII‐L) trichomes in WT and CR‐MYC1 plants under DIC (a, c, e, g) and confocal microscope (b, d, f, h). The upper right insets in the Figure a and e show the close‐up view of the intermediate cell of type VI trichomes. Scale bar: 10 μm. The white arrows in the Figure b and f point to the intermediate cells. Trichomes were stained with propidium iodide (PI) in the Figure b, d, h and f. Scale bar: 25 μm. (i) The diameter of the glandular cells of type VI and VII trichomes in WT and CR‐MYC1 plants. Values are represented as means ± SD (n = 100). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. (j) The percentage of the type VI, VII‐L and VII trichomes in WT and CR‐MYC1 plants based on the morphology of the glandular cells. Three independent lines of CR‐SlMYC1 plants were measured. (k–m) The curvature of type VII trichomes in the WT. The curvature is 0‐30 degree (k), 30–60 degree (l) and 60–90 degree (m) relative to the leaf surface. The images were taken by SEM. Scale bar: 30 μm. (o) The percentage of the small glandular trichomes with different curvatures in WT and CR‐MYC1 plants. Three independent lines of CR‐SlMYC1 were measured. (p) The number of the trichomes on the leaf of WT and CR‐SlMYC1 plants with MeJA treatment. ‘SUM’ on the X‐axis means the sum of all types of trichomes. Values are represented as means ± SD (n = 10). Asterisks indicate significant differences by t‐test: **P < 0.01, *0.01 < P < 0.05. [Colour figure can be viewed at wileyonlinelibrary.com]
In addition to their glandular morphology, type VII trichomes exhibited a slight slant, forming an angle less than 90 degrees relative to the epidermis. In WT, about 15% of the type VII trichomes formed 60‐ to 90‐degree angles, and about 75% formed 30‐ to 60‐degree angles. In CR‐SlMYC1 plants, about 40% of the smaller glandular trichomes (VII‐like plus VII) formed angles of 60 to 90 degrees, and the proportion showing angles of 30 to 60 degrees was reduced to 40% (Figure 7o). In addition to that, the type VII‐L trichomes were all stood straight up (Figure S10). These statistics imply that the additional small glandular trichomes in CR‐SlMYC1 plants were defective type VI trichomes with more but smaller glandular cells and that SlMYC1 is more likely to be involved in regulating the morphology rather than the initiation of type VI trichomes.
We also analysed the number of small glandular trichomes in CR‐MYC1 plants treated with MeJA and found that the glandular morphology of type VI trichomes was not restored (Figure 7p; Figure S14). In addition, the amount of type VII‐like trichomes in CR‐MYC1 plants increased to the same level as in WT when treated with MeJA, suggesting this SlMYC1 regulation was possibly independent of JA.
Finally, we examined the spatiotemporal expression pattern of SlMYC1 using pSlMYC1::GUS transgenic tomatoes. GUS staining showed that SlMYC1 was expressed in the glands and the stalk cells of both type VI and type VII trichomes (Figure S8C). Transcript analyses and in situ assays supported this spatial expression pattern of SlMYC1 (Figure S8D). Interestingly, SlMYC1 expression progressively increased with glandular cell development, with almost no expression at the one‐GC (glandular cell) stage of type VI trichome development (Figure S8E). Therefore, SlMYC1 might not be involved in the initiation of type VI trichomes, but more likely regulates the development of the type VI trichomes, which includes the morphology of stalk cells and intermediate cells, as well as the division, expansion and metabolism of glandular cells at later stages.
Discussion
Trichome formation in Arabidopsis is subject to a complex regulatory network in which multiple factors form a MYB‐bHLH‐WD module to promote trichome specification (Serna and Martin, 2006). In contrast to the unicellular structure of Arabidopsis trichomes, tomato forms seven types of multicellular trichomes (Luckwill, 1943). In particular, four types of tomato trichomes contain a glandular head at their tips, in which a variety of insect‐repellent chemicals are produced. Although a tomato TRY homolog, SlTRY, was shown to inhibit trichome initiation when overexpressed in Arabidopsis, tomato homologs of other trichome regulators identified in Arabidopsis, including SlGL3, have no roles in promoting trichomes in Arabidopsis (Tominaga‐Wada et al., 2013). Given that tomato and Arabidopsis have totally different trichomes, the trichome regulation in these two species is presumably different.
So far, very few regulators of trichome development have been identified in tomato. Moreover, most studies have focused on type I trichomes, partly because their length makes them the easiest type to characterize. For example, woolly (wo) was initially identified from a dominant mutant that promoted type I trichome formation in tomato (Yang et al., 2011). A wo‐interacting transcription factor named Hair was also found to regulate type I trichomes (Chang et al., 2018). Nevertheless, the detailed mechanism of wo function is still far from clear. In addition, tomato produces other types of trichomes, particularly those with glandular cells in which a large number of insect‐repellent metabolites are produced (Kang et al., 2010; Ma et al., 2016). By genetic screen, we identified a loss‐of‐function allele of wo (woW106R ). In this recessive mutant and CRISPR‐Cas9 knockout lines, we found that the wo gene is indispensable for the development of other types of trichomes, including those with glandular cells. The previously reported dominant mutations were located in the SAD domain of the wo gene (P635R) and caused an increase in type I trichomes and a decrease in type VI trichomes (Vendemiatti et al., 2017; Yang et al., 2015; Yang et al., 2011). In contrast, the EMS‐induced mutation woW106R is located in the HD domain of wo, but it also caused a reduction in type VI trichomes. This phenotype is supported by the CRISPR‐Cas9 wo knockout lines, suggesting that mutations within different domains of wo cause different effects on wo function. A recent study reported that the same dominant mutation in tobacco, NbWoV , led to a phenotype similar to that of the tomato mutant (Wu et al., 2020). Surprisingly, overexpression of the wo gene failed to cause the same phenotype, and Nbwo seemed to be inhibited by NbCycB2 while NbWoV was not, suggesting that Nbwo and NbWoV could be functionally distinct (Wu et al., 2020). Thus, mutations within the SAD domain might stabilize wo protein or release it from potential inhibitors, while mutations within the HD domain lead to loss‐of‐function of wo.
Recently, SlMYC1 was reported to regulate the number of type VI trichomes and it was proposed that SlMYC1 controls their initiation (Xu et al., 2018). However, the results of our careful morphological examination, quantification and multiple biochemical analyses bring this hypothesis into question. In CR‐SlMYC1 lines, we found that the apparent reduction in type VI trichomes was likely caused by their decreased size, which transformed them into type VII‐like trichomes. These trichomes retained most of the features of type VI trichomes. Interestingly, however, SlMYC1 forms a regulatory module with wo to promote terpene biosynthesis in type VI glandular cells. Without the recruitment of wo, SlMYC1 alone is not able to associate with TPS promoters. However, the wo gene has a broader expression domain than SlMYC1, and wo mutants have more diverse phenotypes than the CR‐SlMYC1 lines. Thus, wo and SlMYC1 have both overlapping and separate roles in tomato trichomes.
JA has been reported to induce the development of type VI trichomes in tomato (Bosch et al., 2014; Boughton et al., 2005; Li et al., 2004; Xu et al., 2018), but the mechanism behind this induction remains unknown. Here we found that wo was required for JA‐induced type VI trichome formation. Using multiple systems, we demonstrated that wo can specifically interact with SlJAZ2 and that the inhibitory effect of this interaction can be relieved by JA. Consistent with these observations, it was reported previously that overexpression of SlJAZ2 in tomato led to a reduction in the number of trichomes (Yu et al., 2018). Despite the fact that SlMYC1 functions in JA‐mediated terpene biosynthesis, JA‐induced type VI trichome formation seems to be independent of SlMYC1. In wo mutants, the JA‐promoted increase in trichome abundance was significantly suppressed, suggesting that wo and SlMYC1 have distinct roles in tomato trichome formation. After type VI trichome initiation, SlMYC1 is presumably involved in glandular cell division and trichome morphogenesis, as these two types of regulation became aberrant in CR‐SlMYC1 lines. In addition to JA, GA also affects trichomes in a few species. It was found that the DELLA protein, a suppressor of GA signalling, could bind to the WD‐Repeat/bHLH/MYB complex to inhibit trichome initiation in Arabidopsis (Qi et al., 2014). In addition, DELLA was reported to inhibit AtMYC2 during sesquiterpene synthesis (Hong et al., 2012). In cotton, DELLA also interacts with GhHOX3, an HD‐ZIP IV transcription factor, to regulate fibre elongation (Shan et al., 2014). It is likely that DELLA could also interact with the wo‐SlMYC1 module during trichome development in tomato.
The overlap between wo and SlMYC1 expression is in the glandular cells of type VI trichomes. Here we presented multiple lines of evidence supporting the idea that SlMYC1 is recruited by wo to TPS promoters to activate transcription. First, wo directly binds to TPS promoters but alone cannot activate TPS expression. Second, SlMYC1 is sufficient to promote TPS expression but does not interact with TPS promoters. Third, SlMYC1 physically interacts with wo and SlMYC1 fails to induce TPS expression without wo function. Interestingly, however, the interaction between wo and SlMYC1 can be suppressed by JAZ2, and the functions of this regulatory module are greatly enhanced in presence of JA. This model also implies that terpene synthesis and type VI trichome formation are two separate processes. The substantial reduction of terpene levels in woW106 mutants presumably results from a combination of both decreased type VI trichomes and suppressed terpene biosynthesis.
In conclusion, we think that wo is required for MeJA‐induced trichome formation and SlMYC1 plays a key role in the morphological development of the type VI trichomes. In the mature glandular cells of type VI trichomes, SlMYC1 can be recruited by wo to activate the expression of TPS genes for terpene biosynthesis. The wo‐SlMYC1 functional module can be repressed by SlJAZ2 and JA can relieve this repression. In addition to acting together with wo, SlMYC1 also plays wo‐independent roles in glandular cell division and expansion (Figure 8).
Figure 8.
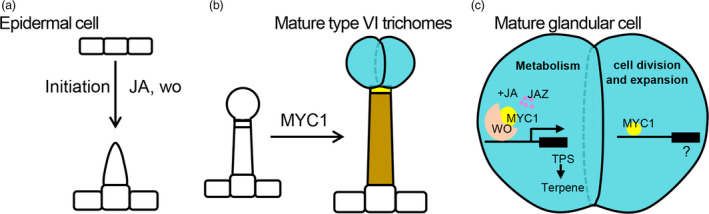
The model depicting the regulatory mechanism of the development and metabolism of tomato trichomes. (a) The trichome initiation requires woolly function, and JA signalling can promote the trichome initiation. woolly is involved in the initiation of almost all types of trichomes with indispensible role in type I, II and III, important role in type IV and V, and partial role in type VI and VII. (b) SlMYC1 regulates the morphology of the glandular cells, intermediate cells and the stalk cells of type VI trichomes. The mature type VI trichomes contain four glandular cells (in blue) where terpenes are mainly biosynthesized. The intermediate cell is shown in yellow, and the stalk cell is shown in brown. (c) The model depicting the regulatory mechanism of the cell division, expansion and metabolism in the glandular cells. wo‐MYC1 module regulates the terpene metabolism, and JA can promote the terpene biosynthesis through the degradation of JAZ2 that represses the wo‐MYC1 module. [Colour figure can be viewed at wileyonlinelibrary.com]
Methods
Plant materials and growth conditions
Tomato (Solanum lycopersicum) cv. Micro‐Tom was used as wild‐type (WT). Seeds of WT and the short hair mutant (TOMJPW3717‐1; name as woW106R) were obtained from TOMATOMA (http://www.tomatoma.nbrp.jp/). Tomato plants were grown in the growth chamber with controlled temperature and light (26 °C under 16‐hour light and 22 °C under 8‐h dark cycles). Nicotiana benthamiana plants were grown in the growth chamber with 22 °C under 16‐h light and 22 °C under 8‐h dark cycles.
Bulked Segregant Analysis (BSA)
Mapping populations were constructed by crossing the short hair mutant and WT. The F2 plants (>200) were used for the phenotype segregation analysis. The genomic DNA of individuals exhibiting mutant‐like phenotypes (>30) and individuals with WT‐like phenotypes (>30) were extracted by CTAB. 2 μg DNA of individuals was mixed to construct two bulks (mutant‐like bulk and WT‐like bulk). Two bulks were sequenced (>30× coverage of the tomato genome) by HiseqXten‐PE150 (Novogene). Trimmed sequences were mapped onto the tomato reference genome. The allelic variant frequencies were analysed and the SNPs with high frequency were identified based on the previous reports (Chang et al., 2019; Garcia et al., 2016).
Molecular biology
For the overexpression of SlMYC1, the full‐length coding sequence of SlMYC1 was amplified using Phanta Max Super‐Fidelity DNA (Vazyme Biotech P505) and cloned into the pHELLSGATE8 vector driven by the cauliflower mosaic virus (CaMV) 35S promoter. To produce mutations in the coding sequence of SlMYC1 and wo, the recombinant pTX vector was designed according to the previous report (Ye et al., 2017). Two target sites were designed on the website (http://skl.scau.edu.cn/targetdesign/) and cloned into the pTX vector using ClonExpress II One Step Cloning Kit (Vazyme Biotech C112‐01/02).
For the promoter‐GUS reporters, about 2500 bp DNA fragment upstream of the examined gene was amplified and cloned into plant binary PMV2 (modified from pHELLSGATE8). The primers used in this experiment were listed in the Table S3.
RNA isolation and real‐time PCR analysis
For RNA extraction, plant tissues were collected and the total RNA was extracted using TransZol (TransGen Biotech ET101‐01). First‐strand cDNA was synthesized from 1mg of the total RNA using HiScript 1st Strand cDNA Synthesis Kit (Vazyme Biotech R111‐01). For qRT‐PCR, AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Q121‐02/03) was used. qRT‐PCR was performed on a CFX384 Real‐Time system (BIO‐RAD) with the cycling program: 5 min 95 °C, 40 cycles of 15 s at 95 °C, 30 s at 55 °C and 15 s at 72 °C. The expression level of target genes was normalized to ACTIN2 gene (Solyc11g005330). Three biological replicates were performed and error bars represent the SD of three biological replicates.
Analysis of trichome morphology and density
The trichomes were visualized either on the stereomicroscope (DFC550 LEICA) or the scanning electron microscopy TM3030Plus (HITACHI JAPAN) at an accelerating voltage of 15 kV. The density of trichomes on the adaxial surface of leaves was determined by counting trichomes on the second leaflet pair of the third leaf in each 4‐week‐old tomato plant (The third leaf is counted in the direction of the new leaf to the old leaf; i.e., the youngest leaf is counted as the first one). For the analysis of trichome density on stems, we chose the stems from the third internodes of 4‐week‐old plants (internodes were counted in the direction from meristematic tissues to hypocotyls. i.e., the internode close to the meristem is the first internode). For each line, about 6‐7 images were taken for the analysis. To image the trichome morphology, the trichomes were dipped in the propidium iodide buffer (200 μg/mL). The samples were then imaged by a Zeiss LSM 880 confocal laser scanning microscope. Error bars represent the SD of the biological replicates.
Harvest of glandular cells of type VI trichomes
The glandular cells of type VI trichomes were harvested by a bead‐beating method from 4‐week‐old plants. The stems and leaves of WT were harvested into the 500 mL glass bottle with 200 mL 70% ethanol and 15 g of 0.75–1 mm glass beads. The bottles were shaken by hand for 2 min. The mixture of glandular cells and other tissues was filtered first through a 350‐μm nylon mesh followed by a 100‐μm mesh after shaking to remove stem and glass beads. Then the supernatant containing the glandular cells of type VI trichomes was transferred to the 50 mL centrifugal tubes. After the centrifugation at 1561 g for 5 min, the glandular cells of type VI trichomes precipitated to the bottom of the centrifugal tubes (Bergau et al., 2016; Schilmiller et al., 2010).
Analysis of RNA‐seq data
For RNA‐seq, young leaflets were collected from 4‐week‐old WT and woW106R mutants. The standard RNA‐sequencing libraries were prepared based on the protocol of the TruSeq Stranded Total RNA Library Prep Kit (Illumina). Paired‐end reads (150 bp) were obtained using an Illumina sequencing system Hiseq 2500 platform (Illumina). Reads were quality‐filtered and the tomato genome SL2.50 (https://solgenomics.net) was used as the reference genome. Reads were aligned to the genome using hisat2 software (http://ccb.jhu.edu/software/hisat2/faq.shtml), and then, transcripts were assembled and estimated for their abundances using cufflinks (http://cole‐trapnell‐lab.github.io/cufflinks/). Differentially expressed genes were determined using cuffdiff (http://cole‐trapnell‐lab.github.io/cufflinks/). Significance was assessed using a threshold of log2 fold change >1, and transcript p‐values were false discovery rate (FDR) (P‐value < 0.01) corrected for multiple tests of significance (Benjamini et al., 2001; Strimmer, 2008). Three biological replicates were conducted. The differentially expressed genes shown in the figure are those showing significant differences in all three biological replicates.
The genes regulated by JA were collected from the previous published data (Du et al., 2017). The co‐regulated genes by JA and woolly were analysis by intersecting the differentially expressed genes in woW106R mutants and the JA treatment. GO enrichment analysis was performed using BiNGO app (https://www.psb.ugent.be/cbd/papers/BiNGO/Home.html) in the Cytoscape software package (http://cytoscape.org). Default parameters were used for all bioinformatics software.
MeJA treatments
Four‐week‐old tomato plants were sprayed with 1 mm MeJA and the control plants were sprayed with water. Leaflets were collected for RNA isolation and qRT‐PCR after 6 h of MeJA treatment.
For the analysis of trichome morphology and density, 4‐week‐old tomato plants were sprayed weekly with 1 mm MeJA and the control plants were sprayed with water. After 30 days of MeJA treatment, the newly formed stems and leaflets were collected for the quantification.
Yeast one‐hybrid assays
The 2500–3000 bp DNA fragment from the promoter of target TPSs was divided into three fragments (700–1000 bp). To avoid breaking up the binding site, we analysed the full‐length promoter by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Based on the result, we divided the putative promoter of TPSs into three overlapped fragments, with about 100 bp overlaps between each two fragments. The 700–1000 bp DNA fragments were amplified and cloned into pLacZi vector. The full‐length coding sequence of woolly and MYC1 was amplified and cloned into pJG4‐5. The recombinant constructs were co‐transformed into yeast strain EGY48 and the transformed yeast cells were incubated on SD/‐Trp‐Ura medium for 3 days at 30 °C. The transformed yeast cells were then transferred to the SD/‐Trp‐Ura + X‐gal medium and incubated for 5 days to test the interaction between the protein and the promoters of TPSs (Zhang et al., 2019).
Biotin‐labelled DNA IP assays
The biotin‐IP experiment was performed as described previously (Wu, 2006). The forward and reverse primers were labelled with biotin. DNA fragments were amplified by PCR and incubated with streptavidin‐agarose bead (Streptavidin Agarose, Catalog Number SA100‐04) for 12 h at 4 °C. The Flag‐wo fusion protein was expressed for the pull‐down assay. Bead‐bound DNA fragments were washed 5 times using PBS buffer and then incubated with total proteins at 4 °C with rotation. The anti‐Flag antibody was used for immunoblotting analysis of Flag‐wo.
Dual‐luciferase (Dual‐LUC) assay
The promoter of TPSs was cloned into the vector pGreen‐0800 to drive the firefly LUC reporter gene. 35S promoter‐driven Renilla (REN) luciferase in the same plasmid was used as the reference to normalize the infection efficiency. The full‐length coding sequence of SlMYC1, JAZ2 and wo was amplified by PCR and cloned into the vector pGreen II 62SK. The vector pGreen II 0800 and pGreen II 62SK were co‐transformed into the mesophyll protoplasts at the ratio of 1:4. The protoplasts were incubated in room temperature for 12 h and then harvested after the centrifugation.
The supernatant of total proteins was used for the transient expression with the Dual‐Luciferase Reporter Assay System (Promega) following the manufacture manual. The fluorescent values of LUC and REN were detected with CYTATION5 image reader (BioTek). The value of LUC was normalized to that of REN. Three biological repeats were measured for each combination.
Yeast two‐hybrid assays
The full‐length coding sequences of tomato JAZs and SlMYC1 were amplified and cloned into the pGBKT7, and the full‐length coding sequences of woolly and SlMYC1 were amplified and cloned into the pGADT7. The recombinant constructs were co‐transformed into yeast strain AH109 to test the interaction. The transformed yeast cells were grown on SD/‐Leu/‐Trp medium in the incubator with 30 °C. The transformed yeast cells were incubated on SD/‐Ade/‐His/‐Leu/‐Trp medium for 5 days to test the protein interactions. The primers used in this experiment were listed in the Table S3.
GUS staining
The young leaves or whole young seedlings of pMYC1:GUS or pwo:GUS transgenic plants were stained with GUS solution (2 mm X‐gal, 100 mm sodium phosphate buffer). Staining was performed overnight at 37 °C in darkness. The samples were washed with the 75% ethanol at room temperature. The samples were imaged by DIC microscopy (Nikon, ECLIPSE Ni‐U).
Bimolecular fluorescence complementation (BiFC)
The full‐length coding sequences of JAZs and SlMYC1 were amplified and cloned into pUC‐SPYNE. The full‐length coding sequences of woolly and SlMYC1 were amplified and cloned into pUC‐SPYCE. The recombinant constructs were transformed into GV3101 and subsequently transiently co‐transformed into 5‐week‐old N. benthamiana leaves. Yellow fluorescent protein signals were imaged after 48 h cultivation by confocal microscopy (LSM 880 Zeiss, Germany). The primers used in this experiment were listed in the Table S3.
Analysis of volatile metabolites
The leaflets were harvested from the 5‐week‐old tomato plants and then immersed in 1 ml of n‐hexane for 1h with gentle shaking at room temperature. The extract solution was then analysed by gas chromatography‐mass with the 7890B GC System (Agilent). Compounds were separated on a capillary HP‐5ms column (Agilent) with a flow rate of 1 ml/min. The initial column temperature was set at 80 °C for 2 min and then increased to 130 °C at a rate of 10 °C/min. After 2 min at 130 °C, the temperature was increased to 190 °C at a rate of 5 °C/min and then increased to 250 °C at a rate of 20 °C/min. All compounds were determined by the comparison of retention times and mass spectra data with those of authentic standards, and with the mass spectra data in the National Institute of Standards and Technology and Wiley libraries (Agilent Technologies, Palo Alto, CA) (Cheng et al., 2007).
Pull‐down assays
To express glutathione S‐transferase (GST)‐MYC1 or maltose‐binding protein (MBP)‐JAZ2, the full‐length coding sequences of SlMYC1 or JAZ2 were amplified by PCR and cloned into the vector pGEX‐4T‐1 or pMAL‐c5X. The recombinant proteins were expressed in Escherichia coli strain Transetta (DE3) with 0.125 mm isopropyl‐b‐D‐thiogalactoside (IPTG). The GST‐SlMYC1 proteins were purified using GST·Bind Resin (Novagen) and MBP‐JAZ2 proteins were purified using the Amylose Resin (NEB).
To express Flag‐wo protein, the full‐length coding sequence of wo was amplified by PCR and cloned into the vector pXSN‐Flag. Flag‐wo protein was expressed in the mesophyll protoplasts according to the previous report (Sang et al., 2007). The protoplasts were then homogenized in the extraction buffer containing 50 mm pH 7.4 Tris‐HCl, 80 mm NaCl, 5% glycerol, 0.1% Tween 20, 1 mm DTT, 1 mm phenylmethylsulphonyl fluoride (PMSF) and 20 μm MG132. The supernatant was collected after the centrifugation (12 000 g at 4 °C). For the protein interaction assay, the total protein was incubated with resin‐bound tagged fusion proteins for 2 h at 4 °C with rotation in the binding buffer containing 50 mm Tris‐HCl pH 7.5, 0.6% Triton and X‐100, 100 mm NaCl. The pellets were washed 5 times, eluted in 5 × SDS loading buffer, and boiled at 95 °C for 10 min before immunoblot. The anti‐MBP and anti‐Flag antibody were used for immunoblot analysis.
LCI assays
The Firefly LCI assay was performed as described previously (Chen et al., 2007). The full‐length coding sequence of wo and SlMYC1 was inserted into the vector pCAMBIA1300‐nLUC. The full‐length coding sequence of wo and JAZ2 was inserted into the vector pCAMBIA1300‐cLUC. All vectors were transformed into agrobacterium strain GV3101. The agrobacterium cells were resuspend in infiltration buffer (10 mm MgCl2, 10 mm MES (pH 5.7), 150 mm acetosyringone). The empty cLUC and nLUC vectors were used as the control. The suspensions were mixed and co‐infiltrated into N. benthamiana. After 2 days, the abaxial side of infiltrated leaves were sprayed with 1 mm luciferin and then kept in darkness for 10 min. The LUC activity was monitored with Tanon‐5200.
Yeast three‐hybrid assays
Yeast three‐hybrid assays were performed using the MATCHMAKER GAL4 Two‐Hybrid System. The full‐length coding sequence of JAZ2 was cloned into the MCS I site of the pBridge vector (Clontech) and the coding sequence of wo was inserted into the MCS II site as the bridge protein. The full‐length coding sequence of SlMYC1 was cloned into the vector pGADT7. The vector pGADT7 and pBridge were co‐transformed into the yeast strain AH109. The transformed yeasts were incubated on the SD/‐Trp/‐Leu medium at 30 °C. After 3 days, the clones were transferred to the SD/‐Trp/‐Leu/‐Met medium and the transfer was repeated 3 times before being transferred to the SD/‐Ade/‐His/‐Leu/‐Trp medium for testing the interaction. Alternatively, the clones were incubated on the plates containing SD/‐Ade/‐His/‐Leu/‐Trp/‐Met to induce the wo expression. All plates were incubated at 30 °C for 4 days.
Spider mite bioassays
The two‐choice assays were performed as described previously (Li et al., 2004). Ten mites were placed within a 1‐cm circle, and the circle was located equidistantly (1 cm) between different single leaflets from 4‐week‐old plants in a 9‐cm petri culture dish. The assays were conducted at 60% relative humidity, and the number of mites was counted after 2 h. Five biological replicates were performed, and error bars represent the SD of five biological replicates.
The procedure of spider mite treatment was previously described (Li et al., 2002). Five spider mites were placed on the fifth leaves of 4‐week‐old tomato plants (usually with 9 leaves). Challenged plants were grown in a growth chamber during the feeding trial. The phenotype was analysed after 5 days. The infected areas with chlorotic lesions were calculated using PhotoShop.
Accession numbers
Sequence data from this article can be found in the Solgenomics databases under the following accession numbers: SlMYC1, Solyc08g005050; woolly, Solyc02g080260; JAZ1, Solyc07g042170; JAZ2, Solyc12g009220; JAZ3, Solyc03g122190; JAZ4, Solyc12g049400; JAZ5, Solyc03g118540; JAZ6, Solyc01g005440; JAZ7, Solyc11g011030; JAZ8, Solyc06g068930; JAZ9, Solyc08g036640; JAZ10, Solyc08g036620; TPS1, Solyc01g105850; TPS5, Solyc01g105890; TPS12, Solyc06g059930; ACTIN2, Solyc11g005330.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
J.C., X.C. and S. W. designed the experiments; B.H., J.C., M.W. and F. Z. performed most of the experiments and analysed the data; M.Y. performed tomato stable transformation; H.X. analysed RNA‐seq data; L.W. assisted in terpene measurement; and J. C., X.C. and S. W. wrote the article.
Supporting information
Figure S1. Verification of the mutation of woolly gene in the woW106R mutant and CR‐wo transgenic plants.
Figure S2. The phenotype of VI glandular trichomes on the fruits of WT, woW106R mutants and CR‐Slwo plants.
Figure S3. Expression pattern of wo.
Figure S4. Relative expression level of TPSs in different materials.
Figure S5. GO Analysis of genes that are up‐regulated by JA and down‐regulated in woolly mutants.
Figure S6. The phenotype of WT and woW106R mutants treated by MeJA.
Figure S7. Relative expression level of JAZs in type VI trichomes (TRI) and leaves (LVS, with all trichomes removed).
Figure S8. Expression pattern of SlMYC1.
Figure S9. Phenotypic comparison between WT, CR‐SlMYC1 and OE‐SlMYC1.
Figure S10. The phenotype of Type VI glandular trichomes on the fruits of WT and CR‐SlMYC1 plants.
Figure S11. Relative expression level of TPSs in the leaves of WT and CR‐SlMYC1 plants with MeJA treatment.
Figure S12. Overexpression (OE) of SlMYC1 enhances pest resistance in tomato.
Figure S13. The morphology of type VII and VII‐like trichomes.
Figure S14. The phenotype of CR‐SlMYC1 plants treated by JA.
Table S1. The candidate genes of short hair mutant.
Table S2. List of the differentially expressed TPSs.
Table S3. Primers used in this paper.
Acknowledgements
This work is supported by the National Key Research and Development Program of China (2018YFD1000800), the grant from the National Natural Science Foundation of China (31722006) and the Key project of Fujian Province (2018NZ0002‐2).
Hua, B. , Chang, J. , Wu, M. , Xu, Z. , Zhang, F. , Yang, M. , Xu, H. , Wang, L.‐J. , Chen, X.‐Y. and Wu, S. (2021) Mediation of JA signalling in glandular trichomes by the woolly/SlMYC1 regulatory module improves pest resistance in tomato. Plant Biotechnol. J., 10.1111/pbi.13473
Corresponding author
References
- Aharoni, A. , Giri, A.P. , Deuerlein, S. , Griepink, F. , de Kogel, W.J. , Verstappen, F.W. , Verhoeven, H.A. et al. (2003) Terpenoid metabolism in wild‐type and transgenic Arabidopsis plants. Plant Cell, 15, 2866–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel, F.D. , Manavella, P.A. , Dezar, C.A. and Chan, R.L. .(2007) The true story of the HD‐Zip family. Trends Plant Sci. 12, 419–426. [DOI] [PubMed] [Google Scholar]
- Balcke, G. , Bennewitz, S. , Bergau, N. , Athmer, B. , Henning, A. , Majovsky, P. , Jimenez‐Gomez, J.M. et al. (2017) Multiomics of tomato glandular trichomes reveals distinct features of central carbon metabolism supporting high productivity of specialized metabolites. Plant Cell, 29(5), 960–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba, P. , Loughner, R. , Wentworth, K. , Nyrop, J.P. , Loeb, G. and Reisch, B. .(2019) A QTL associated with leaf trichome traits has a major influence on the abundance of the predatory mite Typhlodromus pyri in a hybrid grapevine population. Horti, Reas. 6, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou, R. , Fremaux, C. , Deveau, H. , Richards, M. , Boyaval, P. , Moineau, S. , Romero, D.A. et al. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. , Drai, D. , Elmer, G. , Kafkafi, N. & Golani, I. (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res, 125, 278–284. [DOI] [PubMed] [Google Scholar]
- Bergau, N. , Alexander, N.S. , Anja, H. , Balcke, G.U. and Alain, T. (2016) Autofluorescence as a signal to sort developing glandular trichomes by flow cytometry. Front. Plant Sci. 7, 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser, K. , Harper, A. , Welsby, N. , Schauvinhold, I. , Slocombe, S. , Li, Y. , Dixon, R.A. et al. (2009) Divergent regulation of terpenoid metabolism in the trichomes of wild and cultivated tomato species. Plant Physiol. 149, 499–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann, J. , Meyer‐Gauen, G. and Croteau, R. .(1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc. Natl Acad. Sci. USA, 95, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, M. , Wright, L.P. , Gershenzon, J. , Wasternack, C. , Hause, B. , Schaller, A. and Stintzi, A. (2014) Jasmonic acid and its precursor 12‐oxophytodienoic acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiol. 166, 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter, M. , Ruíz‐Rivero, O. , Abdeen, A. and Prat, S. (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Gene Dev. 18, 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughton, A.J. , Hoover, K. and Felton, G.W. (2005) Methyl jasmonate application induces increased densities of glandular trichomes on tomato, Lycopersicon esculentum. J. Chem. Ecol. 31, 2211–2216. [DOI] [PubMed] [Google Scholar]
- Calo, L. , García, I. , Gotor, C. and Romero, L.C. (2006) Leaf hairs influence phytopathogenic fungus infection and confer an increased resistance when expressing a Trichoderma‐1,3‐glucanase. J. Exp. Bot. 57, 3911–3920. [DOI] [PubMed] [Google Scholar]
- Chang, J. , Xu, Z. , Li, M. , Yang, M. , Qin, H. , Yang, J. and Wu, S. (2019) Spatiotemporal cytoskeleton organizations determine morphogenesis of multicellular trichomes in tomato. PLoS Genet. 15, e1008438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J. , Yu, T. , Yang, Q.H. , Li, C.X. , Xiong, C. , Gao, S.H. , Xie, Q.M. et al. (2018) Hair, encoding a single C2H2 zinc finger protein, regulates multicellular trichome formation in tomato. Plant J. 96, 90–102. [DOI] [PubMed] [Google Scholar]
- Chen, F. , Tholl, D. , D’Auria, J.C. , Farooq, A. , Pichersky, E. and Gershenzon, J. (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell, 15, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Zou, Y. , Shang, Y. , Lin, H. , Wang, Y. , Cai, R. et al. (2007) Firefly luciferase complementation imaging assay for protein‐protein interactions in plants. Plant Physiol. 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, A.X. , Xiang, C.Y. , Li, J.X. , Yang, C.Q. , Hu, W.L. , Wang, L.J. , Lou, Y.G. et al. (2007) The rice (E)‐beta‐caryophyllene synthase (OsTPS3) accounts for the major inducible volatile sesquiterpenes. Phytochemistry 68, 1632–1641. [DOI] [PubMed] [Google Scholar]
- Chini, A. , Fonseca, S. , Fernández, G. , Adie, B. , Chico, J.M. , Lorenzo, O. , García‐Casado, G. et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 44, 666–671. [DOI] [PubMed] [Google Scholar]
- Du, M.M. , Zhao, J.H. , David, T.W.T. , Liu, Y.Y. , Deng, L. , Yang, Y.X. , Zhai, Q.Z. et al. (2017) MYC2 Orchestrates a hierarchical transcriptional cascade that regulates jasmonate‐mediated plant immunity in tomato. Plant Cell, 29, 1883–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara, V. , Akhtar, T.A. , Nguyen, T.T. , Spyropoulou, E.A. , Bleeker, P.M. , Schauvinhold, I. , Matsuba, Y. et al. (2011) The tomato terpene synthase gene family. Plant Physiol. 157, 770–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Calvo, P. , Chini, A. , Fernández‐Barbero, G. , Chico, J.M. , Gimenez‐Ibanez, S. , Geerinck, J. , Eeckhout, D. et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell, 23, 701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, S. , Chico, J.M. and Solano, R. (2009) The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12, 539–547. [DOI] [PubMed] [Google Scholar]
- Fridman, E. , Wang, J. , Iijima, Y. , Froehlich, J.E. , Gang, D.R. , Ohlrogge, J. and Pichersky, E. (2005) Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant cell, 17, 1252–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, S.H. , Gao, Y.N. , Xiong, C. , Yu, G. , Chang, J. , Yang, Q.H. , Yang, C.X. et al. (2017) The tomato B‐type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci. 262, 114–103. [DOI] [PubMed] [Google Scholar]
- Garcia, V. , Cécile, B. , Just, D. , Fernandez, L. , Tai, F.W.J. , Mauxion, J.P. et al. (2016) Rapid identification of causal mutations in tomato ems populations via mapping‐by‐sequencing. Nat. Protoc. 11, 2401–2418. [DOI] [PubMed] [Google Scholar]
- Hong, G.J. , Xue, X.Y. , Mao, Y.B. , Wang, L.J. and Chen, X.Y. (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell, 24, 2635–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.H. , Liu, G. , Shi, F. , Jones, A.D. , Beaudry, R.M. and Howe, G.A. (2010) The tomato odorless‐2 mutant is defective in trichome‐based production of diverse specialized metabolites and broad‐spectrum resistance to insect herbivores. Plant Physiol. 154, 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.H. , Mcroberts, J. , Shi, F. , Moreno, J.E. , Jones, A.D. and Howe, G.A. (2014) The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 164, 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.Y. , Williams, M.M. , Loh, Y.T. , Lee, I.G. and Howe, G.A. (2002) Resistance of cultivated tomato to cell content‐feeding herbivores is regulated by the octadecanoid‐signaling pathway. Plant Physiol. 130, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Zhao, Y. , McCaig, B.C. , Wingerd, B.A. , Wang, J. , Whalon, M.E. , Pichersky, E. et al. (2004) The tomato homolog of CORONATINE‐INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate‐signaled defense responses, and glandular trichome development. Plant Cell, 16, 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckwill, L.C. (1943) The genus Lycopersicon: A historical, biological and taxonomic survey of the wild and cultivated tomatoes. PhD thesis (Aberdeen Univ, Aberdeen, UK).
- Ma, D. , Hu, Y. , Yang, C.Q. , Liu, B.L. , Fang, L. , Wan, Q. , Liang, W.H. et al. (2016) Genetic basis for glandular trichome formation in cotton. Nat. Commun. 7, 10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P. , Yang, L. , Esvelt, K.M. , Aach, J. , Guell, M. , DiCarlo, J.E. , Norville, J.E. et al. (2013) RNA‐guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba, Y. , Nguyen, T.T. , Wiegert, K. , Falara, V. , Gonzales‐Vigil, E. , Leong, B. , Schäfer, P. et al. (2013) Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell, 25, 2022–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, E.T. , Kapteyn, J. , Schmidt, A. , Li, C. , Kang, J.H. , Descour, A. , Shi, F. et al. (2011) Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol. 155, 524–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, M. , Katsumata, H. , Abe, M. , Yabe, N. , Komeda, Y. , Yamamoto, K.T. and Takahashi, T. (2006) Characterization of the class IV homeodomain‐leucine zipper gene family in Arabidopsis. Plant Physiol. 141, 1363–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, T. , Huang, H. , Wu, D. , Yan, J. , Qi, Y.J. , Song, Ss and Xie, D. (2014) Arabidopsis DELLA and JAZ proteins bind the WD‐Repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell, 26, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud, C. , Rontein, D. , Onillon, S. , Jabes, F. , Duffe, P. , Giacalone, C. , Thoraval, S. et al. (2009) A novel pathway for sesquiterpene biosynthesis from Z, Z‐farnesyl pyrophosphate in the wild tomato Solanum habrochaites . Plant Cell, 21, 301–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang‐Dong, Y. , Young, H.C. & Jen, S. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc, 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Schie, C.N. , Haring, M.A. and Schuurink, R.C. (2007) Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Mol. Biol. 64, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller, A.L. , Charbonneau, A.L. and Last, R.L. (2012) Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Proc. Natl Acad. Sci. USA, 109, 16377–16382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller, A.L. , Miner, D.P. , Larson, M. , McDowell, E. , Gang, D.R. , Wilkerson, C. and Last, R.L. (2010) Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol. 153, 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller, A.L. , Moghe, G.D. , Fan, P. , Ghosh, B. , Ning, J. , Jones, A.D. and Last, R.L. (2015) Functionally divergent alleles and duplicated loci encoding an acyltransferase contribute to acylsugar metabolite diversity in Solanum trichomes. Plant Cell, 27, 1002–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller, A.L. , Schauvinhold, I. , Larson, M. , Xu, R. , Charbonneau, A.L. , Schmidt, A. , Wilkerson, C. et al. (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl Acad. Sci. USA, 106, 10865–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, F. , Fernández‐CP, Zander M. , Diez, D.M. , Fonseca, S. , Glauser, G. , Lewsey, M.G. , Ecker, J.R. et al. (2013) Arabidopsis basic helix‐loop‐helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell, 25, 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.S. , Joo, J. , Kim, M.J. , Kim, Y.K. , Nahm, B.H. , Song, S.I. , Cheong, J.J. et al. (2011) OsbHLH148, a basic helix‐loop‐helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65, 907–921. [DOI] [PubMed] [Google Scholar]
- Serna, L. and Martin, C. (2006) Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 2006, 274–280. [DOI] [PubMed] [Google Scholar]
- Shan, C.M. , Shangguan, X.X. , Zhao, B. , Zhang, X.F. , Chao, L.M. , Yang, C.Q. , Wang, L.J. et al. (2014) Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun 5, 5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S. , Qi, T. , Huang, H. , Ren, Q. , Wu, D. , Chang, C. , Peng, W. et al. (2011) The Jasmonate‐ZIM domain proteins interact with the R2R3‐MYB transcription factors MYB21 and MYB24 to affect jasmonate‐regulated stamen development in Arabidopsis. Plant Cell, 23, 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulou, E.A. , Haring, M.A. and Schuurink, R.C. (2014) RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genom. 15, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer, K. (2008) drtool: a versatile R package for estimating local and tail area‐based false discovery rates.. Bioinformatics, 24, 1461–1462. [DOI] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G. , Nomura, K. et al. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Tominaga‐Wada, R. , Nukumizu, Y. , Sato, S. and Wada, T. (2013) Control of plant trichome and root‐hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One 8, 54019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van, S.C.C.N. , Haring, M.A. and Schuurink, R.C. (2007) Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Mol. Biol. 64, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliki, F. , Tariq, A.A. , Thuong, T.H.N. , Eleni, A.S. , Petra, M.B. , Ines, S. , Yuki, M. et al. (2011) The Tomato terpene synthase gene family. Plant Physiol. 157, 770–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendemiatti, E. , Zsögön, A. , Silva, Gffe , de Jesus, F.A. , Cutri, L. , Figueiredo, Crf , Tanaka, Fao et al. (2017) Loss of type‐IV glandular trichomes is a heterochronic trait in tomato and can be reverted by promoting juvenility. Plant Sci. 259, 35. [DOI] [PubMed] [Google Scholar]
- Wu, K.K. (2006) Analysis of protein‐DNA binding by streptavidin‐agarose pull down. Methods Mol. Biol. 338, 281–290. [DOI] [PubMed] [Google Scholar]
- Wu, M.L. , Cui, Y.C. , Ge, L. , Cui, L.P. , Xu, Z.C. , Zhang, H.Y. , Wang, Z.J. et al. (2020) NbCycB2 represses Nbwo activity via a negative feedback loop in tobacco trichome development. J. Exp. Bot. 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , van Herwijnen, Z.O. , Dräger, D.B. , Sui, C. , Haring, M.A. and Schuurink, R.C. .(2018) SlMYC11 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell, 30:2988–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y.X. , Stolz, S. , Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007) A downstream mediator in the growthrepression limb of the jasmonate pathway. Plant Cell, 19, 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.X. , Gao, Y.N. , Gao, S.H. , Yu, G. , Xiong, C. , Chang, J. et al. (2015) Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato wov gene in tobacco. BMC Genom. 16, 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.X. , Li, H.X. , Zhang, J.H. , Luo, Z.D. , Gong, P.J. , Zhang, C.J. , Li, J.H. et al. (2011) A regulatory gene induces trichome formation and embryo lethality in tomato. Proc. Natl Acad. Sci. USA, 108, 11836–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J. , Wang, X. , Hu, T.X. , Zhang, F.X. and Ye, Z.B. (2017) An indel in the promoter of ai‐activated malate transporter9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell, 29, 2249–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, Y. , Sano, R. , Wada, T. , Takabayashi, J. and Okada, K. (2009) Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136, 1039–1048. [DOI] [PubMed] [Google Scholar]
- Yu, X. , Chen, G. , Tang, B. , Zhang, J. , Zhou, S. and Hu, Z. (2018) The Jasmonate ZIM‐domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 267, 65–73. [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Zhao, Q.C. , Zeng, D.X. , Xu, J.H. , Zhou, H.G. , Wang, F.L. , Ma, N. et al. (2019) RhMYB108, an R2R3‐MYB transcription factor, is involved in ethylene‐ and JA‐induced petal senescence in rose plants. Horti. Reas. 6, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Verification of the mutation of woolly gene in the woW106R mutant and CR‐wo transgenic plants.
Figure S2. The phenotype of VI glandular trichomes on the fruits of WT, woW106R mutants and CR‐Slwo plants.
Figure S3. Expression pattern of wo.
Figure S4. Relative expression level of TPSs in different materials.
Figure S5. GO Analysis of genes that are up‐regulated by JA and down‐regulated in woolly mutants.
Figure S6. The phenotype of WT and woW106R mutants treated by MeJA.
Figure S7. Relative expression level of JAZs in type VI trichomes (TRI) and leaves (LVS, with all trichomes removed).
Figure S8. Expression pattern of SlMYC1.
Figure S9. Phenotypic comparison between WT, CR‐SlMYC1 and OE‐SlMYC1.
Figure S10. The phenotype of Type VI glandular trichomes on the fruits of WT and CR‐SlMYC1 plants.
Figure S11. Relative expression level of TPSs in the leaves of WT and CR‐SlMYC1 plants with MeJA treatment.
Figure S12. Overexpression (OE) of SlMYC1 enhances pest resistance in tomato.
Figure S13. The morphology of type VII and VII‐like trichomes.
Figure S14. The phenotype of CR‐SlMYC1 plants treated by JA.
Table S1. The candidate genes of short hair mutant.
Table S2. List of the differentially expressed TPSs.
Table S3. Primers used in this paper.


