Abstract
Male sterility is widely used in the production of hybrid seeds in rice, but the use of genic male sterility is limited because of the high labour cost for maintaining male‐sterile lines. Previous studies using T‐DNA insertional mutagenesis demonstrated that disrupting the expression of oxophytodienoic acid reductase 3 (OPR3), which is involved in the jasmonate biosynthesis pathway, results in a kind of male sterility that can be restored to fertility by exogenous jasmonate in Arabidopsis. Here, we created male‐sterile mutations by editing the second and fourth exons of OsOPR7 in rice through clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR‐associated system 9. The induced mutagenesis at these exons resulted in 31.8% and 23.9% male‐sterile plants in the T0 generation, respectively. We screened male‐sterile lines that can be restored to fertility by exogenous methyl jasmonate in the T0, T1 and T2 rice populations and characterized the anther and agronomic traits of the transgenic plants. Results showed the successful generation of male‐sterile lines through the silencing of OsOPR7, the orthologous gene of Arabidopsis OPR3, in a field crop, paving the way for the establishment of a two‐line system for rice hybrid production. The system consists of a male‐sterile line that can be maintained by spraying methyl jasmonate and a restoring line that confers pollen.
Keywords: rice, Oryza sativa, CRISPR, Cas, genome editing, OsOPR7, genic male sterility
Introduction
Rice (Oryza sativa L.) is the most important staple food crop for more than half of the world’s population. Hybrid rice, with a yield that is 20%–30% higher than that of conventional rice, is an important achievement in agriculture and has substantially contributed to the worldwide food supply (Chen and Liu, 2014; Ma and Yuan, 2015). Rice is a self‐pollinating crop, and hybrid seed production is based on male‐sterile lines. Chinese scientists successfully identified male‐sterile genes from wild rice, transferred them to commercial varieties and realized the three‐line system of heterosis utilization as early as 1974 (Ma and Yuan, 2015).
The manipulation of pollination is a key step in rice hybrid breeding. There are two systems, namely the three‐line and the two‐line systems, for rice heterosis utilization. The three‐line system is based on cytoplasmic‐genetic male sterility (CMS) (Huang et al., 2015; Tang et al., 2014). The most popular two‐line system in rice is based on environment‐sensitive nuclear male sterility (EGMS) (Chen et al., 2014; Huang et al., 2014; Yuan, 2014; Zhou et al., 2016). The three‐line system not only is complicated and costly in terms of labour but also requires sophisticated paddy field arrangements. The establishment of a two‐line system on the basis of photoperiod‐ and thermosensitive genic male‐sterile lines (PGMS/TGMS) broadens the utilization of heterosis within and between subspecies. This system increases rice yield by 5%–10% compared with that obtained using a three‐line system. However, two‐line systems are developed under long‐day and short‐day conditions, and the isolation and backcrossing processes normally requires more than 3 years to produce male‐sterile lines. Genic male sterility (GMS) is common in flowering plants, but its application to hybrid production is limited mainly because it cannot propagate pure male‐sterile lines, which are the preconditions for efficient commercial production (Chen et al., 2010; Fan and Zhang, 2018; Li et al., 2016; Wang and Deng, 2018). Previous studies on defective pollen wall 2 (DPW2) (Xu et al., 2017), defective in exine formation protein 1 (OsDEX1) (Yu et al., 2016), ATP‐binding cassette G26 (OsABCG26) (Zhao et al., 2015) and glycerol‐3‐phosphate acyltransferase 3 (OsGPAT3) (Men et al., 2017) discovered various nuclear male‐sterile mutations in rice. No pollen 1 (OsNP1), which encodes glucose–methanol–choline oxidoreductase, is involved in tapetum degeneration and pollen exine formation, and an osnp1 mutant displays normal vegetative growth but complete male sterility that is insensitive to environmental conditions (Chang et al., 2016).
Clustered regularly interspaced short palindromic repeats (CRISPR)‐associated protein 9 (Cas9) is an efficient tool used in rice and other crop breeding for desired traits (Zheng et al., 2016; Lee et al., 2019; Li et al., 2019; Ma et al., 2019; Wang et al., 2019; Wang et al., 2020). OsDGD2β is a digalactosyldiacylglycerol synthase in anther. osdgd2β mutants induced by CRISPR/Cas9 were of a male‐sterile type and characterized by degenerating tapetal cells, shrunken anthers and pollens that are devoid of starch granules (Basnet et al., 2019). Recently, a strategy for effectively establishing a male‐sterile enriched mutant library by knocking out anther‐specific genes using the CRISPR/Cas9 technology was proposed (Ma et al., 2019).
Jasmonates (JAs) play an important role in regulating flower development, especially in anther development (Cai et al., 2014; Gómez et al., 2015; Li e t al., 2018; Xiao et al., 2014; Zhang and Yang, 2014). The loss of function of the genes on the JA biosynthetic pathway resulted in various male‐sterile mutants in Arabidopsis (Ishiguro et al., 2001; Sanders et al., 2000; Stintzi and Browse, 2000; Wasternack and Hause, 2013; Xiao et al., 2014). Particularly, the defective anther dehiscence 1 (dad1) and oxophytodienoate reductase 3 (opr3) mutants displayed malfunction in anther dehiscence, pollen maturation, filament elongation and flower opening. The fertility of such mutants can be restored by applying methyl jasmonate (MeJA) or linolenic acid, a kind of JA precursor (Ishiguro et al., 2001; Stintzi and Browse, 2000). ORR3 catalyses reductive reaction from jasmonoyl‐isoleucine (JA‐Ile) to 12‐oxophytodienoic acid (OPDA). opr3‐1 is a conditional allele conferring incomplete defects on JA‐Ile biosynthesis. By contrast, opr3‐3 is an allele that is responsible for the complete blocking of JA synthesis, and opr3‐3 plants are male‐sterile (Sanders et al., 2000). The rice OsOPR7 gene, which shares a high percentage of nucleotide sequence similarity with AtOPR3 gene, is involved in JA biosynthesis in rice (Oryza sativa L.), and the expression of OsOPR7 in Arabidopsis opr3 mutant restores the fertility of a mutant by regaining JA production (Tani et al., 2008).
In this study, we applied CRISPR/Cas9 to create insertional/deletional mutations at OsOPR7 in rice. The CRISPR/Cas9‐induced osopr7 mutants were male‐sterile, and their fertility was restorable by exogenous MeJA. In addition to current PGMS/TGMS systems, the successful generation of osopr7 male‐sterile lines pave the way for establishing a two‐line system for hybrid production in rice.
Results
Analysis of sequence similarity and structure of the OPR orthologous genes in a variety of plant species
Of the three OPRs in Arabidopsis thaliana, only OPR3 is involved in JA biosynthesis (Breithaupt et al., 2006; Wasternack and Hause, 2018). There are three OPR orthologous genes in tomato, eight in maize, and 10 in rice. Basing on the nucleotide sequences of these OPR orthologous genes, we analysed and compared the structures of the genes and made a phylogenetic tree. As shown in Figure 1a, the 24 OPR orthologs were unevenly distributed among Arabidopsis thaliana (At), Oryza sativa (Os), Zea mays (Zm) and Solanum lycopersicum (Le). The OPRs can be classified into three clades, each with an uneven number of members. AtOPR3 and OsOPR7 are located in Clade III, sharing a high similarity of nucleotide and having a close phylogenetic relationship. AtOPR3 contains four exons and three introns, whereas OsOPR7 contains five exons and four introns. OsOPR7 had long intron sequences and thus had a longer full genomic sequence than AtOPR3. However, AtOPR3 and OsOPR7 had nearly the same coding sequences (Figure 1b).
Figure 1.
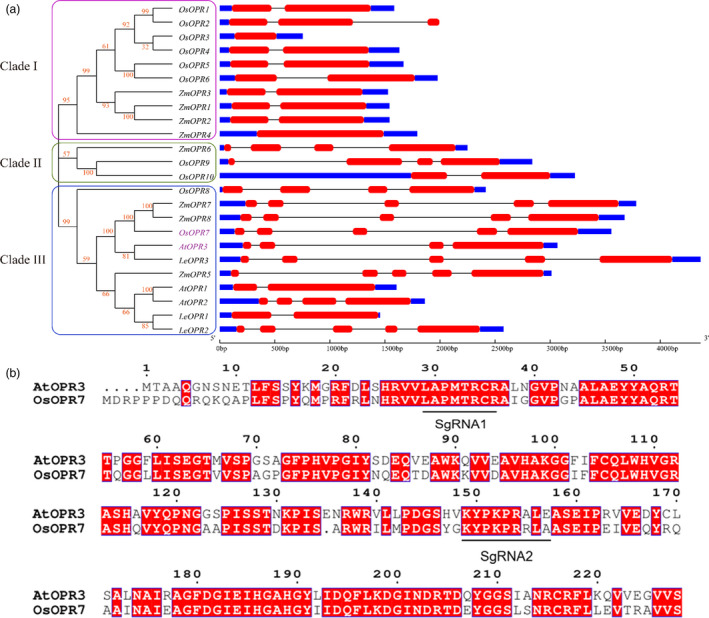
Orthologous OPRs across species. (a) Phylogenetic tree and gene structures of OPRs from various species. Clades I, II and III are indicated in fuchsia, green and light blue lines, respectively. The constructed maximum‐likelihood tree, built using the ClustalW program with 1000 bootstrap replicates, is based on the alignment of 24 OPR coding sequences. The digits in red stand for the bootstrap values. AtOPR3 and OsOPR7 are indicated with purple colour. The right side of the figure displays the gene structures of OPRs from Arabidopsis thaliana (At), Oryza sativa (Os), Zea mays (Zm) and Solanum lycopersicum (Le), which are in the order of their appearance in the phylogenetic tree. Coding sequences (CDS) and untranslated regions (UTR) are shown in the red and blue boxes, respectively. Introns are indicated with horizontal lines. (b) Comparison of the encoded amino acid sequences between AtOPR3 and OsOPR7. The red colour boxes show the amino acids present in the two genes. The underlines indicate the SgRNAs designed. [Colour figure can be viewed at wileyonlinelibrary.com]
Characterization of targeted editions induced by CRISPR/Cas9 in T0 transgenic plants
To knock out the OsOPR7 gene in rice, we transformed ZH11 plants with a VK005‐01 vector. We identified 14 T0 rice transgenic plants from 333 calluses that survived from the hygromycin resistance selection. Eight and six of the identified T0 mutants had nucleotide changes at Exon 2 and Exon 4 of OsOPR7, respectively (Table 1). Sequencing the PCR products of the target sites revealed heterozygous biallelic and homozygous mutations at the targeted regions. Of the eight T0 mutants, which had nucleotide changes at Exon 2 of OsOPR7, five were biallelic genotypes, one was homozygous, and two were heterozygous. Targeted mutagenesis resulted in 13 kinds of allelic variations at Exon 2 of OsOPR7 in addition to allele 0 (a0) of the rice wild type (ZH11). Five alleles (a2, a4, a7, a9 and a13) belonged to the deletional mutant type, and eight alleles (a1, a3, a5, a6, a8, a10, a11 and a12) belonged to the insertional mutant type. No coexistence between deletion and insertion mutations was found after allelic changes at Exon 4 of OsOPR7. Three of the six T0 mutants were biallelic, two were homozygous, and one was heterozygous. Targeted mutagenesis led to a total of nine kinds of allelic variations at Exon 4 of OsOPR7 in addition to the rice ZH11 allele 0 (a0). Six of the alleles (a14, a15, a17, a19, a21 and a22) were deletional, two alleles (a16 and a18) were insertional, and one allele (a20) was co‐existing of deletion–insertion types.
Table 1.
Mutational changes at the target region of OsOPR7 in the T0 transgenic rice
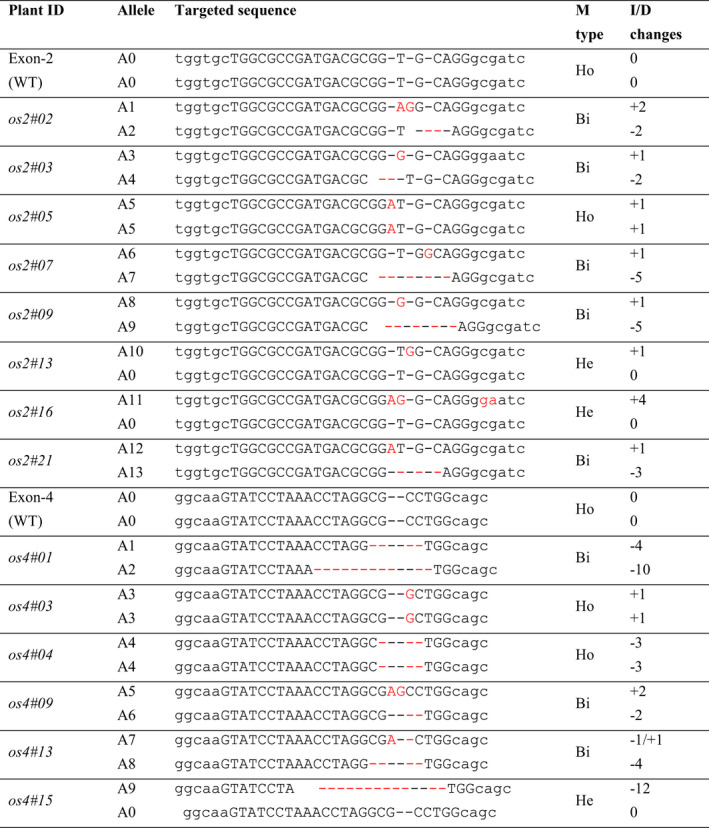
Each row stands for an individual T0 plant or the wild‐type plant. Os2 (or Or4) indicates transgenic plants, in which Exon 2 (or Exon 4) of OsOPR7 gene was targeted. The number after ‘#’ stands for plant ID number. The letters in red colour show the nucleotide changes induced, and the dashes ‘−’ indicate deletion of a nucleotide. Ho, He and Bi represent homozygous alleles, heterozygous alleles and biallelic inducement, respectively. I/D changes mean insertional/deletional number of nucleotides, where ‘+’ or ‘−’ means insertional/deletional changes, respectively.
Each row stands for an individual T0 plant or the wild‐type plant. Os2 (or Or4) indicates transgenic plants, in which Exon 2 (or Exon 4) of OsOPR7 gene was targeted. The number after ‘#’ stands for plant ID number. The letters in red colour show the nucleotide changes induced, and the dashes ‘−’ indicate deletion of a nucleotide. Ho, He and Bi represent homozygous alleles, heterozygous alleles and biallelic inducement, respectively. I/D changes mean insertional/deletional number of nucleotides, where ‘+’ or ‘−’ means insertional/deletional changes, respectively.
Identification of male‐sterile T0 mutants whose fertility can be restored by exogenous MeJA
We compared the floral morphology of the wild type (ZH11) with that of two T0 osopr7 mutants, namely Os2#03 (a3a4) and Os2#09 (a8a9), where Os2 stands for the targeted mutation at Exon 2 and the digits following # represent individuals (Figure 2). Os2#09 and Os2#03 anthers were indehiscent and were parallel. We divided all the seedlings of the generations from T0 to T2 into two parts, as shown in Figure S1, and were used in observing anther sterility and MeJA restoration, respectively. No significant differences were observed between the appearances of the spikelets of the mutants and ZH11 at the beginning of flower opening (Figure 2, left column). However, an apparent difference in spikelet appearance was observed at the fully opening phase of the flowers, when the Os2#03 and Os2#09 anthers were indehiscence in comparison with those of ZH11 (Figure 2, 2nd column from left). Differences inside the flowers were even clearer when pieces of paleae were removed from the flowers (Figure 2, 3rd column from left). Enclosed in red circles in Figure 2 (3rd column from left), a large number of pollen grains were observed on the pistil of ZH11, and a few number of pollen grains were visible on the pistils of the Os2#03 and Os2#09 flowers subjected to MeJA treatment. By contrast, pollen grains were hardly observed on the pistils of Os2#03 and Os2#9 without MeJA treatment.
Figure 2.
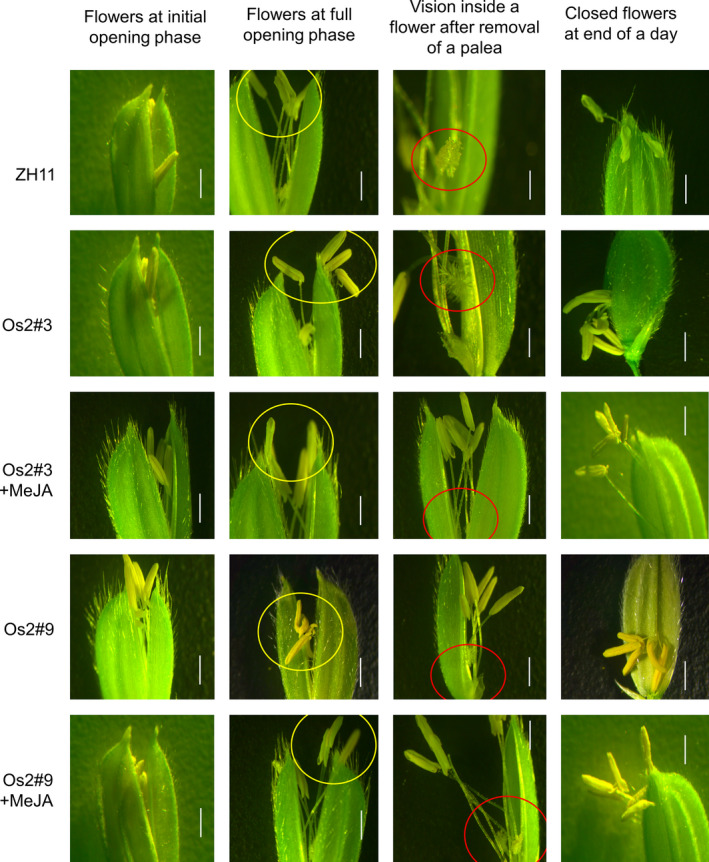
Comparison of floral morphology between ZH11 (control), the T0 osopr7 plant (Os2#3) without the MeJA treatment, and the T0 osopr7 plant (Os2#3 and Os2#9) with the MeJA treatment. The T0 transgenic plants were divided into two parts (Figure S1), which were used for the observation of anther sterility and the MeJA restoration, respectively. The pictures were taken in a same day when the flowers began to open in the morning (the left column), at full opening stage (the two columns in the middle) and at end of the day (the right column). The yellow circles focus on the anthers of the flowers, and the red circles show the pistils with or without pollen grains falling from dehiscence or indehiscence anthers. Scale bars = 1 mm. [Colour figure can be viewed at wileyonlinelibrary.com]
Acquisition of osopr7 T1 and T2 plants and the restoration of their fertility
Given that all the T0 mutants were indehiscent and parallel, we used Os2#09 as an example to observe the T1 and T2 offspring. The Os2#09 half plant without MeJA treatment and the Os2#09 half plant with the MeJA treatment looked similar at the mature stage (Figure 3a). However, their panicles were different; that is, the Os2#09 plant without MeJA treatment had a slim panicle without plump seeds (Figure 3b, right), whereas the Os2#09 plant with MeJA treatment yielded with a considerate amount of seeds on its panicle (Figure 3b, middle), although significantly less than those on the wild‐type panicle (Figure 3b, left). We were able to harvest seeds from the Os2#09 (a8a9) plant with MeJA treatment and grew the seeds for T1 plants because the male fertility of T0 Os2#09 plants was restored through MeJA treatment. The T1 Os2#9 plants (e.g. Os2#09‐3) without MeJA treatment had less pollen grains inside its anther (Figure 3d) compared with ZH11 (Figure 3c) and Os2#09‐3 plants with MeJA (Figure 3e), as indicated by I2‐KI staining results (Figure 3c‐e). We removed a piece of palea from each flower to have a clear vision of the interior parts of the flowers. As shown in Figure 3f‐h, the ZH11 anthers were empty and thus thin, and the ZH11 pistil appeared with visible pollen grains falling from the anthers (Figure 3f). The anthers of the Os2#09‐3 plants with MeJA treatment were dehiscent but still looked thicker than those of ZH11 (Figure 3h). Notably, the anthers of the Os2#09‐3 plant without MeJA treatment were indehiscent, and no pollen grains fell on the pistil (Figure 3g). The expression of OsOPR7 gene in the anthers of ZH11 was 25.6‐fold that in the eight T1 0s2#09 family plants (Figure 4).
Figure 3.
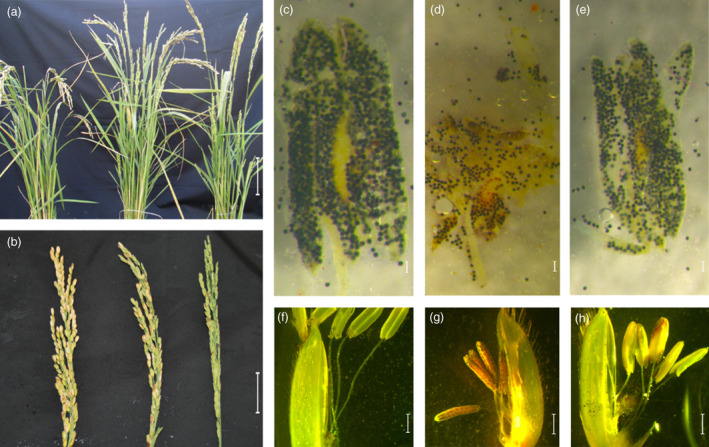
Comparison among between the wild type (ZH11), osopr7 plant without MeJA treatment, and osopr7 plant with MeJA treatment in terms of matured plant appearance, panicles, pollen grains and floral morphology. (a) Matured plant appearance. From left to right: ZH11, the Os2#9 (T0) half plant with MeJA treatment, and T0 Os2#9 half plant with MeJA treatment. (b) Panicles. From left to right: ZH11, T0 Os2#9 half plant with MeJA treatment (20% fertile), and T0 Os2#9 half plant without MeJA treatment (completely sterile). (c)–(e) Pollen grains inside an anther. From left to right: pollen grains from the ZH11 plant, T1 Os2#9‐3 plant, and T1 Os#9‐3 plant treated with MeJA. (f)–(h) Flowers after the removal of paleae. From left to right: pollen grains from a ZH11 plant, T1 Os2#9‐3 plant without MeJA treatment, and T1 Os2#9‐3 plant with MeJA treatment. Scale bars: 10 cm, 5 cm, 1 mm and 0.1 mm for a, b, c–e, and f–h, respectively. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 4.
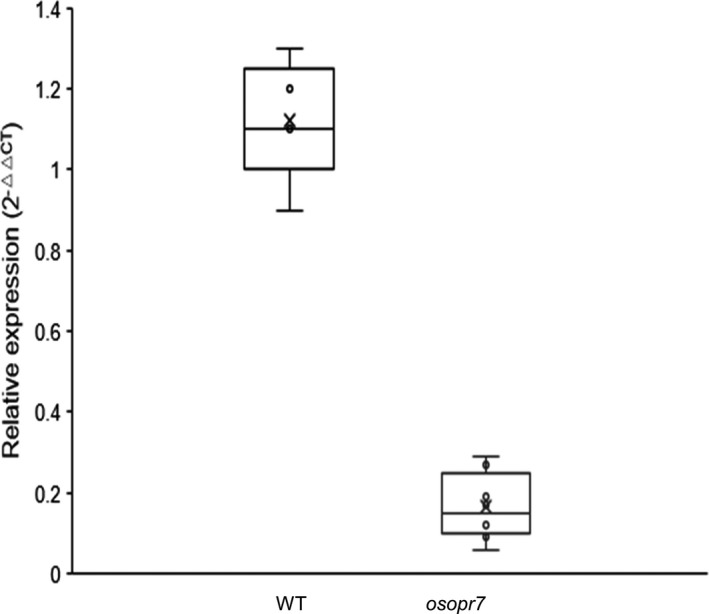
Comparison between WT (ZH11) and T1 osopr7 plants in terms of OsOPR7 expression in anthers. OsACTIN1 was used as internal control. The average value of OsOPR7 expression was based on the data of eight T1 osopr7 plants in the family Os02#‐9.
Characterization of the agronomic traits of Os2#09 T2 plants
We propagated the T1 Os2#09‐3 plants by spraying MeJA and obtained the T2 population. Each T2 Os2#09‐3 half plant with MeJA treatment yielded numerous seeds in contrast with the plants without MeJA treatment, which did not yield any seeds. We compared the MeJA‐treated and untreated plants of two T2 plants (Os2#09‐3‐6 and Os2#09‐3‐11) with the wild‐type control in terms of agronomic traits, such as plant height, flag leaf length, number panicles per plant, number of grains per panicle and rate of seed set. As shown in Table 2, there were no significant differences between T2 plants and the wild type (ZH11) for plant height, flag leaf length and number of panicles per plants. However, significant difference in seed‐setting percentage was observed between the treated and untreated plants and between the T2 plants and wild‐type control. The seed‐setting percentage was 0% for the untreated os2#09‐3‐6 and os2#09‐3‐11 plants and 17.9% and 20.4% for the os2#09‐3‐6 and os2#09‐3‐11 plants, respectively, which were treated with MeJA.
Table 2.
Characterization of some yield‐related agronomic traits of some T2 0s2#09 plants
| Plant ID | Plant height (cm) | Flag leaf length (cm) | Effective panicles per plant | Grain number per panicle | Seed‐setting rate (%) |
|---|---|---|---|---|---|
| ZH11 (CK) | 83.22 ± 0.55 | 32.37 ± 0.52 | 8.08 ± 0.71 | 108.72 ± 3.05 | 92.19 ± 0.57 |
| 0s2#09‐3‐6a | 84.25 ± 0.47 | 33.48 ± 0.64 | 8.23 ± 0.45 | 119.72 ± 6.11 | 0a |
| 0s2#09‐3‐6b | 84.17 ± 0.42 | 32.46 ± 0.34 | 8.21 ± 0.56 | 109.52 ± 4.15 | 17.9b** |
| 0s2#09‐3‐11a | 87.92 ± 0.71* | 34.48 ± 0.45 | 7.84 ± 0.61 | 128.72 ± 3.05 | 0a |
| 0s2#09‐3‐11b | 86.62 ± 0.56* | 34.19 ± 0.57 | 7.91 ± 0.74 | 118.72 ± 3.05 | 20.4b** |
The superscript a or b indicates the plant with or without MeJA treatment. ‘*’ and ‘**’ stand for significant difference between the treatments a and b at statistic levels P < 0.05 and P < 0.01, respectively.
Discussion
Hybrid production requires plants from which no viable pollen is produced. This intentional elimination of male gametophytes can be attained using different methods. A simple way is the emasculation of anthers by hand. A plant with flowers whose anthers had been removed can only serve as a female parent and breeds hybrid seeds after the introduction of pollens from other plants. Emasculation by hand can merely be used in producing hybrids in breeding programmes, and producing hybrid seeds in large quantities through this method is expensive. Thus, this method is commercially impractical. For the economic production of hybrid seeds, a line that cannot produce its own viable pollen grains must be created, and thus, seed formation on such line relies on pollen grains from another genotype.
Various types of male‐sterile lines have been generated, such as cytoplasmic, cytoplasmic‐genic (CMS‐genic) and genic male‐sterile (GMS) lines. To date, the CMS‐genic type is dominant in commercial rice hybrid seed production (Bai et al., 2018; Chang et al., 2016; Chen and Liu, 2014; Fan and Zhang, 2018; Kim and Zhang, 2018). In typical three‐line systems based on CMS‐genic lines, the male sterility of offspring is maternally inherited. The CMS‐genic lines are maintained by crossing it to a sister line, known as the maintainer line, which is genetically identical to CMS‐genic lines but has a normal cytoplasm. Thus, maintainer lines are male fertile. The fertility of CMS‐genic lines can be restored by using restorer lines carrying fertile nuclear genes. A three‐line system is rather complicated and requires sophisticated paddy field arrangements.
In this study, we created a male‐sterile line in rice by knocking out OsOPR7, an orthologous gene of Arabidopsis OPR3 that plays an essential role in the JA biosynthesis pathway (Ishiguro et al., 2001; Tani et al., 2008). The osopr7 plants failed to release pollen grains from mature anthers (Figures 2 and 3g), and a considerable number of the pollen grains were inviable (Figure 3d). The spraying of 500 µm MeJA solution to the osopr7 plants before anthesis restored their fertility (Figure 2, Figure 3a, b, e and h). We proposed a two‐line system for rice hybrid production based on the osopr7 male‐sterile line (Figure 5a‐d). In comparison with other male‐sterile lines, the osopr7 plants have more stable and thorough male sterility because male sterility on these plants does not depend on environmental conditions, such as light and temperature. By contrast, male sterility in some CMS‐genic lines (Huang et al., 2015; Tang et al., 2014) or genic male‐sterile lines (Chen et al., 2014; Huang et al., 2014; Yuan, 2014) does so. Finding a restorer for this two‐line system is easy because nearly all rice fertile genotypes can serve as restorers. However, determining a genotype that can serve as a restorer for some CMS‐genic or genic male‐sterile lines requires tremendous effort.
Figure 5.
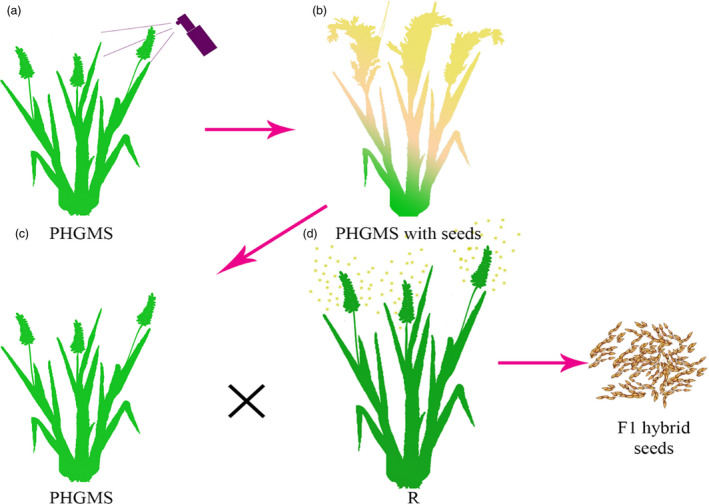
Two‐line system with phytohormone genic male‐sterile (PHGMS) line for hybrid seed production in rice. (a) PHGMS plant that can be restored by spraying MeJA. (b) Male‐sterile plant treated with MeJA restores fertility and sets seeds. (c) osopr7 PHGMS line used as a male‐sterile line in a cross for F1 hybrid seed production. (d) Wild‐type line that serves as pollinator for F1 hybrid seed production. [Colour figure can be viewed at wileyonlinelibrary.com]
Spraying MeJA to rice unlikely causes environmental pollution, and neither the MeJA nor its precursor, linolenic acid, is toxic to humans. Some of MeJA derivatives are commonly used as food additives. Additionally, this two‐line system is affordable; that is, 1 L of 500 µm MeJA solution approximately costs USD 1.5–2.0. Five litres of MeJA solution can be used in spraying rice sparks in a 324‐square‐metre field, producing approximately 33 kg of the seeds of the osopr7 male‐sterile line for the hybrid production sown in 1 ha paddy field. However, spraying MeJA solution to rice sparks requires additional manpower. In developing countries where manpower is not expensive, the economic value of hybrid seeds produced requires manpower input. In developed countries, where manpower is expensive, using drones can be an alternative to manual spraying.
The biological function of OsOPR7 was previously reported (Tani et al., 2008). The focus of the study was on the wounding and drought‐stress responses of the gene. Our effort was to invent a two‐line system for rice hybrid production, which means a significant value in countries depending on rice as staple food. As verified in our study, the malfunction of JA biosynthesis resulted in anther indehiscence in rice. This finding is in accordance with the studies on Arabidopsis (Ishiguro et al., 2001). Moreover, we demonstrated that the loss of function of OsOPR7 enabled inviable pollen grains to be detected through KI‐I2 solution staining (Figure 3). However, the application of MeJA can have some side effects. For example, a previous study on Brassica napus showed that the application of 100 µm MeJA solution resulted in numerous early‐open flowers and a variety of floral organ abnormalities, which were likely due to the combined actions of floral identity genes, such as BnAP1, BnAP2, BnAP3, BnAG1 and BnPI3, as reflected by transcriptional changes in these genes (Pak et al., 2009; Pak et al., 2017). However, this kind of side effect can be controlled by selecting an accurate spraying time and MeJA concentration and dosage. The spray time and concentration that we recommended in the paper (Materials and Methods) did not result in early flowering and significant floral organ abnormality in rice. The negative effects of MeJA spraying on the germination of the harvested seeds and stress responses were not observed.
This two‐line system has several disadvantages. The seed‐setting percentage of osopr7 male‐sterile line, which was approximately 20% after the treatment of exogenous MeJA, was not ideal for maintaining sufficient male‐sterile progeny. Therefore, the system should be further improved by optimizing time slot, spraying frequency, and MeJA concentration before the osopr7 line can be widely used in rice hybrid production.
We performed two parallel experiments to silence OsOPR7. One experiment was designed to edit Exon 2, and the other experiment was designed to target Exon 4 of the gene. The induced mutagenesis at Exon 2 resulted in 13 allelic variations in addition to the wild‐type allele. By contrast, the targeted inducement at Exon 4 caused only nine allelic variations apart from the wild‐type allele (Table 1). Therefore, editing Exon 2 is more effective than editing Exon‐4. In contrast with chemical mutagenesis using ethylmethane sulphonate (EMS), which frequently causes point mutation from guanine to adenine, CRISPR/Cas9‐mediated mutagenesis led to insertional–deletional changes of up to 12 nucleotide fragments causing the frame shift of the coding region. The deletion or insertion at the upper region of a gene might have a greater chance to result in a truncated protein than those at the lower region of a gene. Compared with the CRISPR–Cas technology, EMS‐mediated technology leads to a high number of background mutations. This feature is an obvious shortcoming. Numerous backcrossing are required in the development of an EMS‐mutant line with a desired trait and reduced background mutation load. CRISPR–Cas9 mutagenesis is clearly superior in this matter because a desired mutation can be incorporated into a genome without a background mutation load (Karunarathna et al., 2020). The application of CRISPR–Cas9‐induced mutations in plant breeding is not hampered in countries with less restrictive legislation, such as China, North America and Australia, where CRISPR–Cas9‐mediated plants have not been classified as genetically modified organisms. In this context, we hope that osopr7 male‐sterile lines will be widely used in hybrid seed production in the near future.
Experimental procedures
Plant growth and vector construction for targeted gene mutation
Seeds of Oryza sativa L. cv. Zhonghua11 (ZH11) was sown in soil and kept in a greenhouse under light for 16 h at 28 °C and in darkness for 8 h at 23 °C. The ZH11 rice was cultured in suspension according to our previous description (Guo et al., 2016). The structure of the OsOPR7 gene and the targeted sites for mutagenesis are illustrated in Figure 6a. The vector used for rice transformation was VK005‐01. The components between the right border and left board of VK005‐01 are shown in Figure 6b (Viewsolid Biotech Company Ltd., Beijing, China; https://geneeditorlab.com/). The mpCas9 gene driven by the Ubi promoter and a single‐guide RNA (SgRNA) driven by the rU6 promoter targeting Exon 2 and Exon 4 of OsOPR7 gene (GenBank Accession no AP004707.4, http:// rice.plantbiology.msu.edu/) were connected to each other (Figure 6c). Primers for amplifying DNA fragments from the target sequences in Exon 2 and Exon 4 for the confirmation of the correct mutagenesis of OsOPR7 in rice were designed and synthesized (Sangon Biotech, Shanghai, China; http://www.sangon.com/; Table S1). Competent cells (50 μL; strain DH5α, E. coli) were transformed by heat shock after 5–10 μL of the vector solution was added. E. coli clones containing the designed molecular construct were selected in an LB culture with kanamycin and verified through PCR and nucleotide sequencing. The primer 5‐GATGAAGTGGACGGAAGGAAGGAG‐3 was used according to the protocols for the VK005‐01 vector.
Figure 6.
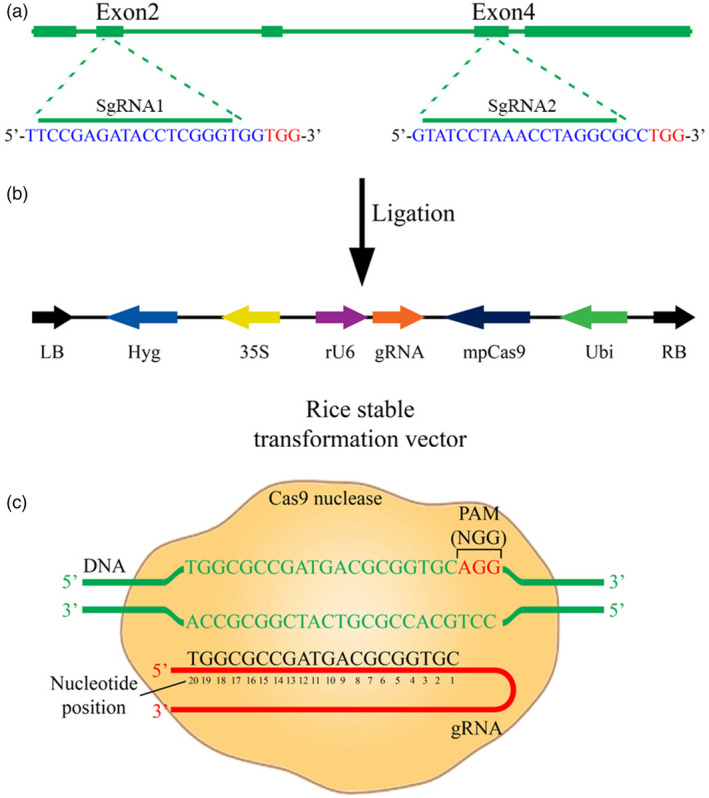
Construction of vectors for induced mutagenesis in rice. (a) Structure of OsOPR7 and targeted sites for mutagenesis in rice. (b) Components between the right and left board of the CRISPR/dpCas9‐OSOPR7 vector. (c) 20 bp target sequences of the gene exon located upstream of the protospacer adjacent motif (PAM) and G‐rich region (NGG) in rice.
Construction of phylogenetic tree and analysis of orthologous OPR genes
The genomic and coding sequences of 24 OPRs were retrieved from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). A maximum‐likelihood tree was constructed based on the alignment of 24 OPR coding sequences with ClustalW program, and 1000 bootstrap replicates were used. The structures of the OPR genes were depicted with Gene Structure Display Server (GDSD 2.0, http://gsds.cbi.pku.edu.cn/; Hu et al., 2015).
Generation of transgenic plants and genotyping for the desired mutants
The transformation of rice calli was carried out according to our previous described method (Guo et al., 2016). Rice calli were selected after they had been cultured on media containing 50 mg/L hygromycin for 4 weeks. Surviving calli were transferred to another media for the regeneration of transgenic plants. After 2–3 months of cultivation, the resulting transgenic seedlings were transferred to a paddy field during the rice growing season. DNA was extracted from each transgenic rice plant through the cetyltrimethyl ammonium bromide (CTAB) method (Rowland and Nguyen, ). All the induced mutants were verified through PCR, and gene‐specific primer pairs (Table S1) were used. Then, the mutants were confirmed by sequencing the PCR products after they were cloned into pMD18T vectors (Takara, Dalian, China). Target genes were detected for mutations by aligning the sequenced fragments with the reference sequences of the respective wild‐type control (ZH11). Heterozygous (wild‐type/single mutation), biallelic (two distinct variants) and homozygous (single mutation/single mutation) mutations were identified by using primers (Table S1) for decoding the mutant allelic sequences of target regions through the degenerate sequence decoding method (Liu et al., 2015; Ma et al., 2015; Xie et al., 2017).
Observation of anther dehiscence and pollen viability
The young osopr7 plants of the T0, T1 and T2 were divided into two plants at the tilling stage. Half of the plants were grown for the observation of anther sterility, and the other half were grown for the observation of MeJA restoration (Figure S1).
The spikelet opening, filament elongation, anther dehiscence and pollen viability of each induced rice mutant were investigated. The spikelets were classified into (i) anther indehiscence type, in which anthers protrude from spikelets, but did not dehisce or dehisce at a delayed time. Pollens were viable, but the number of pollens was lower than normal. Filament elongated normally, and (ii) anther dehiscence type, where anthers dehisce normally and release viable pollen, and filament elongated normally. Pollen viability was examined by KI‐I2 solution staining at room temperature (Xu et al., 2017). Pollen grains were collected from dehiscing anthers and placed on slides, and photographs were obtained using microscopy (Nikon Eclipse 80i).
Restoration of fertility by exogenous MeJA
The anther indehiscence osopr7 mutants were treated with exogenous MeJA for fertility restoration. The wild‐type ZH11 and osopr7 spikes were tagged prior to booting. Three osopr7 panicles were sprayed with 500 µm MeJA solution or sterile water as control. The plants were sprayed three times each day from 8:00 to 10:00 in the morning. The experiment was carried out for 15 days. The number of fertile and sterile spikelet was determined for each tagged spike for the determination of sterile spikelet percentage.
Characterization of agronomic traits of the induced mutants
The ZH11 plants and T0 to T2 rice transgenic plants were grown in paddy fields under normal growth conditions in Pyongyang, DPR of Korea. Plant height, flag leaf length and width, number of productive panicles, panicle length, number of grains per panicle, seed‐setting rate and thousand seed weight of the T0, T1 and T2 plants were measured at the rice maturation stage.
RT‐qPCR analysis
The RNA samples of the leaves of rice transgenic T1 plants were extracted using TRIzol reagent (Invitrogen, California). The housekeeping gene OsACTIN 1 was used as internal control. The target genes were reverse‐transcribed using 1 mg of RNA and a mixture of primers specific to the genes and SgRNA. Gene‐specific and target‐specific primers (Table S1) were used in RT‐qPCR according to the manufacturer’s instruction (iCycler iQ thermocycler manual; Bio‐Rad, Shanghai, China). A SYBR Green kit (Takara, Japan) was used. Comparative quantification of transcripts was performed using the ∆∆CT method (Pak et al., 2009).
Statistical analysis
Baseline and threshold cycles (CT value) were automatically determined using the Bio‐Rad iQ Software (version 3.0). The relative amount of expressed RNA was calculated according to the method of Pak et al. (2009). Data were classified using Win‐Excel and analysed using one‐way analysis of variance (ANOVA), which was performed on SPSS (version 8.0). Comparisons between treatment means were made using Duncan’s multiple range test at a level of P < 0.05.
Competing financial interests
The authors declare no competing financial interests.
Author Contributions
LJ conceived and designed the experiments. HP carried through the experiments in China and DPR Korea and is responsible for plant materials distribution. HW, MT, YK, US and DW assisted HP with experiments and data analysis. LJ and HP wrote the manuscript.
Supporting information
Figure S1 Division of an osopr7 plant.
Table S1 Primers used in the study.
Acknowledgements
The work was mainly sponsored by National Natural Science Foundation of China (Code No. 31671597). The authors also are thankful to supports from Jiangsu Collaborative Innovation Center for Modern Crop Production and State Committee for Scientific Research of DPR Korea, to colleagues at Institute of Rice, Agronomy Academy in DPR Korea for many assistances in field experiments.
Pak, H. , Wang, H. , Kim, Y. , Song, U. , Tu, M. , Wu, D. and Jiang, L. (2021) Creation of male‐sterile lines that can be restored to fertility by exogenous methyl jasmonate for the establishment of a two‐line system for the hybrid production of rice (Oryza sativa L.). Plant Biotechnol J, 10.1111/pbi.13471
Contributor Information
Haksong Pak, Email: lx19101@zju.edu.cn.
Lixi Jiang, Email: jianglx@zju.edu.cn.
References
- Bai, S. , Yu, H. , Wang, B. and Li, J. (2018) Retrospective and perspective of rice breeding in China. J. Genet. Genom. 45, 603–612. [DOI] [PubMed] [Google Scholar]
- Basnet, R. , Hussain, N. and Shu, Q. (2019) OsDGD2β is the sole digalactosyldiacyl glycerol synthase gene highly expressed in anther, and it’s mutation confers male sterility in rice. Rice, 12, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breithaupt, C. , Kurzbauer, R. , Lilie, H. , Schaller, A. , Strassner, J. , Huber, R. , Macheroux, P. and Clausen, T. (2006) Crystal structure of 12‐oxophytodienoate reductase 3 from tomato: Self‐inhibition by dimerization. Proc. Natl. Acad. Sci. USA, 103, 14337–14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Q. , Yuan, Z. , Chen, M. , Yin, C. , Luo, Z. , Zhao, X. , Liang, W. et al. (2014) Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5, 3476. [DOI] [PubMed] [Google Scholar]
- Chang, Z. , Chen, Z. , Wang, N. , Xie, G. , Lu, J. , Yan, W. , Zhou, J. et al. (2016) Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl Acad. Sci. USA, 113, 14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Hu, J. , Zhang, H. and Ding, Y. (2014) DNA methylation changes in photoperiod thermo‐sensitive male sterile rice PA64S under two different conditions. Gene, 537, 143–148. [DOI] [PubMed] [Google Scholar]
- Chen, L. and Liu, Y.G. (2014) Male sterility and fertility restoration in crops. Annual Rev. Plant Biol. 65, 579–606. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Xiao, Y. and Lei, D. (2010) Mechanism of sterility and breeding strategies for Photoperiod/Thermo‐sensitive genic male sterile rice. Rice Sci. 17, 161–167. [Google Scholar]
- Fan, Y. and Zhang, Q. (2018) Genetic and molecular characterization of photoperiod and thermo‐sensitive male sterility in rice. Plant Reprod. 31, 3–14. [DOI] [PubMed] [Google Scholar]
- Gómez, J.F. , Talle, B. and Wilson, Z.A. (2015) Anther and pollen development: a conserved developmental pathway. J. Integr. Plant Biol. 57, 876–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W.L. , Chen, T.L. , Hussain, N. , Zhang, G.P. and Jiang, L.X. (2016) Characterization of salinity tolerance of transgenic rice lines harboring HsCBL8 of wild barkey (Hordeum spontanum) line from Qinghai‐Tibet plateau. Front. Plant Sci. 10.3389/fpls.2016.01678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Jin, J. , Guo, A.‐Y. , Zhang, H. , Luo, J. and Gao, G. (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics, 31, 1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J.‐Z. , E, Z.‐G. , Zhang, H.‐L. and Shu, Q.‐Y . (2014) Workable male sterility systems for hybrid rice: genetics, biochemistry, molecular biology, and utilization. Rice, 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.C. , Yu, C. , Hu, J. , Wang, L. , Dan, Z. , Zhou, W. , He, C. et al. (2015) Pentatricopeptide‐repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility. Proc. Natl. Acad. Sci. USA, 112, 14984–14989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro, S. , Kawai‐Oda, A. , Ueda, J. , Nishida, I. and Okada, K. (2001) The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell, 13, 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.J. and Zhang, D. (2018) Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 23, 53–65. [DOI] [PubMed] [Google Scholar]
- Karunarathna, N.L. , Wang, H.Y. , Harloff, H. , Jiang, L.X. and Jung, C. (2020) Elevating seed oil content in a polypoid crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnol. J.http://doi:10.1111/pbi.13381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.Y. , Chen, Z. , Zhang, C. and Yoon, G.M. (2019) Editing of the OsACS locus alters phosphate deficiency‐induced adaptive responses in rice seedlings. J. Exp. Bot. 70, 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Wang, Y. , Duan, E. , Qi, Q. , Zhou, K. , Lin, Q. , Wang, D. et al. (2018) OPEN GLUME1: a key enzyme reducing the precursor of JA, participates in carbohydrate transport of lodicules during anthesis in rice. Plant Cell Rep. 37, 329–346. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Zhang, D. , Chen, M. , Liang, W. , Wei, J. , Qi, Y. and Yuan, Z. (2016) Development of japonica photo‐sensitive genic male sterile rice lines by editing Carbon Starved Anther using CRISPR/Cas9. J. Genet. Genom. 43, 415–419. [DOI] [PubMed] [Google Scholar]
- Li, S. , Zhang, Y. , Xia, L. and Qi, Y. (2019) CRISPR‐Cas12a enables efficient biallelic gene targeting in rice. Plant Biotechnol. J. 15, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Xie, X. , Ma, X. , Li, Jun , Chen, J. and Li, Y.G. (2015) DSDecode: a web‐based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant, 8, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Chen, L. , Zhu, Q. and Liu, Y.G. (2015) Rapid decoding of sequence‐specific nuclease‐induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol. Plant, 8, 1285–1287. [DOI] [PubMed] [Google Scholar]
- Ma, K. , Han, J. , Hao, Y. , Yang, Z. , Chen, J. , Liu, Y.G. , Zhu, Q. et al. (2019) An effective strategy to establish a male sterility mutant mini‐library by CRISPR/Cas9‐mediated knockout of anther‐specific genes in rice. J. Genet. Genom. 46, 273–275. [DOI] [PubMed] [Google Scholar]
- Ma, G. and Yuan, L. (2015) Hybrid rice achievements, development and prospect in China. J. Integr. Agric. 14, 197–205. [Google Scholar]
- Men, X. , Shi, J. , Liang, W. , Xiao, M. , Shi, J. , Liang, W. , Zhang, Q. et al. (2017) Glycerol‐3‐phosphate acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J. Exp. Bot. 68, 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak, H. , Guo, Y. , Chen, M. , Chen, K. , Li, Y. , Hua, S. , Sums, I. et al. (2009) The effect of exogenous methyl jasmonate on the flowering time, floral organ morphology, and transcript levels of a group of genes implicated in the development of oilseed rape (Brassica napus L.) flowers. Planta, 231, 79–91. [DOI] [PubMed] [Google Scholar]
- Pak, H. , Li, Y. , Kim, H. and Jiang, L. (2017) cDNA‐Amplified fragment length polymorphism analysis reveals differential gene expression induced by exogenous MeJA and GA 3 in Oilseed Rape (Brassica napus L.) flowers. J. Integr. Agric. 16, 47–56. [Google Scholar]
- Sanders, P.M. , Lee, P.Y. , Biesgen, C. , Boone, J.D. , Beals, T.P. , Weiler, E.W. and Goldberg, R.B. (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell, 12, 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A. and Browse, J. (2000) The Arabidopsis male‐sterile mutant, opr3, lacks the 12‐oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA, 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H. , Luo, D. , Zhou, D. , Zhang, Q. , Tian, D. , Zheng, X. , Chen, L. et al. (2014) The rice restorer Rf4 for wild‐abortive cytoplasmic male sterility encodes a mitochondrial localized PPR protein that functions in reduction of WA352 transcripts. Mol. Plant, 7, 1497–1500. [DOI] [PubMed] [Google Scholar]
- Tani, T. , Sobajima, H. , Okada, K. , Chujo, T. , Arimura, S. , Tsutsumi, N. , Nishimura, M. et al. (2008) Identification of the OsOPR7 gene encoding 12‐oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta, 227, 517–526. [DOI] [PubMed] [Google Scholar]
- Wang, H. and Deng, X.W. (2018) Development of the “third‐generation” hybrid Rice in China. Genom. Proteom. Bioinform. 16, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Liu, Q. , Shen, Y. , Hua, Y. , Wang, J. , Lin, J. , Wu, M. et al. (2019) Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 37, 283–286. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wei, S. , He, Y. , Yan, L. , Wang, R. and Zhao, Y. (2020) Synergistic roles of LAX1 and FZP in the development of rice sterile lemma. Crop J. 8, 16–25. [Google Scholar]
- Wasternack, C. and Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. and Hause, B. (2018) A bypass in Jasmonate biosynthesis‐ the OPR3‐ independent formation. Trends Plant Sci. 23, 276–279. [DOI] [PubMed] [Google Scholar]
- Xiao, Y. , Chen, Y. , Charnikhova, T. , Mulder, P.P.J. , Heijmans, J. , Hoogenboom, A. , Agalou, A. et al. (2014) OsJAR1 is required for JA‐regulated floret opening and anther dehiscence in rice. Plant Mol. Biol. 86, 19–33. [DOI] [PubMed] [Google Scholar]
- Xie, X. , Ma, X. , Zhu, Q. , Zeng, D. , Li, G. and Liu, Y.G. (2017) CRISPR‐GE: a convenient software toolkit for CRISPR‐based genome editing. Mol. Plant, 10, 1246–1249. [DOI] [PubMed] [Google Scholar]
- Xu, D. , Shi, J. , Rautengarten, C. , Yang, L. , Qian, X. , Uzair, M. , Zhu, L. et al. (2017) Defective pollen wall 2 (DPW2) encodes an acyl transferase required for rice pollen development. Plant Physiol. 173, 240–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Meng, Z. , Liang, W. , Behera, S. , Kudla, J. , Tucker, M.R. , Luo, Z. et al. (2016) A rice Ca2+ binding protein is required for tapetum function and pollen formation. Plant Physiol. 172, 1772–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L.P. (2014) Development of hybrid rice to ensure food security. Rice Sci. 21, 1–2. [Google Scholar]
- Zhang, D. and Yang, L. (2014) Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 17, 49–55. [DOI] [PubMed] [Google Scholar]
- Zhao, G. , Shi, J. , Liang, W. , Xue, F. , Luo, Q. , Zhu, L. , Qu, G. et al. (2015) Two ATP binding cassette G transporters, rice ATP binding cassette G26 and ATP binding cassette G15, collaboratively regulate rice male reproduction. Plant Physiol. 169, 2064–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. , Yang, S. , Zhang, D. , Zhong, Z. , Tang, X. , Deng, K. , Zhou, J. et al. (2016) Effective screen of CRISPR/Cas9‐induced mutants in rice by single‐strand conformation polymorphism. Plant Cell Rep. 35, 1545–1554. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , He, M. , Li, J. , Chen, L. , Huang, Z. , Zheng, S. , Zhu, L. et al. (2016) Development of commercial thermo‐sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9‐mediated TMS5 editing system. Scientific Rep. 6, 37395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Division of an osopr7 plant.
Table S1 Primers used in the study.


