Wheat (Triticum aestivum L.) is one of the most important cereal crops globally. It is essential to develop new wheat varieties for sustainable agricultural production. In the process of wheat breeding, specific germplasm is an essential factor for developing new varieties. Taigu male‐sterile wheat (TMSW) is a special germplasm discovered in China more than 40 years ago (Deng and Gao, 1982). When Taigu male‐sterile wheat is crossed with normal fertile wheat, 50% of plants are male‐sterile and 50% of the plants are fertile in the F1 generation. Further study revealed that TMSW contains a dominant male‐sterile gene, Ms2, on chromosome 4DS, which controls the complete male sterility of TMSW in heterozygous status (Liu and Deng, 1986). This gene can be easily transferred into different wheat backgrounds for hybridization without artificial emasculation, which is often labour intensive and time consuming, used in traditional breeding programmes.
Further, a dwarf male‐sterile wheat (DMSW) was developed by conventional backcrossing breeding in which Ms2 was closely linked with a dominant dwarf gene Rht10 on chromosome 4DS (Liu and Yang, 1991). In the F1 generation between DMSW and normal fertile wheat, the short plants are male‐sterile, and the tall plants are fertile. This makes it much easier to distinguish male‐sterile plants from fertile plants simply by the plant height in the breeding population. Using TMSW and DMSW, many new varieties have been developed in wheat through a recurrent selection strategy (Zhai and Liu, 2009). Moreover, Ms2 has also been transferred to durum wheat and hexaploid triticale (Ji et al., 1989; Ni et al., 2017).
However, only half of the plants that are fertile in the F1 population between TMSW/DMSW and a normal wheat parent can be selected for new varieties and the other half of the plants which are male‐sterile cannot be used for selection. A few years ago, the Ms2 gene was identified as a reactivated orphan gene due to the insertion of a terminal‐repeat retrotransposon in miniature element in the promoter region of ms2‐D, while the other two Ms2 alleles of ms2‐A and ms2‐B on chromosomes 4AS and 4BS are pseudogene still in TMSW/DMSW (Ni et al., 2017; Xia et al., 2017). Recently, an efficient genetic engineering system called clustered regularly interspaced short palindromic repeats (CRISPR) with CRISPR‐associated protein 9 (CRISPR/Cas9) was developed and subsequently widely applied in plants. To date, many wheat genes have been genetically modified using CRISPR/Cas9 (Wang et al., 2020).
Here, we used Agrobacterium‐mediated CRISPR/Cas9 to edit Ms2 for restoring the male fertility in sterile wheat lines with excellent agronomic and economic traits for breeding purpose. First, two single guide RNAs (g7721 and g9448) were designed to target exons IV and VII in the Ms2 open reading frame (ORF) of Ms2 for CRISPR/Cas9 to increase editing efficiency (Figure 1a). The sgRNAs were synthesized and cloned onto expression vector pWMBX110‐Cas9 (Figure 1b). Next, immature embryos of the hybrid grains from a cross between DMSW material Jimai22 (Ms2ms2/Rht10rht10) and its recurrent parent Jimai22 (ms2ms2/rht10rht10) were infected with the Agrobacterium carrying the constructed vector (Wang et al., 2017). Finally, 133 putative transgenic wheat plants were obtained after co‐cultivation, delay culture and selection culture with phosphinothricin (PPT).
Figure 1.
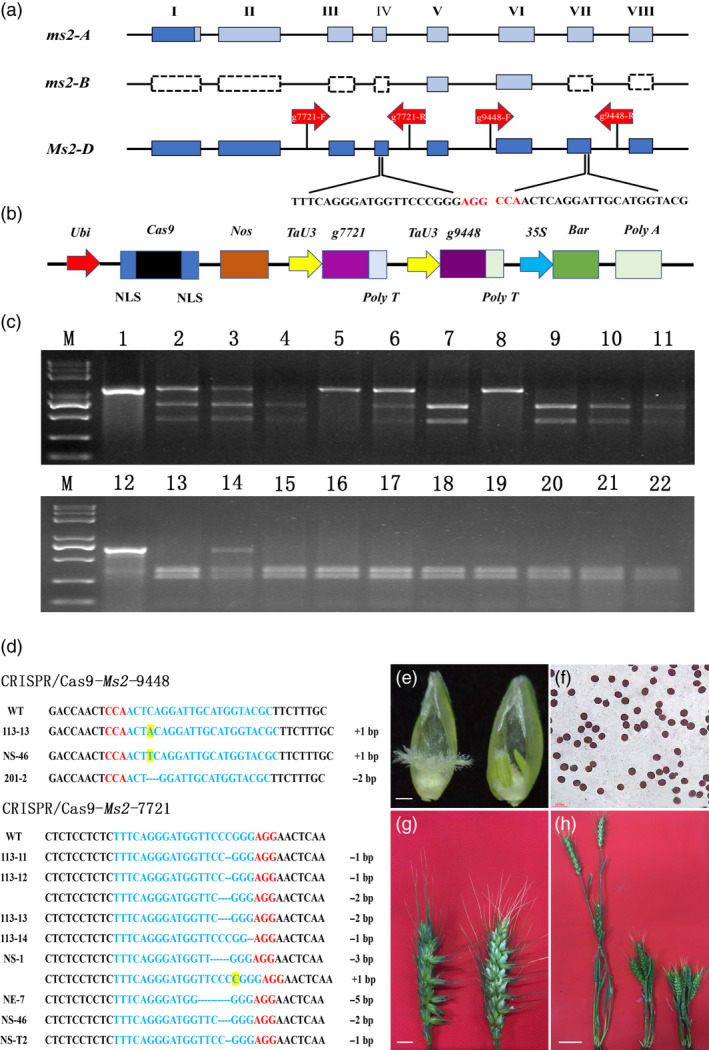
Generation of wheat Ms2 edited mutant plants using CRISPR/Cas9 and their phenotype and genotype analysis. (a) Structure of three alleles of Ms2 in DMSW and the targets designed for editing Ms2‐D; I to VII: the eight exons of Ms2. The dark blue boxes show their ORF; the light blue boxes show the destroyed ORFs; the dashed boxes are the absent exons. The red arrows indicate the detection primers of the two targets; the two base sequences represent the PAM‐guide sequences. The PAM is in red. (b) Linearized structure of vector pWMB110‐Cas9. Ubi: ubiquitin promoter; NLS: nuclear localization signal; Nos: Nos terminator; TaU3: the wheat U3 promoter; 35S: CaMV35S promoter; Bar: phosphinothricin gene. (c) Detection of representative transgenic plants for editing Ms2‐D by PCR/RE. M: DNA marker; 1 and 12: the PCR products of transgenic plants without RE; 2‐11 and 13‐22: the digested PCR products of transgenic plants with SmaI and Hpy188III, respectively. Samples 5 and 8 were homozygous‐edited type, 2, 3, 6 and 14 were heterozygous‐edited type, and 4, 7, 9‐11, 13 and 15‐22 were non‐edited type. (d) Mutant types for target sites 1 and 2 within the sequence of Ms2‐D in T0 edited plants. The PAM is in red and blue letters indicate sgRNA sequences. The dashed lines represent nucleotide deletions, insertions are shaded in yellow, and black frames represent enzymes sites. The net length for insertions or deletions is presented beside the sequences. (e) The floral organs of DMSW (left) and edited plant (right). Scale bar = 100 μm. (f) Pollen viability stained with triphenyltetrazolium chloride (TTC) of the dwarf edited fertile mutants. Scale bar = 0.1 mm. (g) Appearance of spikes in DMSW (left) and dwarf edited fertile plant (right). Scale bar = 1 cm. (h) Whole plant appearance of tall fertile plant (left), DMSW (middle), and dwarf edited fertile plant (right). Scale bar = 5 cm. [Colour figure can be viewed at wileyonlinelibrary.com]
By polymerase chain reaction (PCR) analysis with primers specific to Cas9, all the putative transgenic edited or non‐edited T0 plants showing short plant height were detected. Twelve edited plants were screened out further by PCR/restriction enzyme (RE) analysis using SmaI and Hyp188III and sequencing (Figure 1c,d). Among them, 3 were monoallelic mutants at site g9448, of which 2 had a 1‐bp insertion and the other had a 2‐bp deletion in the target sequence; 9 plants were monoallelic mutants at site g7721, of which 4 had a 1‐bp deletion, 3 had a 2‐bp deletion, one had a 4‐bp deletion, and one had a 5‐bp deletion; 2 were biallelic heterozygous mutants ((−1/‐2) and (+1/−3)); and 2 had simultaneous mutations at the two sites.
The editing efficiency was 8.27% at site g7721, which was significantly higher than that at site g9448 (2.3%) and the two sites simultaneously (1.5%). The total editing efficiency was 9.0%, in which a mutation occurred in one or two loci. Amplification using Rht10 specific primers showed that all T0 edited plants contained Rht10. During spike emergence stage, the edited plants with dwarf phenotype had normal anthers containing viable pollens (Figure 1e,f). During the grain filling period, the edited plants were fertile. As expected, the fertile dwarf mutants showed similar plant height to DMSW (Figure 1g,h).
To detect the mutations inheritance, we planted the edited mutants and analysed T1 plants using PCR/RE assay for the target sites of Ms2 and PCR analysis for Rht10. We found that the mutations on the targeted sites were continuously identified and all mutant plants contained Rht10. In addition, the edited T1 plants showed pistils and normal anthers while the DMSW plants had pistils but no anthers. The edited T1 plants with similar height to DMSW were fertile. Additionally, about one‐fourth of the T1 edited plants with genotype ms2ms2/rht10rht10 were tall due to the segregation of Rht10. The T1 edited plants with genotype ms2ems2e/Rht10Rht10 were not segregated.
All the above results indicated that the fertility of DMSW was completely recovered through editing Ms2 using CRISPR/Cas9. Our results also supported the precise cloning of Ms2 in DMSW (Ni et al., 2017; Xia et al., 2017). But, the editing efficiency in this study is lower than that in our previous report on the editing of wheat TaMTL (Liu et al., 2020). Firstly, DMSW can be only maintained by crossing it with a fertile wheat, and thus the hybrid grains consist of two genotypes Ms2ms2/Rht10rht10 and ms2ms2/rht10rht10 with the same ratio of 50%; but only the grains with Ms2ms2/Rht10rht10 are available materials for editing Ms2 and the grains with ms2ms2/rht10rht10 are not the targeted materials but transformed at the same time because they cannot be distinguished. Secondly, the genotype Ms2ms2/Rht10rht10 controlling the dwarf male‐sterile trait is heterozygous at this locus, and the original ms2 being a pseudogene, the allele of Ms2, might be also edited once it was targeted by the sgRNAs for knocking‐out, which did not affect the fertile phenotype. Thereby, the editing efficiency for Ms2 using DMSW is only one‐fourth of that for other target genes using normal wheat varieties.
In summary, Ms2 was successfully edited and the fertility of DMSW was completely recovered by CRISPR/Cas9. The editing efficiency was 9.0%. The fertile mutants obtained from the male‐sterile lines with excellent agronomic traits, biotic and abiotic resistance and better quality feature can be selected for developing new varieties. This strategy can also be extended to restore the fertility of male‐sterile durum wheat and hexaploid triticale lines containing Ms2 with good performance for the same purposes.
Conflict of interest
No conflict of interest declared.
Author contributions
X.Y. and K.W conceived the project. H.L. constructed plasmid. K.W. and L.D. performed transformation. H.T. and H.L. detected edited plants and performed data analysis. H.T., Y.Z., H.L. and X.Y. made crossing and investigated phenotype. H.T. and X.Y wrote and revised the manuscript.
Acknowledgements
We thank Prof. Yaoguang Liu for providing CRISPR/Cas expression vector. This research was supported by the Ministry of Science and Technology of China for a National Key Research and Development Program (2016YFD0102001), and Ministry of Agriculture and Rural Affairs of China for a National Transgenic Research Program (2016ZX08010004).
Tang, H. , Liu, H. , Zhou, Y. , Liu, H. , Du, L. , Wang, K. and Ye, X. (2021) Fertility recovery of wheat male sterility controlled by Ms2 using CRISPR/Cas9. Plant Biotechnol. J., 10.1111/pbi.13482
Contributor Information
Ke Wang, Email: wangke03@caas.cn.
Xingguo Ye, Email: yexingguo@caas.cn.
References
- Deng, J. and Gao, Z. (1982) Discovery and determination of a dominant male‐sterile gene and its importance in genetics and wheat breeding. Sci Sin. Ser. B. 25, 508–516. [Google Scholar]
- Ji, F.G. , Deng, J.Y. , Li, S.M. and Cheng, C.S. (1989) A primary report on breeding of dominant male‐sterile hexaploid Triticale . Hereditas 11, 1–4. [Google Scholar]
- Liu, B. and Deng, J. (1986) A dominant gene for male sterility in wheat. Plant Breed. 97, 204–209. [Google Scholar]
- Liu, H.Y. , Wang, K. , Jia, Z.M. , Gong, Q. , Lin, Z.S. , Du, L.P. , Pei, X.W. et al (2020) Editing TaMTL gene induces haploid plants efficiently by optimized Agrobacterium‐mediated CRISPR system in wheat. J Exp Bot. 71, 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. and Yang, L. (1991) Breeding of dwarfing‐sterile wheat and its potential values in wheat breeding. Chin Sci Bull. 36, 1562–1564. [Google Scholar]
- Ni, F. , Qi, J. , Hao, Q.Q. , Lyu, B. , Luo, M.C. , Wang, Y. , Chen, F.J. et al (2017) Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nat Commun. 8, 15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Gong, Q. and Ye, X.G. (2020) Recent developments and applications of genetic transformation and genome editing technologies in wheat. Theor Appl Genet. 133, 1603–1622. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Liu, H.Y. , Du, L.P. and Ye, X.G. (2017) Generation of marker‐free transgenic hexaploid wheat via an Agrobacterium‐mediated co‐transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol J. 15, 614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, C. , Zhang, L. , Zou, C. , Gu, Y. , Duan, J. , Zhao, G. , Wu, J. et al (2017) A TRIM insertion in the promoter of Ms2 causes male sterility in wheat. Nat Commun. 8, 15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, H.Q. and Liu, B.H. .(2009) The innovation of dwarf male sterile wheat and its application in wheat breeding. Sci Agric Sin. 42, 4127–4131. [Google Scholar]


