Summary
Alfalfa (Medicago sativa L.) is one of the most important forage crops throughout the world. Maximizing leaf retention during the haymaking process is critical for achieving superior hay quality and maintaining biomass yield. Leaf abscission process affects leaf retention. Previous studies have largely focused on the molecular mechanisms of floral organ, pedicel and seed abscission but scarcely touched on leaf and petiole abscission. This study focuses on leaf and petiole abscission in the model legume Medicago truncatula and its closely related commercial species alfalfa. By analysing the petiolule‐like pulvinus (plp) mutant in M. truncatula at phenotypic level (breakstrength and shaking assays), microscopic level (scanning electron microscopy and cross‐sectional analyses) and molecular level (expression level and expression pattern analyses), we discovered that the loss of function of PLP leads to an absence of abscission zone (AZ) formation and PLP plays an important role in leaflet and petiole AZ differentiation. Microarray analysis indicated that PLP affects abscission process through modulating genes involved in hormonal homeostasis, cell wall remodelling and degradation. Detailed analyses led us to propose a functional model of PLP in regulating leaflet and petiole abscission. Furthermore, we cloned the PLP gene (MsPLP) from alfalfa and produced RNAi transgenic alfalfa plants to down‐regulate the endogenous MsPLP. Down‐regulation of MsPLP results in altered pulvinus structure with increased leaflet breakstrength, thus offering a new approach to decrease leaf loss during alfalfa haymaking process.
Keywords: abscission, alfalfa, forage improvement, Medicago truncatula, Medicago sativa, PETIOLULE‐LIKE PULVINUS (PLP)
Introduction
Alfalfa (Medicago sativa L.) is one of the most widely cultivated forage species in the world and the fourth most valuable crop in the United States after corn, soybean and wheat (Zhang et al., 2005). Alfalfa has high biomass yield, excellent nutritional quality, wide adaptation and nitrogen fixation ability (Putnam et al., 2008). In the United States, it is estimated that more than 95% alfalfa is used for hay production. Leaf loss during the alfalfa haymaking process is considerable, which is in the range of 0.84% to 15.5% of hay dry matter (Pitt, 1990). Based on alfalfa production and price data from USDA National Agricultural Statistics Service (2018 data), haymaking process led to 0.5‐8.6 million tons of leaf loss and economic loss of 0.07 to 1.16 billion dollars in the United States per year. The current methods to decrease mechanical leaf loss include optimization of alfalfa harvesting conditions, mechanical or chemical conditioning and utilization of advanced machinery (Summers & Idowu et al., 2013; Putnam et al., 2008). However, such management practice requires extensive experience and is difficult to control due to unpredictable weather conditions. Based on our observation during alfalfa hay making process, over 95% leaf loss occurred specifically at the leaflet and petiole pulvinus region, which is a joint‐like thickening located at the base of the petiole or petiolule. The weakness of leaflet and petiole pulvinus region might due to the pulvinus structure or the progressive degradation of middle lamella under abscission process at that region. Therefore, strengthening pulvinus structure and manipulating leaflet and petiole abscission are promising targets in alfalfa genetic improvement.
Abscission is a developmental process allowing a plant to shed its unwanted organs in response to various developmental, hormonal or environmental cues (Addicott, 1982; Estornell et al., 2013; Roberts et al., 2002). Abscission occurs at functionally specialized cell layers, called abscission zone (AZ), which is developed at the junction between the leaving organ and the main plant body. AZ is composed of a few layers of small and dense cytoplasm cells (Addicott, 1982; Osborne and Morgan, 1989; Sexton and Roberts, 1982). The abscission process has been divided into four major phases according to studies of floral organ abscission in Arabidopsis thaliana: (i) abscission zone differentiation; (ii) acquisition of the competence to respond to abscission signals; (iii) activation of abscission; and (iv) protective layer formation (Estornell et al., 2013; Kim, 2014; Patharkar and Walker, 2018; Patterson, 2001; Taylor and Whitelaw, 2001). All of these steps are important for the fate of organs that will shed.
The prerequisite of abscission is AZ differentiation; a set of genes participating in this process has been identified in Arabidopsis, tomato and rice (Lewis et al., 2006). In Arabidopsis, BLADE‐ON‐PETIOLE1 (BOP1)/BOP2, ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1), BREVIPEDICELLUS/ KNOTTED‐LIKE FROM ARABIDOPSIS THALIANA1 (BP/KNAT1) and ASYMMETRIC LEAVES 1 (AS1) are critical for floral organ AZ differentiation (Gómez‐Mena and Sablowski, 2008; Gubert et al., 2014; McKim et al., 2008; Shi et al., 2011). Several genes regulating pedicel AZ differentiation have been identified, including JOINTLESS, LATERAL SUPPRESSOR (Ls), MACROCALYX (MC) and SLMBP21 (Liu et al., 2014; Mao et al., 2000; Nakano et al., 2012; Schumacher et al., 1999). In rice, SHATTERING4 (SH4), qSH1, OsSH1 and SHATTERING ABORTION 1 (SHAT1) function as positive regulators to modulate pedicel AZ development, while OsCPL1 represses pedicel abscission zone differentiation (Ji et al., 2010; Konishi et al., 2006; Li et al., 2006; Lin et al., 2012; Zhou et al., 2012b).
After AZ is differentiated, the AZ cells will be staying at a quiescent state, until they perceive abscission‐promoting signals to initiate the abscission process (Nakano et al., 2013). Abscission signals will activate the expression of genes such as cell wall hydrolytic enzymes (e.g. polygalacturonases) to act on structural polysaccharides leading to the hydrolysis of the middle lamella and cell walls of the AZ cells (Estornell et al., 2013). Both ethylene and auxin (IAA) were previously proposed as important signals for abscission activation (Taylor and Whitelaw, 2001). However, ethylene‐insensitive mutants ethylene resistant 1‐1 (etr1‐1) and ethylene‐insensitive 2 (ein2) showed delayed instead of abolished floral organ abscission, indicating that ethylene is not critical for abscission, and thus, auxin has been proposed as a key player in the regulation of abscission process (Patterson and Bleecker, 2004; Taylor and Whitelaw, 2001). Auxin response factors (ARFs) are transcription factors that mediate responses to auxin. In Arabidopsis, arf2 mutant delayed floral organ abscission, while arf1 enhanced arf2 phenotype (Okushima et al., 2005). The mutation of NPH4/ARF7 and ARF19 enhanced arf2 phenotype (Ellis et al., 2005). It has been shown that indoleacetic acid (IAA) signalling is a prerequisite for floral organ shedding in Arabidopsis (Basu et al., 2013). Microarray analysis of gene alterations in response to auxin depletion in tomato and Mirabilis jalapa revealed that acquisition of ethylene sensitivity in leaf and flower AZ is associated with differential expression of auxin‐related genes (Meir et al., 2006; Meir et al., 2010). In tomato, KD1, a KNOTTED1‐LIKE HOMEBOX (KNOX) family gene, was identified to modulate pedicel and petiole abscission by regulating genes involved in the auxin signalling pathway (Ma et al., 2015). However, the transcriptional regulation of hormonal homeostasis in the leaf AZ is still unclear.
To date, a wealth of valuable information has been accumulated based on the studies of floral organ abscission in Arabidopsis, pedicel abscission in tomato and seed shattering in rice. However, our understanding of molecular mechanisms on leaf abscission is still limited. Recently, it has been revealed that both drought‐ and pathogen‐triggered cauline leaf abscission in Arabidopsis might be regulated through salicylic acid signalling (Patharkar and Walker, 2016; Patharkar et al., 2017). HAESA/HAESA‐like 2 (HAE/HSL2), INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), NEVERSHED (NEV) and MAPK KINASE4/5 (MKK4/5) are required for drought‐induced leaf abscission (Patharkar and Walker, 2016). HAESA/HAESA‐like 2 (HAE/HSL2), IDA and NEVERSHED (NEV) are required for the initiation of pathogen‐triggered leaf abscission (Patharkar et al., 2017). BOP1/BOP2 in Arabidopsis and its ortholog NODULE ROOT (NOOT) in Medicago truncatula have been shown to be involved in leaf abscission zone formation (Couzigou et al., 2016; McKim et al., 2008). Recent transcriptome studies of leaf abscission have provided insights into the regulatory mechanism of abscission (Kim et al., 2016; Li et al., 2017; Li et al., 2016; Liao et al., 2016a, 2016b, 2016c).
As a model plant for legume species, Medicago truncatula is also suitable for leaf abscission study. It has compound leaves and its leaflets and petioles abscise at the senescing stage at the base of pulvinus. Over the years, with the development of genomic resources, particularly the generation of large‐scale insertional mutagenesis (Tadege et al., 2005) and the sequencing of the M. truncatula genome (Young et al., 2011), many genes with novel functions have been identified (Chai et al., 2016; Kang et al., 2016; Zhao et al., 2019; Zhou et al., 2019). Leguminous plants open their leaves during the day and close them at night, and this kind of nyctinastic movement is induced by volume change in motor cells in the pulvinus (Ueda and Nakamura, 2007). The PETIOLULE‐LIKE PULVINUS (PLP) gene was identified by forward screening from the retrotransposon‐tagged mutant population of M.truncatula; the loss of function of PLP caused the alteration of pulvini to petiolules and affected the nyctinastic movement of leaflets (Zhou et al., 2012a). In this study, we report novel findings that the loss of function of PLP results in an absence of AZ formation and PLP plays an important role in leaflet and petiole AZ differentiation, thus providing new information on molecular mechanism of leaf and petiole abscission. Furthermore, down‐regulation of the PLP gene (MsPLP) in alfalfa results in the alteration of pulvinus to petiolule‐like structure with increased leaflet breakstrength, offering a new approach to minimize leaf loss during alfalfa haymaking process.
Results
Mutation in PLP abolishes shedding of leaflet and petiole in M. truncatula
The PLP gene has one exon with 579 nucleotides containing a LOB domain (Zhou et al., 2012a). To characterize the role of PLP in leaflet and leaf abscission, three alleles plp‐1 (a Tnt1 inserted after base 178), plp‐2 (a Tnt1 inserted after base 130) and plp‐4 (a Mere1 inserted after base 246) were used for analysis (Figure 1a). The loss of function of PLP altered the leaflet pulvini into petiolule‐like structures (Figure S1a‐d) and the petiole pulvini into petiole‐like structures (Figure S1e‐h).
Figure 1.
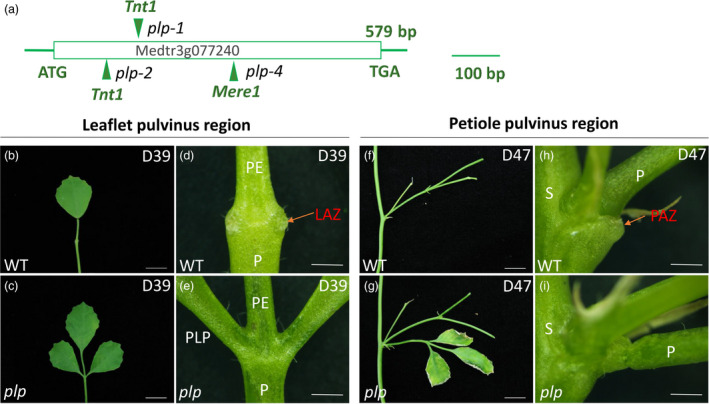
Petiolule‐like pulvinus (plp) mutant of M. truncatula showing defects in leaflet and petiole abscission. (a) Schematic representation of the gene structure of PLP. The arrows indicate the location of Tnt1 and Mere1 retrotransposons in the plp alleles. (b, c) Senescing leaf (39‐day‐old) of wild type and plp mutant. Wild‐type leaflet shed at this stage. (d, e) Close‐up view of leaflet base at leaflet abscission zone (LAZ) in wild type and plp mutant. Red arrow indicates the LAZ in wild type. The leaflet has already abscised from wild type, with the AZ left at the base of pulvinus, while for plp mutant, leaflet is still attached to petiole. (f, g) Senescing leaf (47‐day‐old) with petiole abscission zone. Wild‐type leaf shed at this stage. (h, i) Close‐up view of petiole abscission zone of wild type and plp mutant. Red arrow indicates petiole abscission zone (PAZ). The petiole of wild type has already abscised from stem with the PAZ left at the base of pulvinus, while plp petiole is still attached to stem, although the leaflets are nearly dry. PE, petiolule; P, petiole; PLP: petiolule‐like pulvinus; S, stem. D39, 39‐day‐old; D47, 47‐day‐old. Scale bars: (b, c, f, g) 1 cm; (d, e, h, i) 1 mm. [Colour figure can be viewed at wileyonlinelibrary.com]
In wild‐type M. truncatula, organ detachment started with leaflet at the base of pulvinus (Figure 1b, d) and then followed by petiole at the stem–petiole junction (Figure 1f,h). Leaflet and petiole started to shed at the leaf age of 35 days (67% remaining) and 37 days (87% remaining), respectively (Table S1). The percentage of plants with remaining leaves and petioles decreases with the increase of leaf ages until no leaflets and petioles were left on the plant, when the leaf age reaches 43 and 45 days (Table S1). In contrast, both leaflets and petioles of the plp mutant remain attached to the plant even after 45 days (Table S1). In plp mutants, shedding of leaflet and petiole was abolished even when the whole plant was dead.
The abscission defects of plp were further characterized by shaking assay. After shaking vigorously for 2 min, almost all the senescing leaves of wild type shed, while for plp mutants, most petioles and leaflets were still attached to the plant body (Figure S2a). Markedly reduced leaf shed was observed for plp with respect to wild‐type plants (Figure S2b‐g). The weight of dropped leaves of wild type was 5 times more than that of plp (Figure S2h). The shaking assay further confirmed that plp has a defect in leaflet and petiole abscission at senescing stage.
In order to further characterize the leaflet and petiole abscission defects of plp, a breakstrength assay was performed. The wild‐type leaflet and petiole breakstrength gradually decreased after 33 days old and 35 days old, respectively, indicating a progressive degradation of the middle lamellae in leaflet and petiole abscission zone (Figure 2). However, leaflet and petiole breakstrength of plp showed no obvious change from 33‐ to 39‐day‐old leaf and 35‐ to 39‐day‐old leaf (Figure 2). After 39 days, the leaflet and petiole breakstrength of plp gradually decreased with the increase of leaf ages (Figure 2). In all cases, both leaflet breakstrength and petiole breakstrength of plp mutants were significantly higher than wild type (Figure 2).
Figure 2.
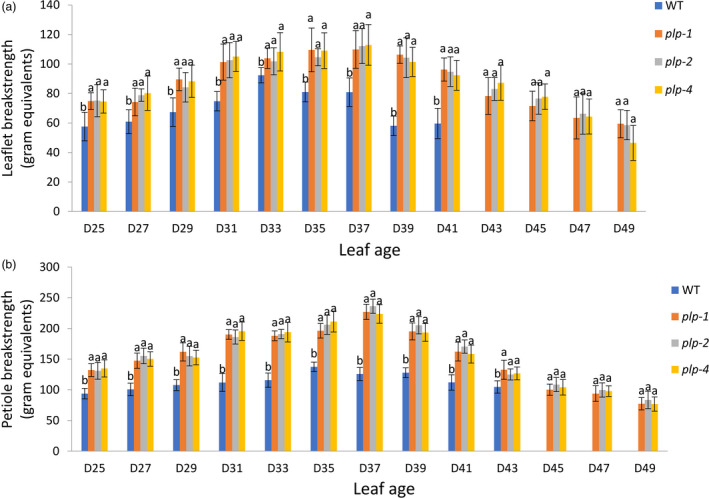
Leaflet and petiole breakstrengths of plp and wild type at different stages in M. truncatula. (a) Leaflet breakstrength measurements of plp and wild type at different stages. Wild‐type leaflets have already dropped when leaf age reached 43 days. (b) Petiole breakstrength measurements of plp and wild type at different stages. Wild‐type petiole has already dropped when leaf age reached 45 days. D25, 25‐day‐old leaf; D27, 27‐day‐old leaf; … D49, 49‐day‐old leaf. Error bars represent SD (n = 8). [Colour figure can be viewed at wileyonlinelibrary.com]
PLP plays an important role in controlling leaflet abscission zone (LAZ) and petiole abscission zone (PAZ) differentiation
To further determine whether normal LAZ and PAZ were developed in plp, we used scanning electron microscopy (SEM) to examine the leaflet pulvinus–petiolule boundary and petiole pulvinus–stem boundary of wild type and plp mutant (Figure 3). In wild type, scars were left after manual removal of leaflet and petiole from the plant. Ruptured cells were observed upon removal of 31‐day‐old leaflet and 35‐day‐old leaflet at the fracture plane (Figure 3b,c). Owing to the weakening of the middle lamellae, removal of 39‐day‐old leaflet only caused very few cell ruptures (Figure 3d). Leaflets were already abscised at 43 days and 47 days, and there was a protective layer of rounded cells formed at the leaflet abscission zone (Figure 3e,f). By contrast, the leaflet fracture surface of plp mutant sample showed no evidence of AZ activation at any age and cells ruptured during leaflet removal (Figure 3g‐k).
Figure 3.
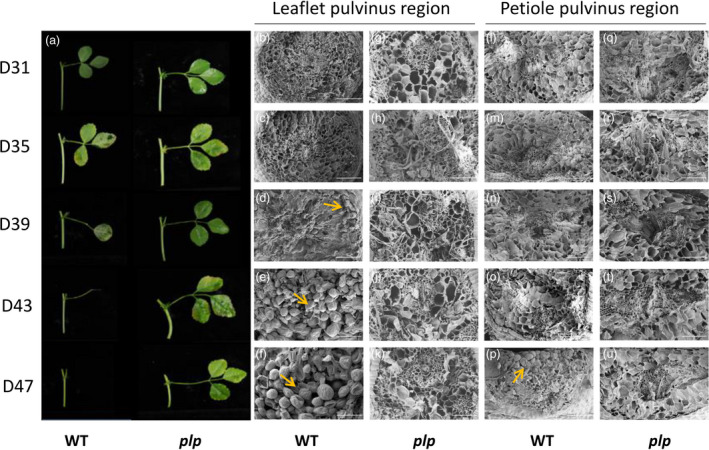
Abscission phenotype and scanning electron micrographs of leaflet and petiole abscission zones in M. truncatula. (a) Leaves of wild type and plp at different ages. In wild‐type plants, 39‐day‐old leaflet started to abscise from the petiolule and petiole of 47‐day‐old leaf had already abscised from stem, while in plp plants, leaflets and petiole did not abscise. (b‐u) Scanning electron micrographs of leaflet and petiole fracture planes in wild type and plp from leaf age of 31 days to 47 days. (b‐f) Leaflet fracture planes of wild type showing progression from broken cells (31‐, 35‐ and 39‐day‐old leaflets) to rounded AZ cells (43‐ and 47‐day‐old leaflets). (g‐k) Leaflet fracture planes of plp mutant showing broken cells (all ages). (l‐p) Petiole fracture planes of wild type showing progression from broken cells (31‐, 35‐, 39‐ and 43‐day‐old leaves) to rounded AZ cells (47‐day‐old leaves). (q‐u) Petiole fracture planes of plp mutant showing broken cells (all ages). D31, 31‐day‐old leaf; D35, 35‐day‐old leaf; D39, 39‐day‐old leaf; D43, 43‐day‐old leaf; D47, 47‐day‐old leaf. Scale bars: 100 μm. [Colour figure can be viewed at wileyonlinelibrary.com]
Broken cells were observed upon removal of petiole (from 31 to 43 days old) in wild type (Figure 3l‐o). Due to the abscission of petiole in 47‐day‐old leaf, the petiole fracture surface of wild type showed a layer composed of spherical elongated cells (Figure 3p). However, in plp mutant, ruptured cells at fractured plane were observed at all leaf ages (Figure 3q‐u).
Phenotype characterization and SEM analysis revealed the important role of PLP in LAZ and PAZ development. To determine more precisely the differences in cellular morphology between wild type and plp, longitudinal sections of leaflet and petiole pulvinus region at 25 and 35 days were examined and compared (Figure 4). The 25‐day‐old leaf (mature leaf) represents the stage before abscission process occurs, whereas the 35‐day‐old leaf (senescing leaf) represents the stage the abscission process was undergoing at both leaflet and petiole abscission zones based on the observation in Table S1. In wild type, a layer of small cytoplasmic dense cells at the junction between pulvinus and petiolule was differentiated at 35 days (Figure 4e, red arrow), and the morphological differences were apparent when compared with the 25‐day‐old section (Figure 4c). The edge of the pulvinus and petiolule junction was detached due to the dissolvement of middle lamella of LAZ cells (Figure 4e). Unlike the wild‐type control, no differentiated small cells were detected in the plp region which is marked by red asterisk indicative of the region corresponding to wild‐type LAZ (Figure 4f). Compared with 25‐day‐old section at petiole pulvinus region, cytoplasmic dense AZ cell layers marked by red arrow were visible between petiole pulvinus and stem in wild type at 35 days (Figure 4k). However, plp did not show such a structure differentiated at the corresponding location marked by a red asterisk at 35‐day‐old leaf (Figure 4l). Therefore, the distinct cell layers that differentiated at the junctions between pulvinus and petiolule and between petiole and stem were absent in plp.
Figure 4.
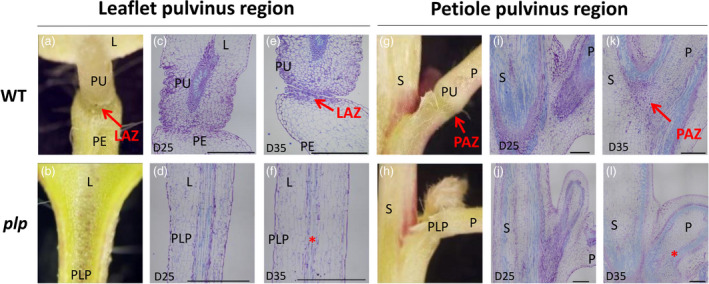
Anatomical comparisons between leaflet abscission zone (LAZ) and petiole abscission zone (PAZ) in wild type and plp. (a, b) Close‐up view of leaflet pulvinus region. (c‐f) Microscopic analysis of longitudinal sections across the LAZ stained by toluidine blue at leaflet pulvinus region. (c, d) Mature leaflet (25‐day‐old) pulvinus region in wild type and plp. No leaflet AZ was formed in either wild type or plp mutant. (e, f) Senescing leaflet (35‐day‐old) pulvinus region in wild type and plp. LAZ of wild type is shown in the picture, and no LAZ is formed in plp mutant. (g, h) Close‐up view of petiole pulvini region. (i‐l) Microscopic analysis of longitudinal sections across the PAZ stained by toluidine blue at the petiole pulvinus region. (i, j) Mature leaf (25‐day‐old) petiole pulvinus region in wild type and plp. No AZ was formed in either wild type or plp. (k, l) Senescing leaf (35‐day‐old) petiole pulvinus region in wild type and plp. Small cytoplasmic cells were observed along the petiole AZ region in wild type, and no AZ cells were observed in plp. D25, 25‐day‐old leaf; D35, 35‐day‐old leaf. Red arrows indicate LAZ and PAZ; red asterisks in (f) and (l) indicate the region where the LAZ and PAZ should be present. Scale bars: (a,b) 5 mm; (g,h) 1 mm; (c‐f,i‐l) 500 μm. L, leaflet; PE, petiolule; PLP: petiolule‐like pulvinus; PU, pulvinus; P, petiole; S, stem. [Colour figure can be viewed at wileyonlinelibrary.com]
PLP expression pattern during the leaf senescing process
The expression pattern of MtPLP in wild‐type plants was analysed by M. truncatula Gene Expression Atlas (Benedito et al., 2008). The expression of MtPLP was detected in almost all the organs at various developmental stages, with a relatively high expression level in vegetative buds and nodules and relatively low levels in seed and leaf (Figure S3).
To determine whether there is a correlation between the PLP expression pattern and leaflet and petiole abscission, we analysed transgenic M. truncatula plants that expressed GUS under the control of PLP promoter. We examined GUS expression during the time course of abscission in three stages as shown in Table S2: stage I, prior to organ separation; stage II, during abscission; and stage III, after abscission when the remaining cells form protective scar tissue. The expression of the proPLP::GUS construct was observed in both leaflet and petiole pulvinus regions and at the base of leaflet–petiolule junction and stem–petiole junction (Figure 5a,e). Prior to organ abscission, the expression of PLP was restricted to the abscission zones, in which the leaflet and petiole had not detached (Figure 5b,f). During abscission, a strong signal was detected at the base of leaflet and petiole pulvinus region (Figure 5c,g). After abscission, at the AZ scars following leaflet and petiole detachment, no GUS signal was detected (Figure 5d,h). Taken together, the proPLP::GUS expression pattern showed that PLP is strongly expressed in the abscission zone prior to abscission and during abscission but not observed after abscission, confirming that PLP contributes to AZ differentiation.
Figure 5.
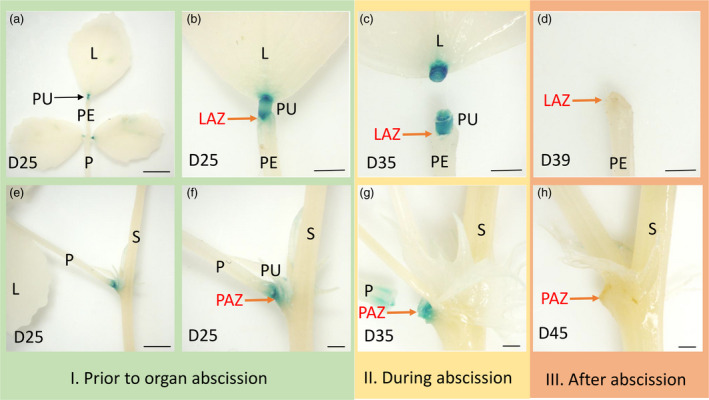
GUS staining of leaflet pulvinus region and petiole pulvinus region at different abscission zone developmental stages of transgenic M. truncatula plants carrying the PLP promoter‐GUS construct. (a, e) GUS expression in the leaflet and petiole pulvinus region. (b, f) Prior to organ abscission, GUS expression was detected at the base of leaflet pulvinus and at the base of leaf pulvinus in 25‐day‐old leaf. (c, g) During organ abscission, GUS activity was detected at the base of 35‐day‐old leaflet and 43‐day‐old leaf petiole. (d, h) After abscission, no GUS expression was observed at the base of leaflet pulvinus or petiole pulvinus after the abscission of leaflet and petiole. L, leaflet; PE, petiolule; PU, pulvinus; P, petiole; S, stem; LAZ, leaflet abscission zone; PAZ, petiole abscission zone. Scale bars: (a, e) 5 mm; (b–d, f–h) 1 mm. [Colour figure can be viewed at wileyonlinelibrary.com]
Gene expression analysis identified genes regulated by PLP in the abscission process
To explore the transcriptional mechanisms underlying the loss of function of AZ development in plp mutants, gene expression in 25‐day‐old leaflet AZ tissues in wild type and three independent mutant lines was analysed using Affymetrix Medicago GeneChips (Affymetrix, CA, USA). The ratio between mutant and wild type above twofold was considered up‐regulated, while below 0.5 was considered down‐regulated. We identified 826 up‐regulated genes and 1216 down‐regulated genes (Tables S3 and S4). The differentially expressed genes were mapped in pathways including photosynthesis, hormone metabolism, signalling, stress, RNA, secondary metabolism, mitochondrial electron transport and DNA pathway (Figure S4a). Genes mapped in the hormone metabolism and cell wall pathways are shown in Figure S4b, c. In the hormone metabolism pathway, 67 auxin‐related genes were down‐regulated, including 53 SMALL AUXIN‐UP RNA (SAUR) like family genes, 4 Gretchen Hagen3 (GH3) genes and 1 AUX/IAA gene. For ethylene‐related genes, 3 ethylene response factors (ERFs), 3 1‐aminocyclopropane‐1‐carboxylic acid synthases (ACSs) and the ethylene‐insensitive 4 (EIN4) were down‐regulated in the plp mutant (Figure S4b). In the cell wall pathway, the expression of 64 cell wall‐related genes were significantly altered in plp mutant, including genes for polygalacturonase (PG), xyloglucan endotransglucosylase/hydrolase (XTH), glycosyl hydrolase (GH), pectin methylesterase (PME) and expansin (EXP) (Figure S4b). Twenty auxin‐related genes were selected for PCR validation (Table S5), and they showed the same expression pattern as microarray data (Figure S5).
Microarray analysis revealed 152 putative abscission‐related transcription factors (TFs). They belong to 23 different families of TFs mainly including bHLH (8.6%), WRKY(8.6%), zinc fingers (8.6%), Aux/IAA family (7.9%), MYB (7.9%) and Homeobox (6.6%), suggesting a complex regulation of leaflet abscission in M. truncatula (Figure S6). In addition, there were several hormone‐related TFs represented by AUX/IAA (7.9%), ARR (4.6%) and ARF (4.6%). We performed a Venn diagram analysis using our identified 152 putative abscission‐related TFs and the 188 soybean abscission‐specific TFs selected by Kim (Kim et al., 2016). Totally 26 overlapped candidates are potentially involved in both soybean and M. truncatula leaf abscission (Figure S7; Table S6). These results suggest that plp affects auxin‐related genes which, in turn, affect LAZ development in M. truncatula.
Expression of the NOOT gene is down‐regulated in plp
In order to examine whether there are any overlaps between published AZ‐related genes and our microarray results, homologous genes of published genes related to AZ development were identified in M. truncatula by blast on phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) (Table. S7). Except for the NOOT gene, none of the genes listed in Table S7 were significantly up‐ or down‐regulated, indicating the regulatory pathway of floral organ abscission in Arabidopsis might be different from that of leaflet and petiole abscission in M. truncatula.
We investigated the relationship between PLP and NOOT. The NOOT gene is involved in leaflet, petiole, petal and seed abscission (Cougizou et al., 2016). For wild type, a 39‐day‐old leaflet was already abscised from petiolule, while a 39‐day‐old leaflet of noot mutant was still attached to petiolule (Figure S8a,b). The 47‐day‐old petiole of wild type had already abscised from stem, while the petiole of plp was still attached to stem tightly (Figure S8a,b). The expression analysis of NOOT in wild type and plp mutants showed that the NOOT gene was significantly down‐regulated for more than twofold compared with wild type (Figure S8e).
Isolation and suppression of MsPLP gene in alfalfa
Because of the high degree of sequence identity between M. truncatula and alfalfa, the alfalfa PLP (MsPLP) gene was cloned using the primers derived from MtPLP. Sequence comparison analysis showed that MtPLP and MsPLP share 97.4% similarity (Figure S9). Analysis of deduced amino acid sequences revealed that MsPLP protein contained 193 amino acids, showing 99% identity to MtPLP (Figure S10). The MsPLP protein is also highly similar to its putative orthologs in Glycin max, Lotus japonicas, Pisum sativum and Trifolium medium, indicating PLP is highly conserved in legume species (Figure S10). The PLP orthologs in various species were obtained by BLASTN through NCBI and Phytozome and used for phylogenetic analysis (Figure S11). Phylogenetic analysis showed that MsPLP and MtPLP were closest to each other and were clustered close to PsAPU, TmELP1 and TpLOB (Figure S11).
To suppress the activity of the endogenous MsPLP in alfalfa, MsPLP‐RNAi vectors (Figure S12) were constructed and introduced into alfalfa by Agrobacterium tumefaciens‐mediated transformation (Figure S13a‐e). Only one plant was selected from each callus, representing an independent line. Fourteen independent transgenic lines were identified by PCR analysis (Figure S13f). Quantitative RT‐PCR analysis revealed that 12 transgenic lines (S1‐S12) had MsPLP expression levels reduced by more than 75% when compared with the wild type (Figure S14a). Three transgenic lines, S1, S2 and S3, were used for further analysis.
MsPLP‐RNAi transgenic alfalfa shows defects in pulvinus development
Plant height and fresh weight showed no significant differences between wild type and transgenic lines, suggesting that knocking down MsPLP had no effects on biomass yield (Figure S14b‐d). The RNAi alfalfa plants showed defects in leaflet and petiole pulvinus development (Figure 6a‐h) similar to the M. truncatula plp mutant.
Figure 6.
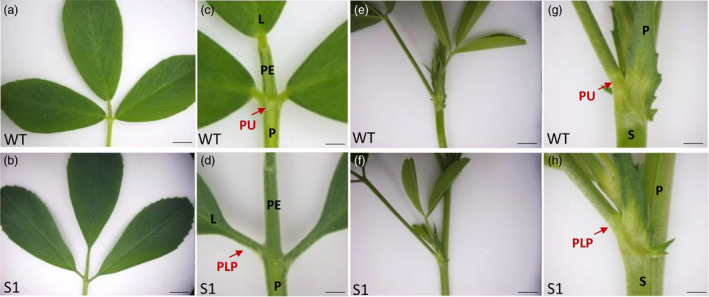
MsPLP‐RNAi transgenic lines showed defects in pulvinus development. (a, b) Leaf of wild type and MsPLP‐RNAi alfalfa. (c, d) Close‐up view of leaflet base in wild type and transgenic alfalfa. Transgenic alfalfa showed defects in pulvinus development, pulvinus was altered to petiolule‐like structure. (e, f) Petiole of wild type and MsPLP‐RNAi alfalfa. (g, h) Close‐up view of petiole base in wild type and transgenic alfalfa. Transgenic alfalfa showed defects in petiole pulvinus development. L, leaflet; PE, petiolule; PU, pulvinus; P, petiole; S, stem; PLP, petiolule‐like pulvinus. Scale bars: (a, b) 1 cm; (c, d) 1 mm; (e, f) 1 cm; (h, 1) 1 mm. [Colour figure can be viewed at wileyonlinelibrary.com]
Suppression of MsPLP strengthens the leaflet and petiole pulvinus region
Leaflet and petiole breakstrengths were measured when plants reached blooming stage, at which alfalfa is normally harvested. In the field, alfalfa plants are generally harvested 10 cm above ground. Therefore, we evaluated the leaflets above the height of 10 cm. Breakstrengths of 3rd leaflets to 12th leaflets were evaluated (Figure 7a). For both terminal and lateral leaflets, leaflet breakstrengths of transgenic lines were significantly higher than that of wild type (Figure 7b,c). For petiole breakstrength, 3rd to 5th petiole and 11th to 12th petiole of the transgenic lines were significantly higher than that of wild type (Figure 7d).
Figure 7.
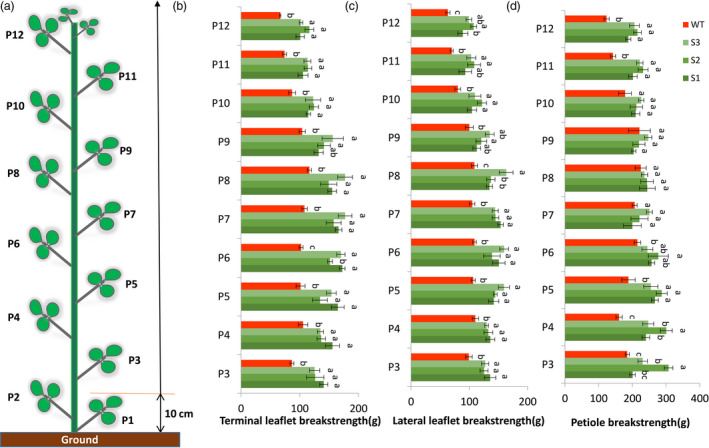
Terminal and lateral leaflet breakstrength assay and petiole breakstrength assay of wild type and transgenic alfalfa. (a) Schematic leaflet distribution on alfalfa stem. The evaluation of leaflet and petiole breakstrength was started from the third leaflet because alfalfa is normally harvested 10 cm above ground under field conditions. (b) Terminal leaflet breakstrength measurements at blooming stage. (c) Lateral leaflet breakstrength measurements at blooming stage. (d) Petiole breakstrength measurements at blooming stage. Error bars indicate SD (n = 5). P3, the 3rd leaflet; P4, the 4th leaflet; … P12, 12th leaflet. [Colour figure can be viewed at wileyonlinelibrary.com]
In order to further characterize whether the pulvinus region was strengthened, the frequency of the breaking positions during breakstrength assay was recorded. In wild type, the terminal leaflet, lateral leaflet and petiole were detached specifically at the pulvinus region during the breakstrength assay, indicating the pulvinus region is the weakness area (Figure S15a,c). In transgenic lines, the detachment of terminal leaflet occurred at three positions, D1: leaflet (8%), D2: petiolule‐like pulvinus (22%) and D3: petiolule (70%); the detachment of lateral leaflet occurred at three positions, D1: leaflet (2%), D2: petiolule‐like pulvinus (57%) and D3: base of petiolule‐like pulvinus (41%); and the detachment of petiole occurred at two positions, D1: petiole‐like pulvinus (95%) and D2: petiole (5%) (Figure S15b,c). The various detachment position demonstrates that the pulvinus region was strengthened by genetically knocking down the PLP gene in alfalfa.
Analysis of nutritive value of MsPLP down‐regulated alfalfa plants
In order to evaluate whether the knockdown of MsPLP will affect alfalfa quality under greenhouse non‐senescing conditions, acid detergent fibre (ADF), neutral detergent fibre (NDF), crude protein (CP), in vitro true dry matter digestibility (IVTDMD), total digestible nutrients (TDN) and relative feed value (RFV) were analysed. No significant change was found in these forage quality parameters between wild type and transgenic lines for the whole plant, leaf and stem (Figure S16a‐c). The results show that down‐regulation of MsPLP has no impact on alfalfa nutritive value when the vegetative plants were gently harvested without leaf loss.
Discussion
PLP is required for AZ differentiation
Abscission zone is a unique anatomical structure which is essential for the abscission process. Several genes involved in Arabidopsis floral organ (BOP1/BOP2, ATH1 and AS1), tomato pedicel (JOINTLESS, MC and SLMBP21) and rice pedicel (SH4, QSH1, SHAT1, OsSH1 and OsCPL1) abscission zone differentiation have been identified (Gómez‐Mena and Sablowski, 2008; Gubert et al., 2014; Ji et al., 2010; Konishi et al., 2006; Li et al., 2006; Lin et al., 2012; Liu et al., 2014; Mao et al., 2000; McKim et al., 2008; Nakano et al., 2012; Zhou et al., 2012b). In M. truncatula, the NOOT gene, a BOP ortholog, has been shown to modulate petal, leaflet and seed abscission zone differentiation (Couzigou et al., 2016). Interestingly, in both plp and noot mutants, simplified stipules were observed, while the pulvinus structure of noot mutant is not modified as the plp mutant, suggesting that the absence of PLP activity modifies the cellular structure of the tissue where leaflet and petiole AZs are differentiated. Until now, the molecular mechanism of leaflet and petiole AZ differentiation is still largely unknown. The present study focused on leaflet and petiole abscission. We discovered that the PLP gene plays an important role in regulating leaflet and petiole abscission zone differentiation in M. truncatula. The loss of function of PLP results in a complete absence of leaflet and petiole abscission during plant senescence. Microscopic observation of cross sections and SEM analysis further confirmed that the plp mutants have no leaflet and petiole abscission zone differentiation, suggesting the importance of PLP in initiating AZ differentiation.
Lateral organ boundary formation and abscission zone development
Night closure of leaflets is a common feature of legumes, and such nyctinastic movement is generated by the pulvinus. It has been proposed that PLP regulates pulvinus establishment through controlling boundary formation for the correct production of compact and convoluted motor cells to mediate leaflet movement (Zhou et al., 2012a). PLP is a LOB domain protein, and it has 75% identity to Arabidopsis ASL4/LOB which belongs to the LBD protein family (Zhou et al., 2012a). LBD proteins play important roles in modulating plant development (pollen, embryo, root, leaf and inflorescence development), hormone response (cytokine, ABA, auxin, JA, gibberellin), anthocyanin and nitrogen metabolism, plant regeneration (callus formation), disease susceptibility, photomorphogenesis, secondary growth and pulvinus development (Majer and Hochholdinger, 2011; Xu et al., 2016). LBDs are specifically expressed at organ boundaries regulating plant lateral organ development (Yang et al., 2016). In wild type, there is a clear boundary region at the base of leaflet and petiole pulvinus, whereas no boundary was formed at the base of leaflet and petiole in the plp mutant (Figure 4). The wild‐type leaflet and petiole AZs were differentiated after the development of leaflet and petiole pulvinus (Figure 4c,e,i,j); thus, they are adventitious AZs according to Addicott (1982). In the plp mutant, the loss of cell fate in leaflet and petiole AZs led to the failure of AZ cell placement and differentiation. These observations indicate that the existence of organ boundaries influences AZ formation. Several genes that are involved in organ boundary formation have been shown to be involved in AZ development including BOP1/2, ATH1, AS1 and HWS (Gómez‐Mena and Sablowski, 2008; González‐Carranza et al., 2007; Gubert et al., 2014; Hepworth et al., 2005; Jun et al., 2010; McKim et al., 2008; Norberg et al., 2005; Xu et al., 2008). Our study provides more evidence to support the potential link between organ boundary formation and AZ development.
In Arabidopsis, BOP1/BOP2 have been demonstrated as the earliest regulators associated with AZ differentiation, which are required for abscission in both wild‐type and 35S::IDA plants (McKim et al., 2008). The homologous genes of BOP in tobacco (Nicotiana tabacum) as well as in a few legume species (Pisum sativum, Lotus japonicas, Lupinus angustifolius and Lupinus luteus) are also necessary for the abscission of lateral organs (Couzigou et al., 2016; Frankowski et al., 2015; Kućko et al., 2019; Wu et al., 2012). In M. truncatula, the noot mutant has no specialized small condensed cell layer along its leaflet and leaf abscission zone region, indicating the role of NOOT gene in regulating the establishment of AZ (Couzigou et al., 2016). In the current study, we found that the NOOT gene was down‐regulated in the plp mutant, suggesting a possible relationship between PLP and NOOT. It has been shown that the LOB domain genes ASYMMETRIC LEAVES2 and LOB were up‐regulated in 35S:BOP and down‐regulated in bop1/2 mutant, therefore indicating a potential link between these genes in regulating lateral organ cell fate and polarity in Arabidopsis (Ha et al., 2007). In addition, the overexpression of LBD15 activated the transcription of BOP1 and BOP2, which were critical in controlling lateral organ development and inflorescence architecture (Zhu et al., 2014). Since PLP is an LBD protein, we propose a potential link between PLP and NOOT in defining the AZ cells at organ boundaries by modulating lateral organ polarity and cell fate. Further research on genetic interactions between PLP and NOOT may provide more details about AZ differentiation.
Transcriptional regulation of hormonal homeostasis, cell wall degradation and remodelling by PLP in the abscission process
The conventional model suggests that auxin gradient plays a central role in abscission (Addicott, 1982). The sensitivity of AZ to ethylene is regulated by the alteration of auxin gradient (Abeles and Rubinstein, 1964; Meir et al., 2006). Previous research on PLP used a DR5rev: GFP auxin response reporter to detect the role of PLP in auxin signalling (Zhou et al., 2012a). A GFP signal was detected specifically in the wild‐type pulvini, while no signal was observed in the plp‐1 mutant (Zhou et al., 2012a). This result was used to confirm the alteration of the unique structure of pulvini in the mutant without realizing its implications in abscission (Zhou et al., 2012a). Here, when abscission is considered, the result also suggests that auxin gradient across AZ was affected due to PLP mutation. Our microarray results showed that 137 genes involved in hormone metabolism were affected by the mutation of PLP. Among them, 70 auxin‐related genes and 12 ethylene‐related genes were significantly affected, indicating a possible role of PLP in modulating abscission process through auxin and ethylene biosynthesis and signalling.
Auxin response factors (ARFs) are transcription factors that mediate responses to auxin. In Arabidopsis, ARF2 was reported to regulate floral organ abscission (Ellis et al., 2005). ARF1 or NPH4/ARF7 and ARF19 were reported to enhance the delayed abscission of arf2 mutant (Ellis et al., 2005; Okushima et al., 2005). The tomato homologue of the Arabidopsis ARF19 was down‐regulated in KD1 antisense transgenic plants with delayed pedicel abscission phenotype (Ma et al., 2015). In tomato, miR160 was found to regulate floral organ abscission and fruit abscission (Damodharan et al., 2016). The sly‐miR160 depletion was associated with the up‐regulation of ARFs including SIARF10A, SIARF10B and SIARF17 which are homologues of Arabidopsis ARF10 and ARF17 (Damodharan et al., 2016). The expression pattern of ARFs during tomato flower pedicel abscission under auxin and ethylene treatment suggested important roles of ARFs in regulating pedicel abscission (Guan et al., 2014). Studies on LBD18, LBD16 and LBD30 revealed the function of LBDs in regulating auxin signalling pathway. LBD18 functions in the initiation and emergence of lateral roots, in conjunction with LBD16, downstream of ARF7 and ARF19 (Lee et al., 2009). JLO (LBD30) is expressed in boundaries and regulates both auxin transport and meristem fate by promoting the expression of the KNOX genes SHOOTMERISTEMLESS (STM) and BP/KNAT1 (Bureau and Simon, 2008). In our study, eight ARFs were affected in plp mutant, including ARF8, ARF10, ARF16 and ARF19, indicating a possible role of PLP in regulating auxin response to control the abscission process.
Abscission is activated after perceiving abscission signals, such as auxin and ethylene in the AZ. The microarray analyses of 25‐day‐old leaflet abscission zone region showed that several genes for cell wall degrading and remodelling factors were down‐regulated in the plp mutant. These include PGs, XTHs, GHs, PMEs and EXPs. The roles of these cell wall modifying enzymes have been associated with abscission in Arabidopsis and tomato (Estornell et al., 2013; Hong et al., 2000; Jiang et al., 2008; Lashbrook and Cai, 2008; Ogawa et al., 2009). In our study, abscission was not yet detected in 25‐day‐old leaflets (Table S1). The expression of PLP gene was detected in both prior and during the abscission process in Figure 5 (a‐c, e‐g), suggesting that PLP might play a role not only in AZ differentiation but also in the competence to respond to abscission signals and the activation of abscission. Both HAE and HSL2 regulate programmed separation of cells during abscission (Cho et al., 2008). Interestingly, AtLOB, the homologous gene of MtPLP in Arabidopsis, is up‐regulated in floral receptacles of the Arabidopsis double mutant hae hsl2, indicating that the AtLOB is closely related to abscission execution (Niederhuth et al., 2013). This reinforces the regulatory role of MtPLP on hormonal‐ and cell wall‐related genes during leaflet abscission. Taken together, the above evidence consistently suggests that PLP affects abscission execution through transcriptional regulation of hormonal homeostasis, cell wall degradation and remodelling. Here we propose a model of PLP in regulating leaflet and petiole abscission (Figure S17).
Promising candidate genes in leaf abscission studies
To analyse genes involved in leaf abscission in soybean, Kim et al. (2016) performed RNA sequencing (RNA‐seq) using RNA isolated from the leaf abscission zones and petioles (Non‐AZ, NAZ) of soybean after treating stem/petiole explants with ethylene for 0, 12, 24, 48 and 72 h. 188 abscission‐specific TFs were identified, which include TFs belonging to homeobox, MYB, Zinc finger, bHLH, AP2, NAC, WRKY, YABBY and ARF families (Kim et al., 2016). In the present study, we identified 26 overlapped candidates that are involved in both soybean and M. truncatula leaf abscission (Figure S7). These genes are promising candidates for future leaf abscission studies (Table S6).
PLP has potential for alfalfa improvement
In the present study, we explored a new way to minimize leaf loss during the haymaking process through genetically modifying alfalfa leaflet and petiole structures. The leaflet pulvinus and petiole pulvinus regions were strengthened with increased breakstrength and decreased detachment frequency by RNAi suppression of MsPLP without negative impacts on biomass yield and nutritive value. The abscission of leaflet and petiole was abolished in transgenic alfalfa. In future studies, a complete knockout of PLP in alfalfa can be achieved by genome editing (Wolabu et al., 2020). A large‐scale field test is needed to evaluate leaf loss and forage quality of the materials using commercial machines under field conditions.
In summary, our study revealed a new function of PLP in regulating AZ development in the model legume M. truncatula and we have successfully applied this knowledge to alfalfa improvement. This study demonstrated that loss of function of PLP resulted in an absence of AZ differentiation. The regulation of abscission by PLP is associated with auxin‐related genes and NOOT. Furthermore, the alteration of pulvinus to petiolule‐like pulvinus has significantly increased the breakstrength of leaflets and petioles in alfalfa, which offers a new approach to decrease leaf loss during the haymaking process. This study demonstrates how knowledge gained from a model plant can be applied to the genetic improvement of a commercial crop.
Experimental procedures
Plant materials and growth conditions
The Medicago truncatula Gaertn. Ecotype R108 was used as the wild type in this study. The source of the three plp alleles (Zhou et al., 2012a) and the noot allele (Couzigou et al., 2016) is described in Table S8. Mutant and wild‐type seeds were scarified with sandpaper and treated at 4°C for 7 days on filter paper and then transferred into soil. An alfalfa genotype, Regen SY‐4D, was used for Agrobacterium tumefaciens‐mediated transformation to produce transgenic plants following the protocol described by Fu et al. (2015). All the plants were grown in Metro‐Mix 830 soil in the greenhouse with the condition 22°C day/20°C night temperature, 16‐h light/8‐h dark photoperiod and 70‐80% relative humidity.
Isolation of MsPLP and creation of MsPLP modified transgenic alfalfa plants
The alfalfa PLP (MsPLP) gene was cloned using the following primers designed based on MtPLP: MsPLPFLF2: ATTGCTTTGTTGCAGGAGAA and MsPLPFLR2: CAAGAGACATAACATAAATAAACCCT. To knockdown the expression of MsPLP in alfalfa, 262‐bp and 205‐bp MsPLP fragments were amplified from alfalfa (genotype Regen SY‐4D) cDNA using primers shown in Table S9. Each fragment was independently cloned into the PENTRTM/ D‐TOPO cloning vector (Invitrogen, Chicago, IL, USA) and transferred into the pANDA35HK vector (Miki & Shimamoto, 2004) by attL × attR recombination reactions (Invitrogen). Sonication‐assisted Agrobacterium‐mediated transformation and tissue culture were performed for transgenic alfalfa generation (Fu et al., 2015). PCR analysis of the regenerated plants was performed using primers from the GUS linker of the pANDA35HK vector: GUS4F: CATGAAGATGCGGACTTACG and GUS5R: ATCCACGCCGTATTCGG.
Protein alignment, phylogenetic analysis and expression pattern of MtPLP
Predicted PLP amino acid sequences from various plant species were obtained by protein blast in GenBank (http://www.ncbi.nlm.nih.gov/) or Phytozome (https://phytozome.jgi.doe.gov/). All sequences were subsequently aligned using the ClusterW by DNASTAR. A phylogenetic tree was built using the neighbour‐joining method by employing the DNASTAR software. The DNA sequence of MtPLP was used for the analysis of expression pattern through BLAST function in Gene Expression Atlas (https://mtgea.noble.org/v2/).
Microscopic examination of leaflet and petiole abscission zone
Leaflet pulvinus region and petiole pulvinus region of M. truncatula mutants and transgenic alfalfa were carefully examined under a dissecting microscope.
In M. truncatula, 25‐day‐old leaves (mature leaves) represent the stage that prior to leaflet abscission and petiole abscission. Thirty five‐day‐old leaves (senescing leaves) represent the stage when leaflet and petiole are under the abscission process. The 25‐day‐old and 35‐day‐old AZ regions of petiole and leaflet were collected and fixed with 4% paraformaldehyde, 2.5% glutaraldehyde and 0.01% (v/v) Triton X‐100 dissolved in PBS (pH7.4) at 4°C for overnight. All specimens were then dehydrated in a series of ethanol and embedded in LR White resin (London Resin Co. Ltd., Berkshire, UK) using gelatin capsules. The resin was polymerized at 60°C for two days. In order to closely examine the abscission zones from wild type and plp mutant, serial 0.25‐micrometre longitudinal sections were cut with a diamond knife with a Leica EM UC7 ultramicrotome (Leica Microsystems GmbH, Vienna, Austria) or RMC MT‐X ultramicrotome (Boeckeler Instruments, Inc., Tucson, Arizona, USA). Semi‐thin sections (0.25 µm in thickness) placed onto the slide glasses (Superfrost Plus; VWR International, LLC, Radnor, Pennsylvania, USA) were stained with 1% (w/v) Toluidine Blue O (with 1% (w/v) sodium borate) for 5 min and observed under a Nikon Microphot‐2 (Nikon Corporation, Tokyo, Japan).
For Zeiss NEON 40 EsB scanning electron microscopy (SEM) examination of leaflet and petiole AZs, leaflets and petioles (31, 35, 39, 43 and 47 days old) were removed at the pulvinus if not yet abscised from the plant in wild type and plp mutant. Samples were fixed, dried and sputter‐coated as reported previously (Gou et al., 2017). The fracture planes on petiolule and stem created by removing leaflets and petiole or naturally abscised were observed.
Shaking assay for abscission
M. truncatula plants were staked in a similar manner in the greenhouse. Both mutant and wild‐type plants at senescence stage (108‐day‐old plant) were shaken vigorously for 2 min by hand. The leaves dropped onto ground were collected and weighed.
Breakstrength assay
Leaflet breakstrength is the force in gram equivalents required to remove a leaflet from the petiolule. Petiole breakstrength is the force to remove a whole leaf from the stem. The leaflet and petiole breakstrengths were measured by using the CT3 Texture Analyzer (BROOKFIELD, MA) in tension mode. Samples were clapped at the same position with dual grip assembly (TA‐DGF001) and extended at a constant speed of 2.0 mm s‐1 until breakage and the breakstrength was recorded. For both wild type and the plp mutants in M. truncatula, we started to mark fully expanded leaves as one‐day‐old leaves when plants were 48 days old. Afterwards, leaf samples at different ages from 25 to 49 days were collected in two‐day intervals for leaflet and petiole breakstrength measurements (eight replicates). For alfalfa materials, we evaluated the terminal, lateral leaflet and petiole breakstrengths from the 3rd to 12th leaflets, which represent leaflets that are normally harvested during alfalfa haymaking process. The breakstrength data were analysed by SPSS software.
β‐Glucuronidase (GUS) staining
To determine whether there is a correlation between the PLP expression pattern and leaflet and petiole abscission, we examined transgenic plants that express β‐glucuronidase (GUS) under the control of PLP promoter. GUS expression was analysed during the time course of abscission in three stages as shown in Table S2: stage I, prior to organ separation; stage II, during abscission; and stage III, after abscission when the remaining cells form a protective scar tissue. At stage II, the organs were separated manually.
RNA extraction, qRT‐PCR and microarray analysis
Pulvinus regions (abscission zones included) of 25‐day‐old leaflets from wild type and three plp alleles of M. truncatula (85‐day‐old plants) and of wild type and 14 transgenic alfalfa lines were collected for RNA isolation, cDNA reverse transcription and qRT‐PCR as previous described (Gou et al., 2019; Zhou et al., 2012a). Primers for quantifying the expression levels of auxin‐related genes and the NOOT gene are listed in Table S10.
For microarray analysis, 10 μg of purified RNA was extracted from the pulvinus region of 25‐day‐old leaves of Medicago truncatula. RNA purification, probe labelling, hybridization, scanning and microarray data analysis were performed following the manufacturer’s instructions (Affymetrix; http:// www.affymetrix.com). Functional enrichments were visualized with MAPMAN (Thimm et al., 2004).
Forage analysis of transgenic alfalfa lines
Three transgenic alfalfa lines S1, S2, S3 and control wild‐type alfalfa plants were grown in the greenhouse and propagated by cuttings. At early blooming stage, plants were harvested and fresh biomass yield was measured immediately. Forage quality was analysed using near‐infrared reflectance spectroscopy (NIRS) as previous described (Gou et al., 2018).
Conflicts of interest
The authors declare no conflict of interest.
Author contribution
ZYW, YZ and JD designed and planned the experiments; JD, SL, MC, CZ, JN, JK, ZW, MB, JS and TZ performed research; JD, YT and LS analysed the data; and JD, YZ and ZYW wrote the manuscript.
Supporting information
Figure S1 Petiolule‐like pulvinus (plp) mutant of M. truncatula shows developmental defects in leaflet and petiole pulvini.
Figure S2 Leaf dropping after shaking assay in plp mutant of M. truncatula.
Figure S3 Expression pattern of MtPLP in wild type, generated from the M. truncatula Gene Expression Atlas.
Figure S4 Differential expression of genes in pulvinus region of 25‐day‐old leaves of wild type and plp mutant of M. truncatula.
Figure S5 Validation of microarray data by qPCR of auxin‐related genes in AZ region of M. truncatula.
Figure S6 Pie chart of leaflet abscission‐related transcription factors (TFs) differentially expressed in M. truncatula during leaflet abscission.
Figure S7 Venn diagram analysis of the overlap of abscission‐specific TFs in soybean and TFs that are differentially expressed in plp mutant of M. truncatula.
Figure S8 Loss of leaflet and petiole abscission phenotype in noot mutant and the expression of NOOT in wild type and plp.
Figure S9 Sequence comparison of MsPLP (M. sativa SY4D) with PLP (M. truncatula R108).
Figure S10 Amino acid alignment of MsPLP (M. sativa SY4D) with PLP (M. truncatula R108) and their orthologs from Arabidopsis thaliana (At),Glycine max (Gm), Lotus japonicas (Lj), Pisum sativum (Ps) and Trifolium medium (Tm).
Figure S11 Phylogenetic analysis of PLP‐related orthologs in different plant species.
Figure S12 Construction of MsPLP‐RNAi binary vectors for alfalfa transformation.
Figure S13 Generation of MsPLP‐RNAi transgenic alfalfa plants.
Figure S14 Molecular and phenotypic characterization of alfalfa MsPLP‐RNAi transgenic lines.
Figure S15 Frequency of detachment occurs at marked positions in both wild type and transgenic alfalfa lines.
Figure S16 Evaluation of nutritive quality of transgenic alfalfa lines.
Figure S17 A proposed model of PLP in regulating leaflet and petiole abscission.
Table S1 Characterization of leaflet and petiole abscission in wild type and plp mutant of M. truncatula.
Table S2 Abscission stages of leaflet and petiole according to leaf ages.
Table S3 List of genes that are up‐regulated in plp mutant.
Table S4 List of genes that are down‐regulated inplp mutant.
Table S5 List of auxin‐related genes that were down‐regulated in LAZ for qPCR validation.
Table S6 List of transcription factors in M. truncatula that overlapped with abscission‐specific TFs in soybean.
Table S7 List of homologous genes of already published genes related to AZ development in M. truncatula.
Table S8 List of M. truncatula lines used in this study.
Table S9 List of primers used for MsPLP‐RNAi vector construction.
Table S10 Primers for qPCR analysis of auxin‐related genes and the NOOT gene.
Acknowledgements
We thank Dr. Million Tadege, Dr. Randy Allen and Dr. Liuling Yan for their valuable suggestions and enthusiastic support on this project. We thank Preston R. Larson at the University of Oklahoma for technical assistance of SEM, Kim Goss for taking care of plants in the greenhouse, and Lishan Yang and Yewei Wang for their help with tissue culture. This research was supported by Noble Research Institute, Oklahoma State University, China Scholarship Council (CSC), National Natural Science Foundation of China (Grant No. 31802116), First Class Grassland Science Discipline Program of Shandong Province, and Science and Technology Foundation of Guizhou Provincial Science and Technology Department (No. LH20177027).
Du, J. , Lu, S. , Chai, M. , Zhou, C. , Sun, L. , Tang, Y. , Nakashima, J. , Kolape, J. , Wen, Z. , Behzadirad, M. , Zhong, T. , Sun, J. , Zhang, Y. and Wang, Z.‐Y. (2021) Functional characterization of PETIOLULE‐LIKE PULVINUS (PLP) gene in abscission zone development in Medicago truncatula and its application to genetic improvement of alfalfa. Plant Biotechnol. J., 10.1111/pbi.13469
Contributor Information
Yunwei Zhang, Email: zwang98_99@yahoo.com.
Zeng‐Yu Wang, Email: zywang@qau.edu.cn.
References
- Abeles, F. and Rubinstein, B. (1964) Regulation of ethylene evolution and leaf abscission by auxin. Plant Physiol. 39, 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addicott, F.T. (1982) Abscission. Berkeley: University of California Press. [Google Scholar]
- Basu, M.M. , González‐Carranza, Z.H. , Azam‐Ali, S. , Tang, S. , Shahid, A.A. and Roberts, J.A. (2013) The manipulation of auxin in the abscission zone cells of Arabidopsis flowers reveals that indoleacetic acid signaling is a prerequisite for organ shedding. Plant Physiol. 162, 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito, V.A. , Torres‐Jerez, I. , Murray, J.D. , Andriankaja, A. , Allen, S. , Kakar, K. , Wandrey, M. et al. (2008) A gene expression atlas of the model legume Medicago truncatula . Plant J. 55, 504–513. [DOI] [PubMed] [Google Scholar]
- Bureau, M. and Simon, R. (2008) JLO regulates embryo patterning and organ initiation by controlling auxin transport. Plant Signal. Behav. 3, 145–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, M. , Zhou, C. , Molina, I. , Fu, C. , Nakashima, J. , Li, G. , Zhang, W. et al. (2016) A Class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl Acad. Sci. USA 113, 6997–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S.K. , Larue, C.T. , Chevalier, D. , Wang, H. , Jinn, T.L. , Zhang, S. and Walker, J.C. (2008) Regulation of floral organ abscission in Arabidopsis thaliana . Proc. Natl Acad. Sci. USA 105, 15629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzigou, J.M. , Magne, K. , Mondy, S. , Cosson, V. , Clements, J. and Ratet, P. (2016) The legume NOOT‐BOP‐COCH‐LIKE genes are conserved regulators of abscission, a major agronomical trait in cultivated crops. New Phytol. 209, 228–240. [DOI] [PubMed] [Google Scholar]
- Damodharan, S. , Zhao, D. and Arazi, T. (2016) A common miRNA 160‐based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 86, 458–471. [DOI] [PubMed] [Google Scholar]
- Ellis, C.M. , Nagpal, P. , Young, J.C. , Hagen, G. , Guilfoyle, T.J. and Reed, J.W. (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana . Development, 132, 4563–4574. [DOI] [PubMed] [Google Scholar]
- Estornell, L.H. , Agustí, J. , Merelo, P. , Talón, M. and Tadeo, F.R. (2013) Elucidating mechanisms underlying organ abscission. Plant Sci. 199, 48–60. [DOI] [PubMed] [Google Scholar]
- Frankowski, K. , Wilmowicz, E. , Kućko, A. , Zienkiewicz, A. , Zienkiewicz, K. and Kopcewicz, J. (2015) Profiling the BLADE‐ON‐PETIOLE gene expression in the abscission zone of generative organs in Lupinus luteus. Acta Physiol. Plant. 37, 220. [DOI] [PubMed] [Google Scholar]
- Fu, C. , Hernandez, T. , Zhou, C. and Wang, Z.‐Y. (2015) Alfalfa (Medicago sativa L.). In Agrobacterium Protocols. ( Wang, K. ed.), pp. 213–221. 3rd, New York: Springer. [Google Scholar]
- Gómez‐Mena, C. and Sablowski, R. (2008) ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell, 20, 2059–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Carranza, Z.H. , Rompa, U. , Peters, J.L. , Bhatt, A.M. , Wagstaff, C. , Stead, A.D. and Roberts, J.A. (2007) HAWAIIAN SKIRT: an F‐box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol. 144, 1370–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou, J. , Debnath, S. , Sun, L. , Flanagan, A. , Tang, Y. , Jiang, Q. , Wen, J. et al. (2018) From model to crop: functional characterization of SPL 8 in M. truncatula led to genetic improvement of biomass yield and abiotic stress tolerance in alfalfa. Plant Biotechnol. J. 16, 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou, J. , Fu, C. , Liu, S. , Tang, C. , Debnath, S. , Flanagan, A. , Ge, Y. et al. (2017) The miR156‐SPL4 module predominantly regulates aerial axillary bud formation and controls shoot architecture. New Phytol. 216, 829–840. [DOI] [PubMed] [Google Scholar]
- Gou, J. , Tang, C. , Chen, N. , Wang, H. , Debnath, S. , Sun, L. , Flanagan, A. et al. (2019) SPL7 and SPL8 represent a novel flowering regulation mechanism in switchgrass. New Phytol. 222, 1610–1623. [DOI] [PubMed] [Google Scholar]
- Guan, X. , Xu, T. , Gao, S. , Qi, M. , Wang, Y. , Liu, X. and Li, T. (2014) Temporal and spatial distribution of auxin response factor genes during tomato flower abscission. J. Plant Growth Regul. 33, 317–327. [Google Scholar]
- Gubert, C.M. , Christy, M.E. , Ward, D.L. , Groner, W.D. and Liljegren, S.J. (2014) ASYMMETRIC LEAVES1 regulates abscission zone placement in Arabidopsis flowers. BMC Plant Biol. 14, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, C.M. , Jun, J.H. , Nam, H.G. and Fletcher, J.C. (2007) BLADE‐ON‐PETIOLE1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial‐abaxial polarity genes. Plant Cell, 19, 1809–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth, S.R. , Zhang, Y. , McKim, S. , Li, X. and Haughn, G.W. (2005) BLADE‐ON‐PETIOLE–dependent signaling controls leaf and floral patterning in Arabidopsis . Plant Cell, 17, 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.B. , Sexton, R. and Tucker, M.L. (2000) Analysis of gene promoters for two tomato polygalacturonases expressed in abscission zones and the stigma. Plant Physiol. 123, 869–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idowu, J. , Grover, K. , Marsalis, M. and Lauriault, L. (2013) Reducing Harvest and Post‐Harvest Losses of Alfalfa and Other Hay. New Mexico State University Circular‐668: 1–5.
- Ji, H. , Kim, S.R. , Kim, Y.H. , Kim, H. , Eun, M.Y. , Jin, I.D. , Cha, Y.S. et al. (2010) Inactivation of the CTD phosphatase‐like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J. 61, 96–106. [DOI] [PubMed] [Google Scholar]
- Jiang, C.Z. , Lu, F. , Imsabai, W. , Meir, S. and Reid, M.S. (2008) Silencing polygalacturonase expression inhibits tomato petiole abscission. J. Exp. Bot. 59, 973–979. [DOI] [PubMed] [Google Scholar]
- Jun, J.H. , Ha, C.M. and Fletcher, J.C. (2010) BLADE‐ON‐PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2 . Plant Cell, 22, 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y. , Li, M. , Sinharoy, S. and Verdier, J. (2016) A snapshot of functional genetic studies in Medicago truncatula . Front. Plant Sci. 7, 1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. .(2014) Four shades of detachment: regulation of floral organ abscission. Plant Signal. Behav. 9, e976154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , Yang, J. , Yang, R. , Sicher, R.C. , Chang, C. and Tucker, M.L. (2016) Transcriptome analysis of soybean leaf abscission identifies transcriptional regulators of organ polarity and cell fate. Front. Plant Sci. 7, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, S. , Izawa, T. , Lin, S.Y. , Ebana, K. , Fukuta, Y. , Sasaki, T. and Yano, M. (2006) An SNP caused loss of seed shattering during rice domestication. Science, 312, 1392–1396. [DOI] [PubMed] [Google Scholar]
- Kućko, A. , Smoliński, D. , Wilmowicz, E. , Florkiewicz, A. and de Dios, Alché J. (2019) Spatio‐temporal localization of LlBOP following early events of floral abscission in yellow lupine. Protoplasma, 5, 1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook, C. C. and Cai, S . (2008) Cell wall remodeling in Arabidopsis stamen abscission zones: temporal aspects of control inferred from transcriptional profiling. Plant Signal. Behav. 3, 733–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.W. , Kim, N.Y. , Lee, D.J. and Kim, J. (2009) LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis . Plant Physiol. 151, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, M.W. , Leslie, M.E. and Liljegren, S.J. (2006) Plant separation: 50 ways to leave your mother. Curr. Opin. Plant Biol. 9, 59–65. [DOI] [PubMed] [Google Scholar]
- Li, M. , Liang, Z. , He, S. , Zeng, Y. , Jing, Y. , Fang, W. , Wu, K. et al. (2017) Genome‐wide identification of leaf abscission associated microRNAs in sugarcane (Saccharum officinarum L.). BMC Genom. 18, 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Liang, Z. , Zeng, Y. , Jing, Y. , Wu, K. , Liang, J. , He, S. et al. (2016) De novo analysis of transcriptome reveals genes associated with leaf abscission in sugarcane (Saccharum officinarum L.). BMC Genom. 17, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Zhou, A. and Sang, T. (2006) Rice domestication by reducing shattering. Science, 311, 1936–1939. [DOI] [PubMed] [Google Scholar]
- Liao, W. , Li, Y. , Yang, Y. , Wang, G. and Peng, M. (2016a) Exposure to various abscission‐promoting treatments suggests substantial ERF subfamily transcription factors involvement in the regulation of cassava leaf abscission. BMC Genom. 17, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, W. , Wang, G. , Li, Y. , Wang, B. , Zhang, P. and Peng, M. (2016b) Reactive oxygen species regulate leaf pulvinus abscission zone cell separation in response to water‐deficit stress in cassava. Sci. Rep. 6, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, W. , Yang, Y. , Li, Y. , Wang, G. and Peng, M. (2016c) Genome‐wide identification of cassava R2R3 MYB family genes related to abscission zone separation after environmental‐stress‐induced abscission. Sci. Rep. 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z. , Li, X. , Shannon, L.M. , Yeh, C.‐T. , Wang, M.L. , Bai, G. , Peng, Z. et al. (2012) Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44, 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Wang, D. , Qin, Z. , Zhang, D. , Yin, L. , Wu, L. , Colasanti, J. et al. (2014) The SEPALLATA MADS‐box protein SLMBP 21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J. 77, 284–296. [DOI] [PubMed] [Google Scholar]
- Ma, C. , Meir, S. , Xiao, L. , Tong, J. , Liu, Q. , Reid, M.S. and Jiang, C.‐Z. (2015) A KNOTTED1‐LIKE HOMEOBOX protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol. 167, 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer, C. and Hochholdinger, F. (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 16, 47–52. [DOI] [PubMed] [Google Scholar]
- Mao, L. , Begum, D. , Chuang, H‐w , Budiman, M.A. , Szymkowiak, E.J. , Irish, E.E. and Wing, R.A. (2000) JOINTLESS is a MADS‐box gene controlling tomato flower abscission zone development. Nature, 406, 910–913. [DOI] [PubMed] [Google Scholar]
- McKim, S.M. , Stenvik, G.‐E. , Butenko, M.A. , Kristiansen, W. , Cho, S.K. , Hepworth, S.R. , Aalen, R.B. et al. (2008) The BLADE‐ON‐PETIOLE genes are essential for abscission zone formation in Arabidopsis . Development, 135, 1537–1546. [DOI] [PubMed] [Google Scholar]
- Meir, S. , Hunter, D.A. , Chen, J.‐C. , Halaly, V. and Reid, M.S. (2006) Molecular changes occurring during acquisition of abscission competence following auxin depletion in Mirabilis jalapa . Plant Physiol. 141, 1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir, S. , Philosoph‐Hadas, S. , Sundaresan, S. , Selvaraj, K.V. , Burd, S. , Ophir, R. , Kochanek, B. et al. (2010) Microarray analysis of the abscission‐related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 154, 1929–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, D. & Shimamoto, K. (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiology, 45, 490–495. [DOI] [PubMed] [Google Scholar]
- Nakano, T. , Fujisawa, M. , Shima, Y. and Ito, Y. (2013) Expression profiling of tomato pre‐abscission pedicels provides insights into abscission zone properties including competence to respond to abscission signals. BMC Plant Biol. 13, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, T. , Kimbara, J. , Fujisawa, M. , Kitagawa, M. , Ihashi, N. , Maeda, H. , Kasumi, T. et al. (2012) MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 158, 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth, C.E. , Patharkar, O.R. and Walker, J.C. .(2013) Transcriptional profiling of the Arabidopsis abscission mutant hae hsl2 by RNA‐Seq. BMC Genom. 14, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg, M. , Holmlund, M. and Nilsson, O. .(2005) The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development, 132, 2203–2213. [DOI] [PubMed] [Google Scholar]
- Ogawa, M. , Kay, P. , Wilson, S. and Swain, S.M. .(2009) ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis . Plant Cell, 21, 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y. , Mitina, I. , Quach, H.L. and Theologis, A. .(2005) AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29–46. [DOI] [PubMed] [Google Scholar]
- Osborne, D.J. and Morgan, P.W. .(1989) Abscission. Crit. Rev. Plant Sci. 8, 103–129. [Google Scholar]
- Patharkar, O.R. , Gassmann, W. and Walker, J.C. .(2017) Leaf shedding as an anti‐bacterial defense in Arabidopsis cauline leaves. PLoS Genet. 13, e1007132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar, O.R. and Walker, J.C. (2016) Core mechanisms regulating developmentally timed and environmentally triggered abscission. Plant Physiol. 172, 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar, O.R. and Walker, J.C. (2018) Advances in abscission signaling. J. Exp. Bot. 69, 733–740. [DOI] [PubMed] [Google Scholar]
- Patterson, S.E. (2001) Cutting loose. Abscission and dehiscence in Arabidopsis . Plant Physiol. 126, 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, S.E. and Bleecker, A.B. (2004) Ethylene‐dependent and independent processes associated with floral organ abscission in Arabidopsis . Plant Physiol. 134, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt, R.E. (1990) Silage and Hay Preservation (NRAES 5): Northeast Regional Agricultural Engineering Service (NRAES).
- Putnam, D. , Robinson, P. and DePeters, E. (2008) Forage quality and testing. In Irrigated Alfalfa Management in Mediterranean and Desert Zones. ( Summers, C. G. & Putnam, D. H. eds.), pp. 2–25. California: University of California Agriculture and Natural Resources Publication 3512. [Google Scholar]
- Roberts, J.A. , Elliott, K.A. and Gonzalez‐Carranza, Z.H. (2002) Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53, 131–158. [DOI] [PubMed] [Google Scholar]
- Schumacher, K. , Schmitt, T. , Rossberg, M. , Schmitz, G. and Theres, K. (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl Acad. Sci. USA, 96, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton, R. and Roberts, J.A. (1982) Cell biology of abscission. Ann. Rev. Plant Physiol. 33, 133–162. [Google Scholar]
- Shi, C.‐L. , Stenvik, G.‐E. , Vie, A.K. , Bones, A.M. , Pautot, V. , Proveniers, M. , Aalen, R.B. et al. (2011) Arabidopsis class I KNOTTED‐like homeobox proteins act downstream in the IDA‐HAE/HSL2 floral abscission signaling pathway. Plant Cell, 23, 2553–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege, M. , Ratet, P. and Mysore, K.S. (2005) Insertional mutagenesis: a Swiss Army knife for functional genomics of Medicago truncatula . Trends Plant Sci. 10, 229–235. [DOI] [PubMed] [Google Scholar]
- Taylor, J.E. and Whitelaw, C.A. (2001) Signals in abscission. New Phytol. 151, 323–340. [Google Scholar]
- Thimm, O. , Bläsing, O. , Gibon, Y. , Nagel, A. , Meyer, S. , Krüger, P. , Selbig, J. et al. (2004) MAPMAN: a user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Ueda, M. and Nakamura, Y. (2007) Chemical basis of plant leaf movement. Plant Cell Physiol. 48, 900–907. [DOI] [PubMed] [Google Scholar]
- Wolabu, T.W. , Cong, L. , Park, J.‐J. , Bao, Q. , Chen, M. , Sun, J. , Xu, B. et al. (2020) Development of a highly efficient multiplex genome editing system in outcrossing tetraploid alfalfa (Medicago sativa). Front. Plant Sci. 11, 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X.M. , Yu, Y. , Han, L.B. , Li, C.L. , Wang, H.Y. , Zhong, N.Q. , Yao, Y. et al. (2012) The tobacco BLADE‐ON‐PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol. 159, 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, B. , Li, Z. , Zhu, Y. , Wang, H. , Ma, H. , Dong, A. and Huang, H. (2008) Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary‐specifying genes to promote sepal and petal development. Plant Physiol. 146, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. , Luo, F. and Hochholdinger, F. (2016) LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci. 21, 159–167. [DOI] [PubMed] [Google Scholar]
- Yang, T. , Fang, G. , He, H. and Chen, J. (2016) Genome‐wide identification, evolutionary analysis and expression profiles of lateral organ boundaries domain gene family in Lotus japonicus and Medicago truncatula . PLoS One, 11, e0161901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, N.D. , Debellé, F. , Oldroyd, G.E. , Geurts, R. , Cannon, S.B. , Udvardi, M.K. , Benedito, V.A. et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature, 480, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.Y. , Broeckling, C.D. , Blancaflor, E.B. , Sledge, M.K. , Sumner, L.W. and Wang, Z.Y. (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain‐containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 42, 689–707. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Liu, R. , Xu, Y. , Wang, M. , Zhang, J. , Bai, M. , Han, C. et al. (2019) AGLF provides C‐function in floral organ identity through transcriptional regulation of AGAMOUS in Medicago truncatula . Proc. Natl. Acad. Sci. USA, 116(11), 5176–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C. , Han, L. , Fu, C. , Chai, M. , Zhang, W. , Li, G. , Tang, Y. et al. (2012a) Identification and characterization of petiolule‐like pulvinus mutants with abolished nyctinastic leaf movement in the model legume Medicago truncatula. New Phytol. 196, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C. , Han, L. , Zhao, Y. , Wang, H. , Nakashima, J. , Tong, J. , Xiao, L. et al. (2019) Transforming compound leaf patterning by manipulating REVOLUTA in Medicago truncatula . Plant J. 100, 562–571. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Lu, D. , Li, C. , Luo, J. , Zhu, B.‐F. , Zhu, J. , Shangguan, Y. et al. (2012b) Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1 . Plant Cell, 24, 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. , Guo, J. , Zhou, C. and Zhu, J. (2014) Ectopic expression of LBD15 affects lateral branch development and secondary cell wall synthesis in Arabidopsis thaliana . Plant Growth Regul. 73, 111–120. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Petiolule‐like pulvinus (plp) mutant of M. truncatula shows developmental defects in leaflet and petiole pulvini.
Figure S2 Leaf dropping after shaking assay in plp mutant of M. truncatula.
Figure S3 Expression pattern of MtPLP in wild type, generated from the M. truncatula Gene Expression Atlas.
Figure S4 Differential expression of genes in pulvinus region of 25‐day‐old leaves of wild type and plp mutant of M. truncatula.
Figure S5 Validation of microarray data by qPCR of auxin‐related genes in AZ region of M. truncatula.
Figure S6 Pie chart of leaflet abscission‐related transcription factors (TFs) differentially expressed in M. truncatula during leaflet abscission.
Figure S7 Venn diagram analysis of the overlap of abscission‐specific TFs in soybean and TFs that are differentially expressed in plp mutant of M. truncatula.
Figure S8 Loss of leaflet and petiole abscission phenotype in noot mutant and the expression of NOOT in wild type and plp.
Figure S9 Sequence comparison of MsPLP (M. sativa SY4D) with PLP (M. truncatula R108).
Figure S10 Amino acid alignment of MsPLP (M. sativa SY4D) with PLP (M. truncatula R108) and their orthologs from Arabidopsis thaliana (At),Glycine max (Gm), Lotus japonicas (Lj), Pisum sativum (Ps) and Trifolium medium (Tm).
Figure S11 Phylogenetic analysis of PLP‐related orthologs in different plant species.
Figure S12 Construction of MsPLP‐RNAi binary vectors for alfalfa transformation.
Figure S13 Generation of MsPLP‐RNAi transgenic alfalfa plants.
Figure S14 Molecular and phenotypic characterization of alfalfa MsPLP‐RNAi transgenic lines.
Figure S15 Frequency of detachment occurs at marked positions in both wild type and transgenic alfalfa lines.
Figure S16 Evaluation of nutritive quality of transgenic alfalfa lines.
Figure S17 A proposed model of PLP in regulating leaflet and petiole abscission.
Table S1 Characterization of leaflet and petiole abscission in wild type and plp mutant of M. truncatula.
Table S2 Abscission stages of leaflet and petiole according to leaf ages.
Table S3 List of genes that are up‐regulated in plp mutant.
Table S4 List of genes that are down‐regulated inplp mutant.
Table S5 List of auxin‐related genes that were down‐regulated in LAZ for qPCR validation.
Table S6 List of transcription factors in M. truncatula that overlapped with abscission‐specific TFs in soybean.
Table S7 List of homologous genes of already published genes related to AZ development in M. truncatula.
Table S8 List of M. truncatula lines used in this study.
Table S9 List of primers used for MsPLP‐RNAi vector construction.
Table S10 Primers for qPCR analysis of auxin‐related genes and the NOOT gene.


