Abstract
Background
The abundance of energy metabolites is intimately interconnected with the activity of chromatin-modifying enzymes in order to guarantee the finely tuned modulation of gene expression in response to cellular energetic status. Metabolism-induced epigenetic gene regulation is a key molecular axis for the maintenance of cellular homeostasis, and its deregulation is associated with several pathological conditions. Nicotinamide N-methyltransferase (NNMT) is a metabolic enzyme that catalyzes the methylation of nicotinamide (NAM) using the universal methyl donor S-adenosyl methionine (SAM), directly linking one-carbon metabolism with a cell's methylation balance and nicotinamide adenine dinucleotide (NAD+) levels. NNMT expression and activity are regulated in a tissue-specific-manner, and the protein can act either physiologically or pathologically depending on its distribution. While NNMT exerts a beneficial effect by regulating lipid parameters in the liver, its expression in adipose tissue correlates with obesity and insulin resistance. NNMT upregulation has been observed in a variety of cancers, and increased NNMT expression has been associated with tumor progression, metastasis and worse clinical outcomes. Accordingly, NNMT represents an appealing druggable target for metabolic disorders as well as oncological and other diseases in which the protein is improperly activated.
Scope of review
This review examines emerging findings concerning the complex NNMT regulatory network and the role of NNMT in both NAD metabolism and cell methylation balance. We extensively describe recent findings concerning the physiological and pathological regulation of NNMT with a specific focus on the function of NNMT in obesity, insulin resistance and other associated metabolic disorders along with its well-accepted role as a cancer-associated metabolic enzyme. Advances in strategies targeting NNMT pathways are also reported, together with current limitations of NNMT inhibitor drugs in clinical use.
Major conclusions
NNMT is emerging as a key point of intersection between cellular metabolism and epigenetic gene regulation, and growing evidence supports its central role in several pathologies. The use of molecules that target NNMT represents a current pharmaceutical challenge for the treatment of several metabolic-related disease as well as in cancer.
Keywords: Nicotinamide N-methyltransferase, Epigenetics, Metabolism, Obesity, Cancer
1. Crosstalk between cellular metabolism and epigenetic regulation
Cells sense nutrient levels by activating complex signaling pathways that, ultimately, regulate gene expression through dynamic changes in chromatin status. Indeed, key energy metabolites, including S-adenosyl methionine (SAM), acetyl-coenzymeA (acetyl-CoA), adenosine triphosphate (ATP), nicotinamide adenine dinucleotide (NAD) and flavin adenine dinucleotide (FAD), act as essential substrates and/or cofactors for the regulation of the enzymes involved in chromatin modifications. The abundance of specific metabolites directly reflects the energetic state of the cell, and, as such, they activate a bio-sensing metabolic mechanism that couples nutrition availability with epigenetic gene regulation in order to guarantee cellular homeostasis [1].
One of the main epigenetic processes regulated by co-substrates generated by cellular metabolism is histone post-translational modifications (PTMs), among which, histone acetylation and methylation have been extensively documented in terms of response to metabolic changes [2]. By influencing chromatin structure, histone PTMs modulate the accessibility of genes and their transcriptional status, thereby providing a link between metabolism and gene regulation. Histone acetylation is regulated, at least in part, by acetyl-CoA, a central metabolite derived from glucose, fatty acid, and amino acid catabolism, which is considered a key indicator of cell metabolic state. Acetyl-CoA is an essential acetyl donor for protein acetylation, particularly histone acetylation carried out by histone acetyltransferases (HATs) [3]. Histone acetylation can be dynamically regulated by endogenous changes in acetyl-CoA, and in fact, it has been demonstrated that high levels of acetyl-CoA contribute to increasing histone acetylation during cellular response to growth factor stimulation and during adipocyte differentiation [4]. In this context, acetyl-CoA connects metabolism to transcriptional regulation by triggering a more permissive chromatin configuration. Another key metabolite involved in the modulation of histone acetylation is nicotinamide adenine dinucleotide (NAD), an essential cofactor involved in the regulation of many major cellular events: NAD catalyzes electron transfer in metabolic redox reactions, including glycolysis, the tricarboxylic acid (TCA) cycle, oxidative phosphorylation, and serine biosynthesis, and the NAD+/NADH redox couple has a central role in energy generation within the cell [5]. NAD+ also serves as a substrate for various enzymes that consume it, such as sirtuins [6]. Sirtuins belong to class III histone deacetylases (HDACs), which differ from other HDACs because they require NAD+ for deacetylase activity. The dependence of sirtuins on NAD+ suggests that this family of enzymes are nutrient-sensing regulators [7] that directly link cell redox state and metabolism to transcriptional regulation. Indeed, sirtuin-compacted chromatin is characterized by histone lysine hypoacetylation and epigenetic silencing [8].
Among the seven characterized mammalian sirtuins (SIRT1–7), SIRT1 and SIRT6 have been proposed to be directly linked to metabolism. Specifically, through mediating the deacetylation of histones H3K9/14 and H3K9/56, respectively, they modulate the expression of genes involved in lipogenesis, glycolysis, gluconeogenesis, and other metabolic pathways [1,[9], [10], [11], [12]]. Glucose has been demonstrated to strongly induce global histone acetylation through a dual mechanism involving acetyl-CoA and NAD+. Indeed, relatively high NADH/NAD+ characterizes favorable energetic conditions, such as the TCA cycle. When glucose is abundant, low NAD+ levels in cells mediates the downregulation of sirtuin HDAC activity. Furthermore, by supplying carbon to the TCA cycle, glucose increases acetyl-CoA, which in turn induces HAT activity [13]. By affecting histone acetylation and deacetylation, the abundance of acetyl-coA and NAD+ modulate chromatin dynamics and gene expression at specific loci, thereby guaranteeing intimate crosstalk between the energetic status of the cell and the epigenome.
Methylation of both DNA sequences and histone residues represents another epigenetic level of gene regulation. While cytosine methylation, catalyzed by DNA methyltransferase enzymes DNMT1, DNMT3a, and DNMT3b, generates 5-methylcytosine (5mC), a major transcriptional repressive mark in many eukaryotes [14], gene regulation mediated by histone methylation is more complex and can be associated with either transcriptional activation or repression, depending on the type of residue and the number of methyl groups added [15]. Generally, methylation of H3K9, H3K27, and H4K20 correlates with a repressed chromatin state, while methylation sites associated with gene activation include H3K4, H33K36, H3K48, and H3K79 [16]. Histone methyltransferases (HMTs) and histone demethylases, respectively, add and remove methyl groups in a highly specific and interconnected manner to regulate chromatin dynamics and gene regulation depending on the biological context [17].
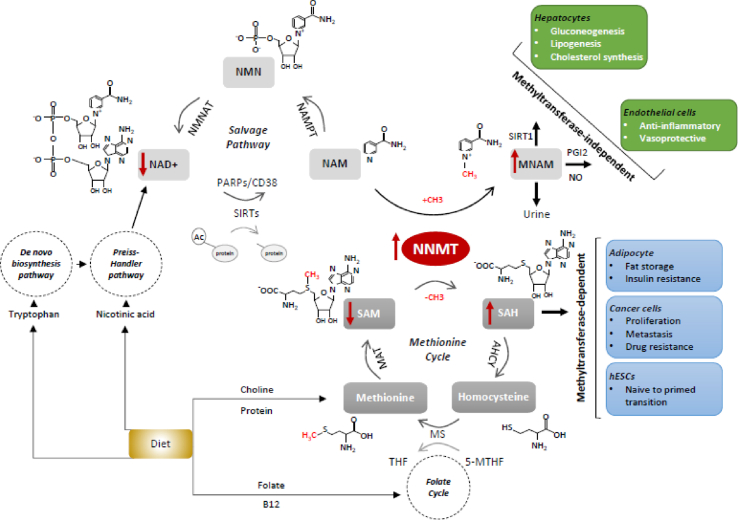
A central methylation pathway in mammalian cells is the one-carbon metabolism, which involves the transfer of methyl groups to various substrates and cofactors within the folate and methionine cycles [18] (Figure 1). The transfer of methyl groups from 5-methyl tetrahydrofolate (5-MTHF) to homocysteine (Hcy) to form methionine links the two cycles. Methionine is then converted to SAM, the universal methyl donor for many biomolecules, including DNA, proteins and small molecule secondary metabolites. Methyltransferase reactions attack the methyl group of SAM to generate the methylated product, with the concomitant formation of S-adenosyl-l-homocysteine (SAH). SAH is further hydrolyzed to Hcy, which is then recycled to methionine by using 5-MTHF as a methyl group donor, thus completing the cycle [19] (Figure1).
Figure 1.
Schematic overview of the main cellular pathways regulated by NNMT. Schematic illustration of the reactions catalyzed by NNMT and the chemical transactions taking place in the methionine cycle and in the NAD+ salvage pathway. NNMT directly links the methionine cycle with the cell's methylation balance and NAD+ levels. In adipose tissue, in cancer cells and in hESCs, NNMT function is principally mediated by its methyltransferase activity (light blue boxes), while NNMT hepatic and endothelial functions are generally methyltransferase-independent (green boxes). Additionally, especially in those tissues in which NAD+ synthesis primarily relies on the salvage pathway. NNMT also regulates NAD + metabolism. Red arrows indicate the most representative variations associated with NNMT overexpression. The structural formulas of the major substrates are shown along with their abbreviated names (in bold). Enzymes catalyzing the reaction steps of the NAD+ salvage pathway and the methionine cycle are also indicated, and abbreviations of the enzymes and substrates not previously mentioned in the text are as follows: AHCY: S-adenosylhomocysteine hydrolase, MS: Methionine synthase, MAT: methionine adenosyltransferase, NAMPT: nicotinamide phosphoribosyltransferase, NMNAT: nicotinamide mononucleotide adenylyltransferase. PARPs and CD38 are other NAD+-consuming enzymes that compete with sirtuins for NAD+ availability. The major metabolic pathways that feed both the methionine and the NAD+ cycles are also illustrated. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Both HMTs and DNA methyltransferases use the SAM cofactor as the methyl group donor. Methionine metabolism has been shown to be sufficient to determine levels of histone methylation through the modulation of SAM and SAH levels. Changes in one-carbon metabolism can modulate gene expression by inducing changes in the histone methylation of key histone marks, such as H3K4me3 [20,21]. Furthermore, to maintain homeostasis in one-carbon metabolism, a feedback mechanism regulates the enzymes that are important in regenerating methionine, whose expression and H3K4me3 signature is modulated in response to decreases in SAM and SAH levels [20]. In this way, cells sense the metabolic status of the one-carbon metabolism through the modulation of the SAM/SAH ratio, which in turn modifies the kinetics of the enzymes that mediate histone methylation and ultimately culminates in gene expression regulation in response to nutrient availability.
By changing DNMTs kinetics, DNA methylation can also be influenced by fluctuations in the nutrients that act as the precursors of SAM, which includes methionine, folate, choline, betaine, and vitamins B2, B6, and B12 (Figure 1). In support of this, numerous studies have reported the effect of nutritional supplementation on global or locus specific alterations in DNA methylation [22]. Theoretically, supplementation of methionine and folate would increase DNA methylation and downregulate gene expression by increasing the SAM/SAH ratio [23]. The activity of both histone- and DNA methyltransferases is inhibited by a high intracellular SAH concentration [24]. As the supply of SAM and the removal of SAH are necessary for DNMT activity, the SAM/SAH ratio has been suggested as a ‘methylation index’ whereby a decrease in this ratio predicts reduced cellular methylation potential. However, it is difficult to extrapolate the direct role of metabolism on DNA methylation, because the mechanisms regulating DNA methylation are very complex and the relationship between methylation pattern and gene expression is tissue and context-dependent [22].
DNA and histone demethylases can also act as molecular transducers of metabolic signals to chromatin. In mammals, DNA methylation in the form of 5-methylcytosine (5mC) can be actively reversed through reactions driven by the ten-eleven translocation enzymes TET1, TET2, and TET3 [25]. The TET enzymes are dioxygenases that use iron (Fe2+) as a cofactor and α -ketoglutarate (α -KG), an essential intermediate in the TCA cycle, as co-substrate, and the availability of either can directly affect reaction kinetics.
In line with this, in the liver of mice, increased α-KG levels, resulting from the administration of glucose, glutamate, or glutamine, correlates with a rapid increase in 5hmC levels [26]. α-KG cofactor is also required for demethylation reactions catalyzed by JmjC-family histone demethylases (JmjCs), while LSD-family histone demethylases (LSD) require FAD to demethylate histone residues [1].
2. Dual role of NNMT in NAD metabolism and methylation balance: physiological and pathological regulation
NNMT is a one-carbon group methyltransferase that catalyzes the transfer of the methyl group from the universal donor SAM to NAM to produce SAH and 1-methylnicotinamide (MNAM) [27] (Figure 1).
Human NNMT maps on chromosome 11 at 11q23.1 and encodes a monomeric cytoplasmic protein of 264 amino acids with a molecular weight of 29.6 kDa (Data reference 1). The NNMT substrate is NAM, a form of vitamin B3 and a precursor of NAD+ [28], a key molecule involved in modulating energy metabolism and a co-substrate for NAD+-dependent sirtuin deacetylases.
Under physiological conditions, NNMT is strongly expressed in the liver. Medium levels of NNMT are present in female tissues (vagina, endometrium, breast, cervix, and ovary), adipose tissue, and muscle, while it is expressed at very low levels in the brain and hematopoietic cells [29]. In several tissues, such as those of the muscles, pancreas, kidney and intestinal tract, as well as adipose tissue, NNMT mRNA and protein levels do not always coincide [30] (Data reference 2), suggesting that NNMT expression could be regulated by complex and tissue-specific mechanisms.
The function of NNMT was originally assigned to the biotransformation and detoxification of multiple drugs and xenobiotic compounds [27] and particularly to the regulation of intracellular levels of NAM through the mediation of its excretion after N-methylation, an irreversible step in the vitamin B3 degradation pathway. In fact, after the methylation of NAM, MNAM is further oxidized in the liver, meaning that it can no longer function as an NAD+ precursor and is excreted in the urine [31].
Growing evidence is currently revealing the involvement of NNMT in several metabolic pathways through both the generation of active metabolites and its influence on the SAM/SAH ratio and cell methylation balance [29](Figure 1). NNMT exerts an apparently contradictory role in liver and in adipose tissue, where its levels correlate with opposing metabolic traits. Specifically, hepatic expression of NNMT improves lipid parameters, while high adipose expression of NNMT correlates with adiposity and insulin resistance [32]. In humans and mice, NNMT expression inversely correlates with total cholesterol, low-density lipoprotein (LDL) cholesterol, triglyceride levels, and other metabolic parameters. Hepatic NNMT overexpression induces a concomitant MNAM change but does not affect either the SAM/SAH ratio or NAD+ content. Its product, MNAM, mediates the beneficial metabolic effects of NNMT by increasing Sirt1 deacetylase protein stability (Figure 1). Proof of this is the fact that the acetylation status of FoxO1, a well-characterized Sirt1 target, is inversely correlated with NNMT expression: NNMT-overexpressing- and MNAM-treated hepatocytes increase Sirt1 stability while decreasing FoxO1 acetylation [33]. In addition, Sirt1 activity upregulates gluconeogenesis and suppresses cholesterol synthesis and lipogenesis [34], and thus, by increasing Sirt1 protein stability, NNMT mediates the regulation of gluconeogenesis and cholesterol synthesis [33]. The products of Sirt1 are deacetylated protein and the two metabolites NAM and acetylated ADP-ribose [32] (Figure 1). If not methylated by NNMT, NAM can be saved and used in the re-synthesis of NAD+ (Figure 1). One model suggests that NAM concomitantly increases Sirt1 activity and stability through NAD+ biosynthesis and NAM methylation, respectively, which are both catalyzed by NNMT [33].
The fact that NNMT hepatic function is not mediated by a methyltransferase activity is consistent with the notion that other methyltransferases are responsible for the transmethylation flux in this tissue. Glycine N-methyltransferase (GNMT) is the most abundant methyltransferase in the mammalian liver [35], where it catalyzes the synthesis of N-methylglycine (sarcosine) from glycine using SAM as the methyl donor. The function of this cycle is to catabolize excess liver SAM in order to maintain a constant SAM/SAH ratio and avoid aberrant methylation reactions in the liver [36]. Evidence of this comes from the fact that GNMT-knockout mice show high SAM content and a large increase in SAM/SAH ratios [37] and also develop steatosis, fibrosis, and hepatocellular carcinoma (HCC) [38]. Individuals with inactivating GNMT mutations have elevated plasma levels of methionine and SAM but a normal concentration of Hcy. These findings indicate that any hepatic reduction in total transmethylation flux caused by the absence of GNMT cannot be compensated for the other methyltransferases that are abundant in the liver [36].
Unlike in the liver, however, in adipose tissue, NNMT is a major methyltransferase, and its expression regulates methyl donor balance and correlates with SAM content and histone methylation [39]. NNMT expression also positively correlates with fat storage and insulin resistance [40] (Figure 1). Mice fed a high-fat diet express high amounts of enzymatically active NNMT, with no parallel change in NNMT activity in the liver. NNMT expression is upregulated in white adipose tissue (WAT) in mouse models of obesity and diabetes, where NNMT knockdown in WAT and liver protects against diet-induced obesity by augmenting the cellular energy expenditure of the adipose tissue [41]. In fact, it has been demonstrated that NNMT inhibition increases the adipose SAM/SAH ratio and modulates histone H3 lysine 4 methylation, with the consequent transcriptional activation of genes involved in the polyamine flux, a major energy metabolic pathway that triggers energy consumption, and, at least in mice, protects against diet-induced obesity. In adipose tissue, where NAD+ synthesis primarily relies on the salvage pathway using nicotinamide, for which NNMT is the only catabolic enzyme, NNMT also regulates NAD+ levels. In WAT, but not in the liver where redundant regulatory pathways for NAD+ metabolism exist, NNMT expression depresses the NAD+ level [41].
Moreover, by modulating the availability of SAM in adipose tissue, NNMT activity can also alter the global DNA methylome and adipocyte function. It has been recently demonstrated that higher NNMT expression is associated with lower global DNA methylation in visceral adipose tissue obtained from morbidly obese patients. Hypomethylated genes in the high-NNMT samples were associated with adipose tissue metabolic pathophysiology. Importantly, decreased NNMT expression following weight loss induced variation in some of the NNMT-related differentially methylated genes [39].
Given the strong connection between obesity and the increased risk of developing insulin resistance and type 2 diabetes, it is not surprising that NNMT expression also plays an important role in these pathologies. Indeed, WAT NNMT expression and circulating levels of MNAM are higher in humans with insulin resistance or type 2 diabetes and correlate with the degree of insulin resistance. In line with this, interventions that improve insulin sensitivity are associated with a decrease in adipose NNMT expression [40]. Furthermore, NNMT could have a central role in hepatic insulin resistance, possibly by regulating the expression of fructose bisphosphatase 1 (Fbp1), a key regulator of the gluconeogenic pathway that is increased in animal models of obesity and insulin resistance [42]. In fact, NNMT knockdown experiments have clearly demonstrated that NNMT is a positive regulator of hepatic gluconeogenesis and that, in vivo, it regulates the conversion of pyruvate to glucose and correlates with the expression of Fbp1 [33].
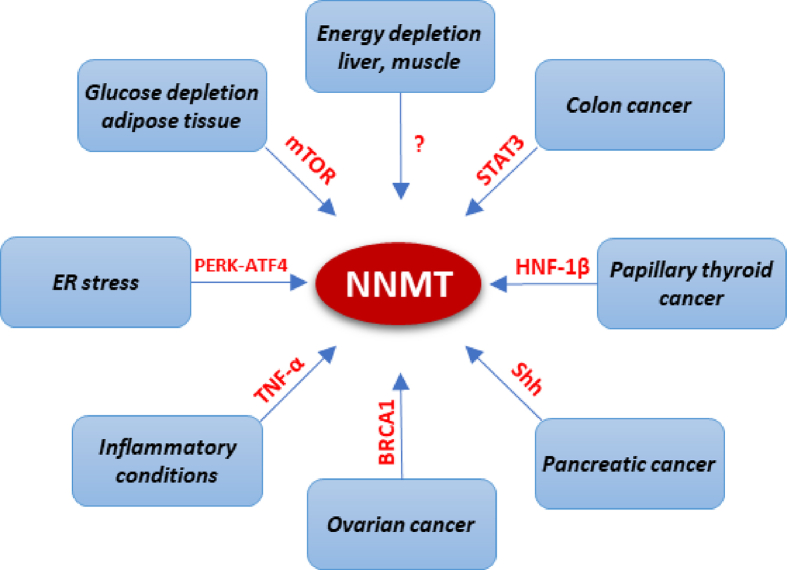
A recent study demonstrated that glucose availability regulates NNMT expression in adipose tissue and suggested that NNMT might function as a sensor for a lack of energy [43]. Additionally, it has been shown that caloric restriction induces an increase in NNMT expression in the human liver [33] and muscle [44] (Figure 2).
Figure 2.
Signaling pathways regulating NNMT expression. Schematic representation of known pathways involved in the regulation of NNMT expression. Various factors influence NNMT regulation in a tissue- and context-dependent manner.
The apparently contradictory role of NNMT in liver and adipose tissue can be explained by the fact that animals have evolutionarily evolved to gain weight and to store energy. In such a scenario, NNMT functions as a ‘fat storage’ molecule in the adipose tissue, while in the liver NNMT allows the organism to cope with the accumulation of body fat produced by episodes of over-nutrition [32].
Adding another layer of complexity is the fact that NNMT expression and activity can be either pathogenic or protective, depending not only on tissue distribution but also on the downstream effectors of NNMT activation. In fact, although highly expressed in normal liver, NNMT upregulation has been associated with several kinds of hepatic diseases. The fact that NNMT is located at the crossroad of the SAM- and NAD+-dependent pathways suggests that this molecule may be involved in alcohol-related liver diseases (ALD) as this pathology has been extensively correlated with both pathways, while supplementation with exogenous SAM, nicotinamide, or nicotinic acid protects mice against ALD [45]. In accordance with this, chronic alcohol consumption upregulates hepatic NNMT expression and activity, while protein inhibition, via both NNMT knockdown and chemical inhibition, protects against alcohol-related fatty liver development in mouse models of ADL by suppressing the de novo lipogenesis pathway in hepatocytes and the liver. This seemingly contradictory role of NNMT in de novo lipogenesis in hepatic physiology and disease remains to be adequately explained. It is possible that in this pathological setting, protein overexpression induces SAM-regulated methylation reactions that are not activated in physiological conditions. Strong support for the pathological role of NNMT in ALD comes from the fact that, in response to chronic alcohol exposure, ER (endoplasmic reticulum) stress, known to play a central role in several metabolic disorders, including alcohol-related fatty liver disease, mainly mediates the upregulation of NNMT via the PERK-ATF4 (protein kinase RNA-like ER kinase-activating transcription factor 4) pathway [45] (Figure 2). Additionally, increased urinary and serum levels of MNAM in patients with cirrhosis are suggestive of the augmented NNMT activity associated with human liver cirrhosis [46], and it has recently been demonstrated that transgenic mice overexpressing NNMT develop fatty liver disease and fibrosis [47]. In this regard, it has been shown that under NAM-replete conditions, typical, for example, of a meat-centric diet, NNMT activation drives the depletion of both NAD+ and SAM and contributes to hepatic steatosis and fibrosis, at least in part, by reducing the methylation of the connective tissue growth factor (CTGF) gene promoter, leading to its transcriptional activation. A possible explanation for the fact that in this setting NNMT may work as a methyltransferase is provided by the observation that, although NNMT is likely not sufficient to change liver methyl donor balance under high-fat diet (HFD), NNMT overexpression in conjunction with high doses of NAM might compete for SAM with other methyltransferase reactions and result in the down regulation of these methyltransferases, meaning that NNMT may therefore play a crucial role in the regulation of the methionine cycle [47].
NNMT pathogenic function has also been described in Parkinson's disease, where protein overexpression has been correlated with neurodegeneration [48].
The cytoprotective effect of NNMT, or its metabolic product MNAM, on the other hand, has been demonstrated in injury and inflammation [29]. NNMT plays a positive role in the muscle fiber repair process as its overexpression in myoblasts elicits significant proliferation and migration activities. Upregulation of NNMT expression in the lungs and skeletal muscle of patients with chronic obstructive pulmonary disease (COPD) has been proposed as an adaptive response designed to improve the regenerative capacities of the muscles in response to injury and protect against oxidative stress [49]. Moreover, the vasoprotective, anti-inflammatory, and anti-thrombotic roles of MNAM are well documented. In inflammatory conditions, such as atherosclerosis, elevated levels of inflammatory cytokines, such as tumor necrosis factor (TNF)-alpha, induce the expression of NNMT [29] (Figure 2) and MNAM, which, through increasing prostacyclin (PGI2) and nitric oxide (NO), improves endothelial function and thereby causes vasorelaxation [50] (Figure 1).
NNMT is a highly polymorphic gene, with more than 10,000 single-nucleotide polymorphisms (SNPs) described across the gene [51] (Data reference 4). The vast majority of them are located in the non-coding region of the gene, and only a few SNPs in the NNMT 5′-flanking region have been associated with differences in its catalytic activity or transcriptional efficiency. Although not associated with changes in NNMT expression, several SNPs in the NNMT gene have been specifically related with human diseases such as hyperhomocysteinemia, congenital heart diseases, abdominal aortic diseases and nonalcoholic steatohepatitis, as well as psychiatric disorders, such as bipolar disorder, epilepsy, and schizophrenia [29,52].
Another well-documented function of NNMT as an epigenetic modulator concerns its regulatory role during the transition from naïve to primed human embryonic stem cells (hESCs) (Figure 1). The switch between these states of pluripotency is associated with changes in gene expression, epigenetics, and metabolic patterns. One well-documented epigenetic change during naïve to primed state transition is the upregulation of the H3K27me3 repressive methylation marker in the promoter of key metabolic genes that regulate this transition [53]. It has been demonstrated that in naïve hESCs, NNMT and MNAM are upregulated and are accompanied by concurrent decreases in SAM levels and H3K27me3 marks. In this scenario, NNMT consumes methyl donors and makes SAM unavailable for other methyltransferases, thereby repressing H3K27me3 marks. NNMT is involved in the maintenance of the naïve state because, by regulating SAM availability, it contributes to the low levels of histone methylation that are typical of the naïve ESC state. During the transition to a primed, more committed, state, the reduction in NNMT levels correlates with increased levels of SAM and H3K27me3 histone methylation marks. Furthermore, it is also known that the resulting repressive chromatin state then controls the metabolism of primed hESCs though the Wnt and the hypoxia-inducible factor (HIF) pathways and a general gene expression shift toward the primed stage [54].
Overall, we can conclude that NNMT is an emblematic protein whose expression and activity are possibly regulated in a tissue- and context-specific manner and which apparently has a contradictory, though as yet not fully understood, role in physiology and disease. The central role of NNMT as a molecular link between metabolic and epigenetic pathways encourages further research to better understand the molecular patterns involved in NNMT regulatory networks.
3. NNMT as a cancer-associated metabolic enzyme
Altered metabolism is a hallmark of cancer. Molecular changes associated with metabolic reprogramming are required to meet the energy demands of cancer cells and to support tumor growth. Metabolic and epigenetic reprogramming are closely interconnected. In fact, metabolic reprogramming, driven by the activation of oncogenes, the inactivation of tumor suppressor genes and alterations in metabolic enzymes, induces the modulation of histone- and DNA-modification enzymes, which in turn regulate the expression of metabolism-associated genes [55]. Several aberrations in metabolic enzymes have been described as promoting tumorigenesis by affecting histone and DNA methylation, among them active mutations in isocitrate dehydrogenase in glioma and acute myeloid leukemia [56], deletion of methylthioadenosine phosphorylase (MTAP) in several cancers [57], and the loss of and increase in glycine N-methyltransferase in, HCC and prostate cancers, respectively [38,58].
There is also growing evidence to suggest that NNMT is deregulated in a wide variety of cancers and that it can be considered a new cancer-associated metabolic enzyme given that it represents a critical node between metabolism and the epigenetic state of cancer cells. NNMT is overexpressed in several tumors, including neuroblastoma, papillary thyroid cancer, bladder, lung, breast, gastric, colorectal, and pancreatic cancers, as well as in renal, oral squamous cells, and ovarian carcinomas [59]. Data from transfection experiments show that NNMT activity promotes the proliferation, migration, and survival of cancer cells [60,61]. Increased expression, rather than activating mutations, is responsible for the oncogenic hyperactivation of NNMT, as supported by the fact that the gene is rarely mutated in different types of cancers (Data reference 3) [62,63] Although only limited information exists regarding the molecular mechanisms involved in the upregulation of NNMT in tumors, it is likely that they act at the transcriptional level as no amplification of the NNMT gene has been reported to date [64].
In colon cancer, enhanced NNMT expression has been correlated with the activation of Signal Transducer And Activator Of Transcription 3 (SAT3) [65]. Hepatocyte nuclear factor-1β (HNF-1β) and sonic hedgehog (Shh) ligand have been shown to have a significant role in modulating NNMT upregulation in papillary thyroid cancer [66] and pancreatic cancer [67], respectively. In ovarian cancer cells, BRCA1 occupies the NNMT promoter and regulates NNMT expression (Figure 2). In this context, BRCA1 knockdown is known to increase NNMT mRNA and protein levels, and NNMT and BRCA1 levels are inversely correlated in high-grade serous ovarian cancer. Importantly, it has been demonstrated that the metabolic reprogramming induced by BRCA1 loss in ovarian cancer cells is mediate by NNMT upregulation [68]. However, it is unlikely that these factors are at play in all NNMT-overexpressing tumors; supporting the idea that NNMT regulation occurs in a cancer-specific manner.
NNMT plausibly exerts its pro-tumorigenic function by controlling the methylation potential of cancer cells (Figure 1). Ulanovskaya et al. [69] demonstrated, through untargeted metabolomics, that elevated NNMT resulted in a reduction in the SAM/SAH ratio and an increase in MNAM levels, accompanied by a concomitant decrease in histone- (but not DNA-) methylation, associated with gene expression changes in several cancer-related genes. Importantly, this work demonstrates that NNMT affects protein methylation by altering SAM and SAH levels rather than through the direct action of its product MNAM. In fact, the authors demonstrated that cancer cells have limited capacity to recover the methylation units that accumulate in MNAM and that this metabolite might act as a “sink” for storing methylation units in cancer cells that overexpress NNMT. Another study highlighted that NNMT-mediated SAM depletion and reduction in methylation potential is the key mechanism regulating cancer-associated fibroblast (CAF) differentiation. Proteomic analysis revealed that NNMT is overexpressed in metastasis-associated stroma and knockdown of the gene reverses cell morphology such that they revert to normal omental fibroblasts. NNMT regulates the expression of CAF markers and pro-tumorigenic cytokines by mediating genome-wide DNA methylation changes and also through the hypomethylation of repressive chromatin marks. As such, these results show that NNMT exerts an additional layer of epigenetic control over histone acetylation by regulating the level of NAD+ and in turn the activity of sirtuin deacetylases [61]. Additionally, the function of NNMT in cellular methylation extends beyond DNA and histones. The tumor suppressor protein phosphatase 2A (PP2A) is regulated by methylation, and several reports have demonstrated that NNMT levels control PP2A activity by influencing its methylation status. By decreasing the methyl donor balance, NNMT-overexpressing cells show a considerable reduction in methylated PP2A, with consequent protein inactivation that in turn promotes the activation of key prosurvival kinases [63,69]. Furthermore, in the liver, NNMT regulates autophagy under nutrient starvation by regulating the degree of PP2A methylation [70].
As with the poor prognosis associated with DNA hypomethylation, NNMT overexpression can be considered a negative prognostic factor for human glioblastoma patients [64]. In fact, NNMT is preferentially expressed by mesenchymal glioblastoma stem cells compared to proneural tumors. Higher NNMT levels have been correlated with a reduced pool of methyl donors and associated with lower DNMT activity, leading to DNA hypomethylation in mesenchymal subtype genes and to accelerate tumor growth. In this context, the upregulation of NNMT induces a shift in the methyl donor pool that promotes the proneural–mesenchymal transition [64].
As with glioblastoma, NNMT may constitute a potential prognostic factor in other cancers as its expression has also been associated with increased tumor size, progression and metastasis. NNMT expression correlates with poor prognosis in a wide range of solid tumors [71]. For example, in pancreatic [72] and gastric cancers [73], high NNMT levels are associated with shorter overall survival and correlate with unfavorable clinic pathological features. Proteomic analysis revealed that NNMT is also highly expressed in ovarian, breast and colon cancer metastasis-associated stroma [61]. NNMT expression progressively increases in the stroma of ovarian neoplasms, and protein overexpression is independently associated with poor prognosis [74]. Moreover, it has been demonstrated that elevated NNMT levels can be used for early diagnosis in some malignant tumors and as a non-invasive biomarker of cancer in body fluids, including serum saliva and urine [[75], [76], [77]].
Drug resistance remains a great challenge for medical oncology, and one of the many mechanisms by which cancer cells develop chemo/radioresistance is through apoptosis evasion [78]. It has been demonstrated that NNMT overexpression significantly inhibits apoptotic cell death and correlates with sensitivity to chemotherapeutic drugs [79]. Several studies suggest that NNMT and MNAM have the potential to increase ATP levels and reduce the production of intracellular reactive oxygen species (ROS), suggesting that they may regulate apoptosis via the mitochondrial-mediated pathway [80]. NNMT increases the resistance of colorectal cancer cells to 5-fluorouracil (5-FU). Indeed, high levels of MNAM reduce ROS production, which consequently inactivates the ASK1-p38 mitogen-activated protein kinase (MAPK) pathway and reduces 5-FU-induced apoptosis [81]. In breast carcinoma, high levels of NNMT correlate with poor survival and unfavorable therapy response in patients who receive chemotherapy. Furthermore, NNMT overexpression has been shown to reduce the sensitivity of breast cancer cells to adriamycin- and paclitaxel -induced apoptosis by increasing SIRT1 stabilization and activity, and there is also evidence that targeting NNMT or downstream SIRT1 could improve the efficacy of breast cancer chemotherapy [82]. NNMT expression has also been associated with radiation resistance in several cancers. It is known that nicotinamide influences radiation sensitivity by inhibiting PARP (Poly (ADP-ribose) polymerase), thus preventing efficient single-strand DNA break repair. In glioblastoma, inhibition of NNMT increases intracellular levels of nicotinamide, leading to radiosensitization [63].
One exception to NNMT overexpression in cancer is in HCC, where, although still abundant, NNMT is significantly downregulated compared to normal adjacent tissues. A plausible mechanistic explanation to this is provided by a recent finding demonstrating that in liver cancer cells, NNMT is a negative autophagy regulator and that in this context its downregulation promotes cancer cell survival [70]. Even so, within HCC samples, higher levels of NNMT positively correlate with tumor stage and shorter disease-free survival. Thus, it may be the case that the liver-specific function of NNMT is lost during progression to HCC and that it perhaps increases in later stage HCC due to the tumor de-differentiation that precedes tumor invasion [83]. In line with this hypothesis, it has been recently demonstrated that NNMT expression is positively correlated with tumor vascular invasion and distant metastasis and thus may possibly represents a prognostic biomarker and therapeutic target in advanced HCC [84]. Interestingly, NNMT and SIRT1 show overlapping roles in hepatic physiology and pathophysiology. In fact, similar to NNMT, SIRT1 plays beneficial roles in the regulation of hepatic lipid metabolism, controlling hepatic oxidative stress and mediating hepatic inflammation and so protecting against the progression of fatty liver diseases [85], whereas in HCC, SIRT1 overexpression promotes cell invasion and migration and enhances the invasive and metastatic potential of HCC [86]. Since NNMT and SIRT1 show similar expression and activation patterns and because in the liver NNMT regulates SITR1 activity and stability, it might be possible that the two proteins cooperate during the progression of HCC and that the function of NNMT might be related with the different roles of SIRT1 in the different stages of the disease. However, further studies are needed to confirm this hypothesis.
4. NNMT inhibitors
Because of its regulatory role in cellular epigenetics and metabolism, together with its well-accepted involvement in several human diseases, NNMT can be considered as a new potential pharmacological target in the treatment of a variety of metabolic disorders, cancers, and other pathologies [87]. Not surprisingly, over the last few years its growing importance has accelerated the development of potent selective molecules that target NNMT. Although still in the early preclinical phase, several NNMT inhibitors have been reported to date, among them methylated quinolines, nicotinamide analogs, covalent inhibitors, and amino-adenosine and alkynyl derived bisubstrate inhibitors [88].
The fact that animals treated with an antisense oligonucleotide that targets NNMT were shown to have reduced weight gain and improvement in insulin sensitivity encouraged the identification of small-molecule NNMT inhibitors (NNMTi) to treat obesity and type 2 diabetes. Specifically, two independent studies recently demonstrated the efficacy of two different NNMTi molecules in treating obesity and related metabolic disorders in preclinical animal models. Kannt et al. reported that treatment with a nicotinamide analog inhibitor of NNMT (JBSNF-00008) caused body weight reduction, improved insulin sensitivity, and normalized glucose tolerance in mice with HFD-induced obesity [89]. Similarly, Neelakantan et al. observed that systemic treatment of diet-induced obese mice with a quinolinium-based NNMTi induced significant loss of body weight and WAT mass, along with concomitant improvements in plasma lipid profile. As in NNMT knockdown experiments, NNMT inhibitor reverses obesity through the modulation of the NAD+ salvage- and SAM-mediated pathways. These observations open up the possibility of using NNMis in combination with other dietary supplements, for example NAD+ precursors, in order to enhance the latter's efficacy in treating metabolic disorders and reducing the adverse effects associated with the high pharmacological doses required [90].
Interestingly, it has been demonstrated that in some tissues, NNMT expression progressively increases with aging. NNMT overexpression may be responsible for the reduction in NAD+, typical of many age-associated pathologies, through modulating the levels of nicotinamide precursors required for NAD+ biosynthesis. One well-documented example is the increased expression of NNMT with aging in muscle tissues [49] and its associated reduction in NAD+. Based on this observation, the NNMT inhibitor 5-amino-1-methylquinolium has been successfully used to accelerate muscle regeneration in an aged mouse muscle injury model. It has been demonstrated that by regulating the NAD+ salvage pathway, NNMT inhibitors affect intracellular NAD+ concentrations, which in turn regulate protein deacetylation by sirtuins, leading to metabolic and transcriptional cell responses that promote myoblast differentiation [91]. However, the possible epigenetic modulation exerted by NNMTi in terms of improving aged skeletal muscles, along with the longitudinal effects of NNMTi treatment on muscle functional outputs still need to be addressed.
As it has been demonstrated that NAD+ repletion can enhance life span in mice [92], it is plausible to think that NNMTis might exert a similar function by modulating the NAD+ salvage pathway. However, the mechanistic implications of NNMTi as an anti-aging intervention still need to be elucidated.
Another line of investigation concerns the therapeutic potential of NNMTis in the management of obesity-induced cardiovascular disease. Multiple studies have established an association between hyperhomocysteinemia and increased risk of ischemic heart disease, stroke, endothelial dysfunction, and thrombosis [93]. Preliminary results suggest that Hcy, the atherogenic product of the NNMT-catalyzed reaction, links NNMT overexpression and cardiovascular diseases. This assumption is supported by the fact that a genome-wide linkage study demonstrated that the NNMT gene is a major genetic determinant of plasma Hcy levels [93], as well as by the observation that in adipose tissue the increased expression of NNMT contributes to the augmented release of plasma Hcy [94].
Additionally, NNMT is emerging as a new molecular target in fatty liver diseases. It has been recently demonstrated that the previously mentioned nicotinamide analog inhibitor of NNMT (JBSNF-00008) protects against alcohol-related fatty liver development in mouse models of ALD by inhibiting hepatic de novo lipogenesis and that chronic alcohol exposure induces ER stress-mediated NNMT upregulation [45]. The authors of the study indicate that as ER stress plays a critical role in several metabolic disorders and that NNMT is highly expressed in metabolic active tissues, ER stress-mediated NNMT upregulation might represent a central axis in these pathologies and targeting NNMT might provide a critical point for therapeutic intervention.
Given its well-documented role in tumor progression, clinical outcome, and drug-sensitivity, NNMT represents an attractive drug target for several neoplasms that are characterized by abnormal NNMT activity. While several studies have demonstrated that NNMT downregulation (through gene knockdown) could be an attractive target in cancer treatment, little information is currently available about NNMTis in cancer management, and future investigations are needed. Recently, the same NNMTi used to treat HFD-induced obesity [90] has been tested in an orthotopic intraperitoneal model of ovarian cancer metastasis and was shown to induce decreased tumor burden, reduce tumor cell proliferation, and increase stromal H3K27 trimethylation [61]. The authors demonstrated that NNMT contributes to neoplasms mainly by regulating the methyl donor balance of cancer cells; thus, NNMT inhibitors may exert their therapeutic benefit by modulating the cancer epigenome [69].
A noteworthy exception to this overexpression in cancers, as mentioned before, is the fact that in the liver, where NNMT is physiologically expressed at high levels, the protein is repressed, at least in the early stages, in the tumor counterpart. A recent report suggested that NNMT depletion contributes to liver cancer progression by promoting autophagy under nutrient starvation, and, as a proof of concept, autophagy inhibitor treatment might represent a possible strategy for treatment of cancers characterized by NNMT repression [70].
Although an ever-increasing number of investigations into the development of strong and specific NNMTi molecules are currently ongoing, further studies are needed to assess the effectiveness and feasibility of NNMT targeting strategies in human settings.
5. Conclusion
Molecular transducers closely interconnect metabolism and epigenetics by activating cell-specific transcriptional networks in response to environmental changes. Among them, NNMT is emerging as a central molecule situated at the crossroads between cellular metabolism and epigenetics, and there is growing evidence to indicate its crucial role in cellular physiology and pathology. Although several signal pathways have been reported to regulate NNMT expression, our understanding of the NNMT regulatory network remains incomplete. It seems possible that different mechanisms, rather than one ubiquitous pattern, regulate NNMT in a tissue- and context-specific manner. Adding further complexity is the fact that both the regulation of NNMT and its function vary as it can act on either the NAD metabolism, the SAM-methylation balance, or both, depending on the tissue involved. It seems clear that NNMT expression is abundant in metabolically active tissue, such as the liver and adipose tissue, as well as in those pathological conditions characterized by an increased metabolic demand, as for example cancer, diabetes and fatty liver disease. Although a growing amount of information is revealing how different transcription factors might regulate NNMT expression in a tissue-specific manner, how these modulations translate into concerted tissue-specific gene regulation is a central question that remains unanswered. It is known that by modulating the SAM/SAH ratio, NNMT levels influence the methylation potential of cells and that the enzyme affects methylation in a site-specific manner rather than impacting all cellular methylation pathways. One hypothesis suggests that this might depend on the relative Km and Ki values of individual methyltransferase enzymes, including NNMT itself, for SAM and SAH, respectively. In fact, it has been demonstrated that methylation events, such as H3K27 and PP2A methylation, controlled by methyltransferases EZH1and EZH2 in the former and LCMT1 in the latter, all of which have higher Km values for SAM (and SAH), tend to be more sensitive to NNMT as compared with methyltransferases, such as CARM1 (H3R17me2a) that show lower Km values [69]. This might also explain how changes in a central metabolic enzyme, like NNMT, which does not directly produce cofactors for epigenetic enzymes but does catalyze a reaction that affects the availability of cofactors (SAM and NAD+) or inhibitors (SAH) of many epigenetic enzymes, may lead to specific gene regulation events. However, further studies are required to elucidate how not only NNMT but also other metabolic enzymes can lead cell type-specific gene expression changes.
As a central metabolic regulator, NNMT is consistently involved in several cellular differentiation processes as they involve dynamic changes in energy demand, depending on the needs of the individual cell types
As an evidence for this, NNMT expression and activity were found to be associated with adipogenesis and myogenesis as well as with the hESC differentiation process [54]. Because dedifferentiation is one of the critical processes in cancer cell transformation and because NNMT expression has been associated with both epithelial- [61] and neuronal [64] mesenchymal transition markers, it is possible that increased levels of NNMT in cancers might shift the cells toward a less differentiated state by decreasing the methylation potential of cancer cells and, in turn, by modulating the expression of the gene that regulates the differentiation process.
However, although in recent years, there has been increased interest in understanding the role of NNMT and its regulation, further investigation is still required to fully elucidate the physiological and pathological functions of NNMT. The use of molecules that can inhibit NNMT function might provide a valid tool to better understand the molecular mechanisms that control the NNMT regulatory network and possibly reverse the pathological conditions associated with its deregulation. Despite several promising NNMTis having been recently developed, they still face technical limitations, such as low cellular permeability and moderate activity at uM levels, which limit their application to cell-based and early preclinical phase studies [87]. Future studies are needed to elucidate inhibitor mechanism-of-action and to evaluate the longitudinal effects of NNMTis in clinical settings in addition to assessing their therapeutic potential.
Funding
This research was funded by the Spanish Association Against Cancer (PROYE18061FERN to M.F.F.), the Health Institute Carlos II (ISCIII/FEDER PI18/01527 to A.F.F. and M.F.F.), the ISCIII-Subdirección General de Evaluación y Fomento de la Investigación (Miguel Servet contract CP11/00131 to A.F.F.), the Plan de Ciencia, Tecnología e Innovación from the Asturias Government cofunding (2018–2022/FEDER to M.F.F.), and FINBA-ISPA (A.R.). The IUOPA is supported by the Obra Social Cajastur-Liberbank, Spain.
Acknowledgments
We sincerely apologize to all colleagues whose work could not be cited because of space constraints. We would like to thank Ronnie Lendrum for editorial assistance and all the members of Cancer Epigenetics and Nanomedicine Laboratory (FINBA-ISPA, IUOPA, CINN–CSIC) for their positive feedback and helpful discussions.
Conflict of interest
None declared.
References
- 1.Etchegaray J.P., Mostoslavsky R. Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Molecular Cell. 2016;62(5):695–711. doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan J., Krautkramer K.A., Feldman J.L., Denu J.M. Metabolic regulation of histone post-translational modifications. ACS Chemical Biology. 2015;10(1):95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sivanand S., Viney I., Wellen K.E. Spatiotemporal control of acetyl-CoA metabolism in chromatin regulation. Trends in Biochemical Sciences. 2018;43(1):61–74. doi: 10.1016/j.tibs.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katsyuba E., Romani M., Hofer D., Auwerx J. NAD+ homeostasis in health and disease. Nature Metabolism. 2020;2(1):9–31. doi: 10.1038/s42255-019-0161-5. [DOI] [PubMed] [Google Scholar]
- 6.Ryu D., Zhang H., Ropelle E.R., Sorrentino V., Mazala D.A., Mouchiroud L. NAD+ repletion improves muscle function in muscular dystrophy and counters global PARylation. Science Translational Medicine. 2016;8(361):361ra139. doi: 10.1126/scitranslmed.aaf5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vassilopoulos A., Fritz K.S., Petersen D.R., Gius D. The human sirtuin family: evolutionary divergences and functions. Human Genomics. 2011;5(5):485–496. doi: 10.1186/1479-7364-5-5-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvanese V., Fraga M.F. SirT1 brings stemness closer to cancer and aging. Aging (Albany NY) 2011;3(2):162–167. doi: 10.18632/aging.100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie Y., Erion D.M., Yuan Z., Dietrich M., Shulman G.I., Horvath T.L. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nature Cell Biology. 2009;11(4):492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker A.K., Yang F., Jiang K., Ji J.Y., Watts J.L., Purushotham A. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes & Development. 2010;24(13):1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 12.Kim H.-S., Xiao C., Wang R.-H., Lahusen T., Xu X., Vassilopoulos A. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metabolism. 2010;12(3):224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donohoe D.R., Bultman S.J. Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. Journal of Cellular Physiology. 2012;227(9):3169–3177. doi: 10.1002/jcp.24054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 15.Roberti A., Valdes A.F., Torrecillas R., Fraga M.F., Fernandez A.F. Epigenetics in cancer therapy and nanomedicine. Clinical Epigenetics. 2019;11(1):81. doi: 10.1186/s13148-019-0675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Experimental & Molecular Medicine. 2017;49(4):e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greer E.L., Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature Reviews Genetics. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ducker G.S., Rabinowitz J.D. One-carbon metabolism in health and disease. Cell Metabolism. 2017;25(1):27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clare C.E., Brassington A.H., Kwong W.Y., Sinclair K.D. One-carbon metabolism: linking nutritional biochemistry to epigenetic programming of long-term development. Annual Review of Animal Biosciences. 2019;7:263–287. doi: 10.1146/annurev-animal-020518-115206. [DOI] [PubMed] [Google Scholar]
- 20.Mentch S.J., Mehrmohamadi M., Huang L., Liu X., Gupta D., Mattocks D. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metabolism. 2015;22(5):861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai Z., Mentch S.J., Gao X., Nichenametla S.N., Locasale J.W. Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nature Communications. 2018;9(1):1955. doi: 10.1038/s41467-018-04426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang N. Role of methionine on epigenetic modification of DNA methylation and gene expression in animals. Animal Nutrition. 2018;4(1):11–16. doi: 10.1016/j.aninu.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadayifci F.Z., Zheng S., Pan Y.X. Molecular mechanisms underlying the link between diet and DNA methylation. International Journal of Molecular Sciences. 2018;19(12) doi: 10.3390/ijms19124055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park L.K., Friso S., Choi S.W. Nutritional influences on epigenetics and age-related disease. Proceedings of the Nutrition Society. 2012;71(1):75–83. doi: 10.1017/S0029665111003302. [DOI] [PubMed] [Google Scholar]
- 25.An J., Rao A., Ko M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Experimental & Molecular Medicine. 2017;49(4) doi: 10.1038/emm.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X., Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nature Reviews Genetics. 2017;18(9):517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 27.Aksoy S., Szumlanski C.L., Weinshilboum R.M. Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. Journal of Biological Chemistry. 1994;269(20):14835–14840. [PubMed] [Google Scholar]
- 28.Makarov M.V., Trammell S.A.J., Migaud M.E. The chemistry of the vitamin B3 metabolome. Biochemical Society Transactions. 2019;47(1):131–147. doi: 10.1042/BST20180420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pissios P. Nicotinamide N-methyltransferase: more than a vitamin B3 clearance enzyme. Trends in Endocrinology and Metabolism. 2017;28(5):340–353. doi: 10.1016/j.tem.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 31.Brachs S., Polack J., Brachs M., Jahn-Hofmann K., Elvert R., Pfenninger A. Genetic nicotinamide N-methyltransferase (Nnmt) deficiency in male mice improves insulin sensitivity in diet-induced obesity but does not affect glucose tolerance. Diabetes. 2019;68(3):527–542. doi: 10.2337/db18-0780. [DOI] [PubMed] [Google Scholar]
- 32.Trammell S.A., Brenner C. NNMT: a bad actor in fat makes good in liver. Cell Metabolism. 2015;22(2):200–201. doi: 10.1016/j.cmet.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong S., Moreno-Navarrete J.M., Wei X., Kikukawa Y., Tzameli I., Prasad D. Nicotinamide N-methyltransferase regulates hepatic nutrient metabolism through Sirt1 protein stabilization. Nature Medicine. 2015;21(8):887–894. doi: 10.1038/nm.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye X., Li M., Hou T., Gao T., Zhu W.-g., Yang Y. Sirtuins in glucose and lipid metabolism. Oncotarget. 2016;8(1):1845–1859. doi: 10.18632/oncotarget.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mudd S.H., Brosnan J.T., Brosnan M.E., Jacobs R.L., Stabler S.P., Allen R.H. Methyl balance and transmethylation fluxes in humans. American Journal of Clinical Nutrition. 2007;85(1):19–25. doi: 10.1093/ajcn/85.1.19. [DOI] [PubMed] [Google Scholar]
- 36.Mato J.M., Martinez-Chantar M.L., Lu S.C. Methionine metabolism and liver disease. Annual Review of Nutrition. 2008;28:273–293. doi: 10.1146/annurev.nutr.28.061807.155438. [DOI] [PubMed] [Google Scholar]
- 37.Luka Z., Capdevila A., Mato J.M., Wagner C. A glycine N-methyltransferase knockout mouse model for humans with deficiency of this enzyme. Transgenic Research. 2006;15(3):393–397. doi: 10.1007/s11248-006-0008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Chantar M.L., Vazquez-Chantada M., Ariz U., Martinez N., Varela M., Luka Z. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47(4):1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crujeiras A.B., Pissios P., Moreno-Navarrete J.M., Diaz-Lagares A., Sandoval J., Gomez A. An epigenetic signature in adipose tissue is linked to nicotinamide N-methyltransferase gene expression. Molecular Nutrition & Food Research. 2018 doi: 10.1002/mnfr.201700933. [DOI] [PubMed] [Google Scholar]
- 40.Kannt A., Pfenninger A., Teichert L., Tonjes A., Dietrich A., Schon M.R. Association of nicotinamide-N-methyltransferase mRNA expression in human adipose tissue and the plasma concentration of its product, 1-methylnicotinamide, with insulin resistance. Diabetologia. 2015;58(4):799–808. doi: 10.1007/s00125-014-3490-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraus D., Yang Q., Kong D., Banks A.S., Zhang L., Rodgers J.T. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508(7495):258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visinoni S., Fam B.C., Blair A., Rantzau C., Lamont B.J., Bouwman R. Increased glucose production in mice overexpressing human fructose-1,6-bisphosphatase in the liver. American Journal of Physiology - Endocrinology And Metabolism. 2008;295(5):E1132–E1141. doi: 10.1152/ajpendo.90552.2008. [DOI] [PubMed] [Google Scholar]
- 43.Ehebauer F., Ghavampour S., Kraus D. Glucose availability regulates nicotinamide N-methyltransferase expression in adipocytes. Life Sciences. 2020;248:117474. doi: 10.1016/j.lfs.2020.117474. [DOI] [PubMed] [Google Scholar]
- 44.Strom K., Morales-Alamo D., Ottosson F., Edlund A., Hjort L., Jorgensen S.W. N(1)-methylnicotinamide is a signalling molecule produced in skeletal muscle coordinating energy metabolism. Scientific Reports. 2018;8(1):3016. doi: 10.1038/s41598-018-21099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song Q., Chen Y., Wang J., Hao L., Huang C., Griffiths A. ER stress-induced upregulation of NNMT contributes to alcohol-related fatty liver development. Journal of Hepatology. 2020;73(4):783–793. doi: 10.1016/j.jhep.2020.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pumpo R., Sarnelli G., Spinella A., Budillon G., Cuomo R. The metabolism of nicotinamide in human liver cirrhosis: a study on N-methylnicotinamide and 2-pyridone-5-carboxamide production. American Journal of Gastroenterology. 2001;96(4):1183–1187. doi: 10.1111/j.1572-0241.2001.03698.x. [DOI] [PubMed] [Google Scholar]
- 47.Komatsu M., Kanda T., Urai H., Kurokochi A., Kitahama R., Shigaki S. NNMT activation can contribute to the development of fatty liver disease by modulating the NAD (+) metabolism. Scientific Reports. 2018;8(1):8637. doi: 10.1038/s41598-018-26882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsons R.B., Smith S.W., Waring R.H., Williams A.C., Ramsden D.B. High expression of nicotinamide N-methyltransferase in patients with idiopathic Parkinson's disease. Neuroscience Letters. 2003;342(1–2):13–16. doi: 10.1016/s0304-3940(03)00218-0. [DOI] [PubMed] [Google Scholar]
- 49.Kim H.C., Mofarrahi M., Vassilakopoulos T., Maltais F., Sigala I., Debigare R. Expression and functional significance of nicotinamide N-methyl transferase in skeletal muscles of patients with chronic obstructive pulmonary disease. American Journal of Respiratory and Critical Care Medicine. 2010;181(8):797–805. doi: 10.1164/rccm.200906-0936OC. [DOI] [PubMed] [Google Scholar]
- 50.Nejabati H.R., Mihanfar A., Pezeshkian M., Fattahi A., Latifi Z., Safaie N. N1-methylnicotinamide (MNAM) as a guardian of cardiovascular system. Journal of Cellular Physiology. 2018;233(10):6386–6394. doi: 10.1002/jcp.26636. [DOI] [PubMed] [Google Scholar]
- 51.Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J. Ensembl 2020. Nucleic Acids Research. 2020 Jan 8;48(D1):D682–D688. doi: 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramsden D.B., Waring R.H., Parsons R.B., Barlow D.J., Williams A.C. Nicotinamide N-methyltransferase: genomic connection to disease. International Journal of Tryptophan Research. 2020;13 doi: 10.1177/1178646920919770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504(7479):282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 54.Sperber H., Mathieu J., Wang Y., Ferreccio A., Hesson J., Xu Z. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nature Cell Biology. 2015;17(12):1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miranda-Goncalves V., Lameirinhas A., Henrique R., Jeronimo C. Metabolism and epigenetic interplay in cancer: regulation and putative therapeutic targets. Frontiers in Genetics. 2018;9:427. doi: 10.3389/fgene.2018.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dang L., Jin S., Su S.M. IDH mutations in glioma and acute myeloid leukemia. Trends in Molecular Medicine. 2010;16(9):387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 57.Bertino J.R., Waud W.R., Parker W.B., Lubin M. Targeting tumors that lack methylthioadenosine phosphorylase (MTAP) activity: current strategies. Cancer Biology & Therapy. 2011;11(7):627–632. doi: 10.4161/cbt.11.7.14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shlomi T., Rabinowitz J.D. Metabolism: cancer mistunes methylation. Nature Chemical Biology. 2013;9(5):293–294. doi: 10.1038/nchembio.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu X.M., Long H. Nicotinamide N-methyltransferase as a potential marker for cancer. Neoplasma. 2018;65(5):656–663. doi: 10.4149/neo_2018_171024N680. [DOI] [PubMed] [Google Scholar]
- 60.Tang S.W., Yang T.C., Lin W.C., Chang W.H., Wang C.C., Lai M.K. Nicotinamide N-methyltransferase induces cellular invasion through activating matrix metalloproteinase-2 expression in clear cell renal cell carcinoma cells. Carcinogenesis. 2011;32(2):138–145. doi: 10.1093/carcin/bgq225. [DOI] [PubMed] [Google Scholar]
- 61.Eckert M.A., Coscia F., Chryplewicz A., Chang J.W., Hernandez K.M., Pan S. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature. 2019;569(7758):723–728. doi: 10.1038/s41586-019-1173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Research. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palanichamy K., Kanji S., Gordon N., Thirumoorthy K., Jacob J.R., Litzenberg K.T. NNMT silencing activates tumor suppressor PP2A, inactivates oncogenic STKs, and inhibits tumor forming ability. Clinical Cancer Research. 2017;23(9):2325–2334. doi: 10.1158/1078-0432.CCR-16-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jung J., Kim L.J., Wang X., Wu Q., Sanvoranart T., Hubert C.G. Nicotinamide metabolism regulates glioblastoma stem cell maintenance. JCI Insight. 2017;2(10) doi: 10.1172/jci.insight.90019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomida M., Ohtake H., Yokota T., Kobayashi Y., Kurosumi M. Stat3 up-regulates expression of nicotinamide N-methyltransferase in human cancer cells. Journal of Cancer Research and Clinical Oncology. 2008;134(5):551–559. doi: 10.1007/s00432-007-0318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu J., Capezzone M., Xu X., Hershman J.M. Activation of nicotinamide N-methyltransferase gene promoter by hepatocyte nuclear factor-1beta in human papillary thyroid cancer cells. Molecular Endocrinology. 2005;19(2):527–539. doi: 10.1210/me.2004-0215. [DOI] [PubMed] [Google Scholar]
- 67.Feldmann G., Habbe N., Dhara S., Bisht S., Alvarez H., Fendrich V. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut. 2008;57(10):1420–1430. doi: 10.1136/gut.2007.148189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanakkanthara A., Kurmi K., Ekstrom T.L., Hou X., Purfeerst E.R., Heinzen E.P. BRCA1 deficiency upregulates NNMT, which reprograms metabolism and sensitizes ovarian cancer cells to mitochondrial metabolic targeting agents. Cancer Research. 2019;79(23):5920–5929. doi: 10.1158/0008-5472.CAN-19-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ulanovskaya O.A., Zuhl A.M., Cravatt B.F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nature Chemical Biology. 2013;9(5):300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shin J.H., Park C.W., Yoon G., Hong S.M., Choi K.Y. NNMT depletion contributes to liver cancer cell survival by enhancing autophagy under nutrient starvation. Oncogenesis. 2018;7(8):58. doi: 10.1038/s41389-018-0064-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S., Qiao L., Yang Z., He C. Prognostic value of nicotinamide N-methyltransferase expression in patients with solid tumors: a systematic review and meta-analysis. Frontiers in Physiology. 2018;9:1407. doi: 10.3389/fphys.2018.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Y., Liu P., Zheng D.H., Wu N., Zhu L., Xing C. Expression profile and prognostic value of NNMT in patients with pancreatic cancer. Oncotarget. 2016;7(15):19975–19981. doi: 10.18632/oncotarget.7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen C., Wang X., Huang X., Yong H., Shen J., Tang Q. Nicotinamide N-methyltransferase: a potential biomarker for worse prognosis in gastric carcinoma. American Journal of Cancer Research. 2016;6(3):649–663. [PMC free article] [PubMed] [Google Scholar]
- 74.Harmankaya I., Akar S., Ugras S., Guler A.H., Ezveci H., Aydogdu M. Nicotinamide N-methyltransferase overexpression may be associated with poor prognosis in ovarian cancer. Journal of Obstetrics and Gynaecology. 2020:1–6. doi: 10.1080/01443615.2020.1732891. [DOI] [PubMed] [Google Scholar]
- 75.You Z., Liu Y., Liu X. Nicotinamide N-methyltransferase enhances the progression of prostate cancer by stabilizing sirtuin 1. Oncology Letters. 2018;15(6):9195–9201. doi: 10.3892/ol.2018.8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sartini D., Muzzonigro G., Milanese G., Pozzi V., Vici A., Morganti S. Upregulation of tissue and urinary nicotinamide N-methyltransferase in bladder cancer: potential for the development of a urine-based diagnostic test. Cell Biochemistry and Biophysics. 2013;65(3):473–483. doi: 10.1007/s12013-012-9451-1. [DOI] [PubMed] [Google Scholar]
- 77.Sartini D., Pozzi V., Renzi E., Morganti S., Rocchetti R., Rubini C. Analysis of tissue and salivary nicotinamide N-methyltransferase in oral squamous cell carcinoma: basis for the development of a noninvasive diagnostic test for early-stage disease. Biological Chemistry. 2012;393(6):505–511. doi: 10.1515/hsz-2012-0112. [DOI] [PubMed] [Google Scholar]
- 78.Mohammad R.M., Muqbil I., Lowe L., Yedjou C., Hsu H.Y., Lin L.T. Broad targeting of resistance to apoptosis in cancer. Seminars in Cancer Biology. 2015;35(Suppl):S78–S103. doi: 10.1016/j.semcancer.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hsu Y.C., Chen H.Y., Yuan S., Yu S.L., Lin C.H., Wu G. Genome-wide analysis of three-way interplay among gene expression, cancer cell invasion and anti-cancer compound sensitivity. BMC Medicine. 2013;11:106. doi: 10.1186/1741-7015-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J., Wang Y., Li G., Yu H., Xie X. Down-regulation of nicotinamide N-methyltransferase induces apoptosis in human breast cancer cells via the mitochondria-mediated pathway. PloS One. 2014;9(2) doi: 10.1371/journal.pone.0089202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie X., Liu H., Wang Y., Zhou Y., Yu H., Li G. Nicotinamide N-methyltransferase enhances resistance to 5-fluorouracil in colorectal cancer cells through inhibition of the ASK1-p38 MAPK pathway. Oncotarget. 2016;7(29):45837–45848. doi: 10.18632/oncotarget.9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y., Zeng J., Wu W., Xie S., Yu H., Li G. Nicotinamide N-methyltransferase enhances chemoresistance in breast cancer through SIRT1 protein stabilization. Breast Cancer Research: BCR. 2019;21(1):64. doi: 10.1186/s13058-019-1150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim J., Hong S.J., Lim E.K., Yu Y.S., Kim S.W., Roh J.H. Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis. Journal of Experimental & Clinical Cancer Research. 2009;28:20. doi: 10.1186/1756-9966-28-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J., You S., Zhang S., Hu Q., Wang F., Chi X. Elevated N-methyltransferase expression induced by hepatic stellate cells contributes to the metastasis of hepatocellular carcinoma via regulation of the CD44v3 isoform. Molecular Oncology. 2019;13(9):1993–2009. doi: 10.1002/1878-0261.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ding R.-B., Bao J., Deng C.-X. Emerging roles of SIRT1 in fatty liver diseases. International Journal of Biological Sciences. 2017;13(7):852–867. doi: 10.7150/ijbs.19370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hao C., Zhu P.-X., Yang X., Han Z.-P., Jiang J.-H., Zong C. Overexpression of SIRT1 promotes metastasis through epithelial-mesenchymal transition in hepatocellular carcinoma. BMC Cancer. 2014;14(1) doi: 10.1186/1471-2407-14-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iyamu I.D., Huang R. Development of fluorescence polarization-based competition assay for nicotinamide N-methyltransferase. Analytical Biochemistry. 2020;604:113833. doi: 10.1016/j.ab.2020.113833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Policarpo R.L., Decultot L., May E., Kuzmic P., Carlson S., Huang D. High-affinity alkynyl bisubstrate inhibitors of nicotinamide N-methyltransferase (NNMT) Journal of Medicinal Chemistry. 2019;62(21):9837–9873. doi: 10.1021/acs.jmedchem.9b01238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kannt A., Rajagopal S., Kadnur S.V., Suresh J., Bhamidipati R.K., Swaminathan S. A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders. Scientific Reports. 2018;8(1):3660. doi: 10.1038/s41598-018-22081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Neelakantan H., Vance V., Wetzel M.D., Wang H.L., McHardy S.F., Finnerty C.C. Selective and membrane-permeable small molecule inhibitors of nicotinamide N-methyltransferase reverse high fat diet-induced obesity in mice. Biochemical Pharmacology. 2018;147:141–152. doi: 10.1016/j.bcp.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neelakantan H., Brightwell C.R., Graber T.G., Maroto R., Wang H.L., McHardy S.F. Small molecule nicotinamide N-methyltransferase inhibitor activates senescent muscle stem cells and improves regenerative capacity of aged skeletal muscle. Biochemical Pharmacology. 2019;163:481–492. doi: 10.1016/j.bcp.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang H., Ryu D., Wu Y., Gariani K., Wang X., Luan P. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352(6292):1436–1443. doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- 93.Souto J.C., Blanco-Vaca F., Soria J.M., Buil A., Almasy L., Ordoñez-Llanos J. A genomewide exploration suggests a new candidate gene at chromosome 11q23 as the major determinant of plasma homocysteine levels: results from the GAIT project. The American Journal of Human Genetics. 2005;76(6):925–933. doi: 10.1086/430409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Riederer M., Erwa W., Zimmermann R., Frank S., Zechner R. Adipose tissue as a source of nicotinamide N-methyltransferase and homocysteine. Atherosclerosis. 2009;204(2):412–417. doi: 10.1016/j.atherosclerosis.2008.09.015. [DOI] [PubMed] [Google Scholar]
Data references
- 1.Atlasgeneticsoncology.org/Geneshttp://atlasgeneticsoncology.org//Genes/NNMTID44506ch11q23.html
- 2.Proteinatlas.orghttps://www.proteinatlas.org/ENSG00000166741-NNMT
- 3.https://cancer.sanger.ac.uk/cosmic/search?q=NNMT
- 4.Ensamble.orghttps://www.ensembl.org/Homo_sapiens/Gene/Variation_Gene/Table?db=core;g=ENSG00000166741;r=11:114257787-114313536