Abstract
Objective
To assess the value of qualitative and quantitative MRI radiomics features for noninvasive prediction of immuno-oncologic characteristics and outcomes of hepatocellular carcinoma (HCC).
Methods
This retrospective, IRB-approved study included 48 patients with HCC (M/F 35/13, mean age 60y) who underwent hepatic resection or transplant within 4 months of abdominal MRI. Qualitative imaging traits, quantitative nontexture related and texture features were assessed in index lesions on contrast-enhanced T1-weighted and diffusion-weighted images. The association of imaging features with immunoprofiling and genomics features was assessed using binary logistic regression and correlation analyses. Binary logistic regression analysis was also employed to analyse the association of radiomics, histopathologic and genomics features with radiological early recurrence of HCC at 12 months.
Results
Qualitative (r = − 0.41–0.40, p < 0.042) and quantitative (r = − 0.52–0.45, p < 0.049) radiomics features correlated with immunohistochemical cell type markers for T-cells (CD3), macrophages (CD68) and endothelial cells (CD31). Radiomics features also correlated with expression of immunotherapy targets PD-L1 at protein level (r = 0.41–0.47, p < 0.029) as well as PD1 and CTLA4 at mRNA expression level (r = − 0.48–0.47, p < 0.037). Finally, radiomics features, including tumour size, showed significant diagnostic performance for assessment of early HCC recurrence (AUC 0.76–0.80, p < 0.043), while immunoprofiling and genomic features did not (p = 0.098–0929).
Conclusions
MRI radiomics features may serve as noninvasive predictors of HCC immuno-oncological characteristics and tumour recurrence and may aid in treatment stratification of HCC patients. These results need prospective validation.
Keywords: Hepatocellular carcinoma, Magnetic resonance imaging, Correlation of data, Genomics, Immunophenotyping
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common neoplasm and third leading cause of cancer-related mortality worldwide [1]. The pathogenesis of HCC is heterogeneous and complex [2]. HCC has a poor prognosis, since it often presents at an advanced stage, at which novel targeted therapies, including tyrosine kinase inhibitors and immune checkpoint inhibitors [3, 4], alone or in combination with locoregional therapy, are the only available therapeutic options. Recently, immunotherapy has shown a 20% response rate in patients with advanced HCC [4], compared to only a 2% response rate reported for multi-kinase inhibitor sorafenib [3].
Due to the marked heterogeneity of HCC and the emerging role of targeted therapies, there is a need for advanced HCC characterisation to appropriately inform patient management and prognostication. The success of immunotherapy is dependent on the tumour’s immune status [5]. Immunoprofiling of tumours is therefore of critical importance in predicting response to immunotherapy [6]. Multiplexed immunohisto-chemistry is a promising advanced technique for assessment of immune cell distribution and localisation in tumours [7]. HCC tumours have also been intensively profiled for genome-wide gene expression, DNA copy number alterations, DNA methylation and gene mutations, which may have a role for the prediction of prognosis, microvascular invasion and treatment response [8]. While immunoprofiling and genomics evaluation are promising for prediction of HCC response to therapy [9], these methods require invasive tissue sampling, specialised equipment and analysis and are limited by sampling bias and possible complications.
HCC is typically diagnosed and staged using noninvasive imaging techniques such as computed tomography (CT) or magnetic resonance imaging (MRI) [10]. Radiomics, a process by which large numbers of features are extracted from digital images, is motivated by the concept that biomedical images contain valuable information reflecting the underlying patho-physiology [11, 12]. Encouraging studies have been published on the potential utility of radiomics for noninvasive characterisation of HCC (reviewed in [13]). Recently, it was shown in a preliminary study that multiparametric MRI histogram features are associated with immunohistochemical HCC markers and gene expression levels of HCC markers and therapeutic targets [14]. In a retrospective analysis, associations of qualitative imaging traits from CT and MRI data with gene signatures of aggressive HCC were observed [15]. While these initial reports on the correlation of each of qualitative, histogram and texture MRI features with several histopathological features and genomics features of HCC are promising, an integrated analysis of the association of MRI radiomics features with HCC immuno-oncological features has not been performed. The development of a comprehensive approach to assess and integrate qualitative and quantitative data obtained from routine MRI could potentially impact the care of patients with HCC.
The objective of our study was to assess the value of qualitative and quantitative MRI radiomics features for noninvasive prediction of immuno-oncologic characteristics and outcomes of HCC.
Materials and methods
Patients
This retrospective HIPAA-compliant single-centre study was approved by our institutional review board with a waiver of informed consent. Our centre’s pathological database was queried between December 2014 and July 2016 using the search term “hepatocellular carcinoma” or “HCC” or “hepatocarcinoma” or “liver cancer”. Inclusion criteria were (a) patients with pathologically proven HCC, (b) patients who underwent abdominal MRI within 4 months before surgery (resection or transplant) for HCC and (c) patients who had not undergone treatment before surgery. Exclusion criteria were (a) lesion size < 10 mm; (b) lack of diffusion-weighted imaging (DWI); (c) technically inadequate MRI exams (due to motion, image artefacts) as evaluated by observers 1 (SL) and 2 (CB), two abdominal radiologists with 8 and 7 years’ experience, respectively; and (d) histopathologic diagnosis other than HCC. A total of 48 patients with HCC (M/F 35/13, mean age 60 years, range 36–77 years) comprised our study population (Fig. 1).
Fig. 1.
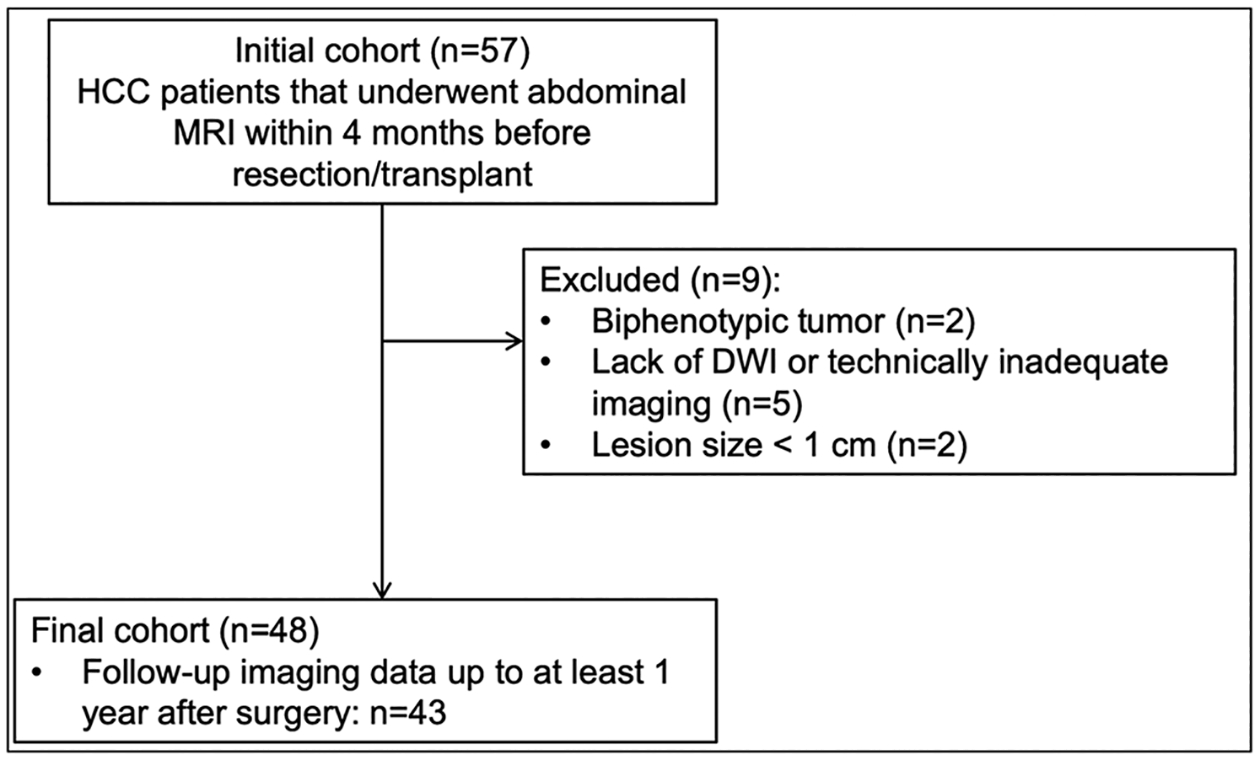
Study flow chart of our single-centre patient population
MRI acquisition
Routine abdominal MRI was performed at either 1.5 T (n = 34) or 3 T (n = 14). All MRI exams included multiphasic contrast-enhanced MRI with liver-specific gadoxetic acid injection (n = 33) or extracellular agent (n = 15), and DWI, among other routine sequences. While DWI b-values varied among MRI scanners, all but one DWI protocol consisted of b50 and b750/800 acquisitions. The DWI data of the patient without b50 and b750/800 data were not analysed, while the other imaging data were kept for analysis. Additional information on the acquisition and used MRI systems can be found in the Supplemental Material.
Qualitative MRI analysis
The qualitative MRI analysis was performed by two abdominal radiologists in consensus (observers 1 and 2). The index HCC lesion (the largest lesion that underwent histopathologic sampling) was identified on MRI. The location (i.e. liver segment) and size of the lesion in the pathological report was used to ensure analysis of the same lesion for the imaging and histopathological and genomics analysis. The observers were aware that patients had HCC, but were blinded to the clinical and pathologic data. The observers categorised each index lesion according to the CT/MRI Liver Imaging Reporting and Data System (LI-RADS) 2018 diagnostic algorithm [16]. In addition, the observers documented the presence/absence of ancillary findings and the qualitative appearance of lesions on conventional sequences (see list of imaging features in Table 1 and representative examples in Supplemental Fig. 1) [15].
Table 1.
Clinical characteristics, imaging features and pathological features assessed in 48 patients with 48 HCCs
| Parameter | Classification | Value |
|---|---|---|
| Demographics | ||
| Age (year) | Mean ± standard deviation (range) | 60 ± 11 (36–77) |
| Sex | M/F | 35/13 |
| Clinical characteristics | ||
| Aetiology of liver disease | - Hepatitis C virus | - 12 |
| - Hepatitis B virus | - 26 | |
| - Nonalcoholic steatohepatitis | - 1 | |
| - Alcohol | - 2 | |
| - Primary sclerosing cholangitis | - 1 | |
| - Unknown | - 6 | |
| Fibrosis stage* | 0/1/2/3/4 | 3/6/10/13/14 |
| Child-Pugh scores | A/B/C | 41/6/1 |
| Model for End-Stage Liver Disease (MELD) score | Mean ± standard deviation (range) | 8.1 ± 2.3 (6–18) |
| Barcelona clinical liver cancer (BCLC) stage | 0/A/B/C | 11/13/22/2 |
| American Joint Committee on Cancer (AJCC) stage** | T1/T2/T3 | 12/27/6 |
| Type of surgery | Resection/transplant | 46/2 |
| Imaging appearance | ||
| Infiltrative pattern | Y/N | 1/47 |
| Multiple lesions | Y/N | 5/43 |
| Extranodular growth | Y/N | 12/36 |
| Macrovascular invasion | Y/N | 2/46 |
| Tumour necrosis | Y/N | 7/41 |
| Tumour haemorrhage | Y/N | 5/43 |
| Tumour fat content | Y/N | 7/41 |
| Mosaic appearance | Y/N | 19/29 |
| Internal arteries | Y/N | 11/37 |
| Capsule | Y/N | 39/9 |
| T2 hyperintensity | Y/N | 44/4 |
| ADC hypointensity*** | Y/N | 20/27 |
| Wash-in/wash-out | Y/N | 41/7 |
| Hepatobiliary phase hypointensity**** | Y/N | 29/4 |
| Quantitative imaging features (nontexture related) | ||
| Tumour size (cm) | Mean ± standard deviation (range) | 4.2 ± 3.3 (1.0–15) |
| ADC mean (10−3 mm2/s) | Mean ± standard deviation | 1.28 ± 0.35 |
| ADC min (10−3 mm2/s) | Mean ± standard deviation | 0.60 ± 0.39 |
| Enhancement ratio early arterial phase (%) | Mean ± standard deviation | 57.0 ± 55.0 |
| Enhancement ratio late arterial phase (%) | Mean ± standard deviation | 97.0 ± 49.4 |
| Enhancement ratio portal venous phase (%) | Mean ± standard deviation | 92.8 ± 40.3 |
| Enhancement ratio late venous phase (%) | Mean ± standard deviation | 77.6 ± 35.4 |
| Enhancement ratio hepatobiliary phase (%) | Mean ± standard deviation | 50.9 ± 37.0 |
| Pathology features | ||
| Lesion size | Mean ± standard deviation (range) | 4.5 ± 4.2 (0.8–21) |
| Grade | Well/moderately/poorly differentiated | 4/31/13 |
| Microvascular invasion | Y/N | 31/17 |
ADC = apparent diffusion coefficient
Fibrosis stage not assessed for 2 patients
AJCC stage not assessed for 3 patients
No ADC map in 1 patient
No HBP in 15 patients
Region of interest analysis
A third observer (observer 3, M.K., a 4th year radiology resident with 1 year of experience in abdominal MRI) performed the quantitative region of interest (ROI) measurements of each index HCC to compute the signal intensity (SI) on contrast-enhanced images (pre-contrast (n = 48), early arterial phase (n = 35), late arterial phase (n = 41), portal venous phase (n = 47), late venous phase (n = 46), hepatobiliary phase (n = 33 in patients who received gadoxetic acid)) and apparent diffusion coefficient (ADC) maps (n = 47) using OsiriX software (version 5.8, Pixmeo). ROIs were placed in a single slice in which the lesion appeared largest for lesions smaller than 3 cm. For lesions with a size larger than 3 cm, the SI was measured on two consecutive slices on which the lesion appeared largest. The entire lesion was covered in the ROI, including areas of necrosis, if present. However, peripheral portions of the lesion were avoided to exclude partial volume effects of adjacent extra-lesional tissue.
Two types of quantitative features were extracted from the ROIs: quantitative nontexture features (for the sake of brevity referred to as quantitative features in the rest of the manuscript) and texture features, as described below.
Quantitative features
Tumour size
The largest diameter of the index lesion on the axial slice on which the lesion appeared largest was recorded.
CE-T1WI
Enhancement ratios (ER) = [(SI post − SI pre)/SI pre] × 100% were computed for all post-contrast phases of contrast-enhanced MRI for each lesion.
Apparent diffusion coefficient analysis
ADC mean and minimum values were calculated for each lesion using monoexponential fit of DWI data from b50 and b750/800 s/mm2.
Texture analysis
Observer 4 (S.J.H., an MRI physicist with 3 years of experience in abdominal MRI) computed Haralick texture features (energy, contrast, correlation, variance, homogeneity, sum average, sum variance, sum entropy, entropy, difference variance, difference entropy, information correlation measures 1 and 2, maximal correlation [17]) in the HCC ROIs drawn on the dynamic multiphasic contrast-enhanced images and ADC maps using MATLAB (version 2016b, MathWorks, Inc.). Before texture analysis, signal values in ROIs were normalised to range between mean ± 3*standard deviation (SD) of the signal values in the ROI [18] and decimated to 16 and 64 discrete bin values. Normalisation was performed to account for inter-patient and inter-scanner differences in MRI signal intensities. Two different grey scales were used to increase the sensitivity of the texture features to both small and large intensity variations in the image.
A total of 218 radiomics features were extracted, consisting of 196 texture features (14 Haralick features × 7 imaging sequences × 2 grey level scales), 14 qualitative features and 8 quantitative features.
Immunoprofiling
Standard clinical histopathologic evaluation of representative H&E stained slides from the 48 HCCs was retrospectively performed in consensus by two pathologists (S.W., with over 25 years’ experience and J.P., with 2 years’ experience). The pathologists assessed tumour grade [19] and presence/absence of microvascular invasion on representative tumour samples.
Immunoprofiling consisted of multiplexed immunohistochemical consecutive staining on single slide (MICSSS) [7] used to sequentially stain endothelial cells (CD31; clone J70, ready-to-use concentration, Ventana Medical Systems), macrophages (CD68; clone KP1, 1:100 dilution, Agilent Dako), T cells (CD3; clone CD3, ready-to-use concentration, Ventana Medical Systems) and cell surface ligand (PD-L1; clone E1L3N, 1:100 dilution, Cell Signaling Technology). Endothelial cells, macrophages and T cells were stained to visualise tissue vasculature, inflammation and immune status, respectively. PD-L1 was chosen as immunotherapy target because of well-established antibodies available for PD-L1 staining [7]. Stained HCC sections were digitally imaged at × 20 magnification using a whole-slide scanner (NanoZoomer C13210–01). Threshold-based segmentation was performed in MATLAB to detect stained pixels [14] (Supplemental Figure 2). Stained tumour fractions were then measured from these segmentations.
Genomics analysis
Isolated total RNA samples were extracted from paraffin-embedded HCC samples of all 48 patients using High Pure RNA Paraffin Kit (Roche). RNA was subjected to the Elements HCC assay (NanoString) as previously reported [20]. Transcriptomic HCC subtypes (S1, S2 and S3) described by Hoshida et al [21] were defined using the nearest template prediction algorithm [20]. Raw transcript count data were normalised by scaling with geometric mean of built-in normalisation genes. The following HCC marker genes and therapeutic targets were profiled: HCC subclass signature genes (IQGAP1, S100A11, RAB31, CD37, POSTN, ARHGDIB, ALOX5AP, LAPTM5, CSPG2, ARPC2, COL2A1, GPC3, AFP, AHCY, TARBP1, ARID3A, FGFR3, SMARCC1, RPS5, EIF4A2, GLYAT, SERPINC1, APOC4, MTHFD1, GPT, HPD, SERPING1, DPAGT1, PCK1, HGD), liver-specific Wnt target (GLUL) [22], stemness markers (EPCAM, KRT19) [23, 24], early HCC markers (BIRC5, HSP70, LYVE1, EZH2) [25, 26], pharmacological target (FGFR4) [27], potentially targetable angiogenesis marker (VEGFA) [28] and targetable immune checkpoints (PD-L1, PD-1, CTLA4) [14, 21].
Outcome analysis
Follow-up imaging data (CT or MRI) was reviewed by observer 5, D.S., a radiologist with 1 year of experience in abdominal MRI. For patients who had follow-up imaging until at least 1 year after surgery, the presence of early HCC recurrence [29] on radiological images was noted.
Statistical analysis
Statistical analysis was performed in MATLAB and SPSS (version 20, IBM Analytics). Logistic regression analysis was used to determine the association of the imaging features (qualitative, nontexture-related quantitative and texture features) with histopathologic grade, microvascular invasion, molecular subtype and clinical outcome. Prior to regression analysis, features were standardised to have zero mean and unit standard deviation. Spearman correlation analysis was employed to determine the correlation of radiomics features with gene expression levels and immunohistochemical stained fractions. p values were adjusted for multiple correlations by using a false discovery rate (FDR) adjustment. An FDR-adjusted p value below 0.05 was considered statistically significant.
Results
Clinical, imaging and pathological characteristics (Table 1)
The majority of the patients were male (35/48 = 72.9%) and had chronic HBV infection (26/48 = 54.2%). Forty-six of the patients underwent partial resection and 2 patients underwent liver transplantation. Pathological evaluation showed that most lesions (44/48 = 91.7%) were moderately or poorly differentiated. Microvascular invasion was observed in 31/48 (64.6%) of the lesions. In terms of imaging, 41/48 HCCs (85.4%) were categorised as LI-RADS 5 and 7/48 (14.6%) as LI-RADS 4. The vast majority of lesions exhibited nodular morphology (47/48 = 97.9%), while one lesion was infiltrative. A capsule was observed in 39 lesions (81.2%). Typical wash-in/was-out enhancement pattern was seen in 41/48 (85.4%) of the lesions.
Association of radiomics features with pathological and immune features
No significant associations were observed between any of the radiomics features and histopathologic grade and microvascular invasion (p = 0.075–1).
Despite the absence of significant association with qualitative histopathologic features, radiomics features were significantly associated with quantitative expression of immune markers CD3, CD31, CD68 and PD-L1 as assessed with MICSSS analysis (Fig. 2, Table 2). Most of the significant correlations were found with expression of CD68 (macrophages), including with several texture features (r 0.33–0.45, p < 0.049) and qualitative features mosaic appearance (r = 0.40, p = 0.023) and wash-in/wash-out enhancement pattern (r = 0.37, p = 0.042). Texture feature variance at the ADC map and the enhancement ratios at the portal and late venous phases were significantly correlated with expression of checkpoint inhibitor PD-L1 (r 0.41–0.47, p < 0.029).
Fig. 2.
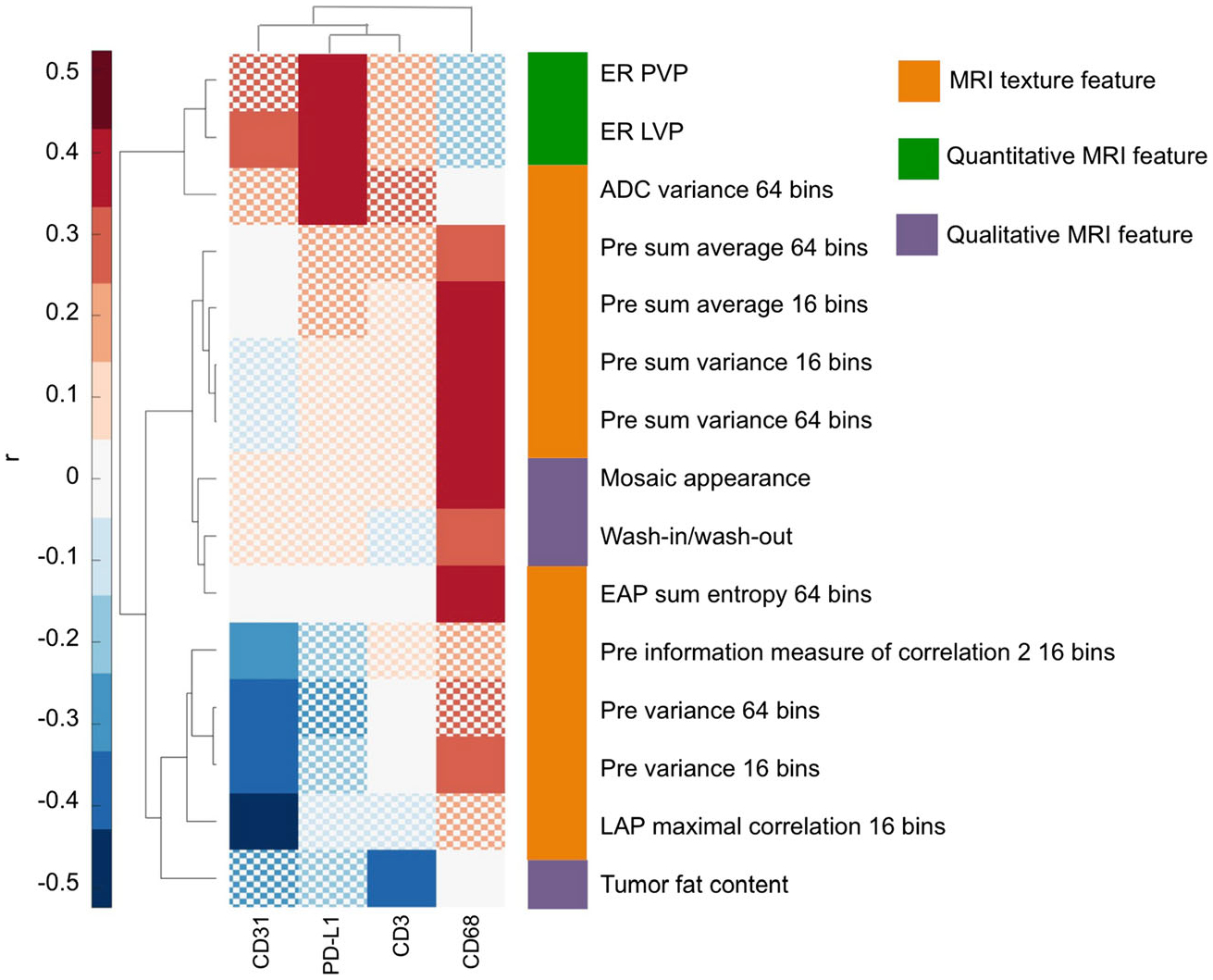
Clustergram of significant correlations, expressed as the Spearman correlation coefficient r, between radiomics features and immunohistochemical markers assessed by multiplex analysis. Only radiomics features and immunohistochemical markers for which a significant association (FDR-adjusted p < 0.05) was observed are shown. Correlations are coloured according to the colour bar shown on the left. Nonsignificant correlations are displayed with a checkerboard pattern. ADC = apparent diffusion coefficient, EAP = early arterial phase, ER = enhancement ratio, LAP = late arterial phase, LVP = late arterial phase, OR = odds ratio, PVP = portal venous phase
Table 2.
Significant correlations of radiomics features with immunoprofiling features in 48 HCCs
| Marker | Radiomics feature | r | FDR-adj p |
|---|---|---|---|
| CD3 | Qualitative features | ||
| Tumour fat content | − 0.41 | 0.019 | |
| CD31 | Texture features | ||
| Pre variance 16 bins | − 0.47 | 0.005 | |
| Pre information measure of correlation 2 16 bins | −0.38 | 0.038 | |
| LAP maximal correlation 16 bins | −0.52 | 0.003 | |
| Pre variance 64 bins | −0.47 | 0.005 | |
| Quantitative features | |||
| ERLVP | 0.37 | 0.023 | |
| CD68 | Texture features | ||
| Pre variance 16 bins | 0.33 | 0.047 | |
| Pre sum average 16 bins | 0.41 | 0.018 | |
| Pre sum variance 16 bins | 0.41 | 0.018 | |
| Pre sum average 64 bins | 0.37 | 0.049 | |
| Pre sum variance 64 bins | 0.42 | 0.017 | |
| EAP sum entropy 64 bins | 0.45 | 0.049 | |
| Qualitative features | |||
| Mosaic appearance | 0.40 | 0.023 | |
| Wash-in/wash-out | 0.37 | 0.042 | |
| PD-L1 | Texture features | ||
| ADC variance 64 bins | 0.41 | 0.029 | |
| Quantitative features | |||
| ERPVP | 0.42 | 0.015 | |
| ER LVP | 0.47 | 0.005 |
Correlation coefficient coefficients (r) resulted from Spearman correlation analysis. p values were adjusted for multiple tests using a false discovery rate (FDR) adjustment
ADC = apparent diffusion coefficient, AUC = area under the curve, EAP = early arterial phase, ER = enhancement ratio, LAP = late arterial phase, LVP = late venous phase, PVP = portal venous phase
Associations of radiomics and genomics features
The distribution of molecular subtypes was as follows: S1, n = 18; S2, n = 9; S3, n = 21. ADC min was the only quantitative feature that showed significant association with molecular subtypes (aggressive subtypes S1/S2 vs. less aggressive subtype S3, odds ratio (OR) = 2.00, p = 0.036, AUC = 0.68). Significant associations between radiomics features and gene expression levels are shown in Fig. 3 and Table 3. Eleven imaging features showed significant associations with 14 gene expression levels (r − 0.61–0.56, p < 0.043). Among these 11 features, there were 9 texture features, 1 quantitative feature and 1 qualitative feature. Most correlations were found with molecular subclass signature genes. The strongest correlation was between texture at the early arterial phase with expression of binding protein TARBP1 (r = − 0.61, p = 0.013). In terms of associations with HCC therapeutic targets, significant correlations were observed between late arterial phase texture features and immunotherapy targets PD-1 and CTLA4 (Fig. 4).
Fig. 3.
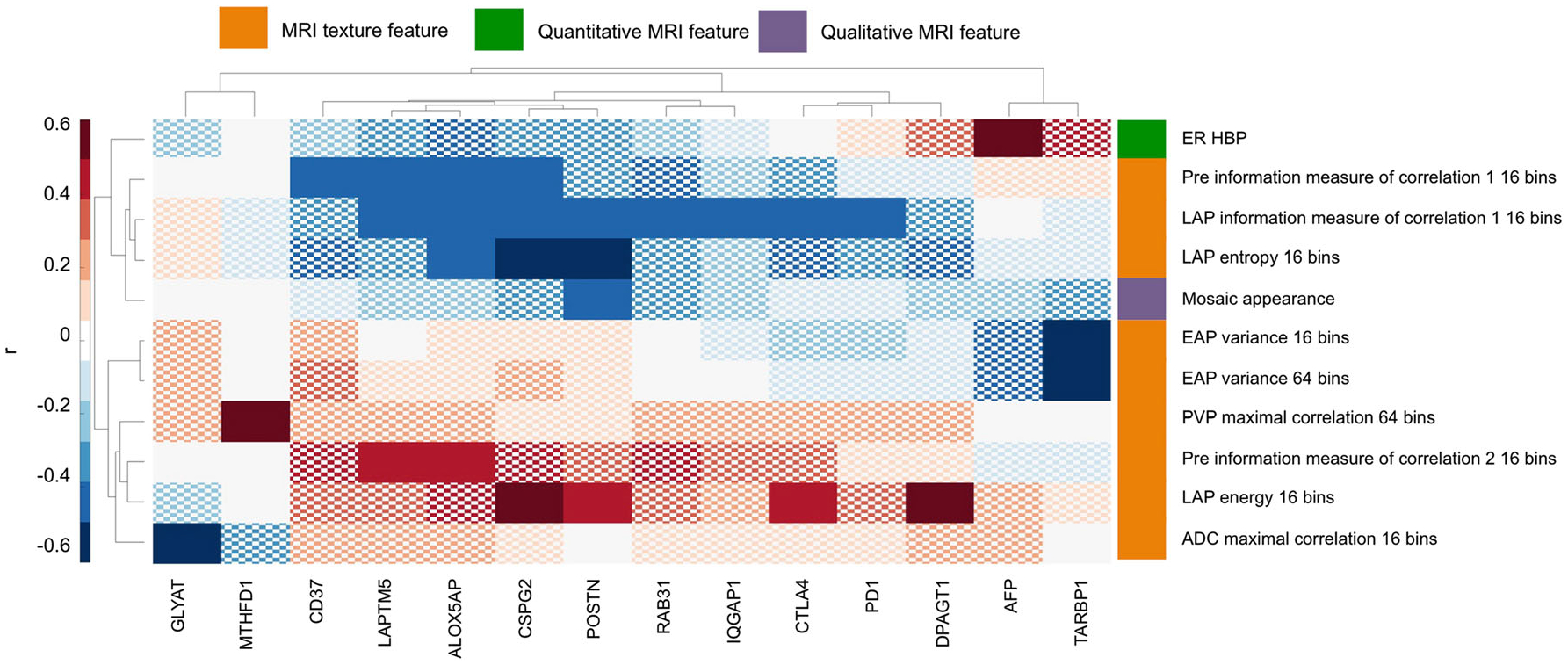
Clustergram of significant correlations, expressed as the Spearman correlation coefficient r, between radiomics features and gene expression levels. Only radiomics features and genes for which a significant association (FDR-adjusted p < 0.05) was observed are shown. Correlations are coloured according to the colour bar shown on the left. Nonsignificant correlations are displayed with a checkerboard pattern. Radiomics features, in particular texture features, showed significant association with gene expression levels of HCC markers and therapeutic targets. ADC = apparent diffusion coefficient, EAP = early arterial phase, ER = enhancement ratio, HBP = hepatobiliary phase, LAP = late arterial phase, PVP = portal venous phase
Table 3.
Significant correlations of radiomics features with gene expression levels in 48 HCCs
| Gene | Feature type | Feature names | r | FDR-adj p |
|---|---|---|---|---|
| IQGAP1 | Texture features | LAP information measure of correlation 116 bins | −0.44 | 0.031 |
| RAB31 | Texture features | LAP information measure of correlation 116 bins | −0.48 | 0.027 |
| CD37 | Texture features | Pre information measure of correlation 116 bins | −0.42 | 0.036 |
| POSTN | Texture features | LAP energy 16 bins | 0.45 | 0.043 |
| LAP entropy 16 bins | −0.55 | 0.011 | ||
| LAP information measure of correlation 116 bins | −0.45 | 0.030 | ||
| Qualitative features | Mosaic appearance | −0.47 | 0.036 | |
| ALOX5AP | Texture features | Pre information measure of correlation 116 bins | −0.46 | 0.028 |
| Pre information measure of correlation 2 16 bins | 0.47 | 0.028 | ||
| LAP entropy 16 bins | −0.46 | 0.039 | ||
| LAP information measure of correlation 116 bins | −0.48 | 0.027 | ||
| LAPTM5 | Texture features | Pre information measure of correlation 116 bins | −0.46 | 0.028 |
| Pre information measure of correlation 2 16 bins | 0.46 | 0.028 | ||
| LAP information measure of correlation 116 bins | −0.42 | 0.036 | ||
| CSPG2 | Texture features | Pre information measure of correlation 116 bins | −0.42 | 0.036 |
| LAP energy 16 bins | 0.50 | 0.028 | ||
| LAP entropy 16 bins | −0.52 | 0.017 | ||
| LAP information measure of correlation 116 bins | −0.46 | 0.030 | ||
| AFP | Quantitative features | ER HBP | 0.56 | 0.025 |
| TARBP1 | Texture features | EAP variance 16 bins | −0.61 | 0.013 |
| EAP variance 64 bins | −0.57 | 0.035 | ||
| GLYAT | Texture features | ADC maximal correlation 16 bins | −0.54 | 0.009 |
| MTHFD1 | Texture features | PVP maximal correlation 64 bins | 0.56 | 0.002 |
| DPAGT1 | Texture features | LAP energy 16 bins | 0.47 | 0.037 |
| PD1 | Texture features | LAP information measure of correlation 116 bins | −0.43 | 0.032 |
| CTLA4 | Texture features | LAP energy 16 bins | 0.47 | 0.037 |
| LAP information measure of correlation 116 bins | −0.48 | 0.027 |
Only genes for which significant correlations were found are shown. Correlation coefficient coefficients (r) resulted from Spearman correlation analysis. P values were adjusted for multiple tests using a false discovery rate (FDR) adjustment
ADC = apparent diffusion coefficient, EAP = early arterial phase, ER = enhancement ratio, HBP = hepatobiliary phase, LAP = late arterial phase, PVP = portal venous phase
Fig. 4.
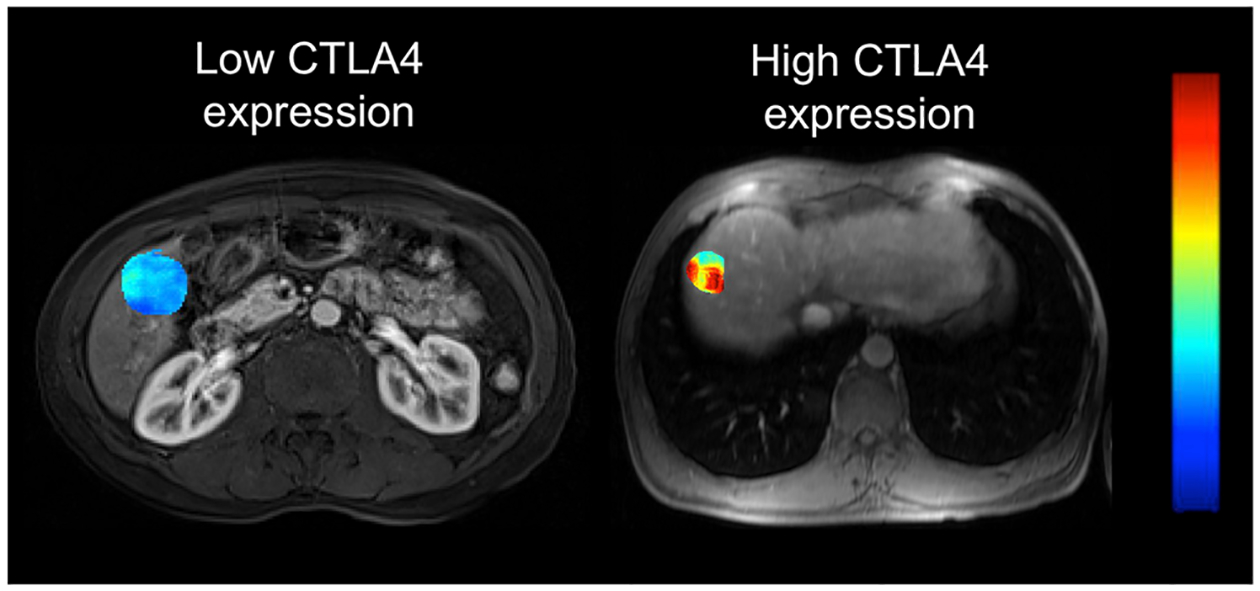
Energy texture maps overlaid on T1-weighted post-contrast image obtained during the late arterial phase image. Left: 59-year-old male HBV patient with a 3.7-cm HCC lesion in right hepatic lobe. Energy texture map overlaid on T1-weighted post-contrast image obtained during the late arterial phase image. Texture feature Energy with 16 bins was 0.025 in the lesion, with corresponding molecular expression of immunotherapy target CTLA4 of 0.44. Right: 51-year-old male HBV patient with a 3.3-cm HCC lesion in right hepatic lobe. Texture feature energy with 16 bins was 0.061 in the lesion, with corresponding molecular expression of immunotherapy target CTLA4 of 0.68. The texture feature maps are coloured according to the scale bar on the right with a range of [0–0.1]
Association of radiomics and immuno-oncologic features with HCC recurrence (Table 4)
Table 4.
Significant associations of imaging features with HCC recurrence within 1 year after resection or transplant in 43 patients
| Feature | AUC (95% CI) | p* | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | OR | p# |
|---|---|---|---|---|---|---|---|---|
| Texture features | ||||||||
| Pre sum entropy 16 bins | 0.80 (0.62–0.79) | <0.001 | 70 | 84 | 58 | 90 | 5.51 | 0.028 |
| Pre entropy 64 bins | 0.77 (0.69–0.96) | 0.002 | 90 | 63 | 43 | 95 | 3.28 | 0.033 |
| LAP information measure of correlation 1 64 bins | 0.77 (0.57–0.97) | 0.043 | 89 | 71 | 50 | 95 | 3.23 | 0.043 |
| LAP information measure of correlation 2 64 bins | 0.77 (0.60–0.93) | 0.009 | 89 | 71 | 50 | 95 | 0.32 | 0.009 |
| LVP information measure of correlation 2 64 bins | 0.75 (0.58–0.91) | 0.019 | 78 | 77 | 50 | 92 | 0.31 | 0.019 |
| HBP information measure of correlation 1 64 bins | 0.78 (0.60–0.95) | 0.016 | 57 | 100 | 100 | 89 | 0.24 | 0.016 |
| Other features | ||||||||
| Size (≥ 5 cm) | 0.76 (0.57–0.59) | 0.004 | 82 | 82 | 54 | 90 | 3.01 | 0.004 |
p value from AUC
p value from logistic regression
Area under the curve (AUC) values from ROC analysis and odds ratios (OR) from logistic regression analysis are shown
AUC = area under the curve; HBP = hepatobiliary phase, LAP = late arterial phase, LVP = late venous phase; NPV = negative predictive value; OR = odds ratio; PPV = positive predictive value
Follow-up imaging data up to at least 1 year after HCC surgery was available for 43/48 (89.6%) patients. In 10 patients (23.3%), early HCC recurrence was detected on imaging within 1 year after surgery. These patients had all previously undergone hepatic resection. The average time to recurrence in those patients was 227 days (range 106–396 days). While the longest time to recurrence was more than 1 year, in that particular patient small enhancing lesions were already visible at the 9 months follow-up imaging exam, indicating that recurrence had occurred within 1 year after resection. Several texture features were significantly associated with recurrence (highest diagnostic performance for pre-contrast texture feature sum entropy with 16 bins; OR = 5.51, p = 0.028, AUC = 0.80). Tumour size ≥ 5 cm was the only nontexture feature that showed significant association with HCC recurrence (OR = 3.01, p = 0.004, AUC = 0.76). None of the immunogenomic features were significantly associated with HCC recurrence (p = 0.098–0.929).
Discussion
In this study, we sought to assess the potential value of MRI-based radiomics analysis for noninvasive prediction of immuno-oncologic characteristics of HCC as well as outcome. There is increasing interest in the characterisation of HCC immune cell microenvironment, since immune cells play a central role in tumour initiation, progression and metastasis [30]. Furthermore, immunologic hallmarks of HCC are increasingly targetable with novel immunotherapies [31]. Early studies have shown that patients who are treated with nivolumab (anti-PD-1) exhibit reasonable objective response and tumour control [4]. In our study, several MRI texture features showed correlation with gene expression of immunotherapy targets CTLA-4 and PD-1. In addition, significant associations of texture features and quantitative enhancement ratios with immunohistochemical PD-L1 expression were identified in our study. These results suggest that MRI radiomics analysis may be used to predict expression of immunotherapy targets in HCC, which may aid in stratification of HCC patients for immunotherapy.
None of the radiomics features showed significant association with pathological grade, in contrast to a previous study [32]. These conflicting results may be due to the fact that we employed statistical correction for multiple testing, which was not performed in the previous study. Microvascular invasion has been identified as predictor of recurrence and poor overall survival [33]. There is increasing interest in noninvasive prediction of microvascular invasion using imaging, both using qualitative [34] and radiomics [35] features. Despite the absence of significant associations of radiomics features with microvascular invasion in our study, combination of multiple MRI features into radiomics signatures may improve the performance of radiomics for prediction of microvascular invasion [35].
Because up to 70% of early-stage HCC patients treated with either curative resection or locoregional therapy will develop tumour recurrence within 5 years [36], we sought to assess the association of imaging features with early tumour recurrence within 1 year of surgery. It has been previously shown that patients with early HCC recurrence have worse prognosis in terms of median survival [29]. Texture features again showed promise for the prediction of recurrence. However, tumour size from MRI at baseline showed similar diagnostic performance. A recent study by Villanueva et al developed prognostic models incorporating clinicopathologic and genomic information in a larger cohort of 287 patients, although imaging was not included in their analysis [23]. Imaging-based prediction of outcome after surgery may have clinical utility. If adverse outcome is predicted by imaging assessment before surgery, neoadjuvant therapies could potentially be employed which may improve outcome [37]. MRI is already routinely performed in HCC patients, making the imaging assessment a cost-effective and noninvasive alternative to tissue sampling.
Our findings of significant associations with radiomics features correspond to previous preliminary studies in which the association of qualitative CT and MRI [15] and quantitative multiparametric MRI histogram features [14] with HCC molecular characteristics was assessed. In the current study, we combined qualitative and quantitative radiomics features to provide a comprehensive overview of associations of radiomics features with HCC pathology and genomics characteristics in a cohort of patients with resected HCC. We also included texture analysis, which was not performed in the previous studies.
Our study is limited by its relatively small sample size, precluding inclusion of a separate validation set. We are currently prospectively recruiting HCC patients to validate the observed associations in our study. In addition, the development of radiomics models was not feasible in our study, since not all radiomics features could be calculated in all patients because of the lack of certain contrast-enhanced MRI phases in several patients. The study was also limited by the heterogeneity of MRI platforms, protocols, sequence parameters and contrast agents, which were not standardised because of the retrospective design of the study. While numerous studies have assessed reproducibility and repeatability of radiomics features extracted from PET and CT images (reviewed in [38]), little is known on the effect of MRI acquisition parameters on radiomics feature robustness. We also did not assess inter-observer variability, although it has been demonstrated that the vast majority of MRI radiomics features in HCC have good-to-excellent inter-observer agreement [39, 40].
MRI, in particular when employing a radiomics approach as supported by our results, can provide comprehensive HCC characterisation that is essential to direct management, predict patient prognosis and potentially enable personalised treatment stratification. Widespread use of advanced immunoprofiling and genomics analysis is hindered by limited resources and financial constraints. MRI can help by providing noninvasive surrogates of advanced immuno-oncological information in patients with HCC.
In conclusion, we observed that texture features, extracted from routine MRI, in conjunction with other qualitative and quantitative imaging features, are associated with HCC immuno-oncological characteristics, and potentially with outcome. Based on these results, we believe that MRI radiomics analysis may potentially be useful for patient-tailored treatment decision-making in HCC patients. These results need prospective validation.
Supplementary Material
Key Points.
MRI radiomics features showed significant associations with immunophenotyping and genomics characteristics of hepatocellular carcinoma.
Radiomics features, including tumour size, showed significant associations with early hepatocellular carcinoma recurrence after resection.
Funding information
This study has received funding from the Research Seed Grant no. RSD1608 from the Radiological Society of North America, and grant U01 CA172320 from the National Cancer Institute and the International Liver Cancer Association.
Abbreviations
- ADC
Apparent diffusion coefficient
- AUC
Area under the curve
- CE
MRI Contrast-enhanced MRI
- CT
Computed tomography
- DWI
Diffusion-weighted imaging
- FDR
False discovery rate
- HCC
Hepatocellular carcinoma
- MICSSS
Multiplexed immunohistochemical consecutive staining on single slide
- MRI
Magnetic resonance imaging
- OR
Odds ratio
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00330-020-06675-2) contains supplementary material, which is available to authorized users.
Guarantor The scientific guarantor of this publication is Bachir Taouli.
Conflict of interest The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry No complex statistical methods were necessary for this paper.
Informed consent Written informed consent was waived by the Institutional Review Board.
Ethical approval Institutional Review Board approval was obtained.
- retrospective
- observational
- performed at one institution
References
- 1.Forner A, Reig M, Bruix J (2018) Hepatocellular carcinoma. Lancet. 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 2.Marrero JA, Kulik LM, Sirlin C et al. (2018) Diagnosis, staging and management of hepatocellular carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 10.1002/hep.29913 [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Ricci S, Mazzaferro V et al. (2008) Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 359:378–390 [DOI] [PubMed] [Google Scholar]
- 4.El-Khoueiry AB, Sangro B, Yau T et al. (2017) Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389:2492–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen DS, Mellman I (2017) Elements of cancer immunity and the cancer-immune set point. Nature 541:321–330 [DOI] [PubMed] [Google Scholar]
- 6.Gnjatic S, Bronte V, Brunet LR et al. (2017) Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer 5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remark R, Merghoub T, Grabe N et al. (2016) In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol 1:aaf6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM (2010) Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis 30: 35–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khemlina G, Ikeda S, Kurzrock R (2017) The biology of hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer 16:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark T, Maximin S, Meier J, Pokharel S, Bhargava P (2015) Hepatocellular carcinoma: review of epidemiology, screening, imaging diagnosis, response assessment, and treatment. Curr Probl Diagn Radiol 44:479–486 [DOI] [PubMed] [Google Scholar]
- 11.Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambin P, Leijenaar RTH, Deist TM et al. (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762 [DOI] [PubMed] [Google Scholar]
- 13.Lewis S, Hectors S, Taouli B (2020) Radiomics of hepatocellular carcinoma. Abdom Radiol (NY). 10.1007/s00261-019-02378-5 [DOI] [PubMed] [Google Scholar]
- 14.Hectors SJ, Wagner M, Bane O et al. (2017) Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Sci Rep 7:2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taouli B, Hoshida Y, Kakite S et al. (2017) Imaging-based surrogate markers of transcriptome subclasses and signatures in hepatocellular carcinoma: preliminary results. Eur Radiol 27:4472–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.American College of Radiology (ACR) (2018) CT/MRI LI-RADS® v2018. https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS/CT-MRI-LI-RADS-v2018
- 17.Haralick RM, Shanmungam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC 3:610–621 [Google Scholar]
- 18.Collewet G, Strzelecki M, Mariette F (2004) Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging 22:81–91 [DOI] [PubMed] [Google Scholar]
- 19.Edmondson HA, Steiner PE (1954) Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer 7:462–503 [DOI] [PubMed] [Google Scholar]
- 20.Tan PS, Nakagawa S, Goossens N et al. (2016) Clinicopathological indices to predict hepatocellular carcinoma molecular classification. Liver Int 36:108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshida Y, Nijman SM, Kobayashi M et al. (2009) Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 69:7385–7392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goossens N, Sun X, Hoshida Y (2015) Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepatol Oncol 2:371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villanueva A, Hoshida Y, Battiston C et al. (2011) Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology 140:1501–1512 e1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamashita T, Forgues M, Wang W et al. (2008) EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res 68:1451–1461 [DOI] [PubMed] [Google Scholar]
- 25.Di Tommaso L, Franchi G, Park YN et al. (2007) Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology 45:725–734 [DOI] [PubMed] [Google Scholar]
- 26.Llovet JM, Chen Y, Wurmbach E et al. (2006) A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology 131:1758–1767 [DOI] [PubMed] [Google Scholar]
- 27.Hagel M, Miduturu C, Sheets M et al. (2015) First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov 5:424–437 [DOI] [PubMed] [Google Scholar]
- 28.Horwitz E, Stein I, Andreozzi M et al. (2014) Human and mouse VEGFA-amplified hepatocellular carcinomas are highly sensitive to sorafenib treatment. Cancer Discov 4:730–743 [DOI] [PubMed] [Google Scholar]
- 29.Poon RT, Fan ST, Ng IO, Lo CM, Liu CL, Wong J (2000) Different risk factors and prognosis for early and late intrahepatic recurrence after resection of hepatocellular carcinoma. Cancer 89:500–507 [PubMed] [Google Scholar]
- 30.Kong LQ, Zhu XD, Xu HX et al. (2013) The clinical significance of the CD163+ and CD68+ macrophages in patients with hepatocellular carcinoma. PLoS One 8:e59771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Wang FS (2019) Clinical immunology and immunotherapy for hepatocellular carcinoma: current progress and challenges. Hepatol Int. 10.1007/s12072-019-09967-y [DOI] [PubMed] [Google Scholar]
- 32.Zhou W, Zhang L, Wang K et al. (2017) Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images. J Magn Reson Imaging 45:1476–1484 [DOI] [PubMed] [Google Scholar]
- 33.Zimmerman MA, Ghobrial RM, Tong MJ et al. (2008) Recurrence of hepatocellular carcinoma following liver transplantation: a review of preoperative and postoperative prognostic indicators. Arch Surg 143:182–188 discussion 188 [DOI] [PubMed] [Google Scholar]
- 34.Renzulli M, Brocchi S, Cucchetti A et al. (2016) Can current preoperative imaging be used to detect microvascular invasion of hepatocellular carcinoma? Radiology 279:432–442 [DOI] [PubMed] [Google Scholar]
- 35.Peng J, Zhang J, Zhang Q, Xu Y, Zhou J, Liu L (2018) A radiomics nomogram for preoperative prediction of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma. Diagn Interv Radiol 24:121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llovet JM, Schwartz M, Mazzaferro V (2005) Resection and liver transplantation for hepatocellular carcinoma. Semin Liver Dis 25: 181–200 [DOI] [PubMed] [Google Scholar]
- 37.Akateh C, Black SM, Conteh L et al. (2019) Neoadjuvant and adjuvant treatment strategies for hepatocellular carcinoma. World J Gastroenterol 25:3704–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Traverso A, Wee L, Dekker A, Gillies R (2018) Repeatability and reproducibility of Radiomic features: a systematic review. Int J Radiat Oncol Biol Phys 102:1143–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Feng S, Wei J et al. (2019) Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol 29:4177–4187 [DOI] [PubMed] [Google Scholar]
- 40.Feng ST, Jia Y, Liao B et al. (2019) Preoperative prediction of microvascular invasion in hepatocellular cancer: a radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur Radiol 29:4648–4659 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


