Abstract
HD-ZIP is a unique type of transcription factor in plants, which are closely linked to the regulation of plant growth and development, the response to abiotic stress, and disease resistance. However, there is little known about the HD-ZIP gene family of pepper. In this study, 40 HD-ZIP family members were analyzed in the pepper genome. The analysis indicated that the introns number of Ca-HD-ZIP varied from 1 to 17; the number of amino acids was between 119 and 841; the theoretical isoelectric point was between 4.54 and 9.85; the molecular weight was between 14.04 and 92.56; most of them were unstable proteins. The phylogenetic tree divided CaHD-ZIP into 4 subfamilies; 40 CaHD-ZIP genes were located on different chromosomes, and all of them contained the motif 1; two pairs of CaHD-ZIP parallel genes of six paralogism genes were fragment duplications which occurred in 58.28~88.24 million years ago. There were multiple pressure-related action elements upstream of the start codon of the HD-Z-IP family. Protein interaction network proved to be coexpression phenomenon between ATML1 (CaH-DZ22, CaHDZ32) and At4g048909 (CaHDZ12, CaHDZ31), and three regions of them were highly homology. The expression level of CaHD-ZIP gene was different with tissues and developmental stages, which suggested that CaHD-ZIP may be involved in biological functions during pepper progress. In addition, Pepper HD-ZIP I and II genes played a major role in salt stress. CaHDZ03, CaHDZ 10, CaHDZ17, CaHDZ25, CaHDZ34, and CaHDZ35 were significantly induced in response to salt stress. Notably, the expression of CaHDZ07, CaHDZ17, CaHDZ26, and CaHDZ30, homologs of Arabidopsis AtHB12 and AtHB7 genes, was significantly upregulated by salt stresses. CaHDZ03 possesses two closely linked ABA action elements, and its expression level increased significantly at 4 h under salt stress. qRT-P-CR and transcription analysis showed that the expression of CaHDZ03 and CaHDZ10 was upregulated under short-term salt stress, but CaHDZ10 was downregulated with long-term salt stress, which provided a theoretical basis for research the function of Ca-HDZIP in response to abiotic stress.
1. Introduction
Plant transcription factors can be divided into 58 families according to the conserved domain and function [1]. Homeobox (HB) protein is a kind of transcription factor closely related to biological growth and development [2]. In 1983, Garber et al. [3] discovers the HB in Drosophila, and then, it is found in invertebrates, vertebrates, fungi, and other high plants. In 1991, Vollbrecht et al. [4] clones the HB gene with the name of Knotted-1 (Kn-1) in maize, and then, the HB gene is cloned in various plants. According to the location, differences, and homology of HD (homeodomain) sequences, plant HB proteins can be subdivided into six major categories: PHD-Finger, Bell, HD-ZIP, WOX, ZF-HD, and KNOX [5]. HD-ZIP transcription factor is a unique transcription factor in plants and plays roles in the growth, development, disease resistance, and abiotic stress [6]. These functions of HB have been reported in mass [7], ferns [8], monocotyledons [9], and dicotyledons [10, 11]. The HD-ZIP transcription factor is mainly composed of two conserved domains, HD (homeodomain) domain and a closely linked LZ (leucine zipper) domain. HD is linked to the specific binding of DNA, and Zip is related to heterodimerization [12]. According to structure and function, HD-ZIP proteins can be divided into four subfamilies: HD-ZIP I, HD-ZIP II, HD-ZIP III, and HD-ZIP IV [13].
HD-ZIP I protein is considered to be involved in the control of plant growth and development and the response to abiotic stress [14]. It is reported that there was a response of HD-ZIP I to acetic acid (ABA) at the transcript level under stress in Arabidopsis [15]. Under NaCl stress, the expressions of Gmhdz51 and Gmhdz 83 of cotton are substantially upregulated at 12 h after treatment [16]. In cotton, GhHB1 (belongs to HD-ZIP I subfamily) may be involved in salt stress and ABA treatment, and the expression of Gh-HB1 increases significantly in early stage of roots, and then decreases sharply, which suggests that HD-ZIP I played an important role in the early development of root [17]. In Arabidopsis, AtHB1 works at a downstream location, and AtPIF1 promotes hypocotyl elongation, particularly in response to short-day photoperiods [18], and mediates the apoptosis of leaf cell [19].
HD-ZIP II subfamily can induce shade avoidance reactions in plants through light signal transduction [20]. For example, when the seeds are stimulated with far-red light in the late germination period, they induce shade avoidance responses [21]. La Rota et al. [22] find that HD-ZIP III is involved in the Arabidopsis sepal development. HD-ZIP is also responsible for regulating plant cell differentiation and participating in the development of apical meristems, embryos, and vascular systems [15]. There are five family members of Arabidopsis HD-ZIP [13], due to differences in domains and expression patterns, each member plays different role [23–25]. The PCN gene in poplar belongs to the HD-ZIP subfamily and has a role in regulating xylem cell differentiation [26]. Transgenic poplars overexpressing PCN exhibited a slow deformation of wood and phloem and upregulation of endogenous hormone expression [28]. HD-ZIP IV subfamily is mainly involved in epithelial cell differentiation and root development [29]. In Arabidopsis thaliana, PDF2, ATML1, and ATHB10 are HD-ZIPIV proteins, which have a regulatory effect on the specific expression of outer cortical cells [30], and the double mutants of PDF2 and ATML1 exhibit epidermal deletion [31]. The OCL1 gene is a member of the maize HD-ZIP subfamily, and the N-terminal amino acid of its START domain plays a decisive role in the activity of this gene; overexpression can lead to delaying in flowering [32].
Pepper is a major vegetable, which is widely cultivated in the world. It is both fresh food, and processing raw material for seasoning, medicine, and cosmetics. It has very significant economic value [33]. The pepper genome has been the sequence in 2014 and can be used for gene prediction and annotation and public use [34, 35]. Several transcription factor families such as DOF, WRKY, AP2/ERF, and NAC in pepper have been studied [36–39]. HD-ZIP family genes have been reported in Arabidopsis, rice, poplar, corn, and other plants [40, 41], but so far, there is no systematic study of peppers HD-ZIP family. Our research analyzed the bioinformatic characteristics of the HD-ZIP gene family of pepper and supported a theoretical basis for studying the function of this gene in response to salt stress.
2. Materials and Methods
2.1. Genome-Wide Identification of HD-ZIP Family Genes in Pepper
We downloaded the pepper HD-ZIP protein sequence (PF00046 and PF02183) from the Pfam database (http://pfam.sanger.ac.uk/) and applied BLAST alignment to the pepper genome database PGP (http://peppergenome.snu.ac.kr/, the protein sequences annotated in CM334 and Zunla-1), the default parameters (Limit Expect Value 1e-5) output data, and the pepper CM334 was identified using the HMMER3.1 software (http://hmmer.org/). For all CaHD-ZIP proteins in the genome, the default output E value is <1 × 10 − 5, and the CaHD-ZIP sequence in the plant transcription factor database is combined to accurately obtain the target sequence [42]. With the help of NCBI-CDD (https://www.ncbi.nlm.nih.gov/ Structure/cdd/wrpsb.cgi) and SMART (http://smart.emblheidelberg.de/smart/save_user_preferences.pl) tools, protein domain identification was performed, and genes without the HD-ZIP main domain were deleted [43]. With the ExPASy-ProtParam tool (https://web.expasy.org/protparam/), the isoelectric point (pI) and molecular weight (MW) of the target sequence are predicted [44]. The online software Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) was used to predict the exon/intron structure of CaHD-ZIP [45].
2.2. Phylogenetic Analysis
To investigate the phylogenetic relationships of the HD-ZIPs among C. annuum L, O. sativa, and A. thaliana, multiple HD-ZIP protein sequences were aligned, and an unrooted phylogenetic tree was constructed in MEGA 5.05 [46]. The phylogenetic tree was constructed using the neighbor-joining (NJ) method. In the phylogenetic tree, group pattern was evaluated with bootstraps (1000 replicates).
2.3. Conserved Motif Analysis
The MEME software (http://meme-suite.org/tools/meme) was used to identify conserved motifs in CaHD-ZIP, the maximum number of motifs was 25, and other variables were the default values.
2.4. Chromosomal Location and Gene Duplication
The protein-coding sequence of CaHD-ZIP was mapped to the pepper genome database using BLASTn, and the gene was displayed on the chromosome by TBtools [47].
Plant gene duplication database (Plant Genome Duplication Database PGDD http://chibba.agtec.uga.edu/duplication/index/locus) was used to identify the gene duplication of pepper HD-ZIP gene [48]. Ka, Ks, and Ka/Ks were estimated using DnaSPV5 [49]. The Ks value of each pair is used to estimate the replication time by the following formula: replication time = Ks/2λ, where λ = 6.1 × 10−9.
2.5. Promoter Element Analysis
The upstream region (2 kb) of the CaHD-ZIP gene is derived from PGD database (http://peppergenome.snu.ac.kr). The promoter elements of CaHD-ZIP family members are predicted by the PLACE [50] (http://www.dna.affrc.go.jp/PLACE), expressed through TBtools.
2.6. Prediction of Protein-Protein Interaction Network
Based on the phylogenetic relationship between the CaHD-ZIP protein and the Arabidopsis HD-ZIP protein, 32 Arabidopsis HD-ZIP proteins which represent the 40 pepper HD-ZIP proteins are uploaded to the string website [51] (https://string-db.org/) to predict protein interactions. The online program run with default parameters.
2.7. Transcriptome Analysis of CaHD-ZIP in Different Tissues
Based on the CM334 RNA-seq [34], the expression patterns of CaHD-ZIP in different stages and tissues were analyzed; the tissues include root, stem, leaf, pericarp (PC), and placenta (PL) at 6, 16, and 25 days postanthesis (DPA), PC and PL at mature green (MG) and at breaker (B) stages, and PC and PL at 5 and 10 days postbreaker (B5 and B10, respectively).
2.8. Plant Cultivation and NaCl Stress
Healthy pepper seeds (Capsicum annuum L. var. conoides (Mill) Irish) were soaked at room temperature for 6-8 h; then, the seeds were placed in a germination box and covered with wet gauze at 28 ± 2°C. The germinated seeds were sown into a pot with substrate (Vpeat : Vvermiculite = 2 : 1). When the sixth leaves appeared, the seedlings were transferred to barral with 8 L distilled water. Each barrel had four peppers. After a week, the seedlings were treated with 100 mM NaCl solution, and those cultivated with distilled water were control. The pH was adjusted to 7.0 using H2SO4 or NaOH. The solution was replaced every three days. After the treatment of 4 h and 58 h, the young leaves were sampled and stored at -80°C until Illumina sequencing. Total RNA was extracted from the pepper leaf with RNA out 1.0 (Tianenze, Beijing, China), and RNA quality was examined by NanoDrop (Thermo Fisher Scientific, Inc.) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). cDNA library construction and Illumina RNA-Seq (HiSeq TM-2500) were carried out by Novo Gene Company (Beijing, China) [52].
2.9. Total RNA Extraction and cDNA Synthesis
After the stress of 4 h and 58 h, total RNA was extracted from pepper leaves with an RNA concentration of 1.0 (Tianenze, Beijing, China). Each group has three biological replicates. Primers were designed according to Premier 6.0 (S5). cDNA was synthesized using the reverse transcriptase MMLV kit (China ABM). Real-time quantification was performed using the CFX Manager (Bio-Rad, USA) and the SYBR Green Real-Time PCR Master Mix (Abm, Canada). The protocol of real-time PCR was as follows: predenaturation at 95°C for 10 minutes, then 40 rounds of amplification at 40°C, 15 s denaturation at 95°C, and annealed at 30°C for 30 s, extended to 72°C and read the plate to record fluorescence data at 65°C. Melting curves were performed at 65°C to 95°C to check the specificity for the amplified products. Each reaction was repeated three times. Pepper ACTIN1 was used as an internal control.
3. Results
3.1. Identification and Structure Analysis of the CaHD-ZIP Gene Family in Pepper
40 CaHD-ZIP target sequences were obtained, which were named with CaHD-ZIP01 to CaHD-ZI-P40 (Table 1). It could be seen from Table 1 that the number of amino acids in each CaHD-ZIP sequence was between 119 and 841; the theoretical isoelectric point was between 4.54 and 9.85; the molecular weight was between 14.04 and 92.56 kD; the instability coefficient results showed that except CaHDZ29 and CaHDZ32, the other 38 CaHD-ZIP family members were unstable proteins (Table 1). The core domain analysis showed that the CaHD-ZIPI subfamily had two domains, HD and LZ. Report to the CaHD-ZIPI, CaHD-ZIPII had an additional N-term. For CaHD-ZIP III and CaHD-ZIP IV subfamilies, in addition to the HD and LZ domains, they had a START domain and a MEKHLA domain (S1A). Analysis of the predicted introns and exons of the pepper HD-ZIP gene revealed that 5 genes (CaHDZ 03, CaHDZ17, CaHDZ23, CaHDZP35, and CaHDZ38) had no introns in the pepper genome. Most CaHD-ZIP genes contained 3-18 exon in the coding DNA sequence. 216 introns had 0 phases, and 1 had 2 phases (S1 B).
Table 1.
Characteristics of CaHD-ZIP genes from Zunla-1 genome and CM334 genome. AA: amino acid; pI: the theoretical isoelectric point of proteins; Mw: the theoretical molecular weight of proteins.
| Gene name | Gene ID | Chromosome or scaffold | No. of AA | pI | MW (kD) | Instability index | NO. of introns |
|---|---|---|---|---|---|---|---|
| CaHDZ01 | CA00g45190 | scaffold1113 | 841 | 5.89 | 92.36 | 52.91 | 17 |
| CaHDZ02 | CA00g57460 | scaffold1298 | 187 | 9.60 | 21.48 | 61.60 | 4 |
| CaHDZ03 | CA01g00300 | 1 | 153 | 9.68 | 17.64 | 62.20 | 0 |
| CaHDZ04 | CA01g05840 | 1 | 164 | 9.50 | 18.89 | 67.79 | 2 |
| CaHDZ05 | CA01g07870 | 1 | 775 | 5.30 | 86.33 | 57.69 | 9 |
| CaHDZ06 | CA01g17370 | 1 | 119 | 9.85 | 14.04 | 75.19 | 1 |
| CaHDZ07 | CA01g19070 | 1 | 835 | 5.86 | 91.07 | 50.23 | 8 |
| CaHDZ08 | CA01g27890 | 1 | 309 | 6.77 | 34.32 | 70.60 | 3 |
| CaHDZ09 | CA02g07270 | 2 | 241 | 9.03 | 27.00 | 48.52 | 2 |
| CaHDZ10 | CA02g08550 | 2 | 279 | 5.79 | 31.69 | 55.49 | 2 |
| CaHDZ11 | CA02g10530 | 2 | 828 | 5.93 | 91.56 | 47.30 | 17 |
| CaHDZ12 | CA02g18040 | 2 | 735 | 5.70 | 81.01 | 41.35 | 9 |
| CaHDZ13 | CA02g24650 | 2 | 272 | 8.40 | 30.28 | 49.85 | 2 |
| CaHDZ14 | CA02g28390 | 2 | 258 | 5.69 | 29.49 | 62.30 | 1 |
| CaHDZ15 | CA02g29480 | 2 | 287 | 4.82 | 32.42 | 74.16 | 3 |
| CaHDZ16 | CA03g16380 | 3 | 820 | 6.07 | 89.03 | 45.35 | 8 |
| CaHDZ17 | CA03g20530 | 3 | 135 | 9.81 | 16.29 | 55.67 | 0 |
| CaHDZ18 | CA03g23110 | 3 | 452 | 5.56 | 50.94 | 48.73 | 5 |
| CaHDZ19 | CA03g28010 | 3 | 322 | 5.03 | 37.08 | 52.26 | 2 |
| CaHDZ20 | CA03g34750 | 3 | 774 | 6.71 | 86.78 | 53.61 | 10 |
| CaHDZ21 | CA03g35060 | 3 | 838 | 6.06 | 92.05 | 47.62 | 17 |
| CaHDZ22 | CA04g03990 | 4 | 728 | 5.53 | 80.14 | 42.32 | 9 |
| CaHDZ23 | CA04g15670 | 4 | 131 | 8.56 | 15.48 | 55.19 | 0 |
| CaHDZ24 | CA04g16620 | 4 | 194 | 8.86 | 22.42 | 59.32 | 2 |
| CaHDZ25 | CA05g13390 | 5 | 284 | 4.54 | 32.23 | 50.11 | 1 |
| CaHDZ26 | CA06g08530 | 6 | 224 | 5.17 | 25.98 | 48.36 | 1 |
| CaHDZ27 | CA06g12600 | 6 | 313 | 8.13 | 35.61 | 59.61 | 3 |
| CaHDZ28 | CA06g20810 | 6 | 734 | 6.08 | 80.87 | 52.29 | 9 |
| CaHDZ29 | CA07g14020 | 7 | 211 | 9.35 | 24.26 | 37.29 | 2 |
| CaHDZ30 | CA08g08650 | 8 | 243 | 5.27 | 27.63 | 58.24 | 1 |
| CaHDZ31 | CA09g07360 | 9 | 745 | 5.48 | 82.68 | 41.08 | 9 |
| CaHDZ32 | CA10g05400 | 10 | 734 | 5.66 | 80.87 | 39.58 | 9 |
| CaHDZ33 | CA10g19210 | 10 | 321 | 8.88 | 36.14 | 58.46 | 3 |
| CaHDZ34 | CA11g05100 | 11 | 311 | 5.73 | 35.61 | 62.75 | 2 |
| CaHDZ35 | CA11g05650 | 11 | 151 | 6.33 | 17.80 | 46.65 | 0 |
| CaHDZ36 | CA11g18960 | 11 | 841 | 5.84 | 92.13 | 51.57 | 17 |
| CaHDZ37 | CA12g00830 | 12 | 716 | 5.90 | 81.57 | 45.82 | 8 |
| CaHDZ38 | CA12g03310 | 12 | 152 | 6.13 | 17.83 | 49.39 | 0 |
| CaHDZ39 | CA12g13110 | 12 | 837 | 6.04 | 92.56 | 47.05 | 17 |
| CaHDZ40 | Capana09g002322∗ | 9 | 245 | 7.77 | 28.43 | 54.10 | 2 |
3.2. Phylogenetic Analysis of CaHD-ZIP Genes
In order to examine the phylogenetic relationship between the 32 HD-ZIP transcription factors in pepper and the known members of other plants, we created a rootless development tree between Arabidopsis, rice, and pepper and implemented it in the MEGA 6 software. The phylogenetic tree implied that there are four groups of HD-ZIP, which is similar to previous studies on sesame, poplar, and corn [40, 41, 53]. A number of HD-ZIP I to HD-ZIP IV members of pepper are 14, 10, 5, and 11. The tree also showed that most CaHD-ZIP proteins move closer to members of Arabidopsis thaliana than members from rice (Figure 1). For example, in the third group, CaHD-ZIP 11 was clustered with AtPHV and AtPHB, while Oshox 32 and Oshox 33 in the third group are clustered in a single clade. These results offered an important basis for the prediction of the function of pepper HD-ZIP protein.
Figure 1.
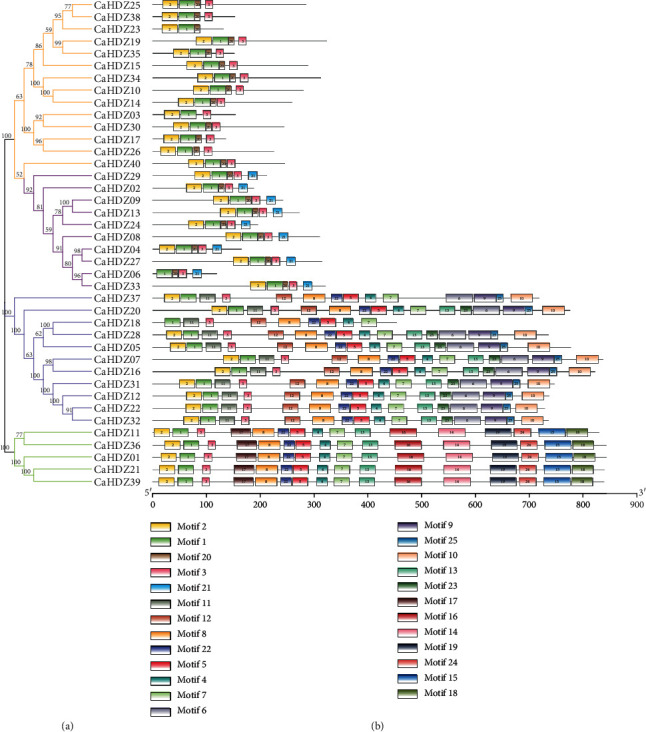
Unrooted neighbor-joining phylogenetic tree and conserved motif analysis of CaHD-ZIP proteins. (a) The phylogenetic tree was generated based on the protein sequences of CaHD-ZIP proteins. (b) Conserved motif analysis of CaHD-ZIP proteins. Different color boxes represent different types of motifs.
3.3. Analysis of Conserved Motifs of the CaHD-ZIP Gene Family
The conserved motif of CaHD-ZIP protein was further analyzed using the meme software. The software detected a total of 25 motifs in the 40 CaHD-ZIP proteins, which were designated 1 to 25 (Figure 1 and S2). As expected, all identified CaHD-ZIP proteins (except CaHDZ06, 18) contained the LZ domain (motif 3) and the HD domain (motifs 1 and 2). A START field (topic 8) was found in the members of the third and fourth groups but was not found in the first and second groups. Among the third category, Motif15 and Motif18 were found to correspond to the Mekhla domains e. In addition to these, new functional motifs, some domains with unknown functions were also discovered, for example, Motif 6, 9, 10, 11, 12, 23, and 25 (only detected members of the fourth group) and Motif 14, 17, and 19 (only found in members of the third group). The results also indicated that members of the same group of CaHD-ZIP usually had similar motifs and therefore may have similar function.
3.4. Chromosomal Locations and Syntenic Analysis
40 CaHD-ZIP genes were located on twelve chromosomes, 7 CaHD-ZIP genes located on chromosome 02 (CaHDZ09, 10, 11, 12, 13, 14 and CaHDZ15), 6 CaHD-ZIP genes were located on the long arms of chromosome 01 and 03, chromosomes 4, 6, 11, and 12 included 3 genes, respectively, and chromosomes 09 and 10 had two genes, respectively, each of chromosome of 5, 7, and 8 had one CaHD-ZIP gene, and chromosome 0 had two genes of CaHDZ01 and CaHDZ02 (Figure 2).
Figure 2.
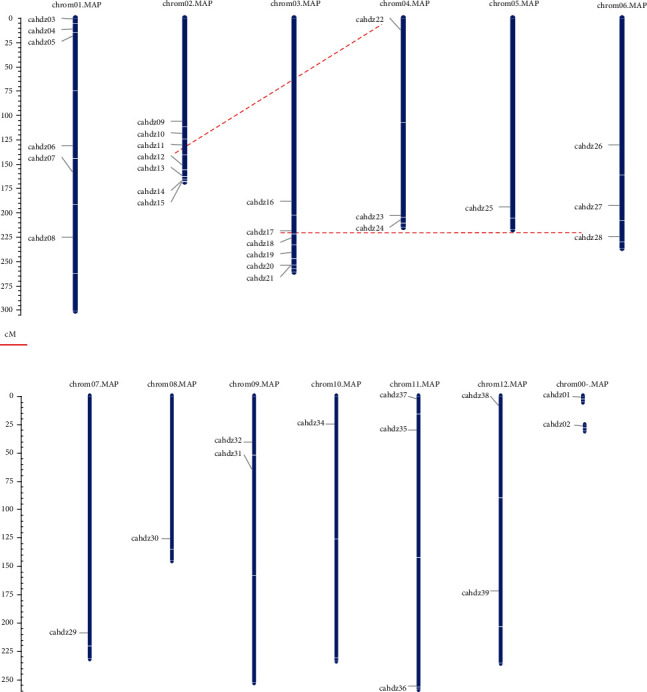
Chromosomal locations and duplication events of CaHD-ZIP genes of pepper. Locations of CaHD-ZIP were based on physical locations. The numbers on the top represent the chromosome number. The chromosome 00 (Chr00) means two different scaffolds containing unassigned CaHD-ZIP to any of the 12 pepper chromosomes. Red lines indicate 2 pairs of paralogism genome duplication events.
Analysis of HD-ZIP genes duplication of pepper showed that two pairs of paralogism genes (CaH-DZ18 and CaHDZ28 and CaHDZ12 and CaHDZ22) were fragment replication. The nonsynonymous substitution rate (Ka), the synonymous substitution rate (Ks), and the Ka/Ks ratio were shown in S3. It was reported that CaHD-ZIP's two-segment replication happened between 58.28 million and 88.24 million years ago. The Ka/Ks values of the two replication pairs were lower than 0.3, which indicated that there were no significant functional differences between these CaHD-ZIP genes after the replication event.
3.5. Promoter Analysis of the CaHD-ZIP Proteins
To further explore the possible regulation mechanism of CaHD-ZIP under various pressure, the case element in the promoter sequence of CaHD-ZIP gene was studied. The cis-elements are divided into four main subgroups: stress-response, hormone responsiveness, photosensitivity, and MYB binding sites [54] (Figure 3, S6). In our study of 32 CaHD-ZIP genes, 19 of them possessed acetic acid response elements and 13 possessed low-temperature response originals. In addition, we got a large number of light-responsive cis-elements in CaHD-ZIP, especially in CaHDZ13 gene, 19 MYB binding sites, and 12 defense stress-response elements. It was useful to noting that the wound response element was only found in three members (CaHDZ05, CaHD06, and CaHD18), and hypoxia-specific induction was only found in two members (CaHDZ06 and CaHDZ36). CaHDZ03 has two closely connected defensive stress response and ABA action elements.
Figure 3.
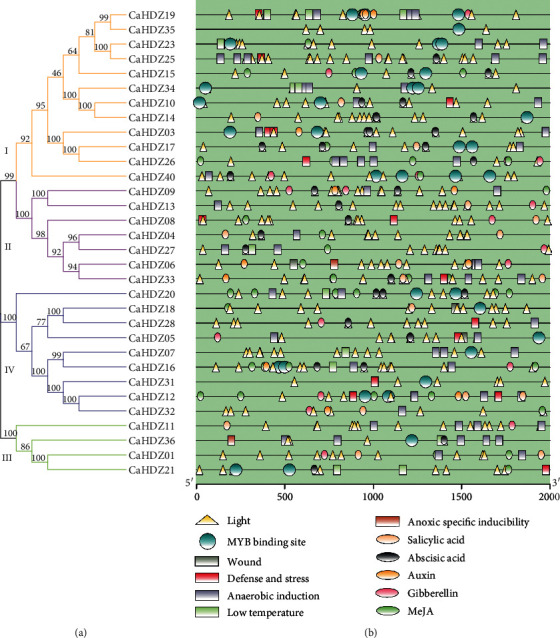
Cis analysis of CaHD-ZIP promoter related to stress response. Different cis-elements with the same or similar functions are shown in the same shape and color.
3.6. Prediction of Protein-Protein Interaction Network
In order to further understand the interaction of pepper HD-ZIP proteins, an interactive network based on Arabidopsis orthodoxy was established using STRING. The results showed that there was just one pair of ATML1 (CaHDZ22, 32) and At4g048909 (CaHDZ12, 31). There was a coexpression phenomenon, and the protein homology between the two was high. It has been confirmed by the protein two-hybrid test on Arabidopsis [55]. Three high homology regions were verified, one of them was located on HB-2 (CaHDZ27), and it had high homology to HAT22 (CaHDZ9, 13, 24, 29), HAT14 (CaHDZ06, 33), and HAT3 (CaHDZ08). The second region was centered on HB5 (CaHDZ19, 23), and it had high homology with HB6 (CaHDZ15, 25), HB16 (CaHDZ35, 38), and ATHB-7 (CaHDZ03, 17, 26, 30). The last area was a high correlation with HDG11 (CaHDZ28) and HDG4 (CaHDZ05). The relationship of other HD-ZIPs had yet to be explored (Figure 4).
Figure 4.
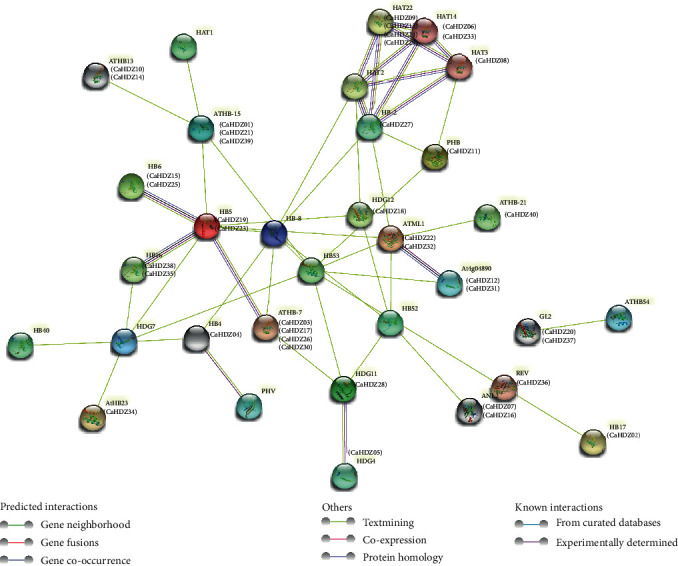
Predicted protein-protein interaction network of CaHD-ZIP proteins. The colors of the line indicate different data sources.
3.7. Expression Analysis of CaHD-ZIP Genes in Different Tissues and Development Stages
To study the role of CaHD-ZIP in pepper growth and development, we cited publicly available RNA-seq data from 5 tissues (root, stem, leaf, peel, and placenta) to generate CaHD-ZIPs heat map of transcription patterns, which included seven developmental stages of the peel and placenta (Figure 5(a), S5). The expression profile of each CaHD-ZIP gene revealed various patterns in different organs and stages. CaHDZ 04, 13, 17, 18, 24, 28, 37 had higher expression levels in all tissues; CaHDZ 01, 14, 30 had higher expression levels in roots, stems, peels, and placenta; CaHDZ11, 25 had higher expression in leaf and placentas. The expression level of CaHDZ35, 26, 05, 32 in each tissue was relatively low. The expression levels varied with the developmental stages significantly, for example, CaHDZ14 was highly expressed in PC-6DPA, PL-6DPA, and PL-16DPA, but hardly expressed in PC-B10 and PL-B5; the expression of CaHDZ30 in PC-6DPA was the highest. The expression of CaHDZ29 in PL (including PL-16DPA, PL-25DPA, PL-MG, PL-B, PL-B5, and PL-B10) was higher than that of other periods. The expression level of CaHDZ27 was higher throughout the PL period. The expression level of CaHDZ20 in PC-B, PC-B5, and PC-B10 was relatively low PL period. These results suggested that CaHD-ZIP may be involved in biological functions of pepper development.
Figure 5.
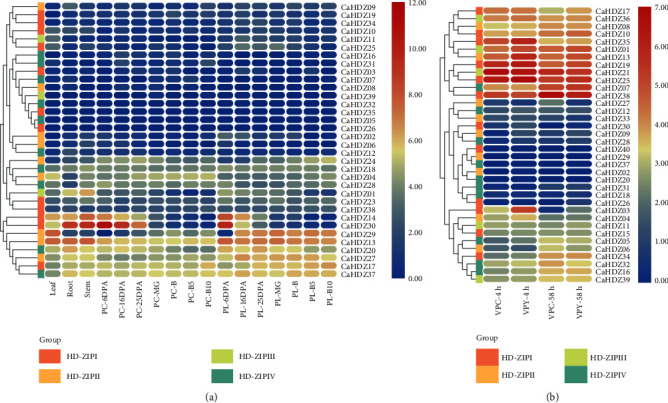
(a) Pepper HD-ZIPgenes expression of different tissues and developmental stages. Raw data were taken from RNA-seq data of CM334. The tissues included root, stem, leaf, pericarp (PC), and placenta (PL) at 6, 16, and 25 days postanthesis (DPA), PC and PL at mature green (MG) and at breaker (B) stages, PC and PL at 5 (B5) and 10 (B10) days after postbreaker, respectively. (b) The heat map of CaHD-ZIP genes under NaCl treatment (VPC represents CK, VPY represents NaCl stress). Blue and red colors represent relatively low and high expression (log 2 RPKM value).
3.8. CaHD-ZIP Expression Profile under Salt Stress
The expression pattern of CaHD-ZIP transcription factor under salt stress was obtained from the transcription data (Figure 5(b), S4). It was noted that CaHDZ 01, CaHDZ04, CaHDZ 09, CaHDZ32, CaH-DZ13, CaHDZ19, and CaHDZ35 had higher expression levels after 4 h of salt stress, and CaHDZ 03, CaH-DZ11, and CaHDZ17 had higher expression after 58 h of salt stress. Expression levels of CaHDZ 06 and Ca-HDZ27 were lower than those of the control after 4 h and 58 h of salt stress. It was useful to note that the expression level of CaHDZ03 increased significantly after 4 h of salt stress.
3.9. Expression Analysis of CaHD-ZIP Genes in Response to Salt Stresses
Based on the differential expression data of the transcription, 9 genes were selected in different subfamilies, and qRT-PCR was used to find out the role of the gene under salt stress (Figure 6). The expression levels of CaHDZ03, CaHDZ04, CaHDZ25, CaHDZ32, CaHDZ35, and CaHDZ39 were higher than that of the control after 4 h salt stress; the expression levels of CaHDZ03, CaHDZ04, CaHDZ10, C-aHDZ21, and CaHDZ33 decreased after 4 h of salt stress. The expression of the CaHDZ21, CaHDZ33, and CaHDZ32 genes decreased after 58 h of salt stress. CaHDZ03 and CaHDZ10 can be induced to increase the expression level under short-term salt stress and decreased expression level under long-term salt stress. These results suggested that these genes may play a significant role in peppers response to salt stress.
Figure 6.
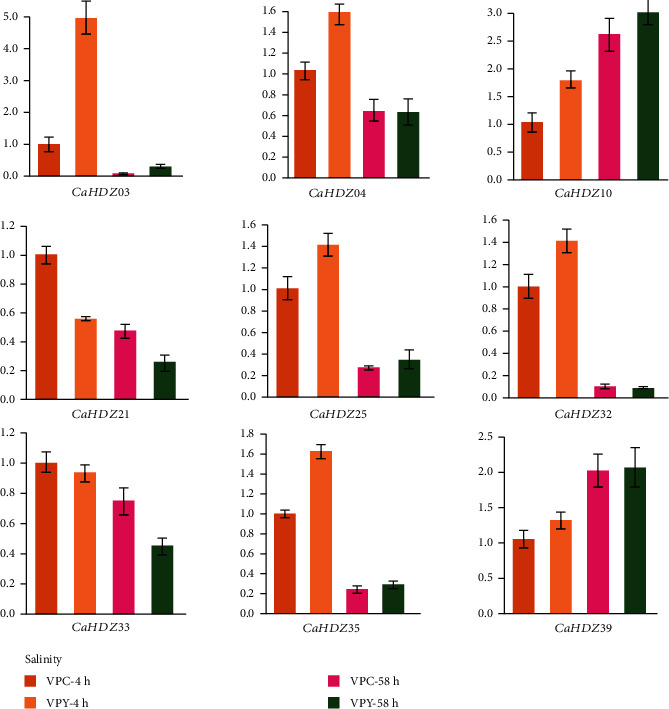
The heat map of the CaHD-ZIP genes under NaCl treatment (VPC represents CK, VPY represents NaCl stress). Expression patterns of 9 CaHDZ genes under salt stress. Salt stress (100 mM NaCl) was carried out on pepper seedlings at the six-leaf stage. The relative expression level of CaHDZ gene was analyzed by qRT-PCR, using sesame VPC4h gene as the internal control.
4. Discussion
HD-ZIP, as a plant-specific transcription factor, is closely linked to abiotic stress. HD-ZIP transcription factors are widely disseminated in different plants. HD-ZIP genes are highly conserved, but their proteins are different. This study got 40 HD-ZIP transcription factors; those factors contain a highly conserved domain, but most members are unstable proteins. We analyzed 217 introns, 216 of them have phase 0, only one of them has phase 2, and all the introns were available on both sides of the exon. This phase was called symmetrical exons. Based on the early intron hypothesis [56], excessive phase 0 introns and symmetrical exons can effectively promote exon shuffling by avoiding the interruption of the open reading frame, which could accelerate the recombination and exchange of protein domains [57]. There were a large number of phase 0 introns and symmetric exons of CaHD-ZIP genes in our study. It was corresponding with the statement and indicates that exon shuffling may be playing an important role in the evolution.
The phylogenetic tree was constructed using HDZ proteins of pepper, rice, and Arabidopsis. HDZ proteins of pepper can be divided into four categories (HD-ZIP I-IV), and the number of HD-ZIP I, II, III, and IV are 14, 10, and 5, respectively, 11, and 17, 10, 5, and 16 in Arabidopsis [58]; 12, 9, 4, and 7 in physic nut [59]; 11, 7, 5, and 8 in grape [60]; 16, 10, 9, and 10 in sesame [53]. These results suggested that the HD-ZIPI protein was the most abundant type of pepper HD-ZIP transcription factor, and the amount of HD-ZIPIII protein in pepper was the same as that of most plants. In addition, the phylogenetic tree also showed that most CaHDZ proteins were closer to members of Arabidopsis thaliana than members from rice [59] (Figure 7. Motif analysis also demonstrated that CaHDZ protein motifs distributed differently with subfamilies, and genes of the same family had similar structure and function (Figure 1, S2), which provided a powerful guarantee for the evolution of pepper. Gene duplication is the highest evolutionary mechanism that helps plants adapt to various environmental stresses [15]. Compared with 41 pairs of soybean [16] and 10 pairs of cassava [61] paralogism HD-ZIP genes, there were only two pairs of pepper, CaHZ18 and CaHDZ28 and CaHDZ12 and CaHDZ22; those genes distributed on different chromosomes. Paralogous genes originated from the partial duplication of CaHD-ZIP between chromosomes. Replication of two fragments occurred between 5828 and 88.24 million years ago. The genomes of most polyploid plants contain a large number of repetitive chromosome segments, so the probability of partial repetitions is significantly higher than that of tandem repeats and translocations [62]. It speculated that the evolution of the HD-ZIP family of pepper was slow.
Figure 7.
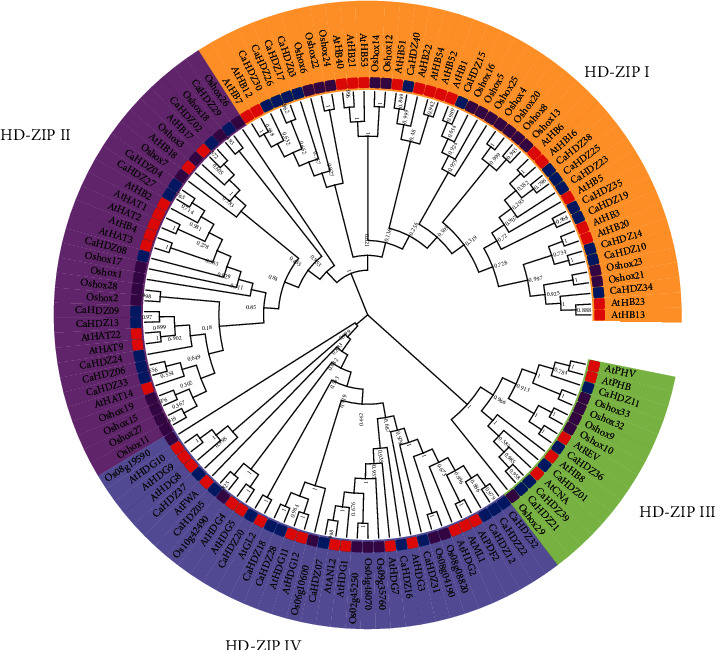
Phylogenetic trees of 132 HD-ZIP proteins in pepper (blue square), Arabidopsis (red square), and rice (purple square). The HD-ZIP gene is divided into four subfamilies (groups I, II, III, and IV, with yellow, purple, green, and blue branches, respectively).
More and more evidence shows that the HD-ZIP gene is associated with plant growth and development [16, 40, 53]. For example, AtHB2 regulates the shade response of red light/far red light, which is linked to the formation of lateral roots. In sesame, HD-ZIP I, II, and III are widely expressed in all tissues [53], and similar phenomena had been found in pepper, but the number of CaHDZ genes expressed was less than that of sesame. In addition, we also found that the CaHD-ZIPIV gene in pepper showed a clear tissue-specific expression pattern (Figure 5). For instance, CaHDZ28 was hardly expressed in roots but was expressed in other tissues; the expression of CaHDZ12 was opposite to that of CaHDZ28. HD-ZIP IV gene is highly expressed in young leaves and flowers of tomato [63]. These results indicated that different subgroups of HD-ZIP genes have distinct functions. Members of the CaHD-ZIP family have multiple pressure-related action elements upstream of the start codon. These case elements are divided into four main subgroups: stress-response, hormone responsiveness, photosensitivity, and MYB binding site [54]. ABA response element, low-temperature response element (LTRE), and dehydration response element (Dre) are the main transcription factors regulating ABA signal transduction and participate in salt and drought stress [64]. CaHD-ZIP gene contained ABA signal transduction, and the greater the number of related action elements, the higher the gene expression level under salt stress. A lot of evidence shows that HD-ZIP I protein is involved in developmental reprogramming, c and coping with environmental pressure [65]. For example, AtHB7 and AtHB12 can act as negative feedback on the effect of ABA signal in plants under water shortage [65, 66]. Some of the HD-ZIP I genes in pepper, such as CaHDZ03, 10, 17, 25, 34, and 35, were significantly induced in response to salt stress. Notably, the expression of CaHDZ07, 17, 26, and 30, homologs of Arabidopsis AtHB12 and AtHB7 genes, was markedly upregulated by salinity stresses, indicating that these genes may regulate drought and salt tolerance through an ABA-dependent pathway. In particular, CaHDZ03 possessed two closely linked defense stress responses and ABA action elements, and its expression level increased significantly at 4 h under salt stress. The correlation analysis of qRT-PCR (Figure 6) and transcription data (S4) showed that the expression of CaHDZ03 gene can be upregulated under short-term salt stress. In addition, we found that the defense stress-response may also be related to those gene expressions which contain defense stress-response elements. CaHDZ03, CaHDZ19, CaHDZ25, and CaHDZ26 had higher expressions than other genes under salt stress; CaHDZ10 was increased under short-term salt stress but decreased under long-term salt stress.
5. Conclusion
In the present study, we identified 40 HD-ZIP genes in pepper, including four types, which were unevenly distributed on 12 chromosomes. Syntenic analysis showed that CaHDZ18, CaHDZ28, CaHDZ12, and CaHDZ22 are fragment duplication. Two fragment duplications occurred at 58.28~88.24 million years ago. There were multiple upstream of the start codon of HD-ZIP family members. There was coexpression between ATML1 (CaHDZ22, CaHDZ32) and At4g048909 (CaHDZ12, CaHDZ31), and there were three regions with high homology of them. Expressions of the CaHD-ZIP gene ranged with plant tissue and developmental stage. The HD-ZIPI responded more significantly to salt stress than other subfamilies.
Acknowledgments
This work was partially supported by the National Natural Science Foundation of China (31860548 and 32060676), the Shihezi University Breeding Project (KX0301).
Data Availability
The attached table contains all the data used to fund the results of this study. Transcription data under salt stress has not been uploaded to the NCBI because this article has not been published, but S4 contains the data needed for this article. The qRT-PCR data used to support the findings of this study are included within the supplementary information file (Table S7).
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Supplementary Materials
Table S1: the main domain and gene structure information of pepper HD-ZIP protein sequence.
Table S2: conserved motifs of CaHD-ZIP family proteins, used to analyze the conserved sequences of pepper.
Table S3: the Ka/Ks ratios for CaHD-ZIPproteins which calculate using DnaSPV5.
Table S4: transcriptome data (including raw data for analysis of tissue and expression under salt stress).
Table S5: the specific primer for qRT-PCRof each CaHD-ZIP gene.
Table S6: pepper HD-ZIP gene promoter function element.
Table S7: correlation between HD-ZIP gene RNA-seq data and qRT-PCR of 9 pepper.
References
- 1.Jin J., Zhang H., Kong L., Gao G., Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research. 2013;42(D1):D1182–D1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schena M., Davis R. W. HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(9):3894–3898. doi: 10.1073/pnas.89.9.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garber R. L., Kuroiwa A., Gehring W. J. Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila. The EMBO Journal. 1983;2(11):2027–2036. doi: 10.1002/j.1460-2075.1983.tb01696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vollbrecht E., Veit B., Sinha N., Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350(6315):241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- 5.Ariel F. D., Manavella P. A., Dezar C. A., Chan R. L. The true story of the HD-ZIP family. Trends in Plant Science. 2007;12(9):419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Sen S., Chakraborty J., Ghosh P., Basu D., Das S. Chickpea WRKY70 regulates the expression of a homeodomain-leucine zipper (HD-ZIP) I transcription factor CaHDZ12, which confers abiotic stress tolerance in transgenic tobacco and chickpea. Plant and Cell Physiology. 2017;58(11):1934–1952. doi: 10.1093/pcp/pcx126. [DOI] [PubMed] [Google Scholar]
- 7.Sakakibara K., Nishiyama T., Kato M., Hasebe M. Isolation of homeodomain–leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain–leucine zipper genes in land plants. Molecular Biology and Evolution. 2001;18(4):491–502. doi: 10.1093/oxfordjournals.molbev.a003828. [DOI] [PubMed] [Google Scholar]
- 8.Aso K., Kato M., Banks J. A., Hasebe M. Characterization of homeodomain-leucine zipper genes in the fern Ceratopteris richardii and the evolution of the homeodomain-leucine zipper gene family in vascular plants. Molecular Biology and Evolution. 1999;16(4):544–552. doi: 10.1093/oxfordjournals.molbev.a026135. [DOI] [PubMed] [Google Scholar]
- 9.Meijer A. H., Scarpella E., Dijk E. L., et al. Transcriptional repression by Oshox1, a novel homeodomain leucine zipper protein from rice. The Plant Journal. 1997;11(2):263–276. doi: 10.1046/j.1365-313X.1997.11020263.x. [DOI] [PubMed] [Google Scholar]
- 10.Deng X., Phillips J., Bräutigam A., et al. A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses. Plant Molecular Biology. 2006;61(3):469–489. doi: 10.1007/s11103-006-0023-x. [DOI] [PubMed] [Google Scholar]
- 11.Mattsson J., Söderman E., Svenson M., Borkird C., Engström P. A new homeobox-leucine zipper gene from Arabidopsis thaliana. Plant Molecular Biology. 1992;18(5):1019–1022. doi: 10.1007/BF00019223. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura M., Katsumata H., Abe M., et al. Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiology. 2006;141(4):1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sessa G., Steindler C., Morelli G., Ruberti I. The Arabidopsis ATHB-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Molecular Biology. 1998;38(4):609–622. doi: 10.1023/A:1006016319613. [DOI] [PubMed] [Google Scholar]
- 14.Mao H., Yu L., Li Z., Liu H., Han R. Molecular evolution and gene expression differences within the HD-ZIP transcription factor family of Zea mays L. Genetica. 2016;144(2):243–257. doi: 10.1007/s10709-016-9896-z. [DOI] [PubMed] [Google Scholar]
- 15.Henriksson E., Olsson A. S. B., Johannesson H., et al. Homeodomain Leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiology. 2005;139(1):509–518. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X., Chen Z., Zhao H., Zhao Y., Cheng B., Xiang Y. Genome-wide analysis of soybean HD-ZIP gene family and expression profiling under salinity and drought treatments. PLoS One. 2014;9(2, article e87156) doi: 10.1371/journal.pone.0087156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni Y., Wang X., Li D., Wu Y., Xu W., Li X. Novel cotton homeobox gene and its expression profiling in root development and in response to stresses and phytohor-mones. Acta Biochimica et Biophysica Sinica. 2008;40(1):78–84. doi: 10.1111/j.1745-7270.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- 18.Capella M., Ribone P. A., Arce A. L., Chan R. L. Arabidopsis thaliana HomeoBox 1 (AtHB1), a homedomain-leucine zipper I (HD-ZIP I) transcription factor, is regulated by PHYTOCHROME-INTERACTING FACTOR 1 to promote hypocotyl elongation. The New Phytologist. 2015;207(3):669–682. doi: 10.1111/nph.13401. [DOI] [PubMed] [Google Scholar]
- 19.Aoyama T., Dong C. H., Wu Y., et al. Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell. 1995;7(11):1773–1785. doi: 10.1105/tpc.7.11.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turchi L., Baima S., Morelli G., Ruberti I. Interplay of HD-ZIP II and III transcription factors in auxin-regulated plant development. Journal of Experimental Botany. 2015;66(16):5043–5053. doi: 10.1093/jxb/erv174. [DOI] [PubMed] [Google Scholar]
- 21.Ilegems M., Douet V., Meylan-Bettex M., et al. Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development. 2010;137(6):975–984. doi: 10.1242/dev.047662. [DOI] [PubMed] [Google Scholar]
- 22.La Rota C., Chopard J., Das P., et al. A data-driven integrative model of sepal primordium polarityin Arabidopsis. The Plant Cell. 2012;23(12):4318–4333. doi: 10.1105/tpc.111.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prigge M. J., Otsuga D., Alonso J. M., Ecker J. R., Drews G. N., Clark S. E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. The Plant Cell. 2005;17(1):61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green K. A., Prigge M. J., Katzman R. B., Clark S. E. CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. The Plant Cell. 2005;17(3):691–704. doi: 10.1105/tpc.104.026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McConnell J. R., Emery J., Eshed Y., Bao N., Bowman J., Barton M. K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411(6838):709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 26.Crewdson E. Molecular Investigations into Cambium Differentiation in Tree Stems. Melbourne School of Land & Environment Forest & Ecosystem Science; 2010. [Google Scholar]
- 27.Du J., Miura E., Robischon M., Martinez C., Groover A. The Populus Class III HD ZIP Transcription Factor POPCORONA affects cell differentiation during secondary growth of woody stems. PloS One. 2011;6(2, article e17458) doi: 10.1371/journal.pone.0017458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J., Jung J. H., Reyes J. L., et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. The Plant Journal. 2005;42(1):84–94. doi: 10.1111/j.1365-313X.2005.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vernoud V., Laigle G., Rozier F., Meeley R. B., Perez P., Rogowsky P. M. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. The Plant Journal. 2009;59(6):883–894. doi: 10.1111/j.1365-313X.2009.03916.x. [DOI] [PubMed] [Google Scholar]
- 30.Kamata N., Sugihara A., Komeda Y., Takahashi T. Allele-specific effects of PDF2 on floral morphology in Arabidopsis thaliana. Plant Signaling & Behavior. 2013;8(12, article e27417) doi: 10.4161/psb.27417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javelle M., Klein-Cosson C., Vernoud V., et al. Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: preferential expression in the epidermis. Plant Physiology. 2011;157(2):790–803. doi: 10.1104/pp.111.182147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Depège-Fargeix N., Javelle M., Chambrier P., et al. Functional characterization of the HD-ZIP IV transcription factor OCL1 from maize. Journal of Experimental Botany. 2011;62(1):293–305. doi: 10.1093/jxb/erq267. [DOI] [PubMed] [Google Scholar]
- 33.Fillion J., Sauve F., Selwyn J. Multiresidue method for the determination of residues of 251 pesticides in fruits and vegetables by gas chromatography/mass spectrometry and liquid chromatography with fluorescence detection. Journal of AOAC International. 2000;83(3):698–713. doi: 10.1093/jaoac/83.3.698. [DOI] [PubMed] [Google Scholar]
- 34.Kim S., Park M., Yeom S.-I., et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nature Genetics. 2014;46(3):270–278. doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- 35.Qin C., Yu C., Shen Y., et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proceedings of the National Academy of Sciences. 2014;111(14):5135–5140. doi: 10.1073/pnas.1400975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Z., Cheng J., Cui J., et al. Genome-wide identification and expression profile of Dof transcription factor gene family in pepper (Capsicum annuum L) Frontiers in Plant Science. 2016;7(574) doi: 10.3389/fpls.2016.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diao W.-P., Snyder J. C., Wang S.-B., et al. Genome-wide identification and expression analysis of WRKY gene family in Capsicum annuum L. Frontiers in Plant Science. 2016;7(211, article 1727) doi: 10.3389/fpls.2016.00211. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Jin J. H., Wang M., Zhang H. X., et al. Genome-wide identification of the AP2/ERF transcription factor family in pepper (Capsicum annuum L) Genome. 2018;61(9):663–674. doi: 10.1139/gen-2018-0036. [DOI] [PubMed] [Google Scholar]
- 39.Diao W., Snyder J., Wang S., et al. Genome-wide analyses of the NAC transcription factor gene damily in pepper (Capsicum annuum L.): chromosome location, phylogeny, structure, expression patterns, cis-elements in the promoter, and interaction network. International Journal of Molecular Sciences. 2018;19(4, article 1028) doi: 10.3390/ijms19041028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y., Zhou Y., Jiang H., et al. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-ZIP gene family in maize. PLoS One. 2011;6(12, article e28488) doi: 10.1371/journal.pone.0028488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu R., Chi X., Chai G., et al. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine Zipper gene family in poplar (Populus trichocarpa) PLoS One. 2012;7(2, article e31149) doi: 10.1371/journal.pone.0031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo M., Liu J.-H., Ma X., Zhai Y.-F., Gong Z.-H., Lu M.-H. Genome-wide analysis of the Hsp 70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Science. 2016;252:246–256. doi: 10.1016/j.plantsci.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Letunic I., Copley R. R., Schmidt S., et al. SMART 4.0: towards genomic data integration. Nucleic Acids Research. 2004;32(1):142–144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Research. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo A. Y., Zhu Q. H., Chen X., Luo J. C. GSDS: a gene structure display server. Hereditas. 2007;29(8):1023–1026. [PubMed] [Google Scholar]
- 46.Horton P., Park K.-J., Obayashi T., et al. WoLF PSORT: protein localization predictor. Nucleic Acids Research. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C., Xia R., Chen H., He Y. TBtools: an Integrative toolkit developed for interactive analyses of big biological data. Molecular Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 48.Tang H., Wang X., Bowers J. E., Ming R., Alam M., Paterson A. H. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Research. 2008;18(12):1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Padmalatha K. V., Dhandapani G., Kanakachari M., et al. Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Molecular Biology. 2012;78(3):223–246. doi: 10.1007/s11103-011-9857-y. [DOI] [PubMed] [Google Scholar]
- 50.Lescot M., Déhais P., Thijs G., et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong S., Joung J. G., Zheng Y., et al. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harbor Protocols. 2011;2011(12):940–949. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 53.Wei M., Liu A., Zhang Y., et al. Genome-wide characterization and expression analysis of the HD-ZIP gene family in response to drought and salinity stresses in sesame. BMC Genomics. 2019;20(1):p. 748. doi: 10.1186/s12864-019-6091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu X., Hao C., Cheng Z.-M., Zhong Y. Genome-wide identification, characterization, and expression analysis of the grapevine superoxide dismutase (SOD) family. International Journal of Genomics. 2019;20:13. doi: 10.1155/2019/7350414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rombolá-Caldentey B., Rueda-Romero P., Iglesias-Fernández R., Carbonero P., Oñate-Sánchez L. Arabidopsis DELLA and two HD-ZIP transcription factors regulate GA signaling in the epidermis through the L1 box cis-element. The Plant Cell. 2014;26(7):2905–2919. doi: 10.1105/tpc.114.127647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilbert W. Cold Spring Harbor symposia on quantitative biology. Vol. 52. Cold Spring Harbor Laboratory Press; 1987. The exon theory of genes; pp. 901–905. [DOI] [PubMed] [Google Scholar]
- 57.Patthy L. Intron-dependent evolution: preferred types of exons and introns. FEBS Letters. 1987;214(1):1–7. doi: 10.1016/0014-5793(87)80002-9. [DOI] [PubMed] [Google Scholar]
- 58.Mukherjee K., Brocchieri L., Bürglin T. R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Molecular Biology and Evolution. 2009;26(12):2775–2794. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Y., Wang J., Bao X., et al. Genome-wide identification and expression profile of HD-ZIP genes in physic nut and functional analysis of the JcHDZ16 gene in transgenic rice. BMC Plant Biology. 2019;19(1):p. 298. doi: 10.1186/s12870-019-1920-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang H., Jin J., Liu H., et al. Genome-wide analysis of HD-ZIP genes in grape (Vitis vinifera) Tree Genetics & Genomes. 2015;11(1):p. 827. [Google Scholar]
- 61.Ding Z., Fu L., Yan Y., et al. Genome-wide characterization and expression profiling of HD-ZIP gene family related to abiotic stress in cassava. PLoS One. 2017;12(3, article e0173043) doi: 10.1371/journal.pone.0173043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cannon S. B., Mitra A., Baumgarten A., Young N. D., May G. The roles of segmental and tandem gene duplication in theevolution of large gene families in Arabidopsis thaliana. BMC Plant Biology. 2004;4:p. 10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao Y., Gao S., Xiong C., et al. Comprehensive analysis and expression profile of the homeodomain leucine Zipper IV transcription factor family in tomato. Plant Physiology and Biochemistry. 2015;96:141–153. doi: 10.1016/j.plaphy.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida T., Fujita Y., Sayama H., et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. The Plant Journal. 2010;61(4):672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 65.Gong S., Ding Y., Hu S., Ding L., Chen Z., Zhu C. The role of HD-ZIP class I transcription factors in plant response to abiotic stresses. Physiologia Plantarum. 2019;167:516–525. doi: 10.1111/ppl.12965. [DOI] [PubMed] [Google Scholar]
- 66.Valdes A. E., Overnas E., Johansson H., Rada-Iglesias A., Engstrom P. The homeodomain-leucine zipper (HD-ZIP) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Molecular Biology. 2012;80(4-5):405–418. doi: 10.1007/s11103-012-9956-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: the main domain and gene structure information of pepper HD-ZIP protein sequence.
Table S2: conserved motifs of CaHD-ZIP family proteins, used to analyze the conserved sequences of pepper.
Table S3: the Ka/Ks ratios for CaHD-ZIPproteins which calculate using DnaSPV5.
Table S4: transcriptome data (including raw data for analysis of tissue and expression under salt stress).
Table S5: the specific primer for qRT-PCRof each CaHD-ZIP gene.
Table S6: pepper HD-ZIP gene promoter function element.
Table S7: correlation between HD-ZIP gene RNA-seq data and qRT-PCR of 9 pepper.
Data Availability Statement
The attached table contains all the data used to fund the results of this study. Transcription data under salt stress has not been uploaded to the NCBI because this article has not been published, but S4 contains the data needed for this article. The qRT-PCR data used to support the findings of this study are included within the supplementary information file (Table S7).


