Abstract
SARS-CoV-2 main protease is one of the major targets in drug development efforts against Covid-19. Even though several structures were reported to date, its dynamics is not understood well. In particular, impact of dimerization and ligand binding on the dynamics is an important issue to investigate. In this study, we performed molecular dynamics simulations of SARS-CoV and SARS-CoV-2 main proteases to investigate influence of dimerization on the dynamics by modeling monomeric and dimeric apo and holo forms. The dimerization causes an organization of the interdomain dynamics as well as some local structural changes. Moreover, we investigated impact of a peptide mimetic (N3) on the dynamics of SARS-CoV and SARS-CoV-2 Mpro. The ligand binding to the dimeric forms causes some key local changes at the dimer interface and it causes an allosteric interaction between the active sites of two protomers. Our results support the idea that only one protomer is active on SARS-CoV-2 due to this allosteric interaction. Additionally, we analyzed the molecular dynamics trajectories from graph theoretical perspective and found that the most influential residues – as measured by eigenvector centrality – are a group of residues in active site and dimeric interface of the protease. This study may form a bridge between what we know about the dynamics of SARS-CoV and SARS-CoV-2 Mpro. We think that enlightening allosteric communication of the active sites and the role of dimerization in SARS-CoV-2 Mpro can contribute to development of novel drugs against this global health problem as well as other similar proteases.
Communicated by Ramaswamy H. Sarma
Keywords: Allostery, Covid-19, dimerization, eigenvector centrality, linear mutual information, main protease (3C-like protease), molecular dynamics, N3, SARS-CoV, SARS-CoV-2
Introduction
Covid-19 is a global health problem that has been affecting the entire world since the end of 2019 (China Novel Coronavirus Investigating and Research Team, 2020). About 66 million individuals were diagnosed with Covid-19 and the disease caused death of about 1.5 million people as of 8 December 2020 (https://covid19.who.int/). Genome of epidemiological agent of Covid-19, namely SARS-CoV-2 virus, contains two proteases that help it replicate in host cells. These proteases are 3C-like protease and papain like protease. 3C-like protease, also known as main protease (Mpro), is a major drug target since human beings do not have any homolog of it.
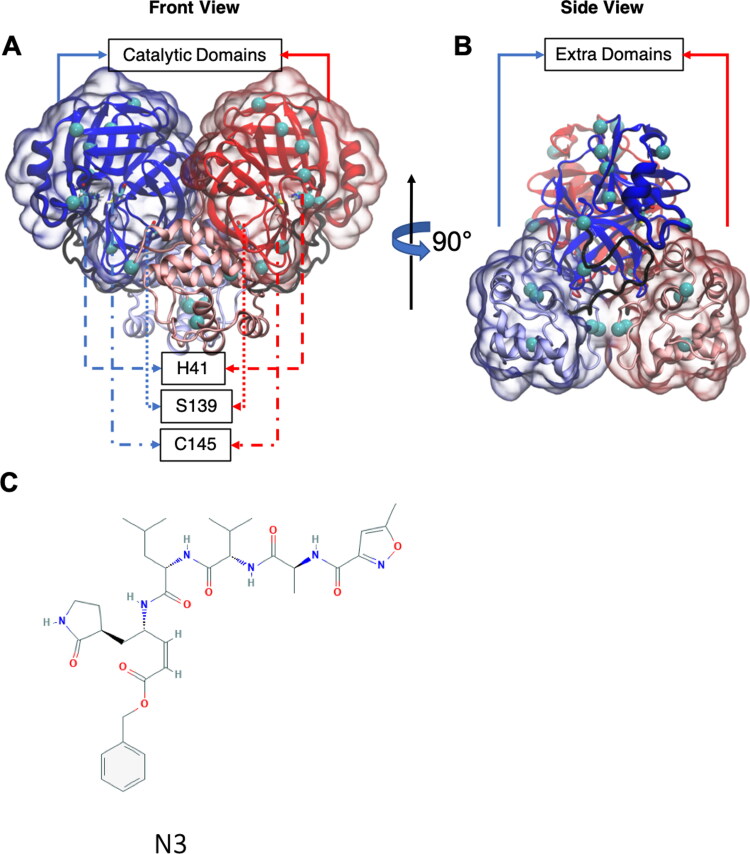
SARS-CoV-2 Mpro is a homodimer and each protomer contains 306 residues. Each protomer has two major domains: catalytic domain (residues 1 to 180) and extra-domain (residues 200 to 306). Active site residues H41 and C145 are sandwiched between two subdomains of the catalytic domain. The major domains are connected with a flexible region of 19 amino acids and the domains are oriented in an approximately tetragonal molecular geometry (Figure 1(A,B)). Even though SARS-CoV Mpro is not different structurally from the SARS-CoV-2 Mpro, there are 14 mutations at various locations (See Figure 1 and Supporting Information Figure S1).
Figure 1.
SARS-CoV-2 Mpro given in New Cartoon representation. Coloring is according to chain IDs (or protomers): A, blue; B, red. Each protomer contains two major domains. Catalytic domain and extra domain were highlighted with transparent surface representation. The residues connecting the catalytic and extra domains is shown in black tube representation. Residue 139 and catalytic dyad residues (H41 and C145) are presented in Licorice representation. SARS-CoV Mpro differs from SARS-CoV-2 Mpro at the mutation sites highlighted as cyan VDW spheres. (A) Front view. (B) Side view. (C) 2D structure of N3. All protein visualizations were produced with VMD (Humphrey et al., 1996).
Several experimental structures of SARS-CoV-2 Mpro were reported to date but most of the structures, even the ones with mutations, are not significantly different (> 3 Å RMSD) from each other. Even though we have a lot of structural information about SARS-CoV-2 Mpro, the role of its dynamics and role of dimerization is not well-understood. Understanding impact of dimerization on the dynamics of SARS-CoV-2 Mpro can open new venues for potential therapeutics (Goyal & Goyal, 2020). In fact, several experimental and computational studies were conducted to understand influence of dimerization on SARS-CoV Mpro (Chen et al., 2006, 2008; Shi et al., 2008; Shi & Song, 2006; Wei et al., 2006; Zheng et al., 2007). However, our knowledge on impact dimerization on structure and dynamics of SARS-CoV-2 Mpro is still quite limited.
Another issue that may have a potential to open new therapeutic venues is investigation of allosteric interactions and their relations to the dynamics of proteases (Gunasekaran et al., 2004; Lee, 2015; Sheik Amamuddy, Veldman, et al., 2020). Indeed, there are some studies on allosteric interactions of SARS-CoV Mpro and their relation to its dynamics (Barrila et al., 2006; Lim et al., 2014, 2019). On the other hand, there is only a single coarse-grained elastic network model study that investigate allosteric interactions SARS-CoV-2 Mpro (Dubanevics & McLeish, 2021). As we will show later, dynamically driven allosteric interactions may exist between side chain atoms as well as representative atoms like Cα atoms. Therefore, an atomic level investigation can reveal valuable information in this front.
Molecular dynamics (MD) simulation method is an established computational method to understand dynamics of proteins in different temporal (from picoseconds to microseconds) and spatial domains (from individual atoms to multiple subunits; Karplus & McCammon, 2002; Adcock & McCammon, 2006). For SARS-CoV-2, several MD studies were conducted to investigate impact of the dynamics but most of them were short simulations aiming to identify potential compounds against SARS-CoV-2 (Alamri et al., 2020; Alazmi & Motwalli, 2020; Ghosh et al., 2020; Khan et al., 2020; Kumar et al., 2020; Liang et al., 2020; Tahir Ul Qamar et al., 2020). The dynamics of SARS-CoV and SARS-CoV-2 Mpro need to be enlightened further from different perspectives. For example, how dimerization can affect the dynamics is an important and open research question to investigate because mutations can render several inhibitors targeting only active sites (Kurt Yilmaz et al., 2016). There is only a single computational study focused on the impact of the dimerization SARS-CoV-2 Mpro. Though quite informative, that study lacks a comparison of the dynamics of SARS-CoV Mpro and SARS-CoV-2 Mpro from the dimerization perspective (Suarez & Diaz, 2020). Additionally, impact of peptide or ligand binding on the dynamics is another issue that is worthy of further research. Enlightening these issues and revealing allosteric interactions can have a broad impact not just on Covid-19 but on all viral diseases that utilize similar proteases and mechanisms for replication. More importantly, it is necessary to compare the dynamical similarities and differences between SARS-CoV and SARS-CoV-2 to see how much they are similar in terms of their dynamics. In this way, we can form a bridge between SARS-CoV and SARS-CoV-2 Mpro dynamics. As a result, we can see which information on SARS-CoV Mpro dynamics is transferrable to SARS-CoV-2 Mpro.
Due to the reasons explained in the previous paragraphs, we performed atomistic MD simulations of SARS-CoV Mpro and SARS-CoV-2 Mpro to understand the impact of dimerization and ligand binding on the dynamics and structures. We used a peptide mimetic called N3 to investigate impact of ligand binding (Figure 1(C)). This peptide mimetic was shown experimentally to bind several proteases including SARS-CoV and SARS-CoV-2 (Cui et al., 2019; Jin et al., 2020; Li et al., 2007; Wang et al., 2016, 2017; Xue et al., 2007).
We performed the following simulations to do a systematic investigation: At first, we performed one microsecond-long MD simulations of monomeric and dimeric apo forms of SARS-CoV and SARS-CoV-2 Mpro. Then, we carried out one microsecond-long MD simulations of the monomeric and the dimeric forms with N3 to analyze how ligation affects the dynamics of SARS-CoV and SARS-CoV-2 Mpro. After these simulations, we performed a local analysis of catalytic dyad residues (H41 and C145) and residue 139, which is a key residue at the interface of the dimers. Finally, we analyzed linear correlations with linear mutual information (LMI) to identify impact of dimerization. We utilized the LMI maps to build a network with graph theory and tried to identify residues sensitive to effector binding using eigenvector centrality obtained from this network.
Materials and methods
Materials
PDB structure 6y2e was used to model apo state of SARS-CoV-2 Mpro (Zhang et al., 2020). PDB structure 6lu7, which contains also the coordinates of N3, was used as starting structure to investigate dynamics of the holo state of SARS-CoV-2 Mpro (Jin et al., 2020). For SARS-CoV Mpro, PDB structure 1q2w was used to model apo state and 2hob was used to model N3 bound state of SARS-CoV Mpro (Xue et al., 2007). All PDB structures contained only a monomer in the asymmetric unit. The biological unit and the symmetry operations therein were used to obtain the dimers. The first chain and the second chain were named as protomer A and protomer B in all simulations, respectively (Figure 1).
Methods
Parameterization of N3
Antechamber (Wang et al., 2006) was used to calculate the AM1 BCC partial charges (Jakalian et al., 2002) of N3 with GAFF (Wang et al., 2004) and the topology file of N3 for GROMACS was obtained with Acpype tool (Sousa da Silva & Vranken, 2012).
Molecular dynamics simulations
Both monomeric and dimeric systems were solvated in a rhombic dodecahedral box with periodic boundary conditions and a minimum distance of 12 Ȧ between the solvent and the box. The sodium and chloride ions were added to neutralize the net charge of the systems at a concentration 0.15 M. TIP3P water model was used to model the interactions of water molecules and AMBER99SB-ILDN force field was preferred for the proteins in all simulations (Lindorff-Larsen et al., 2010; Jorgensen et al., 1983). System compositions for all simulations are given in Supporting Information Table S1.
All MD simulations were performed with GROMACS 2018 (Pall et al., 2015; Pronk et al., 2013; Van Der Spoel et al., 2005). After minimizations, the systems were equilibrated in NVT ensemble for 1 ns. After 1 ns-long NPT equilibrations, 1 microsecond-long production runs in identical conditions to the NPT run were carried out. The pressure was set to 1 bar and the simulation temperature was set to 298.15 K. The pressure was controlled with Berendsen barostat while the temperature was controlled with velocity rescaling algorithm during NPT simulations (Saint-Martin et al., 2004). Particle mesh Ewald method was used to model the electrostatic interactions (Darden et al., 1993; Essmann et al., 1995). The bond lengths and angle of water molecules were constrained with SETTLE algorithm and LINCS algorithm was used to constraint the hydrogen bonds (Hess, 2008; Hess et al., 1997; Miyamoto & Kollman, 1992). Time-step was 2 fs in all simulations and the trajectories were sampled every 200 ps. Production runs were also under isobaric-isothermal ensemble but Parrinello-Rahman barostat (Parrinello & Rahman, 1981) was utilized during the production runs. All simulations were triplicated with a different initial random seed to investigate reproducibility of the results.
Linear mutual information
LMI is a parameter widely used to investigate linear correlations of protein fluctuations. Unlike Pearson correlations, it does not suffer from the angular dependency problem (Lange & Grubmuller, 2006). To calculate LMI between residues i and j, one has to calculate and where Here, Ri denotes the position vector of atom i. Inserting Ci, Cj and Cij into the following equation
gives us pairwise LMI. LMI between a residue pair is 0 when there is no correlation, and it is 1 when they are completely correlated. g_correlation program developed by Lange and Grubmuller was used to calculate pairwise LMI (Lange & Grubmuller, 2006).
Graph theory and eigenvalue centrality
In recent years, dynamical cross-correlations (DCC), linear and generalized mutual information have been widely used to identify key residues that can affect function of biological macromolecules upon mutation or effector binding (Negre et al., 2018; Penkler et al., 2018; Sethi et al., 2009; Van Wart et al., 2014). According to the application of this theory to protein dynamics, all Cα atoms (or any other selected atom type) form the nodes of a graph. The edges correspond to pairwise interactions between the nodes and a weight (importance) can be assigned to each edge. The weights can be Euclidean distances, hydrogen bonds, covalent bonds or DCCs, LMIs and so forth (Di Paola et al., 2013). Since we want to analyze importance of the dynamics, LMIij (in fact, –log(|LMIij|) obtained from the trajectories was assigned as edge weights between residues i and j. Only the residues above a certain LMI (≥0.3) and distance threshold (≤7 Å) are considered interacting with each other, namely forming an edge. We should note that similar threshold values were used in other studies as well (Lim et al., 2014; Sethi et al., 2009; Van Wart et al., 2014; Sheik Amamuddy, Verkhivker, et al., 2020; Botello-Smith & Luo, 2019).
In such a network, degree of a node is sum of its edge weights. Therefore, the nodes that have a high coupling to the surrounding residues have a high degree. Degree of a node by itself does not imply importance of a residue. To determine if a residue is influential, it is important to consider the degree of the nodes it is connected as well. For example, the people with high number of friends are the ones with a high degree in a social network. However, only having several friends that have a high degree can make an individual an influential person. In this sense, eigenvector centrality is a measure used to quantify the influence of a node in a network (Zaki & Meira, 2014). Eigenvector centrality was used before to assess key residues to effector binding in IGPS (Negre et al., 2018). Motivated by the IGPS results, we calculated eigenvector centrality of each residue in the dynamics network of SARS-CoV-2 and SARS-CoV Mpro in order to analyze only the dimeric states. We utilized NetworkX (https://networkx.github.io/) and correlationplus (https://github.com/tekpinar/correlationplus) Python packages for eigenvector centrality calculations.
Results and discussion
Stability of the proteases in the simulations
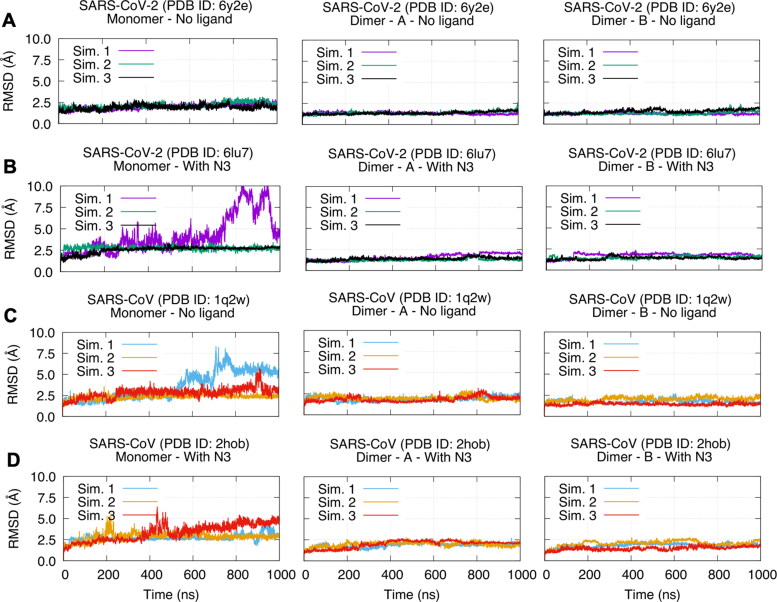
We plotted Cα RMSD of all simulations in Figure 2. In addition, time evolutions of all simulations are presented in Supporting Information Movies S1–S8 from front and back viewpoints. In general, all dimeric forms are more stable than the monomeric forms regardless of the ligand binding state (compare the left columns with the middle and the right columns in Figure 2). In two simulations of the monomeric forms, we can see that RMSD can go over 7.5 Å. The first simulation of the monomeric holo SARS-CoV-2 Mpro shows a conformational change about 700 ns. This conformational change is reorientation of extra-domain compared to the catalytic domain (Figure 2(B) and Supporting Information Movie S2). In addition, we can also observe a reorientation of the extra-domain in the monomeric apo SARS-CoV Mpro (Figure 2(C) and Supporting Information Movie S3). Even though the domain reorientations are not repeated in all replica simulations of the monomers, this gives us a hint that a change in interdomain flexibility is an essential impact of dimerization both for SARS-CoV and SARS-CoV-2 Mpro. The most flexible regions in the dimers are terminal residues, particularly C terminal (see RMSF plots in Supporting Information Figure S2). On the other hand, active site residues H41 and C145 are located at positions of the lowest flexibility regardless of their oligomerization state. Low flexibility character of active site residues was reported before for other proteins as well (Yuan et al., 2003). Even though individual trajectories show different characters, SARS-CoV and SARS-CoV-2 Mpro display similar stability/flexibility behavior, namely, the monomers have higher flexibility and the dimers are relatively more stable.
Figure 2.
RMSD plots of SARS-CoV and SARS-CoV-2 Mpro in monomeric (left column) and dimeric forms (middle and right columns) for all simulations. (A) SARS-CoV-2 – Mpro apo (PDB ID: 6y2e). (B) SARS-CoV-2 – Mpro holo (PDB ID: 6lu7). (C) SARS-CoV – Mpro apo (PDB ID: 1q2w). (D) SARS-CoV – Mpro holo (PDB ID: 2hob).
Impact of dimerization on SARS-CoV and SARS-CoV-2 Mpro
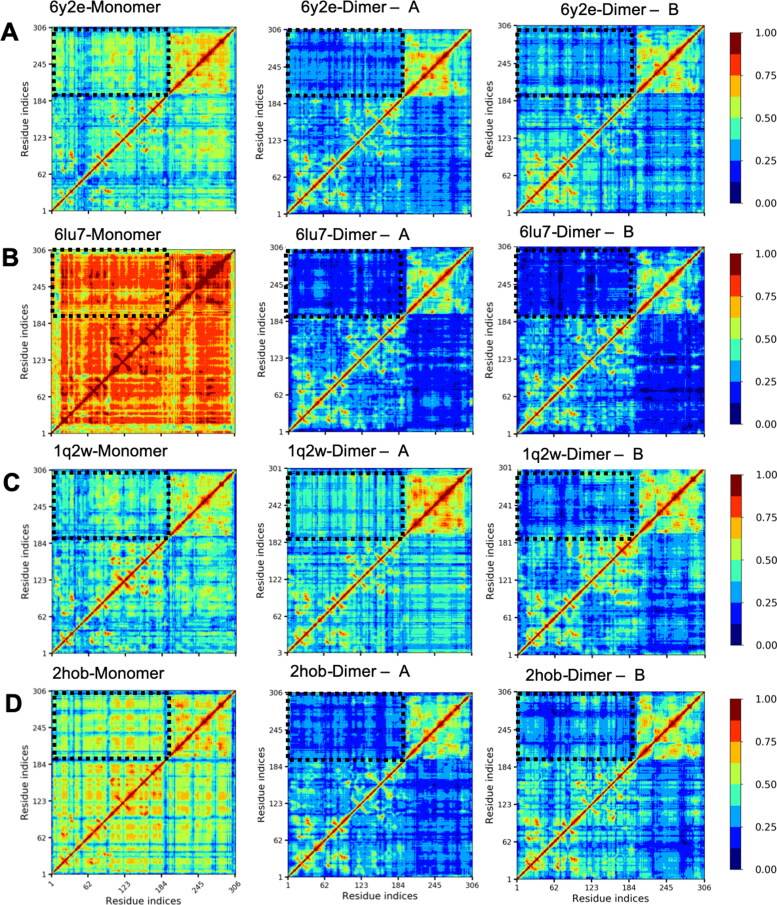
Even though the RMSD and RMSF plots showed some differences in individual trajectories, they were not sufficient to reach a conclusion about major differences between monomeric and dimeric forms because the replica simulations were not showing identical patterns. Due to this reason, we measured inter-residue linear correlations with LMI. All apo and holo monomers show highly disorganized intrachain correlations and the domain borders are indistinguishable between the catalytic domain and the extra domain. When dimerized, the most important contribution of dimerization appears in establishing correlations of the domains into distinguishable regions (Figure 3(A,B), the left column vs. the middle and the right columns). Even upper and lower parts of the catalytic domain become distinguishable upon dimerization. Dimerization organizes correlations into clearly distinguishable domains and interdomain correlations reduce significantly upon dimerization. The catalytic domain and the extra domain patterns still have high intradomain correlations even though interdomain correlations reduced noticeably. In short, a reorganization of LMI maps and a reduction of interdomain LMI are hallmarks of the dimerization in SARS-CoV-2 Mpro. We performed the same analysis for SARS-CoV Mpro as well and the reduction of interdomain correlations is clearly observable there as well (Figure 2(C,D)). This behavior does not change upon N3 binding. The results are not different, and the patterns are similar in the second and the third simulation set as well (Supporting Information Figures S3 and S4). This indicates that dimerization serves to almost identical function in both SARS-CoV and SARS-CoV-2 and LMI parameter by itself cannot show impact of N3 binding.
Figure 3.
Intrachain LMI maps of SARS-CoV and SARS-CoV-2 Mpro in monomeric (left column) and dimeric forms (middle and right columns) for the first simulation set. (A) SARS-CoV-2 – Mpro apo (PDB ID: 6y2e). (B) SARS-CoV-2 – Mpro holo (PDB ID: 6lu7). (C) SARS-CoV – Mpro apo (PDB ID: 1q2w). (D) SARS-CoV – Mpro holo (PDB ID: 2hob). Black dashed rectangles highlight interdomain LMI.
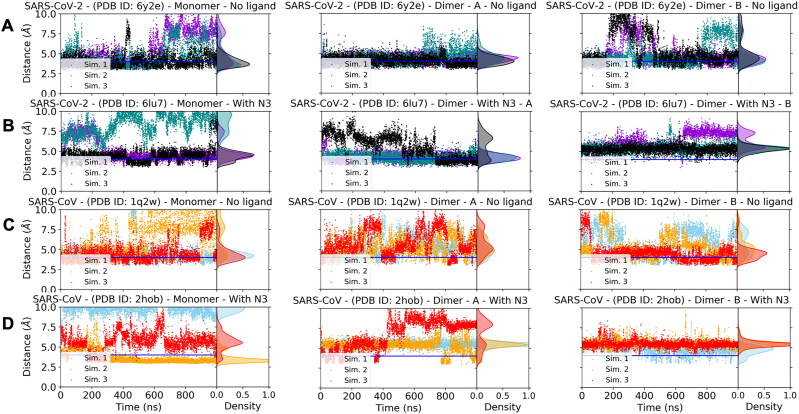
Figure 4.
Distance between SG atom of C145 and NE2 atom of H41 in apo and holo states of SARS-CoV and SARS-CoV-2 Mpro. In addition to the time evolution of the distances, density of distance states is given in each plot as Gaussian-like shaded areas. The monomers are in the left column, the protomer A is in the middle column, and the protomer B is in the right column. Horizontal blue line indicates 4 Å distance. (A) SARS-CoV-2 Mpro apo state (PDB ID: 6y2e). (B) SARS-CoV-2 Mpro holo state (PDB ID: 6lu7). (C) SARS-CoV Mpro apo state (PDB ID: 1q2w). (D) SARS-CoV-2 Mpro holo state (PDB ID: 2hob).
Key local changes upon N3 binding and allosteric interaction between the active sites
Dynamics of catalytic dyad
As explained above, the dimerization stabilizes interdomain correlations. However, impact of N3 binding on the dynamics is not visible in the LMI maps. Therefore, we wondered if N3 binding can make some key local changes in the catalytic dyads of the monomeric and the dimeric states. Several studies showed that the distance between SG atom of C145 and NE2 atom of H41 should be ≤ 4 Å for catalytic competency of SARS-CoV Mpro (Chen et al., 2005, 2006, 2008; Lim et al., 2014; Pang, 2004; Shi et al., 2011; Tan et al., 2005; Zheng et al., 2007). Therefore, we wondered if that is the case for SARS-CoV-2 Mpro as well and analyzed how this distance between the catalytic dyad residues evolved in our simulations for monomeric/dimeric and apo/holo states of both SARS-CoV and SARS-CoV-2 Mpro (Figure 4). In all monomeric forms (the left column in Figure 4(A–D)), the distance is spread to a wider distance range and it can go up to 10 Å. In general, the dimerization reduces the distribution of the distances to a smaller density range (2.5–7.5 Å). When we check impact of N3 binding in dimeric forms, it is interesting to see that the distance can be about 4 Å even in apo states. On the other hand, only one chain can have ≤ 4 Å distance in the holo dimeric states. This result shows clearly that there is an allosteric interaction between two active sites both in SARS-CoV and SARS-CoV-2 Mpro even though the allosteric effect is more visible in the SARS-CoV-2 Mpro. In addition, since it was shown that only one monomer is active in SARS-CoV Mpro, our simulations suggest this assumption can be valid for SARS-CoV-2 Mpro (Chen et al., 2006).
Dynamics of an interface residue
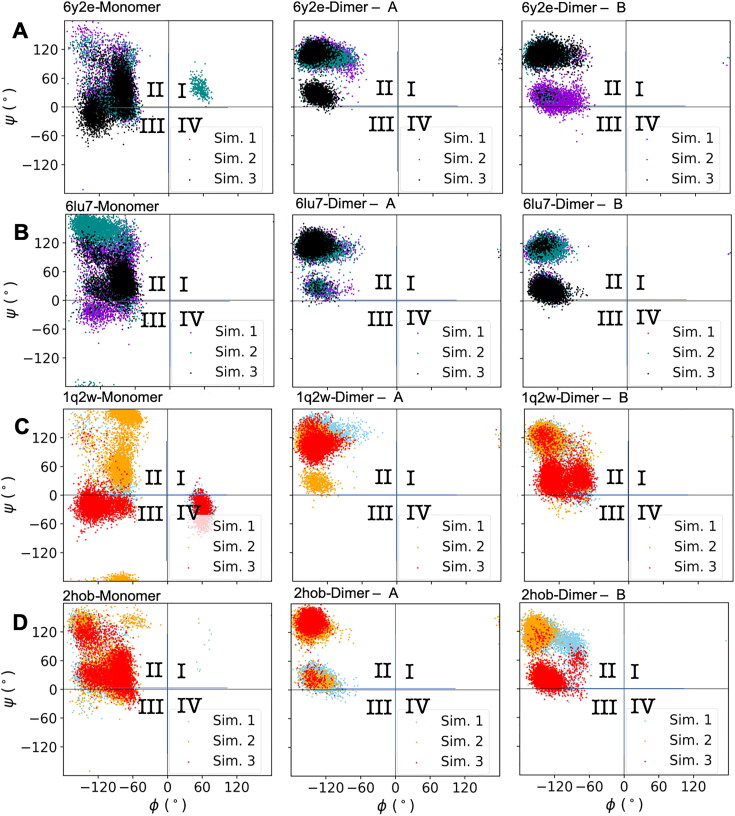
Residue 139 is at the interface of two protomers (See Figure 1 for the details) and it is not far from the catalytic dyad. Previous studies pointed that a mutation on this residue of SARS-CoV Mpro can have a debilitating effect on the catalytic activity (Barrila et al., 2006; Bacha et al., 2004). Due to this reason, we wondered if we can quantify the differences between apo/holo and monomer/dimer states of SARS-CoV and SARS-CoV-2 Mpro. We studied ψ – ϕ distribution of this residue for all states investigated in this study (Figure 5). We divided the dihedral surface into four quarters to simplify the analysis. In general, ψ – ϕ values are concentrated in the quarter II of the dihedral surface for all cases. For monomeric states, namely the left column, we observe that the distribution is spread into the other quarters as well. For only SARS-CoV-2 Mpro, we can observe a slight difference in the weight of populated states between apo and holo dimeric states (see the middle and the right columns of Figure 5(A,B)). The values are more localized in the N3 bound state into two approximately circular regions within quarter II. A previous study reported that not only residue 139 but also the adjacent two residues (residue 140 and 141) also have an impact on catalytic activity of SARS-CoV Mpro (Li et al., 2016). Even though we analyzed them as well, we did not observe a significant difference between apo/holo and monomeric/dimeric states on ψ – ϕ surface for them. Our results suggest that if residue 139 is mutated, it may have a crippling effect on catalytic activity of SARS-CoV-2 Mpro as it was observed in SARS-CoV Mpro. However, this study suggests that the impact of residue 139 on catalytic activity is a secondary impact via coupling of dimerization to the catalytic activity.
Figure 5.
ψ - ϕ angles of residue 139 in SARS-CoV and SARS-CoV-2 Mpro. The dihedral surface was divided into four quarters: I, II, III, IV. (A) SARS-CoV-2 Mpro apo state (PDB ID: 6y2e). (B) SARS-CoV-2 Mpro holo state (PDB ID: 6lu7). (C) SARS-CoV Mpro apo state (PDB ID: 1q2w). (D) SARS-CoV-2 Mpro holo state (PDB ID: 2hob).
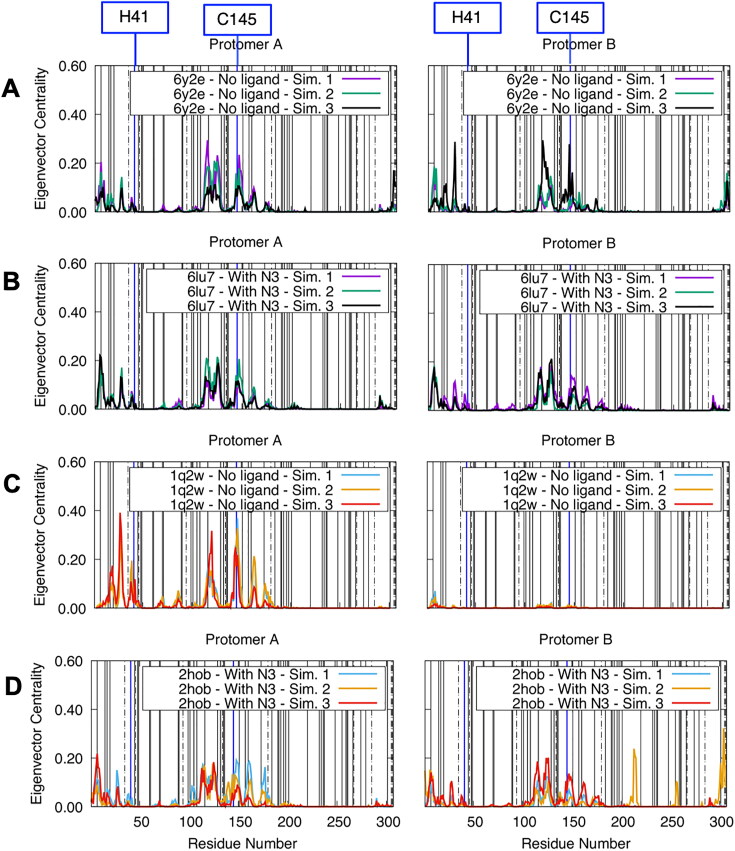
Dynamically influential residues are located in the active sites and dimeric interface
Residues with high eigenvector centrality are influential residues and they have high susceptibility to effector binding (Negre et al., 2018; Zaki & Meira, 2014). Therefore, we plotted eigenvector centrality per residue for dimeric states in Figure 6. Almost all residues with high eigenvector centrality are within the catalytic domain or at the protomer interface and this is true for both of the protomers (Supporting Information Figure S5). We can see only a few residues that have high eigenvector centrality in the extra domain, particularly in C-terminal for holo SARS-CoV Mpro (Supporting Information Figure S5D). Dynamically influential residues are located approximately in the same positions both in SARS-CoV and SARS-CoV-2 Mpro. We recalculated eigenvector centrality with distance threshold values of 8–10 Å and we observed that the values may slightly increase or decrease but the locations of dynamically influential residues are robust (see the same plot for 10 Å distance threshold in Supporting Information Figure S6).
Figure 6.
Residue-wise eigenvector centralities of SARS-CoV and SARS-CoV-2 Mpro in only dimeric forms (left column: protomer A, right column: protomer B) with 7 Å distance threshold and 0.3 LMI value threshold. (A) SARS-CoV-2 Mpro apo state (PDB ID: 6y2e). (B) SARS-CoV-2 Mpro holo state (PDB ID: 6lu7). (C) SARS-CoV Mpro apo state (PDB ID: 1q2w). (D) SARS-CoV-2 Mpro holo state (PDB ID: 2hob). Solid vertical lines show locations of early mutations in GISAID database and a mutational analysis of dynamics study (Sheik Amamuddy, Verkhivker, et al., 2020). Locations of catalytic dyad residues were highlighted with blue vertical lines. Dot-dashed lines indicate locations of mutations between SARS-CoV and SARS-CoV-2 Mpro.
In the eigenvector centrality plot, a peak in the N-terminal is clearly visible in all of the simulations. Several experimental studies on SARS-CoV Mpro show importance of these residues in enzymatic activity. Two studies on SARS-CoV Mpro reported that N-terminal residues are indispensable for catalytic activity (Chen et al., 2005; Zhong et al., 2008). In addition, Chen et al. reported that mutation of Gly-11 can also disrupt dimerization (Chen et al., 2008). Hsu et al. showed that truncation of residue 4 and the following three residues has a strong impact on both enzymatic activity and dissociation constant of the dimers. The region 144–152 also contains one of the highest peaks. C145, which is one of the catalytic dyad residues, is one of the residues in this group. Moreover, residue S147 in this region also have high eigenvector centrality and it was shown that, though far away from the interface, this residue has a debilitating effect on catalytic activity when mutated to alanine. Interestingly, H41 does not have a high centrality by itself while the surrounding residues have it. The difference might stem from differences of the functional role of C145 versus H41. While C145 have a dynamic/chemical role, H41 serves as a base for nucleophilic attack, therefore, might only have a chemical role. In conclusion, previous experimental studies on SARS-CoV Mpro support the idea that the high eigenvector centrality residues identified here are sensitive to effector binding and mutations.
Fifty early stage mutations of SARS-CoV-2 Mpro reported in GISAID database (Elbe & Buckland-Merrett, 2017) and analyzed by Sheik Amamuddy, Verkhivker, et al. (2020) are marked as vertical solid lines in Figure 6. Similarly, when mutations of SARS-CoV Mpro are also considered (dot-dashed lines), we observe that most of the early state mutations are in low eigenvector centrality locations. However, residue 7 and residue 116 have relatively high eigenvector centralities. Both an experimental and a computational study showed that a mutation in this residue can impact the dynamics and enzymatic activity of SARS-CoV and SARS-CoV-2 Mpro (Wei et al., 2006; Sheik Amamuddy, Verkhivker, et al., 2020). Similarly, a mutation in residue 116 caused a noticeably large difference in Cα RMSD. Since the mutations reported in GISAID are from clinical isolates, we must still assume that the mutated forms are still functional. In addition, A7V and A116V mutations are only mutations between amino acids of similar physicochemical properties. Therefore, we think that high eigenvector centrality may point to the active site residues that may have a disruptive influence on the protease function via dynamics when mutated to amino acids of only different physicochemical properties. However, we put a note of caution that the observation here should not be interpreted as a correlation between eigenvector centrality and a possibility of future mutations.
Conclusions
On the basis of our results, we conclude that SARS-CoV Mpro and SARS-CoV-2 Mpro display similar dynamics. Impact of dimerization can be observed in the organization of LMI into clearly distinguishable domains and a reduction in the interdomain correlations both for SARS-CoV and SARS-CoV-2 Mpro. In addition, the dimerization alters distribution of ψ – ϕ angles for residue 139. Another important local effect is in the catalytic dyad. N3 binding to the dimers shows an allosteric effect and causes only a single protomer to be active at a time. We do not know yet if this allosteric effect is a general feature of all ligands or if it specific to N3 peptide mimetic. If it is a general characteristic, it can contribute significantly to the drug design efforts against Covid-19. Finally, we identified some influential residues that are sensitive to effector binding and/or mutations. We believe that the results in this study sheds light in the dynamics of SARS-CoV-2 Mpro and they can contribute to the drug/vaccine design efforts against Covid-19 or similarly functioning proteases.
Authors’ contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supplementary Material
Glossary
Abbreviations
- COVID-19
Coronavirus Disease 2019
- LMI
Linear Mutual Information
- Mpro
main protease
- MD
Molecular Dynamics
- N3
N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N∼1∼-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINA MIDE
- SARS-CoV
Severe Acute Respiratory Syndrome Coronavirus
Funding Statement
This work was granted access to the Jean-Zay HPC resources of IDRIS under the allocation 2020-AP010711656 made by GENCI. Also, we utilized GPU-based computing resources of Inception program (Investissement d’Avenir grant [grant no. ANR-16-CONV-0005]. MT thanks to Programme PAUSE of College de France for their support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Adcock, S. A., & McCammon, J. A. (2006). Molecular dynamics: Survey of methods for simulating the activity of proteins. Chemical Reviews, 106(5), 1589–1615. 10.1021/cr040426m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamri, M. A., Tahir Ul Qamar, M., Mirza, M. U., Bhadane, R., Alqahtani, S. M., Muneer, I., Froeyen, M., & Salo-Ahen, O. M. H. (2020). Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CL(pro). Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1782768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazmi, M., & Motwalli, O. (2020). In silico virtual screening, characterization, docking and molecular dynamics studies of crucial SARS-CoV-2 proteins. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1803965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacha, U., Barrila, J., Velazquez-Campoy, A., Leavitt, S. A., & Freire, E. (2004). Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry, 43(17), 4906–4912. 10.1021/bi0361766 [DOI] [PubMed] [Google Scholar]
- Barrila, J., Bacha, U., & Freire, E. (2006). Long-range cooperative interactions modulate dimerization in SARS 3CLpro. Biochemistry, 45(50), 14908–14916. 10.1021/bi0616302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botello-Smith, W. M., & Luo, Y. (2019). Robust determination of protein allosteric signaling pathways. Journal of Chemical Theory and Computation, 15(4), 2116–2126. 10.1021/acs.jctc.8b01197 [DOI] [PubMed] [Google Scholar]
- Chen, H., Wei, P., Huang, C., Tan, L., Liu, Y., & Lai, L. (2006). Only one protomer is active in the dimer of SARS 3C-like proteinase. The Journal of Biological Chemistry, 281(20), 13894–13898. 10.1074/jbc.M510745200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Chen, L., Tan, J., Chen, J., Du, L., Sun, T., Shen, J., Chen, K., Jiang, H., & Shen, X. (2005). Severe acute respiratory syndrome coronavirus 3C-like proteinase N terminus is indispensable for proteolytic activity but not for enzyme dimerization. Biochemical and thermodynamic investigation in conjunction with molecular dynamics simulations. The Journal of Biological Chemistry, 280(1), 164–173. 10.1074/jbc.M408211200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Hu, T., Zhang, J., Chen, J., Chen, K., Ding, J., Jiang, H., & Shen, X. (2008). Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: Crystal structure with molecular dynamics simulations. The Journal of Biological Chemistry, 283(1), 554–564. 10.1074/jbc.M705240200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, W., Cui, S., Chen, C., Chen, X., Wang, Z., Yang, H., & Zhang, L. (2019). The crystal structure of main protease from mouse hepatitis virus A59 in complex with an inhibitor. Biochemical and Biophysical Research Communications, 511(4), 794–799. 10.1016/j.bbrc.2019.02.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald - an N.Log(N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Di Paola, L., De Ruvo, M., Paci, P., Santoni, D., & Giuliani, A. (2013). Protein contact networks: An emerging paradigm in chemistry. Chemical Reviews, 113(3), 1598–1613. 10.1021/cr3002356 [DOI] [PubMed] [Google Scholar]
- Dubanevics, I., & McLeish, T. C. B. (2021). Computational analysis of dynamic allostery and control in the SARS-CoV-2 main protease. Journal of the Royal Society Interface, 18, 20200591. 10.1098/rsif.2020.0591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe, S., & Buckland-Merrett, G. (2017). Data, disease and diplomacy: GISAID's innovative contribution to global health. Global Challengers, 1(1), 33–46. 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Ghosh, R., Chakraborty, A., Biswas, A., & Chowdhuri, S. (2020). Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1802347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal, B., & Goyal, D. (2020). Targeting the dimerization of the main protease of coronaviruses: A potential broad-spectrum therapeutic strategy. ACS Combinatorial Science, 22(6), 297–305. 10.1021/acscombsci.0c00058 [DOI] [PubMed] [Google Scholar]
- Gunasekaran, K., Ma, B., & Nussinov, R. (2004). Is allostery an intrinsic property of all dynamic proteins? Proteins, 57(3), 433–443. 10.1002/prot.20232 [DOI] [PubMed] [Google Scholar]
- Hess, B. (2008). P-LINCS: A parallel linear constraint solver for molecular simulation. Journal of Chemical Theory and Computation, 4(1), 116–122. 10.1021/ct700200b [DOI] [PubMed] [Google Scholar]
- Hess, B., Bekker, H., Berendsen, H. J. C., & Fraaije, J. G. E. M. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. [DOI] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jakalian, A., Jack, D. B., & Bayly, C. I. (2002). Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. Journal of Computational Chemistry, 23(16), 1623–1641. 10.1002/jcc.10128 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of M(pro) from COVID-19 virus and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Karplus, M., & McCammon, J. A. (2002). Molecular dynamics simulations of biomolecules. Nature Structural Biology, 9(9), 646–652. 10.1038/nsb0902-646 [DOI] [PubMed] [Google Scholar]
- Khan, M. T., Ali, A., Wang, Q., Irfan, M., Khan, A., Zeb, M. T., Zhang, Y. J., Chinnasamy, S., & Wei, D. Q. (2020). Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2-a molecular dynamic study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1769733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Sharma, P. P., Shankar, U., Kumar, D., Joshi, S. K., Pena, L., Durvasula, R., Kumar, A., Kempaiah, P., & Al, P. (2020). Discovery of new hydroxyethylamine analogs against 3CL(pro) protein target of SARS-CoV-2: Molecular docking, molecular dynamics simulation, and structure-activity relationship studies. Journal of Chemical Information and Modeling, 60(12), 5754-5770. 10.1021/acs.jcim.0c00326 [DOI] [PubMed] [Google Scholar]
- Kurt Yilmaz, N., Swanstrom, R., & Schiffer, C. A. (2016). Improving viral protease inhibitors to counter drug resistance. Trends in Microbiology, 24(7), 547–557. 10.1016/j.tim.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, O. F., & Grubmuller, H. (2006). Generalized correlation for biomolecular dynamics. Proteins, 62(4), 1053–1061. 10.1002/prot.20784 [DOI] [PubMed] [Google Scholar]
- Lee, A. L. (2015). Contrasting roles of dynamics in protein allostery: NMR and structural studies of CheY and the third PDZ domain from PSD-95. Biophysical Reviews, 7(2), 217–226. 10.1007/s12551-015-0169-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Teng, X., Qi, Y., Tang, B., Shi, H., Ma, X., & Lai, L. (2016). Conformational flexibility of a short loop near the active site of the SARS-3CLpro is essential to maintain catalytic activity. Scientific Reports, 6, 20918. 10.1038/srep20918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Shen, W., Liao, M., & Bartlam, M. (2007). Preliminary crystallographic analysis of avian infectious bronchitis virus main protease. Acta Crystallographica. Section F, Structural Biology and Crystallization Communications, 63(Pt 1), 24–26. 10.1107/S1744309106052341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, J., Pitsillou, E., Karagiannis, C., Darmawan, K. K., Ng, K., Hung, A., & Karagiannis, T. C. (2020). Interaction of the prototypical α-ketoamide inhibitor with the SARS-CoV-2 main protease active site in silico: Molecular dynamic simulations highlight the stability of the ligand-protein complex. Computational Biology and Chemistry, 87, 107292. 10.1016/j.compbiolchem.2020.107292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L., Gupta, G., Roy, A., Kang, J., Srivastava, S., Shi, J., & Song, J. (2019). Structurally- and dynamically-driven allostery of the chymotrypsin-like proteases of SARS, Dengue and Zika viruses. Progress in Biophysics and Molecular Biology, 143, 52–66. 10.1016/j.pbiomolbio.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L., Shi, J., Mu, Y., & Song, J. (2014). Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A mutations on the extra domain. PLoS One, 9(7), e101941. 10.1371/journal.pone.0101941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen, K., Piana, S., Palmo, K., Maragakis, P., Klepeis, J. L., Dror, R. O., & Shaw, D. E. (2010). Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins, 78(8), 1950–1958. 10.1002/prot.22711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, S., & Kollman, P. A. (1992). Settle - An analytical version of the SHAKE and RATTLE algorithm for rigid water models. Journal of Computational Chemistry, 13(8), 952–962. 10.1002/jcc.540130805 [DOI] [Google Scholar]
- Negre, C. F. A., Morzan, U. N., Hendrickson, H. P., Pal, R., Lisi, G. P., Loria, J. P., Rivalta, I., Ho, J., & Batista, V. S. (2018). Eigenvector centrality for characterization of protein allosteric pathways. Proceedings of the National Academy of Sciences of the United States of America, 115(52), E12201–E12208. 10.1073/pnas.1810452115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall, S., Abraham, M. J., Kutzner, C., Hess, B., & Lindahl, E. (2015). Tackling Exascale software challenges in molecular dynamics simulations with GROMACS. Lecture Notes in Computer Science, 8759, 3. [Google Scholar]
- Pang, Y. P. (2004). Three-dimensional model of a substrate-bound SARS chymotrypsin-like cysteine proteinase predicted by multiple molecular dynamics simulations: Catalytic efficiency regulated by substrate binding. Proteins, 57(4), 747–757. 10.1002/prot.20249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello, M., & Rahman, A. (1981). Polymorphic transitions in single-crystals - A new molecular-dynamics method. Journal of Applied Physics, 52(12), 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Penkler, D. L., Atilgan, C., & Tastan Bishop, O. (2018). Allosteric modulation of human Hsp90α conformational dynamics. Journal of Chemical Information and Modeling, 58(2), 383–404. 10.1021/acs.jcim.7b00630 [DOI] [PubMed] [Google Scholar]
- Pronk, S., Páll, S., Schulz, R., Larsson, P., Bjelkmar, P., Apostolov, R., Shirts, M. R., Smith, J. C., Kasson, P. M., van der Spoel, D., Hess, B., & Lindahl, E. (2013). GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics (Oxford, England), 29(7), 845–854. 10.1093/bioinformatics/btt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Martin, H., Hess, B., & Berendsen, H. J. (2004). An application of flexible constraints in Monte Carlo simulations of the isobaric–isothermal ensemble of liquid water and ice Ih with the polarizable and flexible mobile charge densities in harmonic oscillators model. The Journal of Chemical Physics, 120(23), 11133–11143. 10.1063/1.1747927 [DOI] [PubMed] [Google Scholar]
- Sethi, A., Eargle, J., Black, A. A., & Luthey-Schulten, Z. (2009). Dynamical networks in tRNA:protein complexes. Proceedings of the National Academy of Sciences of the United States of America, 106(16), 6620–6625. 10.1073/pnas.0810961106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik Amamuddy, O., Veldman, W., Manyumwa, C., Khairallah, A., Agajanian, S., Oluyemi, O., Verkhivker, G., & Tastan Bishop, O. (2020). Integrated computational approaches and tools for allosteric drug discovery. International Journal of Molecular Sciences. 21(3), 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik Amamuddy, O., Verkhivker, G. M., & Tastan Bishop, O. (2020). Impact of early pandemic stage mutations on molecular dynamics of SARS-CoV-2 M(pro). Journal of Chemical Information and Modeling, 60(10), 5080-5102. 10.1021/acs.jcim.0c00634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., & Song, J. (2006). The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. The FEBS Journal, 273(5), 1035–1045. 10.1111/j.1742-4658.2006.05130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Han, N., Lim, L., Lua, S., Sivaraman, J., Wang, L., Mu, Y., & Song, J. (2011). Dynamically-driven inactivation of the catalytic machinery of the SARS 3C-like protease by the N214A mutation on the extra domain. PLoS Computational Biology, 7(2), e1001084. 10.1371/journal.pcbi.1001084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Sivaraman, J., & Song, J. (2008). Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. Journal of Virology, 82(9), 4620–4629. 10.1128/JVI.02680-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa da Silva, A., & Vranken, W. (2012). ACPYPE - AnteChamber PYthon Parser interfacE. BMC Research Notes, 5(1), 367. 10.1186/1756-0500-5-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez, D., & Diaz, N. (2020). SARS-CoV-2 main protease: A molecular dynamics study. Journal of Chemical Information and Modeling, 60(12), 5815-5831. 10.1021/acs.jcim.0c00575 [DOI] [PubMed] [Google Scholar]
- Tahir Ul Qamar, M., Alqahtani, S. M., Alamri, M. A., & Chen, L. L. (2020). Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. Journal of Pharmaceutical Analysis, 10(4), 313–319. 10.1016/j.jpha.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J., Verschueren, K. H. G., Anand, K., Shen, J., Yang, M., Xu, Y., Rao, Z., Bigalke, J., Heisen, B., Mesters, J. R., Chen, K., Shen, X., Jiang, H., & Hilgenfeld, R. (2005). pH-dependent conformational flexibility of the SARS-CoV main proteinase (M(pro)) dimer: Molecular dynamics simulations and multiple X-ray structure analyses. Journal of Molecular Biology, 354(1), 25–40. 10.1016/j.jmb.2005.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Van Wart, A. T., Durrant, J., Votapka, L., & Amaro, R. E. (2014). Weighted implementation of suboptimal paths (WISP): An optimized algorithm and tool for dynamical network analysis. Journal of Chemical Theory and Computation, 10(2), 511–517. 10.1021/ct4008603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Chen, C., Liu, X., Yang, K., Xu, X., & Yang, H. (2016). Crystal structure of feline infectious peritonitis virus main protease in complex with synergetic dual inhibitors. Journal of Virology, 90(4), 1910–1917. 10.1128/JVI.02685-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Chen, C., Yang, K., Xu, Y., Liu, X., Gao, F., Liu, H., Chen, X., Zhao, Q., Liu, X., Cai, Y., & Yang, H. (2017). Michael acceptor-based peptidomimetic inhibitor of main protease from porcine epidemic diarrhea virus. Journal of Medicinal Chemistry, 60(7), 3212–3216. 10.1021/acs.jmedchem.7b00103 [DOI] [PubMed] [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2006). Automatic atom type and bond type perception in molecular mechanical calculations. Journal of Molecular Graphics & Modelling, 25(2), 247–260. 10.1016/j.jmgm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- Wei, P., Fan, K., Chen, H., Ma, L., Huang, C., Tan, L., Xi, D., Li, C., Liu, Y., Cao, A., & Lai, L. (2006). The N-terminal octapeptide acts as a dimerization inhibitor of SARS coronavirus 3C-like proteinase. Biochemical and Biophysical Research Communications, 339(3), 865–872. 10.1016/j.bbrc.2005.11.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, X., Yang, H., Shen, W., Zhao, Q., Li, J., Yang, K., Chen, C., Jin, Y., Bartlam, M., & Rao, Z. (2007). Production of authentic SARS-CoV M(pro) with enhanced activity: Application as a novel tag-cleavage endopeptidase for protein overproduction. Journal of Molecular Biology, 366(3), 965–975. 10.1016/j.jmb.2006.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Z., Zhao, J., & Wang, Z. X. (2003). Flexibility analysis of enzyme active sites by crystallographic temperature factors. Protein Engineering, 16(2), 109–114. 10.1093/proeng/gzg014 [DOI] [PubMed] [Google Scholar]
- Zaki, M. J., & Meira, W. (2014). Data mining and analysis: Fundamental concepts and algorithms. New York: Cambridge University Press. [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, K., Ma, G., Zhou, J., Zen, M., Zhao, W., Jiang, Y., Yu, Q., & Feng, J. (2007). Insight into the activity of SARS main protease: Molecular dynamics study of dimeric and monomeric form of enzyme. Proteins, 66(2), 467–479. 10.1002/prot.21160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, N., Zhang, S. N., Zou, P., Chen, J. X., Kang, X., Li, Z., Liang, C., Jin, C. W., & Xia, B. (2008). Without its N-finger, the main protease of severe acute respiratory syndrome coronavirus can form a novel dimer through its C-terminal domain. Journal of Virology, 82(9), 4227–4234. 10.1128/JVI.02612-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W., China Novel Coronavirus Investigating and Research Team (2020). A Novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.