Abstract
Background
Culicoides imicola (Diptera: Ceratopogonidae) is an important Afrotropical and Palearctic vector of disease, transmitting viruses of animal health and economic significance including African horse sickness and bluetongue viruses. Maternally inherited symbiotic bacteria (endosymbionts) of arthropods can alter the frequency of COI (cytochrome c oxidase subunit I) mitochondrial haplotypes (mitotypes) in a population, masking the true patterns of host movement and gene flow. Thus, this study aimed to assess the mtDNA structure of C. imicola in relation to infection with Candidatus Cardinum hertigii (Bacteroides), a common endosymbiont of Culicoides spp.
Methods
Using haplotype network analysis, COI Sanger sequences from Cardinium-infected and -uninfected C. imicola individuals were first compared in a population from South Africa. The network was then extended to include mitotypes from a geographic range where Cardinium infection has previously been investigated.
Results
The mitotype network of the South African population demonstrated the presence of two broad mitotype groups. All Cardinium-infected specimens fell into one group (Fisher’s exact test, P = 0.00071) demonstrating a linkage disequilibrium between endosymbiont and mitochondria. Furthermore, by extending this haplotype network to include other C. imicola populations from the Mediterranean basin, we revealed mitotype variation between the Eastern and Western Mediterranean basins (EMB and WMB) mirrored Cardinium-infection heterogeneity.
Conclusions
These observations suggest that the linkage disequilibrium of Cardinium and mitochondria reflects endosymbiont gene flow within the Mediterranean basin but may not assist in elucidating host gene flow. Subsequently, we urge caution on the single usage of the COI marker to determine population structure and movement in C. imicola and instead suggest the complementary utilisation of additional molecular markers.
Graphical Abstract
Keywords: Cardinium, Endosymbiont, Symbiosis, COI, Culicoides
Background
Culicoides spp. (Diptera: Ceratopogonidae) are a group of tiny biting midges responsible for the spread of several pathogens to animals and humans, including African horse sickness, Schmallenberg, bluetongue and Oropouche viruses. An important vector species, Culicoides imicola, was initially considered to be limited to Afrotropical regions but recent genetic analysis suggests this species has been present in the Mediterranean basin since the Late Pleistocene or Early Holocene [1]. The movement of C. imicola at the northern fringes of the Mediterranean basin is likely driven by wind events which have led to recurrent incursions of the species into mainland Europe [2]. Thus, the tracking of C. imicola through population genetics and climatic modelling is important in establishing where potential future outbreaks of midge-borne viruses may occur.
Mitochondrial DNA (mtDNA) markers have previously been used in phylogeographic studies to detect biodiversity and dispersal events of C. imicola populations [1–3]. However, mtDNA-based inferences can be confounded by the linkage disequilibrium (co-inheritance) of heritable bacteria (endosymbionts) and host mtDNA. As endosymbionts enter a naïve population, selective sweeps of the bacteria can occur and with them the homogenisation of linked mtDNA haplotypes, leading to the apparent absence of biodiversity [4]. Conversely, variation in endosymbiont presence over space can lead to a perceived genetic population structure when none exists. Subsequently, inferences from these studies can only be interpreted in the context of any history of infection with endosymbiotic bacteria.
In this study, we examine the relationship between mtDNA in C. imicola and the heritable endosymbiont Candidatus Cardinium hertigii (Bacteroidetes). Cardinium is present in several biting midge species, including C. imicola, although Cardinium’s significance in Culicoides biology is unknown [5–7]. As Cardinium infection could have implications for inferring movement of C. imicola into naïve geographic areas through mtDNA marker analysis, we analysed associations between mitochondrial haplotypes (mitotypes) and Cardinium infection in this important vector species.
Methods
One hundred and six C. imicola individuals were collected between 2015 and 2016 using light traps from two countries: France (location: Corsica) and South Africa (location: Onderstepoort). Amplification of the host COI gene [8] and the Cardinium Gyrase B (GyrB) gene [7] was undertaken by conventional PCR assay. DNA extracts which passed quality control through successful COI amplification but were negative for GyrB were then screened with a more sensitive nested PCR assay (cycling conditions and primer sequence information in Additional file 1: Table S1). PCR assays consisted of a total of 15 μl per well, comprised of 7.5 μl GoTaq® Hot Start Polymerase (Promega), 5.1 μl nuclease free water, 0.45 μl forward and reverse primers (concentration 10 pmol/μl) and 1.5 μl DNA template. PCR products were separated on 1% agarose gels stained with Midori Green Nucleic Acid Staining Solution (Nippon Genetics Europe). COI amplicons identified by gel electrophoresis were purified enzymatically (ExoSAP) before being sent for sequencing through the Sanger method (GATC Biotech AG, Konstanz, Germany). Chromatograms were then visually assessed for correct base calls. Accession numbers for COI sequences generated in this study are LR877434–LR877461.
We then investigated possible associations of the endosymbiont Cardinium with C. imicola mitotype distribution through haplotype network analysis. To this end, haplotype networks were constructed, with analysis and visualisation undertaken using the TCS haplotype network algorithm generated in PopART v1 [9]. The networks included C. imicola mitotypes from this study (South Africa and Corsica) as well as mitotypes detected from the Iberian Peninsula and Israel ([3]; Portugal n = 12, Israel n = 29; accession numbers: AF078098–AF078100, AF080531-AF080535, AJ549393–AJ549426). The Iberian population is known to be free from Cardinium, whereas that of Israel is known to carry Cardinium [5, 6]. To investigate associations between endosymbiont and mitotype, the mtDNA haplotype group and Cardinium infection status of C. imicola individuals within and between populations were compared using Fisher’s exact test (significance cut-off P < 0.001) in RStudio version 1.3 [10]. A Group was defined as a cluster of mitotypes with no more than one SNP difference between at least one other mitotype within the cluster.
Results
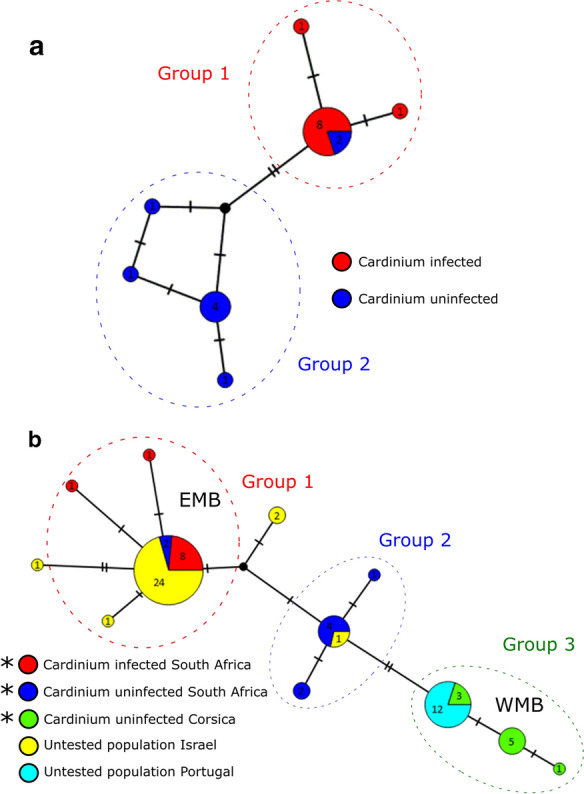
To confirm individuals in this study were not cryptic species of C. imicola missed by morphological identification, a distance estimation of mtDNA barcodes was assessed giving a minimum identity of > 98%, consistent with all individuals belonging to the same species. Cardinium infection was observed in the South African population (10/33 individuals) but not in Corsican populations (0/73 individuals) (Table 1). The additional nested screening revealed no evidence of low-titre infections. A C. imicola mitotype network (Fig. 1a) of the single South African population, containing both Cardinium-infected and uninfected individuals, showed the presence of seven mtDNA haplotypes split into two broad mitotype groups. Cardinium infection status and these general mitotype groupings were associated, with all Cardinium-infected specimens falling in one group (Fisher’s exact test, P = 0.00071 [OR: Infinity; CI 3.27–Infinity]).
Table 1.
Cardinium screening results using conventional and nested GyrB PCR assays
| Species | Collection site | Year | Number of Cardinium-positive individuals by conventional PCR | Number of Cardinium-positive individuals by nested PCR |
|---|---|---|---|---|
| C. imicola | Corsica (Site 1) | 2015 | 0/46 | 0/46 |
| Corsica (Site 2) | 0/27 | 0/27 | ||
| South Africa (Onderstepoort) | 2016 | 10/33 | 10/33 |
Fig. 1.
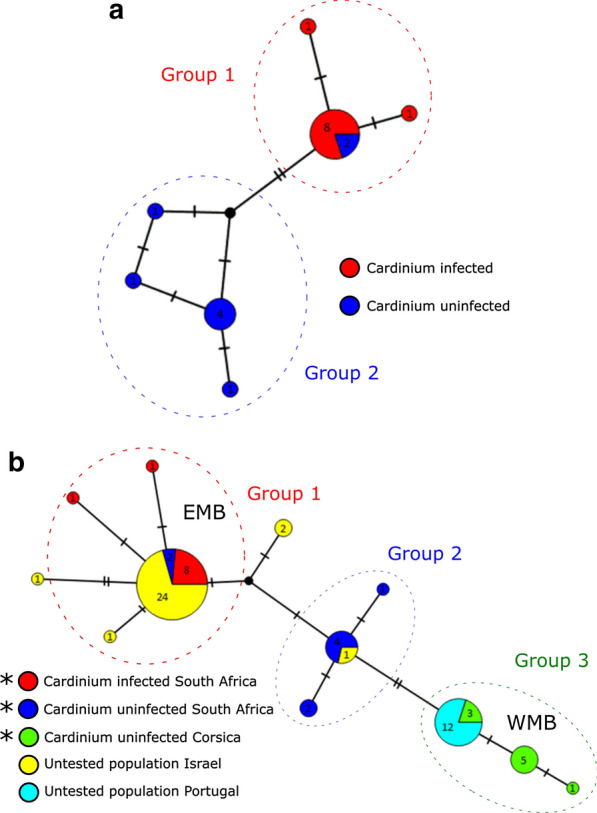
Culicoides imicola mtDNA haplotype networks a mtDNA haplotype network of Cardinium-infected and -uninfected Culicoides imicola from a single site in Onderstepoort, South Africa; based on a 517 bp COI sequence. b mtDNA haplotype network of Culicoides imicola from sites spanning South Africa and the Mediterranean basin; based on a 451 bp COI sequence. Haplotype networks were generated using the TCS network algorithm in PopART v1.7 (9). Numbers within circles represent the numbers of individuals designated to each haplotype. The numbers of substitutions separating haplotypes are indicated by dashes. A group is defined as a cluster of mitotypes with no more than one SNP difference between at least one other mitotype within the cluster. EMB Eastern Mediterranean Basin, WMB Western Mediterranean Basin. *Indicates mtDNA haplotypes generated in this study
Extending the haplotype network to include C. imicola populations from the Western Mediterranean Basin (WMB; Corsica and Portugal) and the Eastern Mediterranean Basin (EMB; Israel) led to 12 haplotypes being observed with 0–1.77% range of divergence over a 451 bp region. Further to this, 24/29 Israeli (EMB) haplotypes clustered within the main Cardinium-infected mitotype observed in South Africa, whereas the Portuguese (WMB) haplotypes all clustered within an uninfected Corsican mitotype (Fig. 1b). Grouping mitotypes of known infection status within one SNP of others within a cluster defined three groups, and again all Cardinium-infected specimens fell in one group (Fisher’s exact test, P < 0.0001 [OR: NA; CI NA]).
Discussion
Insect endosymbionts are generally considered in the context of driving diverse and extreme host phenotypes, such as reproductive manipulations and virus blocking [11, 12]. However, their role in confounding interpretations of mtDNA structure is also of importance [4]. As both endosymbiont and mitochondria are maternally inherited together, a selective sweep of Cardinium is likely to have led to our observed concordance of C. imicola mitotype and Cardinium (Fig. 1a). Null models of endosymbiont-mtDNA dynamics predict three phases: first, the symbiont sweep, during which the endosymbiont-associated mitotype increases in frequency and the infected and uninfected individuals have different associated mitotypes. This is followed by the mitotype of infected individuals becoming increasingly represented in uninfected individuals where the endosymbiont fails to transmit from mother to offspring. In the final phase, mutation re-establishes diversity in the mitotype pool. Although a structure is observed between infection status and mtDNA haplotype, the presence of two uninfected individuals in the main infected haplotype in C. imicola from South Africa is consistent with an endosymbiont showing imperfect vertical transmission, generating uninfected individuals from lineages with the dominant Cardinium-infected mitotype.
Although the infection status of the Portuguese and Israeli individuals included in the extended haplotype network (Fig. 1b) has not been directly assessed, the recent Pagès [6] screening for Cardinium found no indication of C. imicola infection in the Iberian Peninsula, and we observed no infections in our Corsican population. Despite this, others [5] have observed the endosymbiont to be common in Israel, indicating infection heterogeneity exists between the EMB and WMB. In light of this, mitotype variation between the EMB and WMB mirrors infection heterogeneity between C. imicola populations from the EMB and WMB. Thus, the mtDNA structure may be explained by the dispersal of Cardinium-infected individuals exclusively into the EMB.
The matrilineal subdivision between Cardinium-infected and -uninfected C. imicola (Fig. 1b) is similar to observations by previous studies [3, 13] which noted a remarkable divergence between C. imicola mitotypes from the EMB and WMB. This observation suggests that the linkage disequilibrium of Cardinium and mitochondria reflects endosymbiont gene flow within the Mediterranean basin but may not assist in elucidating host gene flow. For example, if dispersal events occur involving Cardinium-infected C. imicola, selective sweeps of the endosymbiont may erase any previous biodiversity in this marker. This is reminiscent of similar patterns observed in other insects with endosymbiont infections: the mosquito vector, Culex pipiens [14], the parasitoid wasp, Nasonia vitripennis [15], and the ladybird, Adalia bipunctata [16]. In all these cases, the frequency of mtDNA haplotypes are more closely associated with the endosymbionts in a population rather than geography.
Conclusions
The haplotype networks produced in this study (Fig. 1) suggest Cardinium infection in Culicoides is another example where the presence of an endosymbiont impacts mtDNA-based inference of population history. Previous work had indicated Israeli and Southern Africa samples grouped together on analysis of mtDNA variation but were distinct on analysis of microsatellite variation [1]. We conclude that association of Cardinium and mtDNA provides a likely explanation for this discordance, and these data support the general importance of using multiple loci (e.g. microsatellites and nuclear markers) in phylogeographic analysis.
Supplementary Information
Additional file 1: Table S1. Primer attributes and PCR conditions.
Acknowledgements
The authors acknowledge the Agricultural Research Council-Onderstepoort Veterinary Research, Pretoria, South Africa, for allowing the undertaking of field work at their site.
Abbreviations
- EMB
Eastern Mediterranean Basin
- WMB
Western Mediterranean Basin
- COI
Cytcochrome c oxidase subunit I
Authors’ contributions
JP, GDDH, SS and MB and assisted in the conception and design of the study. Field work was undertaken by JP, CG and GV. Laboratory work was undertaken by JP. Analyses and interpretation of the data were undertaken by JP, GDDH, MB, SS, CG and GV, as well as drafting of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a BBSRC Doctoral Training Partnership studentship (BB/M011186/1) awarded to JP and a Marie Curie Individual Fellowship (H2020-MSCA-IF-2014) grant awarded to SS.
Availability of data and materials
Accession numbers for COI sequences generated in this study are LR877434–LR877461.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/1/2021
A Correction to this paper has been published: 10.1186/s13071-021-04653-1
Contributor Information
Jack Pilgrim, Email: jack.pilgrim@liverpool.ac.uk.
Stefanos Siozios, Email: siozioss@liverpool.ac.uk.
Matthew Baylis, Email: baylism@liverpool.ac.uk.
Gert Venter, Email: venterjgert@gmail.com.
Claire Garros, Email: claire.garros@cirad.fr.
Gregory D. D. Hurst, Email: g.hurst@liverpool.ac.uk
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-020-04568-3.
References
- 1.Jacquet S, Garros C, Lombaert E, Walton C, Restrepo J, Allene X, et al. Colonization of the Mediterranean basin by the vector biting midge species Culicoides imicola: an old story. Mol Ecol. 2015;24:5707–5725. doi: 10.1111/mec.13422. [DOI] [PubMed] [Google Scholar]
- 2.Jacquet S, Huber K, Pagès N, Talavera S, Burgin LE, Carpenter S, et al. Range expansion of the Bluetongue vector, Culicoides imicola, in continental France likely due to rare wind-transport events. Sci Rep. 2016;6:27247. doi: 10.1038/srep27247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dallas JF, Cruickshank RH, Linton YM, Nolan DV, Patakakis M, Braverman Y, et al. Phylogenetic status and matrilineal structure of the biting midge, Culicoides imicola, in Portugal Rhodes and Israel. Med Vet Entomol. 2003;17:379–387. doi: 10.1111/j.1365-2915.2003.00454.x. [DOI] [PubMed] [Google Scholar]
- 4.Hurst GDD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc Biol Sci. 2005;272:1525–1534. doi: 10.1098/rspb.2005.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morag N, Klement E, Saroya Y, Lensky I, Gottlieb Y. Prevalence of the symbiont Cardinium in Culicoides (Diptera: Ceratopogonidae) vector species is associated with land surface temperature. FASEB J. 2012;26:4025–4034. doi: 10.1096/fj.12-210419. [DOI] [PubMed] [Google Scholar]
- 6.Pagès N, Muñoz-Muñoz F, Verdún M, Pujol N, Talavera S. First detection of Wolbachia-infected Culicoides (Diptera: Ceratopogonidae) in Europe: Wolbachia and Cardinium infection across Culicoides communities revealed in Spain. Parasit Vectors. 2017;10:582. doi: 10.1186/s13071-017-2486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis SE, Rice A, Hurst GDD, Baylis M. First detection of endosymbiotic bacteria in biting midges Culicoides pulicaris and Culicoides punctatus, important Palaearctic vectors of bluetongue virus. Med Vet Entomol. 2014;28:453–456. doi: 10.1111/mve.12055. [DOI] [PubMed] [Google Scholar]
- 8.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 9.Leigh JW, Bryant D. Popart: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 10.RStudio Team. RStudio: Integrated Development for R. [Internet] RStudio, Inc., Boston, MA; 2020. http://www.rstudio.com.
- 11.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 12.Bourtzis K, Dobson SL, Xi Z, Rasgon JL, Calvitti M, Moreira LA, et al. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop. 2014;132:150S–163S. doi: 10.1016/j.actatropica.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Calvo JH, Calvete C, Martinez-Royo A, Estrada R, Miranda MA, Borras D, et al. Variations in the mitochondrial cytochrome c oxidase subunit I gene indicate northward expanding populations of Culicoides imicola in Spain. Bull Entomol Res. 2009;99:583–591. doi: 10.1017/S0007485309006622. [DOI] [PubMed] [Google Scholar]
- 14.Rasgon JL, Cornel AJ, Scott TW. Evolutionary history of a mosquito endosymbiont revealed through mitochondrial hitchhiking. Proc R Soc B Biol Sci. 2006;273:1603–1611. doi: 10.1098/rspb.2006.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raychoudhury R, Grillenberger BK, Gadau J, Bijlsma R, van de Zande L, Werren JH, et al. Phylogeography of Nasonia vitripennis (Hymenoptera) indicates a mitochondrial–Wolbachia sweep in North America. Heredity. 2010;104:318–326. doi: 10.1038/hdy.2009.160. [DOI] [PubMed] [Google Scholar]
- 16.Schulenburg JHGVD, Hurst GDD, Tetzlaff D, Booth GE, Zakharov IA, Majerus MEN. History of infection with different male-killing bacteria in the two-spot ladybird beetle Adalia bipunctata revealed through mitochondrial DNA sequence analysis. Genetics. 2002;160:1075-86. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primer attributes and PCR conditions.
Data Availability Statement
Accession numbers for COI sequences generated in this study are LR877434–LR877461.


