Abstract
Bone formation in the craniofacial complex is regulated by cranial neural crest (CNC) and mesoderm-derived cells. Different elements of the developing skull, face, mandible, maxilla (jaws) and nasal bones are regulated by an array of transcription factors, signaling molecules and microRNAs (miRs). miRs are molecular modulators of these factors and act to restrict their expression in a temporal-spatial mechanism. miRs control the different genetic pathways that form the craniofacial complex. By understanding how miRs function in vivo during development they can be adapted to regenerate and repair craniofacial genetic anomalies as well as bone diseases and defects due to traumatic injuries. This review will highlight some of the new miR technologies and functions that form new bone or inhibit bone regeneration.
Keywords: microRNA therapeutic, microRNA Inhibitor system (PMIS), Bone repair, Bone regeneration, microRNA mouse models, Bone development
Graphical Abstract
INTRODUCTION
Bone formation during early development can be by two processes;1) intramembranous bone formation or 2) endochondral ossification [1–3]. Cranial neural crest (CNC) cells migrate from the neural tube into the anterior region of the skull to form some of the bones and cartilages of the face and anterior skull. The cartilages and bones in the posterior region of the skull are derived from prechordal mesoderm cells. These cells condense and either differentiate into osteoblasts (which directly form bone by intramembranous bone formation or differentiate into chondrocytes (which form cartilage and bones by endochondral bone formation). Thus, CNC-derived cells involved in intramembranous bone formation form the anterior region of the skull, face, mandible and maxilla. The mesoderm-derived cells form the posterior regions of the skull by endochondral ossification [3–7].
The molecular mechanisms that control craniofacial growth that gives rise to the different vertebrate head sizes and morphology include signaling and growth factors [3,8–19]. In particular, the Wnt, Fgf, Bmp, Shh, and Tgf-β signaling pathways control the early patterning and growth of the craniofacial skeleton by regulating the migration, proliferation, differentiation and transformation of cells derived from the mesoderm and cranial neural crest [20–29]. These factors and pathways interact and intersect to control development of the brain and skull [7,9,10,13,14,30–32]. Furthermore, early signals by these pathways especially the Wnt pathway emanating from the pharyngeal endoderm and epithelium, appear to regulate patterning of the developing skeleton [19,33–35]. Tissue-tissue interactions that give rise to cell fate decisions are fundamental to the development of head structures, especially for the patterning and morphogenesis of craniofacial organs including teeth [36–38]. These early developmental cues drive the morphogenesis and patterning of perinatal craniofacial tissues. Hippo signaling has also been shown to regulate prenatal and post-natal craniofacial development and growth [39,40]. Recently, microRNAs (miRs) have emerged as new modulators of craniofacial bone development and maintenance.
miRs are short non-coding RNA molecules approximately 22 nucleotides long. miRs bind to complementary targets on the 3’ untranslated region (UTR) of messenger RNAs (mRNAs), attenuating mRNA translation via either mRNA strand degradation or sequestration [41]. Through this mechanism, miRs play a broad role in the regulation of mRNA translation and have been demonstrated to play a significant part in an array of biological processes [42–44].
As we understand the mechanisms and functions of miRs they act as global modulators of multiple gene expression pathways. It is not simply one miR regulates one gene, network and/or process. In our research we find that miRs are regulators of one or more genetic pathways controlling specific tissue, organ or cellular processes. The temporal and spatial levels of miR expression change either moderately or dramatically depending on the tissue during development. Almost all cells will express low levels of specific miRs, but there appears to be a threshold of miR expression for them to be effective modulators of cellular processes. The complexity of miR biogenesis, processing and degradation during developmental time points mediates the fine-tuning of gene expression.
There are many cell-based studies on the role of miRs in osteogenesis that have identified several genetic pathways for the differentiation of cells into osteoblasts and osteocytes. We have found that in vitro based miR expression and profiling differ from the in vivo state. The process of using osteogenic differentiation media in different types of cell cultures can result in miR expression profiles that are different from in vivo bone development. Many cell types can be manipulated to express miRs using osteogenic media and many miRs can induce the bone-forming program in cells under osteogenic conditions. This review will focus on miR mouse and rat models for craniofacial bone development, regeneration and repair.
I. NEW TECHNOLOGY TO INHIBIT MICRORNAS IN VIVO AND AS THERAPEUTIC REAGENTS
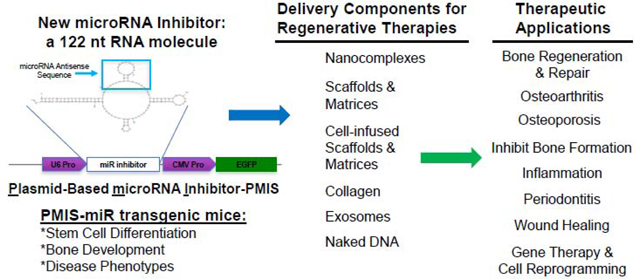
Plasmid-Based microRNA Inhibitor System (PMIS)
The problems with current miR inhibition methods are that chemically modified anti-miR oligonucleotides (AMOs) and locked-nucleic acids (LNAs) bind miRs transiently and inefficiently, do not remain in dividing cells and require repeated large doses of oligos in cells to be effective and they have severe off-target effects. In previous preclinical and clinical trials with LNAs, antagomirs, AMOs and other miR inhibitors they were toxic, non-specific and caused multiple adverse effects in animals and humans. The sponges and decoys also suffer from a lack of stability, inefficient binding of the miR, lack of specificity and require toxic delivery systems.
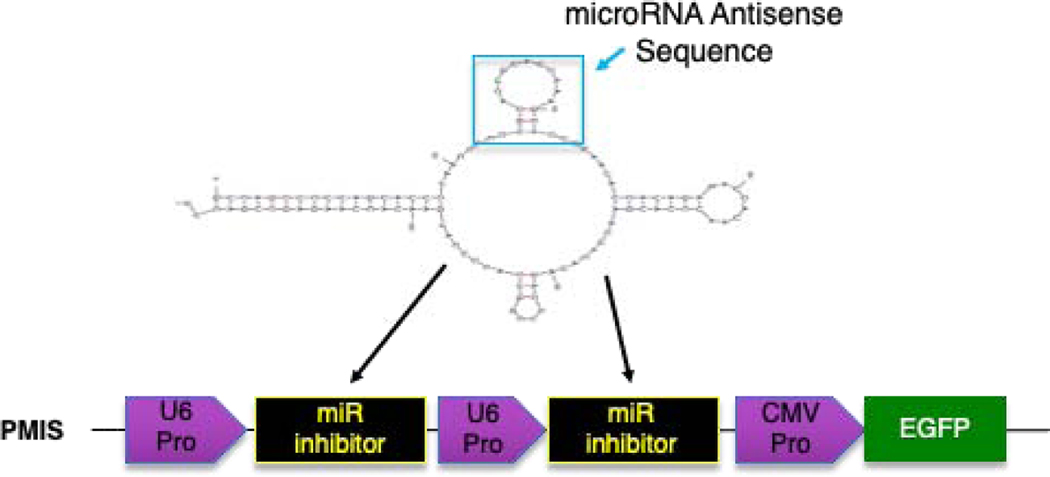
To circumvent these major problems, we developed a new method of miR inhibition, the Plasmid-Based miRNA Inhibition System (PMIS), to allow for the simultaneous knockdown of homologous miR families in vitro and in vivo [45–47]. The PMIS inhibitor is composed of native, unmodified nucleic acids that enables the development of stably expressing cells (lentivirus) and animal models for the study of genome-wide miR family inhibition to identify miR targets and cellular processes. The PMIS can be used to knockdown miRs during embryonic development to determine their effect on stem cells, cell proliferation and differentiation as well as developmental processes. The PMIS system represents a major paradigm shift in miR biotechnology that allows researchers to finally dissect the role of identical miRs expressed in clusters and on multiple chromosomes. The PMIS can distinguish between and differentially inhibit miRs with only one nucleotide change in their seed sequence. The unique structure of the PMIS-miR complex is bound by factors of the RNA-Induced Silencing Complex (RISC), making it very stable, efficient, with a high specificity and affinity for specific miRs and is not toxic in animals and cells (Fig. 1)[45]. The PMIS transcript is expressed using the U6 Polymerase III promoter to control PMIS-miR transcript expression levels in the cell. We found that using the CMV Polymerase II promoter that PMIS-miR expression levels were exceedingly high and interfered with cellular processes. The U6 promoter expresses PMIS-miR constructs at lower levels with increased functionality. Furthermore, we engineered the PMIS miR inhibitor to bind to the RISC proteins and it is extremely stable in the cell. This miR inhibitor works in transgenic animals (in vivo) as well as xenograft animal models without adverse side effects, it has great promise as a therapeutic molecule in clinical applications including bone repair and regeneration.
Figure 1. Schematic representation of the PMIS. A U6 promoter drives expression of the PMIS transcript.
The inhibitor complex contains a stem loop that comprises the antisense miR of choice. The 21–22 nucleotide antisense sequence of each miR is cloned into this site in the stem loop. EGFP expression is used to determine expression. More than one miR inhibitor can be constructed in the same plasmid.
We are currently working on inducible PMIS-miR constructs, but we find that the specificity of the PMIS-miR is very efficient and while it is expressed in every cell, not every cell expresses the targeted miR, so only those cells/tissues with high levels of targeted miR expression are affected during development. Furthermore, not every cell/tissue expresses the same genes at high levels, thus we see differential gene expression with and without inhibiting specific miRs. Clearly, we observe other cell/tissue developmental defects in our PMIS-miR mice depending on the miR that is inhibited and specific genes targeted by the miR. It has been reported many times that conditional ablation of a miR cluster for tissue-specific knockouts have no effect on development. This is because the tissue specific Cre knocks out the miR too late in development to observe a defect. The specificity of the PMIS system during embryonic development provides a new mechanism to study miR function, especially at time points that miRs play critical roles in developmental processes.
II. FUNCTIONS OF MICRORNAS IN CRANIOFACIAL BONE FORMATION AND DEVELOPMENT
The tissue-specific Dicer, DGCR8 and Ago2 knockouts in mice have shown miRs are involved in bone development [48–54] and the developing tooth [48,55,56]. miRs are post-transcriptional regulators that repress gene expression by regulating the translation of mRNA and promoting the degradation of targeted mRNAs and are thought to regulate tooth and bone development by repressing the transcription factors and signaling factors involved in these processes [57].
Murine models for miR-17–92 regulation of bone formation
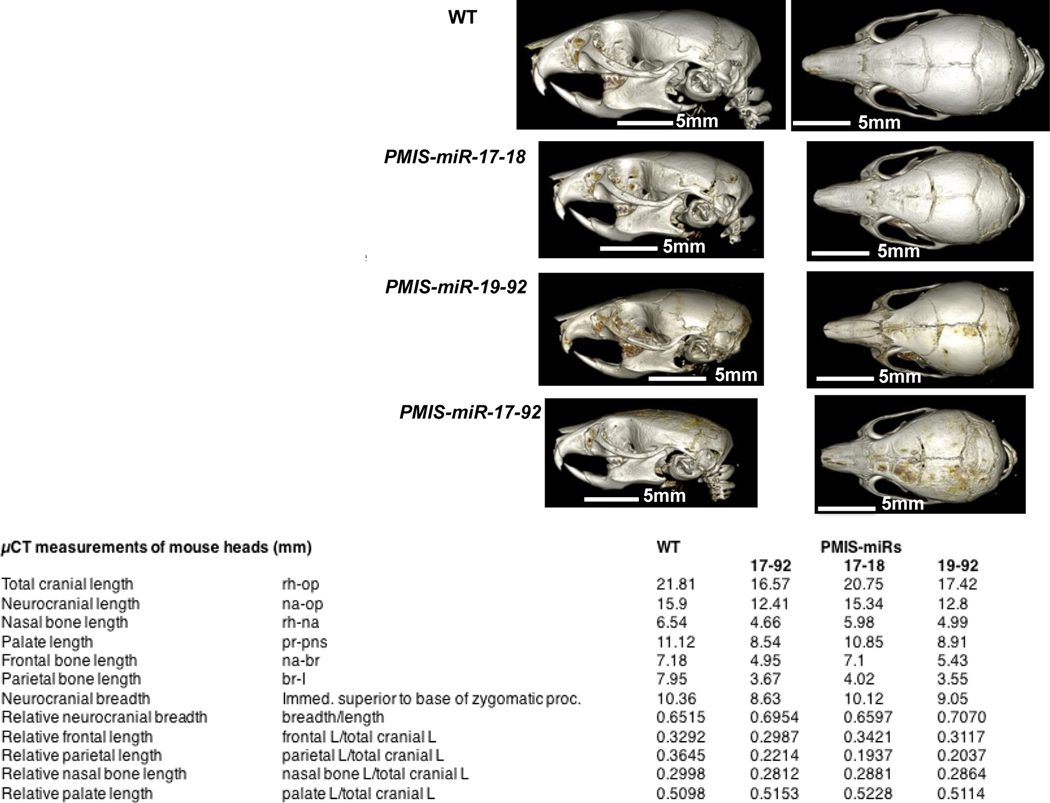
miRs come in a variety of genomic contexts, both intragenic and intergenic [58,59], and can be isolated or grouped into polycistronic clusters, as is the case with the widely-studied miR cluster, miR-17–92 [60,61]. miR-17–92 is a cluster of six highly conserved miRs from four different families located on chromosome 13 in humans and chromosome 14 in mice [62]. However, identical miRs to the miR-17–92 cluster are also expressed on chromosomes X and 7 in humans and chromosomes X and 5 in mice [45]. Recent studies have implicated the miR-17–92 cluster in the development of orofacial and craniofacial defects [63,64]. We knocked down all three clusters on these chromosomes using PMIS-miR-17–18, PMIS-miR-19–92 and PMIS-miR-17–92 mice to dissect the functions of identical miRs within the miR-17–92 clusters and found each contributed to craniofacial bone development in different aspects of growth [46]. Craniofacial defects were identified in all of the PMIS transgenic mice at three weeks of age by High-Resolution X-Ray Microtomograph scans (uCT scans). Specific measurements from the uCT scans identified defects in total cranial length (rhinion to opisthion, rh-op), neurocranial length (nasion to opisthion, na-op), nasal bone length (rhinion to nasion, rh-na), palate length (prosthion to anterior nasal spine, pr-pns), frontal bone length (nasion to bregma, na-br), and parietal bone length (bregma to lambda, br-l). We report defects in the width and length of cranial structures in all PMIS mice compared to WT (Fig. 2) [46]. Other defects identified included suture, mandibular condyle growth defects and microcephaly [46].
Figure 2. miRs within the miR-17–92 cluster differentially regulate craniofacial development.
WT, PMIS-miR-17–18, PMIS-miR-19–92 and PMIS-miR-17–92, 3 week-old heads were analyzed by μCT. In-depth measurements were obtained for different aspects of craniofacial growth. Quantitative measurements of total cranial length and breadth of the PMIS transgenic mice are shown compared to WT, N=3.
Murine models for miR-26b regulation of bone formation
Recent studies have reported that miR-26b functions as an odontogenic regulator [65,66]. A miR-26b transgenic over-expression (OE) mouse is similar to the Lef-1 general knockout mouse as miR-26b regulates Lef-1 expression [65]. These mice lack teeth, including molars and incisors, and full body uCT images reveal skeletal and bone defects in these mice. The cranial base and cranial breath measurements are essentially identical in the miR-26b OE and WT mice. However, the miR-26b OE mice have a shorter nasal bone, snout, frontal bone, parietal bone, cranial breath, and cranial base length compared to WT mice. The cranial base angle and ramus height are also decreased in the miR-26b OE mice compared to WT mice. The miR-26B OE mice have a decreased mandibular length compared to WT mice [65]. It appears that miR-26b targets Lef-1 and Wnt signaling to control bone growth and formation.
Murine models for miR-200 regulation of bone formation
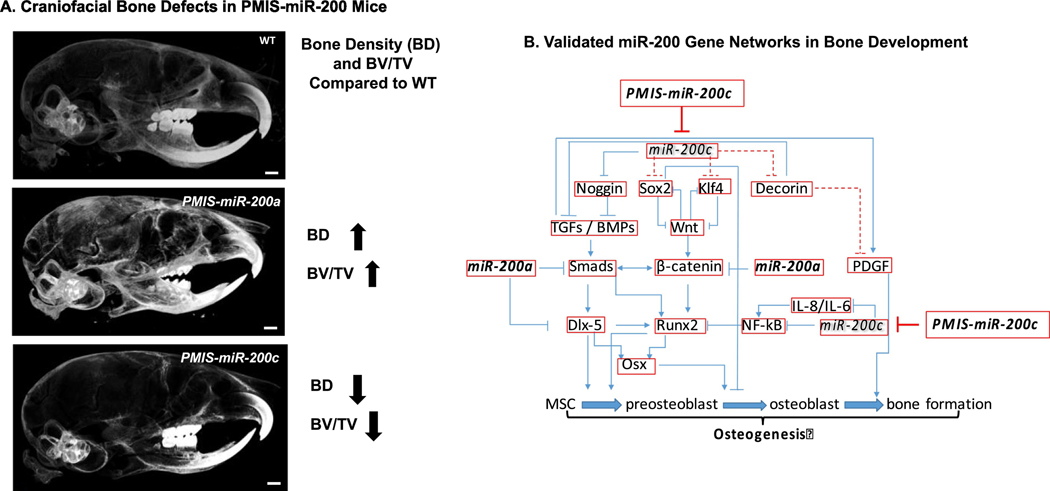
The miR-200c/141 knockout mice have defects in mandibular bone formation (a decrease in alveolar bone) linked to a Noggin regulatory pathway and Bmper expression [66]. miR-200c directly targets and inhibits Noggin expression (a Bmp inhibitor) resulting in an increase in Bmp activity and cell differentiation [66]. However, the lack of miR-200c increases Noggin expression and decreases Bmp activity resulting in a bone developmental defect in these mice. The PMIS-miR-200c mice also have a decrease in ossification of craniofacial bones, maxilla/palatine bones and facial bone [47]. The molecular mechanism of miR-200c in osteogenic differentiation includes a Sox2-mediated Wnt signaling and Klf4 pathway. Conversely, the inhibition of miR-200a (PMIS-miR-200a) in mice results in an increase in bone density (Fig. 3). Through multiple validation experiments and gene expression analyses we have identified multiple miR-200 regulated pathways (Fig. 3). These in vivo results demonstrate the effect the miR-200 family has on gene regulatory pathways, gene networks and cellular processes.
Figure 3. The in vivo role of the miR-200 family in craniofacial bone development.
A. Bone density in 4 week old WT, PMIS-miR-200a and PMIS-miR-200c mice heads. BD, bone density; BV/TV, bone volume/tissue volume. B. Gene networks involved in bone formation regulated by the miR-200 family.
Murine models for miR-23–24-27 regulation of bone formation
The miR-23–24-27 cluster has been shown to regulate osteoblast differentiation through several genetic pathways. A report using a novel miR-23a cluster knockdown mouse model demonstrated a role for this cluster in maintaining stage-specific HoxA factor expression during osteogenesis [67]. HoxA5 and HoxA11 are targets of the miR-23a cluster and these Hox genes regulate bone-specific gene expression through chromatin modifications. The miR-23a cluster knockdown increased cortical and trabecular bone mass [67]. These researchers used an inducible anti-miR-23a CI knock-in mouse to show regulation of the PRC2 complex and repressive H3K27me3 deposition contrasting with BAF-linked activating H3K27ac chromatin modifications promote osteoblast differentiation by this miR cluster [67]. Other researchers have used miR decoys for miR-23a, miR-27a and miR-24–2 to knockdown these miRs in mice to show regulation of osteocyte differentiation [68]. The knockdown of miR-23a or miR-27a decreased osteocytes but knockdown of miR-24–2 had no effect. In contrast over-expression of the miR-23a cluster in mice demonstrated low bone mass associated with a decrease in osteoblasts but an increase in osteocytes [68]. It was suggested that the miR-23a cluster regulates osteocyte differentiation by modulating the TGF-ß signaling pathway.
Murine models for miR-34 regulation of bone formation
A miR-34c over-expression (OE) mouse demonstrated that miR-34c can regulate bone remodeling by affecting both osteoblasts and osteoclasts leading to increased bone resorption [69]. The defect in osteoblast differentiation by miR-34c OE was due to targeting Satb2 and Runx2. However, miR-34c also targets Notch signaling in osteoblasts and miR-34c regulates bone homeostasis by regulating multiple targets in osteoblasts and osteoclasts [69].
Murine models for miR-214 regulation of bone formation
miR-214 OE under the control of the osteoclast specific Acp5 promoter in mice demonstrated a role for this miR in bone formation. The increase in miR-214 decreased Pten levels, increased osteoclast activity and reduced bone mineral density [70]. The expression of miR-214 was upregulated in the process of m-CSF and RANKL induced osteoclastogenesis, thus inhibition of miR-214 levels reduced osteoclastogenesis.
Murine models for miR-335 regulation of bone formation
A transgenic mouse over-expressing miR-335 using the osterix promoter revealed higher bone mass and increased bone formation in the transgenic mouse compared to wildtype [71]. Mechanistically in these mice Runx2 and Osx were upregulated, while the Wnt antagonist Dickkopf-1 was down-regulated. The researchers suggest that application of miR-335 modified bone marrow stem cells could be used in craniofacial bone regeneration [71].
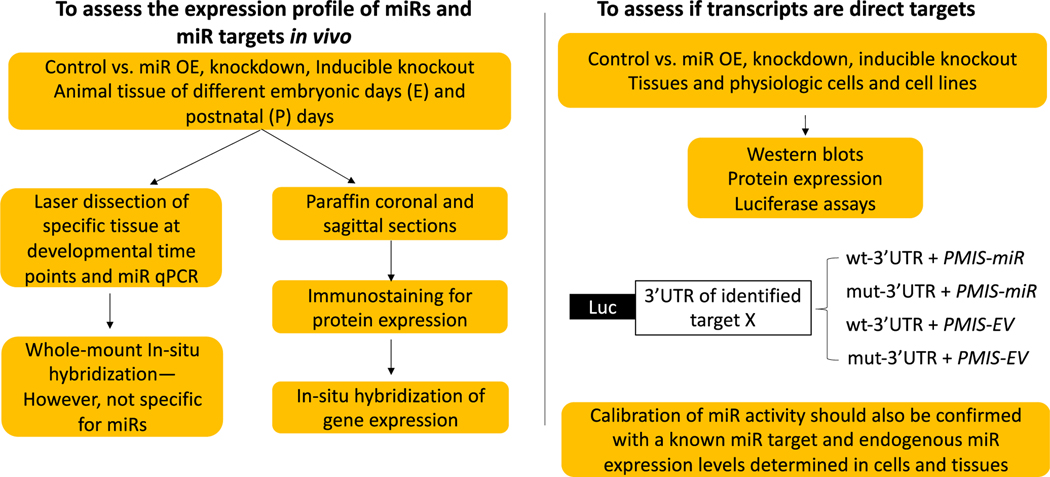
A common theme in these critical mouse models is that several miRs can regulate identical and intersecting pathways required for bone formation and bone remodeling. The use of animal models for studying miR regulation of bone formation is critical to understanding how miRs affect osteogenesis. We follow a validation process to determine miR targets in mouse tissues and cell-based assays (Fig. 4). The role of miRs in bone homeostasis will be discussed in the following section.
Figure 4. Validation Workflow of in vivo RNA-Seq., scRNA-Seq. or miR Microarray potential targets (compare Control vs miR knockdown, miR OE or inducible (conditional) miR inactivation).
• Note- In situ hybridization for gene and miR transcripts may not yield accurate information as miRs and miR targets may not be degraded upon miR inhibition or over expression (OE), respectively.
III. THE ROLES OF MIRS IN INFLAMMATION, BONE MAINTENANCE, REGENERATION AND REPAIR
miRs in bone homeostasis: a balance of formation and resorption
miRs are actively involved in the bone homeostasis by regulating the balance of osteogenesis and osteoclastogenesis. This regulation has been investigated using different cell lines and in vivo models, including bone homeostasis under estrogen deficiency, mechanical loading and corticosteroid stimulation. miRs regulate bone loss by directly and indirectly modulating the inflammation that subsequently activates osteoclastogenesis through the RANKL-RANK-OPG system. miRs may directly regulate the RANKL/OPG balance to adjust osteoclastogenesis. The upregulation and activation of RANKL accelerates the osteoclastic differentiation and bone resorption, while downregulation of RANKL diminishes osteoclastogenesis. For example, miR-29a and miR-146a regulate the RANKL-RANK-OPG system by directly targeting RANKL and Smad4, resulting in the inhibition of bone resorption in ovariectomized (OVX) mice [72,73]. Inhibition of these miRs are shown to relatively improve bone mass by overcoming osteoclastogenesis [74]. miRs also modulate osteoblastogenesis by directly targeting osteogenic signal pathways to inhibit bone formation. miR-139, −145, −26a, −451, and −100 have been found to target osteogenic signals, including Wnt/β-catenin, Notch, BMP/Smads, and FGF-21, which accelerating osteoporosis in the OVX murine models [75–81]. Similarly, some miRs upregulated the activities of Wnt-β/catenin and Bmp/Smads and promoted osteogenic differentiation by targeting the inhibitory regulator of these pathways. For examples, miR-199a [82,83], miR-219a [84], and miR-208a [85] have been reported to improve osteogenesis and bone formation by directly targeting an adipogenesis promoter, TET2 (Tet Methylcytosine Dioxygenase 2), Rorβ (retinoic acid-related orphan receptor), a transcriptional enhancer of inhibitor HBP1 of the Wnt pathway, and a Bmp ligand Activin A receptor type I (ACVR1). In addition, miR-23b [86] and miR-103a [87] directly target the osteogenic transcription factor, Runx2, to inhibit bone formation, which accelerates osteoporosis in the OVX model.
Anti-inflammatory Therapeutic Potential of miRs in Periodontitis and Bone Loss
The exaggerative inflammatory response plays key roles in the pathogenesis of periodontitis, including activated alveolar bone loss and inhibited bone formation. Thus, by manipulating the miRs that actively participate in periodontitis, an epigenetic approach is an innovative therapeutic tool for periodontitis by fine-tuning the inflammatory response. Although numerous miRs are involved in the pathogenic progress of periodontitis, and both miR-146a and miR-200c have been reported to negatively feedback to IL-6. miR-146a also negatively affects IL-1β and TNF-a, and Hey2 expression [88]. However, miR-146a has been reported to be upregulated in gingival tissue of periodontitis and in gingival cells under LPS stimulation, which activates inflammation [89]. In the mouse model of P. gingivalis (PG) infection-induced experimental periodontitis, injection of a miR-146a mimic exhibited protective function on periodontitis associated bone loss [90]. In comparison to miR-146a, although it has smaller variation in periodontitis patients, miR-200c was found to be significantly down-regulated in the gingival tissues of periodontitis patients [91]. miR-200c reduced IL-8 expression by targeting the inhibitor of nuclear factor kappa B kinase subunit beta targeting (IKBKB) in the NF-kB signal pathway [92]. Our previous studies have confirmed that miR-200c directly targets the 3’UTRs of IL-6, IL-8 and CCL-5 [93]. Reporter gene analysis showed that miR-200c targets the 3’UTR of the interferon related developmental regulator1 (Ifrd1). We have observed that a local application of plasmid DNA encoding miR-200c effectively suppressed IL-6, IL-8, Ifrd1, and NF-kB in rat gingival tissues. In a rat model of periodontitis induced by PG-LPS, we observed that the local application of miR-200c significantly down regulated the proinflammatory cytokines and alveolar bone loss stimulated by of LPS injection.
Therapeutic function of miRs in bone healing and regeneration.
Several approaches have been attempted to explore the therapeutic function of miRs on enhancing bone formation and regeneration. The majority of the reported studies of miR-driven bone regeneration were cell-based. Both mesenchymal stromal cells from bone marrow (BMSCs) and adipose tissues (ADSCs) have served as progenitor cells of bone forming cells. Either mimics and/or antagomir inhibitors of specific miRs were transfected into BMSCs or ADSCs to increase osteogenic differentiation. The mimics of miR-375 [94,95], miR-101 [57], miR-450b [96], miR-135 [97], miR-218 [98], miR-335 [99,100], miR-199a [83], and miR-99a [101] have reported to regenerate bone tissue by targeting the inhibitors or antagonist of osteogenic signaling BMP, IGF, and WNT, including IGFBP3, EZH2 (the enhancer of zeste homolog 2), Hoxa2, DKK1, HIF1a, and KDM6B. In addition, the antagomirs or inhibitors of miRs that target osteogenic differentiation also have shown the osteogenic capabilities and bone regeneration after transfection into BMSCs or ADSCs. The antagomirs to miR-133a [102], miR-137 [103,104], and miR-31 [105,106] improve Runx2 and Bmp signaling in the progenitor cells. The antagomirs of miR-146a (74), miR-21 [107], and miR-221 [108] increase osteogenic differentiation by rescuing Smad signaling. Inhibitors of miR-138 [109–111] and miR-124 [112] also demonstrated the enhancement of osteogenic differentiation by regulating FAK and Dlx signaling. Cells transfected with miR mimics and antagomirs also exhibited the capabilities to regenerate bone tissues at ectopic sites of murine models and restore calvarial bone defects and long bone fractures at murine models.
Section III Summary
| miRs | genes/pathways targeted | effect on bone growth/regeneration |
|---|---|---|
| miRs-29a, −146a | RANKL-RANK-OPG | inhibition of bone resorption (OVX model) |
| miRs-139, −145, −26a, −451 and −100 | osteogenic signals | increase osteoporosis (OVX model) |
| miRs-199a, −219a, −208a | osteogenic signals | improve osteogenesis |
| miRs-23b, −103a | RUNX2 | inhibit bone formation (OVX model) |
| miRs-146a, −200c | inflammation, IL6, IL-1B, TNF-a | inhibition of pro-inflammatory cytokines |
| miR-200c | inflammation, IL-6, IL-8, CCL-5 | inhibition of pro-inflammatory cytokines |
| miRs-375, −101, −450b, −135, −218, −335, −199a, −99a | BMP, IGF, WNT | regenerate bone tissue, increase osteogenic differentiation |
| miRs-133a, −137, −31 | RUNX2, BMP | bone progenitor cell differentiation |
| miRs-146a, −21,−221 | SMADs | osteogenic differentiation |
| miRs-138, −124 | FAK, DLX | increase osteogenic differentiation |
IV. MICRORNA DELIVERY APPROACHES FOR THERAPEUTIC TREATMENTS
General strategies of miR-based therapeutic approaches
Depending on the expression strategy of the target miR, the miR therapies are divided into two categories: 1) miR activation or gain-of-function therapy which up-regulates exogenous miR expression and 2) miR inhibition or loss-of-function therapy which down-regulates endogenous miR expression [113,114]. Overexpression of the target miR also can be achieved through the delivery of either miR mimics that are double-strand oligonucleotides or miR-expressing viral vectors. The double-stranded miR mimics are designed either to target a single mRNA or a group of multiple miR units target different mRNAs [115,116]. Viral vector-mediated miR over expression enables the sustained generation of miRs, which is especially attractive for regenerative medicine applications. Instead of using a viral vector for over expressing miRs, our group recently reported that plasmid DNA-mediated sustained expression of miR-200c was a viable and translational way for craniofacial and periodontal bone regeneration [117–119]. In contrast to the miR activation therapy, the miR inhibition therapy aims to block the endogenous miR. So far, the most straightforward technique is to use anti-miR oligonucleotides and several different methods have been developed to inhibit miR functions. The major limitation of the anti-miR oligonucleotides is their short half-life because of the susceptibility to degradation. Therefore, the oligonucleotide has a transient effect because its concentration decreases quickly with cell division [113]. Our newly developed plasmid-based miR inhibitor system (PMIS) can largely address this challenge by sustained production of miR inhibitor molecules to inhibit miR family members in both cells and mice [120].
Chemically modified anti-miRs
Tremendous efforts have been reported to develop chemical modification techniques that improve the performance of anti-miRs (synthesized oligonucleotides) through enhancing cellular uptake, binding affinity, and nuclease resistance [114]. For example, phosphorothioate linkage is the most commonly used internucleotide modification on synthesized oligonucleotides. This modification delays plasma clearance and enhances cellular uptake through the nonspecific binding to serum albumin and membrane proteins [121,122]. AntagomiRs containing a terminal phosphorothioate linkage, a cholesterol conjugation, and 2’-O-methyl-modified ribose sugars (2-OMe), are the first miR inhibitors reported to work in many different tissues in mammals [123]. By using this technique, some specific antagomiRs have been developed to modulate bone mass through systemic injection methods. One group reported that the blocking of miR-148a by antagomiR-148a through tail vein injection significantly increased bone formation while suppressing bone resorption in ovariectomized (OVX) mice [124]. Similarly, hind limb-unloading-induced bone loss in mice was partly prevented by injection of antagomiR-103a since miR-103 was a mechanosensitive miR in bone tissue [87]. Moreover, aging-related bone loss was mitigated in rats through the injection of antagomiR-31a-5p into the femoral bone marrow cavity [125]. Although antagomiRs have shown encouraging results in several different tissues and animal models, the safety concerns caused by high dose requirements prevent their applications [113,114]. Additionally, other chemical modification strategies have been reported with promising applications, e.g., peptide nucleic acids (PNAs) [126] and locked nucleic acids (LNAs) [127].
Viral vectors
The high transfection efficiency and sustained gene expression of viral vectors make them the most effective techniques so far for miR delivery both in vitro and in vivo. Retrovirus, lentivirus, adeno-associated virus (AAV), and baculovirus are the most frequently used viral vectors for gene therapy [128–130]. Retroviruses carrying two copies of the single-strand RNA genome can stably integrate into the host chromosomes, guaranteeing long-term expression of inserted therapeutic genes [131]. However, the inability of transducing nondividing cells and the safety concern caused by genome integration at an undesired location significantly impede the application of retrovirus vectors in regenerative medicine [114]. Lentivirus is a subgroup of retrovirus is a more promising vector for in vivo miR delivery because it can not only transfect nondividing cells [130] but also prefer to integrate within introns of active transcriptional units, thereby reducing the potential to cause insertional oncogenesis [132,133]. Therefore, lentivirus-mediated miR delivery has been used to improve bone formation in many reports. For example, lentivirus-mediated miR-29a over expression can protect rats against glucocorticoid-Induced bone loss and fragility [134]. Lentivirus- mediated miR- 26a over expression in bone marrow mesenchymal stem cells (BMSCs) significantly improved murine cranial bone regeneration [135]. Additionally, lentivirus-mediated anti-miR-31 to inhibit the expression of miR-31 in BMSCs also can repair critical-sized calvarial defects in rats [105]. AAV vectors are emerging as the most promising gene therapy tool for clinical applications because they have several advantages: 1) small size (only ~4.7kb DNA genome; 2) infection of both dividing and nondividing cells; 3) nonpathogenicity in humans, and; 4) relatively low immunogenicity [113,114,129,136]. A recent study reported that either systemic or direct joint administration of an rAAV9 vector carrying an artificial-miR that targets shn3 (rAAV9-amiR-shn3) in mice markedly improved bone formation via augmenting osteoblast activity [137]. Baculoviruses are insect viruses that can transfect a wide variety of mammalian stem cells at high efficiency [138]. Importantly, these baculovirus vectors are relatively safe for clinical applications because they lack the capabilities of replication and gene integration in mammalian cells [139]. One study reported the aberrant elevated expression of miR-214 in the BMSCs isolated from OVX rats. To suppress the miR levels, the authors constructed hybrid baculovirus vectors expressing miR sponges to bind miR-214. Notably, transplantation of the miR-214 sponges in OVX-BMSCs effectively healed the critical-size bone defects and ameliorated the bone quality in OVX rats while the OVX-BMSCs ectopically expressing Bmp2 failed to heal the defects [140].
Lipid-based delivery system
Lipid-based nanocarriers are the most commonly used nonviral gene delivery systems in vitro. Besides numerous in vitro studies, lipid-mediated miR delivery is also feasible for in vivo application based on recent findings. One study reported that tail vein injection of a miR-451a mimic containing liposome (Invivofectamine® 3.0) significantly enhanced osteoblastogenesis, reversed OVX-induced bone loss, and improved bone strength [141].
However, so far most of the lipid-mediated miR applications are relying on in vitro stem cells (e.g., BMSCs) transfection first and then transplantation of these miR-expressing cells in vivo [142]. These “ex-situ” strategies instead of direct administration of liposome/miR to “in situ” bone defects are largely because the nonspecific distribution and low stability of liposomes in tissue fluid will severely limit their efficacy in vivo [114,142,143]. To address this challenge, one seminal study developed an innovative bone targeting delivery system comprising 1,2-dioleoyl-3-trimethylammonium-propane-based (DOTAP) liposome linked with (AspSerSer)6, which selectively bind to osteoblast-mediated mineralizing nodules and amorphous calcium phosphate [144]. Through systemic injection of this (AspSerSer) 6-liposome-containing casein kinase-2-interacting protein-1 (Plekho1) siRNA, bone formation, bone microarchitecture, and bone mass were all significantly improved in both healthy and osteoporotic rats [144]. Soon after, this exciting bone formation surface-targeting delivery system, (AspSerSer)6-liposome, was successfully applied to deliver antagomiR-214 [145] and agomiR-33–5p [146] to improve bone formation/mass in OVX [145] and unloading [146] mice models, respectively. In addition to targeting bone formation surface, a novel eight repeating sequences of aspartate (D-Asp8) peptide-conjugated DOTAP-based liposome was developed to deliver miR to bone resorption surfaces [147] because of the ability of D-Asp8 for selectively binding to highly crystallized hydroxyapatite (HA), which is the characteristic of bone resorption surfaces [148]. They demonstrated that D-Asp8 conjugation promoted the enrichment of antagomir-148a/liposome after intravenous injection and the subsequent down-regulation of miR-148a expression in osteoclasts in vivo, resulting in significantly reduced bone resorption in OVX-induced osteoporotic mice [147].
Polymer-based delivery system
Numerous cationic natural and synthetic polymers have been widely studied and shown great promise for both plasmid DNA and RNA gene delivery [149]. Compared to lipid vectors, one obvious advantage of polymer-based delivery systems is that they are more flexible and versatile through variation in polymer molecular weight, structure, composition, and conjugation [113]. Among the currently reported polymer-based vectors, high molecular weight branched polyethyleneimine (PEI, 25KD) is still the gold standard and has been most widely used in both preclinical studies and clinical trials because of its relatively high nucleic acid transfer efficiency. Recently, we successfully delivered plasmid DNA containing miR-200c into primary human periodontal ligament fibroblasts and bone marrow MSCs. miR-200c delivered using PEI effectively inhibited IL-6, IL-8, and CCL-5 in periodontal ligament fibroblasts and enhanced osteogenic differentiation of human bone marrow MSCs in vitro [119]. However, the significant safety concern derived from PEI’s high cytotoxicity and non-degradability prevent its applications in regenerative medicine [150]. Many efforts have been reported to reduce PEI’s cytotoxicity while improving the gene transfer efficiency. One strategy is to crosslink low molecular weight PEI via a disulfide linkage [149]. Another promising strategy is to combine with other inorganic nanoparticles. For example, a PEI-capped gold nanoparticle (AuNPs) was developed for miR-29b delivery. These AuNPs not only didn’t show obvious cytotoxicity on hMSCs but also they were more efficient in improving osteoblastic differentiation compared to Lipofectamine RNAi MAX/miR-29b complexes [151]. Similarly, PEI combined with iron oxide magnetic nanoparticles (MNPs) indicated moderate cytotoxicity and high uptake efficiency (~75%). MNPs-PEI provided a long-last effect in delivering miR-335 into hMSCs in vitro [152].
Chitosan (CS) is one of the most studied natural polymeric gene carriers derived from partial deacetylation of chitin [153,154]. Chitosan-based gene carriers are especially attractive for regenerative medicine because of its high positive charge, excellent biodegradability, favorable biocompatibility, low toxicity, low cost, and low immunogenicity [153–155]. Additionally, chitosan has been widely studied in bone tissue engineering either as drug/gene carriers or scaffolds because of the pro-osteoblastic activity and inherent antimicrobial ability [156,157]. In one study, chitosan nanoparticles cross-linked by tripolyphosphate (TPP) were used to deliver miR-199a-5p agomiR plasmid. Their data indicated that the chitosan nanoparticles could stably overexpress miR-199a-5p in hMSCs in vitro and promoted both ectopic bone formation after implantation in vivo and bone defects repair delivered by hydrogel [158]. Chitosan/TPP/Hyaluronic Acid (HA) nanoparticles were also developed to deliver antimiR-138 to rat bone marrow mesenchymal stem cells (rMSCs) [159]. These composite nanoparticles showed a high transfection efficiency (~70%) and significantly promoted rMSCs osteogenic differentiation in vitro. Moreover, some in vivo studies proved that it was feasible to effectively prevent the bone loss in OVX mice by systemic administration of miR-34a mimic [160], or miR-182 inhibitor [161] using TPP/chitosan nanoparticles.
Inorganic nanoparticle-based delivery system
Calcium phosphates (CaP) have been used as gene carriers for decades through the way of DNA-calcium phosphate co-precipitation to introduce plasmid DNA into many cell types [162–164]. Among the gene vectors being considered to date, CaP nanoparticle is one of the most promising materials for dental and bone tissue regeneration applications by virtue of the excellent osteoconductivity, biocompatibility, and biodegradability [165]. Besides DNA, e.g., pDNA encoding bone morphogenetic protein-2 (Bmp-2) [166], CaP nanoparticles also have successfully delivered miRs [167] and siRNAs [168] as well. The excellent osteoconductivity and biocompatibility of CaP nanoparticles as non-viral vectors are especially advantageous for bone and dental tissue engineering applications even though their transfection efficacy needs to be significantly improved compared to viral vectors [166,169].
Inspired by the rationale of co-precipitation of Ca(2+) with DNA, nano-sized CaCO(3)/DNA co-precipitates were also developed for gene delivery because of its high biocompatibility and inducible biodegradability [170]. Our unpublished data indicated that the CaCO3/PS has much higher (at least two times) efficiency than PEI (25KD) at the same culture condition with 10% serum presence to deliver plasmid DNA (eGFP) to mouse cranial osteoblasts with significantly lower cytotoxicity. Moreover, as an alternative to CaPO4-based biomaterials, CaCO3-based biomaterials have shown some excellent properties, e.g., biocompatibility, biodegradation, bioactivity, and osteoconductivity for bone implantation and regeneration [171,172]. Therefore, CaCO3-based materials show great promise for bone tissue engineering applications including as a non-viral gene delivery vector and scaffold materials.
Mesoporous silica nanoparticles (e.g., MCM-41 and SBA-15 MSNs), are emerging as a multifunctional drug delivery carriers because they are cable of absorbing/encapsulating large amounts of bioactive molecules through the hundreds of empty channels with a honeycomb-like porous structure (mesopores). MSNs have some unique features, e.g., large pore volume (~0.9 cm3/g), tunable pore size (2–10 nm), high surface area (~900 m2/g), good chemical and thermal stability, good biocompatibility, excellent surface functionality, which are all advantageous for various controlled release applications [173,174]. In addition to pDNA, one study indicated that MSNs could efficiently deliver siRNA, pDNA, and small anti-tumor drugs into cells without significant cytotoxicity when the surfaces of MSNs were noncovalent coated with PEI (10KD not 25 KD) [175]. More recently, a new technique was developed for functionalization of MSN surface with PEI through disulfide bonds which can achieve lysosomal delivery of chemotherapy drug (doxorubicin) and intracellular delivery of miR-145 [176].
Section IV Summary
| miRs | delivery approach/type of molecule | effect on bone growth/regeneration |
|---|---|---|
| miR-200c | viral vector/miR OE | bone regeneration |
| miR-148a | antagomirs/modified oligos | bone regeneration |
| miR-103a | antagomirs/modified oligos | inhibition of bone loss |
| miR-31a | antagomirs/modified oligos | inhibition of bone loss |
| miR-29a | viral vector/miR OE | protect against bone loss |
| miR-26a | viral vector/miR OE | improved bone regeneration |
| miR-31 | viral vector/miR inhibition | increased bone formation |
| miR-214 | viral vector/miR inhibition | increased bone formation |
| miR-451a | liposome/miR mimic | enhanced osteoblastogenesis |
| mlRs-214, −33 | modified liposome/antagomir | improved bone formation |
| miR-148a | modified liposome/antagomir | reduced bone resorption (OVX) |
| miR-200c | polymer-based/miR OE | enhanced osteogenic differentiation |
| miR-199a | chitosan/antagomir | promoted ectoptic bone formation |
| miR-138 | chitosan/antimir | increased osteogenic differentiation |
| miRs-34a, −182 | chitosan/mimic-inhibitor | prevent bone loss (OVX) |
V. SCAFFOLD-BASED LOCAL MIR DELIVERY SYSTEM
Stem cells for gene therapy
Currently, there are two gene therapy strategies in bone tissue engineering. The first one is a stem cell-based gene delivery which has been used in most reported pDNA or miR-based gene therapy studies. The second one is direct gene therapy or acellular gene delivery which delivers the therapeutics via viral or nonviral vectors to the injury sites to promote bone healing by recruiting endogenous reparative cells without using exogenous stem cells [177,178]. Compared to embryonic stem (ES) cells and induced pluripotent stem cells (iPSCs), which are pluripotential, the multipotent adult stem cells, especially several types of mesenchymal stem cells (MSCs), are the most widely used in dental and bone tissue engineering due to the ethical and legal controversies with ES and iPSCs [179–181]. Almost half-century ago, bone marrow-derived MSCs (BMSCs) were identified and isolated from murine femur bones as an adherent fibroblast-like population with the capacity to self-renew and differentiate into at least three cell lineages, including bone, fat, and cartilage [182]. Since then, non–marrow tissue-derived MSCs were isolated and identified in almost all tissues, e.g., adipose tissue, placenta, umbilical cord blood, dermis, and orofacial tissue [181,183] while most of the current studies are still using BMSCs in miR-based bone tissue engineering. However, it should be noted that craniofacial bones originate from two sources: most are of cranial neural crest origin while the parietal bones arise from the paraxial mesoderm. Therefore, BMSCs harvested from craniofacial bones exhibit distinct properties from long bone MSCs although overall, they are very similar. For example, craniofacial BMSCs are growing more rapidly with higher levels of alkaline phosphatase in cell culture, and forming more compact bone and less bone marrow upon transplantation in vivo [179–181]. While the BMSCs or other type of stem cells have been used for gene delivery and bone tissue engineering, very few studies have discussed if the origination of MSCs could affect tissue regeneration. Therefore, we need to investigate if it is appropriate to use long-bone derived MSCs to repair cranial bone defects as done previously in many cases. Moreover, too many procedures including stem cell isolation, culture, characterization, and gene transfection in vitro add extra safety concern and complexity that will prevent these approaches from clinical applications. Therefore, cell-free direct gene delivery is a more straightforward and translational strategy for regenerative medicine while it requires developing robust delivery system for local and sustained release of therapeutic molecules to promote, recruit and reprogram endogenous stem cells for tissue regeneration.
Scaffolds for gene delivery and regenerative medicine
In addition to stem cells, scaffolds are another critical component for the success of tissue engineering including miR-meditated bone and dental tissue regeneration [113,114,142,143]. Compared to systemic or bolus delivery, scaffold-based delivery provides more controllable, sustained, and local release of miRs to minimize the off-target side effects or immune reaction [113]. One basic rationale for developing a tissue engineering scaffold/biomaterial is to at least partially mimic the structure and functions of the targeting natural extracellular matrix (ECM), e.g., the bone matrix for bone tissue engineering. Ideally, these synthesized biomimetic biomaterials should not only provide cells with transient and sufficient physical support, e.g., topological and mechanical cues, via its three-dimensional (3D) structure but also instructional chemical signals, e.g., cytokines, growth factors, or miRs to guide these implanted exogenous cells or recruited endogenous cells to proliferate and differentiate until they produce native ECM [184,185]. Accordingly, collagen/gelatin, calcium phosphate/hydroxyapatite, and their composite materials, including hydrogels and 3D scaffolds, are the most used biomaterials for bone and dental tissue engineering because of the similar chemical components to the bone matrix. Additionally, some other natural and synthetic polymers, e.g., chitosan, alginate, polyethylene glycol (PEG), poly (lactic acid) (PLA), poly (glycolic acid) (PGA), and their copolymer poly (lactic-co-glycolic acid) (PLGA), poly(hydroxyl butyrate) (PHB), and poly (caprolactone) (PCL), are also widely used for drug delivery and bone tissue engineering largely because of their excellent biocompatibility and flexibility in composition and fabrication [184,185].
Hydrogels fabricated by either natural or synthetic polymers are attractive for tissue engineering applications because they are highly hydrated and similar to natural ECM. Therefore, a hydrogel can easily encapsulate cells and drugs in a mild condition and mix with them evenly for controlled release through tuning the degree of crosslinking. Moreover, injectable hydrogels are flexible and fit different geometrical deformities with minimal invasiveness which is especially advantageous for craniofacial bone tissue engineering applications [113]. Recently, an innovative PEG hydrogel was synthesized for simultaneous encapsulation of hMSCs and delivery of siNoggin and/or miR-20a. These locally and sustainably released RNAs significantly promoted osteogenic differentiation of hMSCs in vitro and subsequent bone repair in critical-sized rat calvarial defects after transplantation in vivo [186]. In another study, one commercialized hydrogel named HyStem-HP contained thiol-modified hyaluronic acid, gelatin, and heparin was used to deliver miR-26a for successful repair of mouse critical-sized calvarial bone defects by targeting both angiogenesis and osteogenesis [187]. Instead of relying on transplantation of exogenous stem cells for carrying miRs, one study reported that they developed a cell-free nanoparticle/hydrogel composite system for bone regeneration. Both stromal cell-derived factor-1α (SDF-1α) and chitosan/tripolyphosphate/hyaluronic acid/anti-miR-138 nanoparticles were incorporated in chitosan/β-sodium glycerol phosphate hydrogel which sustained release to promote rat critical-sized calvarial bone regeneration by recruiting and differentiating the endogenous stem cells [188].
Another type of biomimetic scaffold/biomaterials that are promising for miR-based regenerative medicine are 3D nanofibrous scaffolds since they closely mimic the nanofiber structure of collagens, the main organic component of ECM. These biomimetic nanofibrous scaffolds have been widely studied in many types of tissues, e.g., skin, vascular, cartilage, bone, and nerve [185,189,190]. The porous nanofibrous scaffolds with the high surface area are valuable because they not only provide a 3D matrix for cell growth and nutrient transport but also enable a high dose of drug loading and local sustained release. Importantly, the topography of nanofiber could directly influence the migration/differentiation of stem cells [185,189,190] and facilitate endocytosis mediated gene transfer through modulating cell-matrix interactions [191,192]. To date, several techniques including electrospinning, phase separation, and self-assembly have been investigated to prepare nanofibrous scaffolds. Among these reported methods, the electrospinning technique has been most widely studied in many fields because it is facile and versatile [193–195]. Numerous studies have proved that electrospun nanofibers are versatile biomaterials for both viral and non-viral vector-mediated gene delivery, e.g., pDNA and siRNA, which were summarized in a recent review [196]. Additionally, a bilayer vascular scaffold was specially prepared via emulsion electrospinning of PEG-b-PLA-co-PCL (PELCL) and dual-power electrospinning of PCL and gelatin. The inner layer of PELCL loaded with complexes of miR-126 in REDV peptide-modified trimethyl chitosan-g-PEG, modulated the response of vascular endothelial cells (VEC), while the outer layer of PCL/gelatin provided the mechanical stability. The bilayer vascular scaffold loaded with miR-126 complexes were capable of accelerating VEC proliferation and improving endothelialization in vivo [197].
The major limitation of most electrospun scaffolds is the lack of macropores for cell growth and tissue formation due to their morphological structure of overlaid nanofiber mats [198]. Thus, it is critical to develop innovative strategies for producing biologically and clinically relevant 3D electrospun nanofibrous scaffolds with desired structural properties. To tackle this significant technical challenge, we and our collaborators have developed an innovative technique of thermally induced nanofiber self-agglomeration (TISA). The 3D electrospun PCL or PCL/PLA blend nanofibrous scaffolds are prepared using a TISA technique having high porosity of >95% as well as interconnected and hierarchically structured pores with sizes from sub-micrometers to ~300 μm. Compared to other 3D porous scaffolds, our 3D TISA nanofibrous scaffolds had higher elasticity, which enable them with press-fit ability suitable for irregular-shaped defects. Importantly, they favorably supported the growth and osteoblastic differentiation of hMSCs in vitro and both ectopic and cranial bone regeneration in vivo [199–202]. Another technique for the preparation of 3D nanofibrous scaffolds in our group is the thermally induced phase separation (TIPS) method with the porogen leaching technique (TIPS&P) [203–208]. One main advantage of these TIPS&P nanofibrous scaffolds fabricated either by PLLA or gelatin is that the macropore structure is well-defined with high interconnectivity and relatively strong mechanical properties. These biodegradable biomimetic 3D nanofibrous scaffolds are especially intriguing for bone tissue engineering with pro-osteoblastic activity and potent drug delivery capacity combined with versatile nanoparticles as we previously reported [209–216]. Furthermore, our group developed an innovative porogen-free TIPS technique for fabrication of 3D porous scaffolds with interconnected, hierarchically structured macropores, and biomimetic nanostructures. The porosity, pore size, and mechanical properties of these porous microspheres-aggregated 3D PCL scaffolds were highly tailorable and showed bone-like apatite forming ability and multiple drug loading and sustained release capacity. Importantly, the 3D nanofibrous scaffolds fabricated by TIPS&P combined with PLGA microspheres were successfully used for locally and controlled release of miR-26a polyplexes for heathy and osteoporotic cranial bone regeneration. This innovative two-stage release and cell-free strategy showed strong bone regenerative ability by harnessing endogenous reparative stem cells and signaling pathways [81]. Moreover, a new PLLA nanofibrous spongy microsphere-delivery system was fabricated and used as an injectable scaffold for both MSN-mediated fast release of IL-2/TGF-β and PLGA microsphere-mediated slow release of miR-10a/polyplexes. In a mouse model of periodontitis, this injectable and biomolecule-delivering system effectively rescued the periodontal bone loss through modulating regulatory T cells mediated immune response [217].
Perspective for biomaterials-based miRNA therapy
As we discussed above, most current miR-based bone regeneration studies rely on ex vivo stem cell transfection before their in vivo transplantation. On one side, this reminds us that stem cells play an important role in tissue repair by either differentiating the target cells or producing trophic and immunomodulatory factors [218]. On the other side, this also indicates that current conventional gene delivery vectors/scaffold systems are not efficient and safe enough to promote sufficient endogenous tissue regeneration using the direct gene therapy strategy. The cell-free strategy is less complicated, more straightforward, and translational than a cell-dependent strategy. This significant challenge urges the development of innovative biomimetic tools for translational miR-based therapy. The exosome is emerging as a promising natural nanoparticle produced by all types of cells for gene delivery with excellent transfection efficiency, biocompatibility, and particular cell/tissue targeting ability. Importantly, exosomes similar to liposomes are capable of being engineered with extra targeting and functional moieties in addition to loading both small miR and large pDNA for customized drug delivery [219–221]. Aptamer, often termed ‘chemical antibodies’, developed by the groundbreaking technology systematic evolution of ligand exponential enrichment (SELEX), is another exciting cell targeting tool for gene therapy [222,223], and have shown promising functions in osteoblasts or MSCs-targeting delivery of nanoparticles/siRNA or miR for successfully improving bone formation in vivo [224,225].
Section V Summary
| miRs | scaffold type/cells | effect on bone growth/regeneration |
|---|---|---|
| miR-20a | PEG hydrogel/hMSCs | promoted osteogenic differentiation |
| mlR-26a | HyStemHP/exogenous stem cells | repair calvarial bone defects |
| anti-miR-138 | chitosan nanoparticles/hydrogel | bone regeneration/endogenous cells |
| miR-126 | PELCL/PCL/gelatin | improved endothelialization |
| miR-26a | PLGA nanosphere/3D PLLA scaffold | bone regeneration |
| miR-10a | PLGA microsphere | rescue bone loss |
VI. CURRENT RESEARCH
Although genetically miR modified BMSCs and ADSCs have exhibited the therapeutic capabilities to generate bone tissue and facilitate bone formation and regeneration. The limited source of the cells and the biosafety of manipulating gene transfection in vitro sets up a huge barrier for their application in the clinic. Thus, the cell-free gene delivery of miRs has the advantage to provide a promising approach for bone regeneration. In comparison to the cell-based approaches, there are relatively few studies that use cell-free gene therapy of miRs for bone regeneration. The mature oligo miR-26a has been attempted to be delivered by nonviral nanoparticles for sustained release from PLGA microspheres [81]. miR-26a was reported to activate Wnt/β-catenin by targeting sclerostin domain containing 1 (SOSTDC1)[79]. The system has been demonstrated to successfully regenerate calvarial bone in a mouse model of critical-sized defects. miR-200c directly targets Noggin, an antagonist of Bmp signaling [66]. Our previous studies have demonstrated that miR-200c also directly targets Sox2 and Klf4 and activated Wnt signaling activities [117]. The expression of miR-200c was upregulated during in vitro osteogenic differentiation of human BMSCs. Inhibition of miR-200c in vivo using the PMIS system has exhibited significant downregulation of craniofacial bone development and the osteogenic differentiation of human BMSCs. We were surprised to observe that the naked plasmid encoding miR-200c can safely and effectively increase the transfection of miR-200c in vitro and in vivo. By over expressing miR-200c we effectively promoted the osteogenic differentiation of human BMSCs in vitro. Furthermore, the plasmid miR-200c incorporated collagen sponges has been shown to significantly improve bone formation and restore different sized calvarial defects in rats [117]. Conversely, miR-200a OE delivered to skull sutures inhibited suture fusion and in PMIS-miR-200a mice calvarial bone development and bone density were increased in the transgenic mice (Fig. 3).
Our group has developed new technology and methods to inhibit miRs both in vivo and in vitro for therapeutic applications that can rapidly and efficiently repair and regenerate bone using several of the delivery methods outlined in this review. The PMIS system can be applied to also inhibit bone formation dependent on the miR that is targeted for inhibition. Experiments in dogs, rats and mice demonstrate that the PMIS is not toxic and highly effective. This is an exciting time for miR therapeutic research and their ability to regenerate bone and tissues.
Highlights.
microRNAs regulate genetic pathways that form bone and inhibit bone regeneration
New microRNA inhibition technology (PMIS) proven in transgenic mice models
microRNAs regulate bone formation, inflammation, osteoporosis and periodontitis
New microRNA inhibition technology (PMIS) more efficacious than oligonucleotides
microRNA gene therapy approaches are compared to stem cell-based scaffolds
ACKNOWLEDGMENTS
We thank members of the Amendt, Sun and Hong laboratories for helpful discussions. We thank the Roy J. Carver Trust for funding the uCT scanner. The following NIH grant mechanisms contributed to this work: NIH grants DE027569; EB025873; DE028527 to BAA; 5T90DE023520; NIH DE026433 to LH; NIH DE029159 to HS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Berendsen AD and Olsen BR (2015) Bone Development. Bone, 80, 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Endo T. (2015) Molecular mechanisms of skeletal muscle development, regeneration, and osteogenic conversion. Bone, 80, 2–13. [DOI] [PubMed] [Google Scholar]
- [3].Chai Y and Maxson RE Jr. (2006) Recent advances in craniofacial morphogenesis. Dev. Dyn, 235, 2353–2375. [DOI] [PubMed] [Google Scholar]
- [4].Shah NM, Groves AK and Anderson DJ (1996) Alternative Neural Crest Cell Fates Are Instructively Promoted by TGFb Superfamily Members. Cell, 85, 331–343. [DOI] [PubMed] [Google Scholar]
- [5].LaBonne C and Bronner-Fraser M. (1999) Molecular mechanisms of neural crest formation. Annual Rev. of Cell and Dev. Biol, 15, 81–112. [DOI] [PubMed] [Google Scholar]
- [6].LeDouarin NM (2004) The avian embryo as a model to study the development of the neural crest: a long and still ongoing story Mech. Dev, 121, 1089–1102. [DOI] [PubMed] [Google Scholar]
- [7].Wilkie AOM and Morriss-Kay GM (2001) Genetics of Craniofacial Development and Malformation. Nat. Rev. Genet, 2, 458–468. [DOI] [PubMed] [Google Scholar]
- [8].Gritli-Linde A. (2008) The etiopathogenesis of cleft lip and cleft palate: usefulness and caveats of mouse models. Curr. Top. Dev. Biol, 84, 37–138. [DOI] [PubMed] [Google Scholar]
- [9].Hu D and Marcucio RS (2009) A SHH-responsive signaling cente in the forebrain regualtes craniofacial morphogenesis via the facial ectoderm. Development, 136, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Young NM, Chong HJ, Hu D, Hallgrímsson B and Marcucio RS (2010) Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development, 137, 3405–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eames BF and Schneider RA (2008) The genesis of cartilage size and shape during development and evolution. Development, 135, 3947–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Solem RC, Eames BF, Tokita M and Schneider RA (2011) Mesenchymal and mechanical mechanisms of secondary cartilage induction. Dev. Biol, 356, 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Parsons TE, Schmidt EJ, Boughner JC, Jamniczky HA, Marcucio RS and Hallgrimsson B. (2011) Epigenetic integration of the developing brain and face. Dev. Dyn, 240, 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Marcucio RS, Young NM, Hu D and Hallgrimsson B. (2011) Mechanisms that underlie co-variation of the brain and face. Genesis, 49, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hutchinson EF, Kieser JA and Kramer B. (2014) Morphometric growth relationships of the immatrue human mandible and tongue. Eur. J. Oral Sci, 122, 181–189. [DOI] [PubMed] [Google Scholar]
- [16].Depew MJ and Compagnucci C. (2008) Tweaking the hinge and caps: testing a model of the organization of jaws. J. Exp. Zoolog. B Mol. Dev. Evol, 310, 315–335. [DOI] [PubMed] [Google Scholar]
- [17].Minoux M and Rijli FM (2010) Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development, 137, 2605–2621. [DOI] [PubMed] [Google Scholar]
- [18].Medeiros DM and Crump JG (2012) New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev. Biol, 371, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Reid BS, Yang H, Melvin VS, Taketo MM and T. W. (2011) Ectodermal Wnt/b-catenin signaling shapes the mouse face. Dev. Biol, 349, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cox TC (2004) Taking it to the max: the genetic and developmental mechanisms coordinating midfacial morphogenesis and dysmorphology. Clin. Genet, 65, 163–176. [DOI] [PubMed] [Google Scholar]
- [21].Cordero DR, Brugmann S, Chu Y, Bajpai R, Jame M and Helms JA (2011) Cranial neural crest cells on the move: their roles in craniofacial development. Am. J. Med. Genet. A, 155, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bush JO and Soriano P. (2010) Ephrin-B1 forward signaling regualtes craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries. Genes & Dev, 24, 2068–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lan Y and Jiang R. (2009) Sonic hedgehog signaling regulates recipricol epithelial-mesenchymal interactions controlling palatal outgrowth. Development, 136, 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I and Rice DP (2004) Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Invest, 113, 1692–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang Z, Song Y, Zhao X, Zhang X, Fermin C and Chen Y. (2002) Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development, 129, 4135–4146. [DOI] [PubMed] [Google Scholar]
- [26].Bush JO and Jiang R. (2012) Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development, 139, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Twigg SR, Kan R, Babbs C, Bochukova EG, Robertson SP, Wall SA, Morriss-Kay GM and Wilkie AO (2004) Mutations of ephrin-B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome. Proc Natl Acad Sci U S A, 101, 8652–8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sasaki T, Ito Y, Xu X, Han J, Bringas J,P,, Maeda T, Slavkin HC, Grosschedl R and Chai Y. (2005) LEF1 is a critical epithelial survival factor during tooth morphogenesis. Dev. Biol, 278, 130–143. [DOI] [PubMed] [Google Scholar]
- [29].He F, Xiong W, Wang Y, Matsui M, Yu X, Chai Y, Klingensmith J and Chen Y. (2010) Modulation of BMP signaling by Noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev Biol, 347, 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Szabo-Rogers HL, Geetha-Loganathan P, Whiting CJ, Nimmagadda S, Fu K and Richman JM (2009) Novel skeletogenic patterning roles for the olfactory pit. Development, 136, 219–229. [DOI] [PubMed] [Google Scholar]
- [31].Sasaki T, Ito S, Bringas P, Chou S, Urata MM, Slavkin H and Chai Y. (2006) TGFb-mediated FGF signaling is crucial for regualting cranial neural crest cell proliferation during frontal bone development. Development, 133, 371–381. [DOI] [PubMed] [Google Scholar]
- [32].Hammond P, McKee S, Suttie M, Allanson J, Cobben JM, Maas SM, Quarrell O, Smith AC, Lewis S, Tassabehji M. et al. (2014) Opposite effects on facial morphology due to gene dosage sensitivity. Hum. Genet, 133, 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Alexander C, Piloto S, Le Pabic P and Schilling TF (2014) Wnt signaling interacts with bmp and edn1 to regualte dorsal-ventral patterning and growth of the craniofacial skeleton. PLoS Genet.. 10, e1004479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, Fuerer C, Clevers H, Nusse R and Helms JA (2007) Wnt signaling mediates regional specification in the vertebrate face. Development, 134, 3283–3295. [DOI] [PubMed] [Google Scholar]
- [35].Wang Y, Song L and Zhou CJ (2011) The canonical Wnt/b-catenin signaling pathway regulates Fgf signaling for early facial development. Dev. Biol, 349, 250–260. [DOI] [PubMed] [Google Scholar]
- [36].Wang X-P, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S and Thesleff I. (2004) Follistatin Regulates Enamel Patterning in Mouse Incisors by Asymmetrically Inhibiting BMP Signaling and Ameloblast Differentiation. Dev. Cell, 7, 719–730. [DOI] [PubMed] [Google Scholar]
- [37].Tummers M and Thesleff I. (2003) Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development, 130, 1049–1057. [DOI] [PubMed] [Google Scholar]
- [38].Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong CM, Schimmang T and Thesleff I. (2007) An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol, 5, e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sun Z, da Fontoura CSG, Moreno M, Holton NE, Sweat M, Sweat Y, Lee MK, Arbon J, Bidlack FB, Thedens DR et al. (2018) FoxO6 regulates Hippo signaling and growth of the craniofacial complex. PLoS Genet, 14, e1007675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang J, Xiao Y, Hsu CW, Martinez-Traverso IM, Zhang M, Bai Y, Ishii M, Maxson RE, Olson EN, Dickinson ME et al. (2016) Yap and Taz play a crucial role in neural crest-derived craniofacial development. Development, 143, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bartel DP (2004) MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- [42].Wienholds E and Plasterk RHA (2005) MicroRNA function in animal development. FEBS Letters, 579, 5911–5922. [DOI] [PubMed] [Google Scholar]
- [43].Alvarez-Garcia I and Miska EA (2005) MicroRNA functions in animal development and human disease. Development, 132, 4653–4662. [DOI] [PubMed] [Google Scholar]
- [44].Iorio MV and Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med, 4, 143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cao H, Yu W, Li X, Wang J, Gao S, Holton NE, Eliason S, Sharp T and Amendt BA (2016) A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene Therapy, 23, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ries RJ, Yu W, Holton N, Cao H and Amendt BA (2017) Inhibition of the miR-17–92 Cluster Separates Stages of Palatogenesis. J. Dent. Res, 96, 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Akkouch A, Eliason S, Sweat ME, Romero-Bustillos M, Zhu M, Qian F, Amendt BA and Hong L. (2019) Enhancement of MicroRNA-200c on Osteogenic Differentiation and Bone Regeneration by Targeting Sox2-Mediated Wnt Signaling and Klf4. Hum Gene Ther, doi: 10.1089/hum.2019.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cao H, Wang J, Li X, Florez S, Huang Z, Venugopalan SR, Elangovan S, Skobe Z, Margolis HC, Martin JF et al. (2010) MicroRNAs play a critical role in tooth development. J. Dent. Res, 89, 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gaur T, Hussain S, Mudhasani R, Parulkar I, Colby JL, Frederick D, Kream BE, van Wijnen AJ, Stein JL, Stein GS et al. (2010) Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse Dev. Biol, 340, 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M and Kronenberg HM (2008) Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A, 105, 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sugatani T and Hruska KA (2009) Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem, 284, 4667–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T, Kato S, Miyasaka N, Ezura Y and Noda M. (2010) Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem, 109, 866–875. [DOI] [PubMed] [Google Scholar]
- [53].Sugatani T, Vacher J and Hruska KA (2011) A microRNA expression signature of osteoclastogenesis. Blood, 117, 3648–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sugatani T, Hildreth B.E.r., Toribio RE, Malluche HH and Hruska KA (2014) Expression of DGCR8-dependent microRNAs is indispensable for osteoclastic development and bone-resorbing activity. J Cell Biochem, 115, 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Michon F, Tummers M, Kyyronen M, Frilander MJ and Thesleff I. (2010) Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev Biol, 340, 355–368. [DOI] [PubMed] [Google Scholar]
- [56].Oommen S, Otsuka-Tanaka Y, Imam N, Kawasaki M, Kawasaki K, Jalani-Ghazani F, Anderegg A, Awatramani R, Hindges R, Sharpe PT et al. (2012) Distinct roles of microRNAs in epithelium and mesenchyme during tooth development. Dev. Dyn, 241, 1465–1472. [DOI] [PubMed] [Google Scholar]
- [57].Wang H, Meng Y, Cui Q, Qin F, Yang H, Chen Y, Cheng Y, Shi J and Guo Y. (2016) MiR-101 Targets the EZH2/Wnt/beta-Catenin the Pathway to Promote the Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells. Sci Reports, 6, 36988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ballarino M, Pagano F, Girardi E, Morlando M, Cacchiarelli D, Marchioni M, Proudfoot NJ and Bozzoni I. (2009) Coupled RNA Processing and Transcription of Intergenic Primary MicroRNAs. Mol Cell Biol, 29, 5632–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Godnic I, Zorc M, Jevsinek Skok D, Calin GA, Horvat S, Dovc P, Kovac M and Kunej T. (2013) Genome-Wide and Species-Wide In Silico Screening for Intragenic MicroRNAs in Human, Mouse and Chicken. PLOS ONE, 8, e65165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y and Takahashi T. (2005) A Polycistronic MicroRNA Cluster, miR-17–92, Is Overexpressed in Human Lung Cancers and Enhances Cell Proliferation. Cancer Res, 65, 9628–9632. [DOI] [PubMed] [Google Scholar]
- [61].Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR et al. (2008) Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17 92 Family of miRNA Clusters. Cell, 132, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Concepcion CP, Bonetti C and Ventura A. (2012) The miR-17–92 family of microRNA clusters in development and disease. Cancer J (Sudbury, Mass.), 18, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang J, Bai Y, Li H, Greene SB, Klysik E, Yu W, Schwartz RJ, Williams TJ and Martin JF (2013) MicroRNA-17–92, a Direct Ap-2α Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting. PLOS Genet., 9, e1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cao H, Yu W, Li X, Wang J, Gao S, Holton NE, Eliason S, Sharp T and Amendt BA (2016) A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene Ther, 23, 527–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Eliason S, Sharp T, Sweat M, Sweat YY and Amendt BA (2020) Ectodermal Organ Development Is Regulated by a microRNA-26b-Lef-1-Wnt Signaling Axis. Frontiers Physiol., 11, 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Cao HJ, Jheon A, Li X, Sun Z, Wang JB, Florez S, Zhang ZC, McManus MT, Klein OD and Amendt BA (2013) The Pitx2:miR-200c/141: noggin pathway regulates Bmp signaling and ameloblast differentiation. Development, 140, 3348–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Godfrey TC, Wildman BJ, Beloti MM, Kemper AG, Ferraz EP, Roy B, Rehan M, Afreen LH, Kim E, Lengner CJ et al. (2018) The microRNA-23a cluster regulates the developmental HoxA cluster function during osteoblast differentiation. J Biol Chem, 293, 17646–17660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zeng HC, Bae Y, Dawson BC, Chen Y, Bertin T, Munivez E, Campeau PM, Tao J, Chen R and Lee BH (2017) MicroRNA miR-23a cluster promotes osteocyte differentiation by regulating TGF-β signalling in osteoblasts. Nat Commun, 8, 15000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J and Lee BH (2012) miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet, 21, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhao C, Sun W, Zhang P, Ling S, Li Y, Zhao D, Peng J, Wang A, Li Q, Song J. et al. (2015) miR-214 promotes osteoclastogenesis by targeting Pten/PI3k/Akt pathway. RNA Biol, 12, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhang L, Tang Y, Zhu X, Tu T, Sui L, Han Q, Yu L, Meng S, Zheng L, Valverde P. et al. (2017) Overexpression of MiR-335–5p Promotes Bone Formation and Regeneration in Mice. J Bone Miner Res, 32, 2466–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lian WS, Ko JY, Chen YS, Ke HJ, Hsieh CK, Kuo CW, Wang SY, Huang BW, Tseng JG and Wang FS (2019) MicroRNA-29a represses osteoclast formation and protects against osteoporosis by regulating PCAF-mediated RANKL and CXCL12. Cell Death Dis, 10, 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhao J, Huang M, Zhang X, Xu J, Hu G, Zhao X, Cui P and Zhang X. (2019) MiR-146a Deletion Protects From Bone Loss in OVX Mice by Suppressing RANKL/OPG and M-CSF in Bone Microenvironment. J Bone Miner Res, 34, 2149–2161. [DOI] [PubMed] [Google Scholar]
- [74].Xie Q, Wei W, Ruan J, Ding Y, Zhuang A, Bi X, Sun H, Gu P, Wang Z and Fan X. (2017) Effects of miR-146a on the osteogenesis of adipose-derived mesenchymal stem cells and bone regeneration. Sci Reports, 7, 42840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Feng Y, Wan P, Yin L and Lou X. (2020) The Inhibition of MicroRNA-139–5p Promoted Osteoporosis of Bone Marrow-Derived Mesenchymal Stem Cells by Targeting Wnt/Beta-Catenin Signaling Pathway by NOTCH1. J Microbiol Biotechnol, 30, 448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yu FY, Xie CQ, Sun JT, Peng W and Huang XW (2018) Overexpressed miR-145 inhibits osteoclastogenesis in RANKL-induced bone marrow-derived macrophages and ovariectomized mice by regulation of Smad3. Life Sci, 202, 11–20. [DOI] [PubMed] [Google Scholar]
- [77].Fukuda T, Ochi H, Sunamura S, Haiden A, Bando W, Inose H, Okawa A, Asou Y and Takeda S. (2015) MicroRNA-145 regulates osteoblastic differentiation by targeting the transcription factor Cbfb. FEBS Lett, 589, 3302–3308. [DOI] [PubMed] [Google Scholar]
- [78].Liu X, Zhu W, Wang L, Wu J, Ding F and Song Y. (2019) miR-145–5p suppresses osteogenic differentiation of adipose-derived stem cells by targeting semaphorin 3A. In Vitro Cell Dev Biol Anim, 55, 189–202. [DOI] [PubMed] [Google Scholar]
- [79].Sun L, Li Z, Xue H, Ma T, Ren C, Li M, Lu Y, Sun H and Zhang K. (2019) MiR-26a promotes fracture healing of nonunion rats possibly by targeting SOSTDC1 and further activating Wnt/beta-catenin signaling pathway. Mol Cell Biochem, 460, 165–173. [DOI] [PubMed] [Google Scholar]
- [80].Li Y, Fan L, Liu S, Liu W, Zhang H, Zhou T, Wu D, Yang P, Shen L, Chen J. et al. (2013) The promotion of bone regeneration through positive regulation of angiogenic-osteogenic coupling using microRNA-26a. Biomaterials, 34, 5048–5058. [DOI] [PubMed] [Google Scholar]
- [81].Zhang X, Li Y, Chen YE, Chen J and Ma PX (2016) Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects. Nat Commun, 7, 10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Qi XB, Jia B, Wang W, Xu GH, Guo JC, Li X and Liu JN (2020) Role of miR-199a-5p in osteoblast differentiation by targeting TET2. Gene, 726, 144193. [DOI] [PubMed] [Google Scholar]
- [83].Chen X, Gu S, Chen BF, Shen WL, Yin Z, Xu GW, Hu JJ, Zhu T, Li G, Wan C. et al. (2015) Nanoparticle delivery of stable miR-199a-5p agomir improves the osteogenesis of human mesenchymal stem cells via the HIF1a pathway. Biomaterials, 53, 239–250. [DOI] [PubMed] [Google Scholar]
- [84].Aquino-Martinez R, Farr JN, Weivoda MM, Negley BA, Onken JL, Thicke BS, Fulcer MM, Fraser DG, van Wijnen AJ, Khosla S. et al. (2019) miR-219a-5p Regulates Rorbeta During Osteoblast Differentiation and in Age-related Bone Loss. J Bone Miner Res, 34, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Arfat Y, Basra MAR, Shahzad M, Majeed K, Mahmood N and Munir H. (2018) miR-208a-3p Suppresses Osteoblast Differentiation and Inhibits Bone Formation by Targeting ACVR1. Molecular therapy. Nucleic acids, 11, 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Deng L, Hu G, Jin L, Wang C and Niu H. (2018) Involvement of microRNA-23b in TNF-alpha-reduced BMSC osteogenic differentiation via targeting runx2. J Bone Miner Metab, 36, 648–660. [DOI] [PubMed] [Google Scholar]
- [87].Zuo B, Zhu J, Li J, Wang C, Zhao X, Cai G, Li Z, Peng J, Wang P, Shen C. et al. (2015) microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J Bone Miner Res, 30, 330–345. [DOI] [PubMed] [Google Scholar]
- [88].Lina S, Lihong Q, Di Y, Bo Y, Xiaolin L and Jing M. (2019) microRNA-146a and Hey2 form a mutual negative feedback loop to regulate the inflammatory response in chronic apical periodontitis. J Cell Biochem, 120, 645–657. [DOI] [PubMed] [Google Scholar]
- [89].Motedayyen H, Ghotloo S, Saffari M, Sattari M and Amid R. (2015) Evaluation of MicroRNA-146a and Its Targets in Gingival Tissues of Patients With Chronic Periodontitis. J Periodontol, 86, 1380–1385. [DOI] [PubMed] [Google Scholar]
- [90].Jiang S, Hu Y, Deng S, Deng J, Yu X, Huang G, Kawai T and Han X. (2018) miR-146a regulates inflammatory cytokine production in Porphyromonas gingivalis lipopolysaccharide-stimulated B cells by targeting IRAK1 but not TRAF6. Biochim Biophys Acta Mol Basis Dis, 1864, 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Stoecklin-Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M and Papapanou PN (2012) MicroRNAs and their target genes in gingival tissues. J Dent Res, 91, 934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Chuang TD and Khorram O. (2014) miR-200c regulates IL8 expression by targeting IKBKB: a potential mediator of inflammation in leiomyoma pathogenesis. PLoS One, 9, e95370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Hong L, Sharp T, Khorsand B, Fischer C, Eliason S, Salem A, Akkouch A, Brogden K and Amendt BA (2016) MicroRNA-200c Represses IL-6, IL-8, and CCL-5 Expression and Enhances Osteogenic Differentiation. Plos One, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Chen S, Tang Y, Liu Y, Zhang P, Lv L, Zhang X, Jia L and Zhou Y. (2019) Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif, 52, e12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Chen S, Zheng Y, Zhang S, Jia L and Zhou Y. (2017) Promotion Effects of miR-375 on the Osteogenic Differentiation of Human Adipose-Derived Mesenchymal Stem Cells. Stem Cell Reports, 8, 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Fan L, Fan J, Liu Y, Li T, Xu H, Yang Y, Deng L, Li H and Zhao RC (2018) miR-450b Promotes Osteogenic Differentiation In Vitro and Enhances Bone Formation In Vivo by Targeting BMP3. Stem Cells Dev, 27, 600–611. [DOI] [PubMed] [Google Scholar]
- [97].Xie Q, Wang Z, Zhou H, Yu Z, Huang Y, Sun H, Bi X, Wang Y, Shi W, Gu P. et al. (2016) The role of miR-135-modified adipose-derived mesenchymal stem cells in bone regeneration. Biomaterials, 75, 279–294. [DOI] [PubMed] [Google Scholar]
- [98].Shi L, Feng L, Liu Y, Duan JQ, Lin WP, Zhang JF and Li G. (2018) MicroRNA-218 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells and Accelerates Bone Fracture Healing. Calcif Tissue Int, 103, 227–236. [DOI] [PubMed] [Google Scholar]
- [99].Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J and Chen J. (2011) Effects of miR-335–5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res, 26, 1953–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zhang L, Tang Y, Zhu X, Tu T, Sui L, Han Q, Yu L, Meng S, Zheng L, Valverde P. et al. (2017) Overexpression of MiR-335–5p Promotes Bone Formation and Regeneration in Mice. J Bone Miner Res, 32, 2466–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Tang Y, Zhang L, Tu T, Li Y, Murray D, Tu Q and Chen JJ (2018) MicroRNA-99a is a novel regulator of KDM6B-mediated osteogenic differentiation of BMSCs. J Cell Mol Med, 22, 2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Mencia Castano I, Curtin CM, Duffy GP and O’Brien FJ (2016) Next generation bone tissue engineering: non-viral miR-133a inhibition using collagen-nanohydroxyapatite scaffolds rapidly enhances osteogenesis. Sci Reports, 6, 27941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zhang H, Yan T, Liu Z, Wang J, Lu Y, Li D and Liang W. (2018) MicroRNA-137 is negatively associated with clinical outcome and regulates tumor development through EZH2 in cervical cancer. J Cell Biochem, 119, 938–947. [DOI] [PubMed] [Google Scholar]
- [104].Kong L, Zuo R, Wang M, Wang W, Xu J, Chai Y, Guan J and Kang Q. (2020) Silencing MicroRNA-137–3p, which Targets RUNX2 and CXCL12 Prevents Steroid-induced Osteonecrosis of the Femoral Head by Facilitating Osteogenesis and Angiogenesis. Int J Biol Sci, 16, 655–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Deng Y, Bi X, Zhou H, You Z, Wang Y, Gu P and Fan X. (2014) Repair of critical-sized bone defects with anti-miR-31-expressing bone marrow stromal stem cells and poly(glycerol sebacate) scaffolds. Eur Cell Mater, 27, 13–24; discussion 24–15. [DOI] [PubMed] [Google Scholar]
- [106].Deng Y, Zhou H, Zou D, Xie Q, Bi X, Gu P and Fan X. (2013) The role of miR-31-modified adipose tissue-derived stem cells in repairing rat critical-sized calvarial defects. Biomaterials, 34, 6717–6728. [DOI] [PubMed] [Google Scholar]
- [107].Li X, Guo L, Liu Y, Su Y, Xie Y, Du J, Zhou J, Ding G, Wang H, Bai Y. et al. (2017) MicroRNA-21 promotes osteogenesis of bone marrow mesenchymal stem cells via the Smad7-Smad1/5/8-Runx2 pathway. Biochem Biophys Res Commun, 493, 928–933. [DOI] [PubMed] [Google Scholar]
- [108].Sadeghi M, Bakhshandeh B, Dehghan MM, Mehrnia MR and Khojasteh A. (2016) Functional synergy of anti-mir221 and nanohydroxyapatite scaffold in bone tissue engineering of rat skull. J Mater Sci Mater Med, 27, 132. [DOI] [PubMed] [Google Scholar]
- [109].Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S and Kassem M. (2011) MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A, 108, 6139–6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yan J, Zhang C, Zhao Y, Cao C, Wu K, Zhao L and Zhang Y. (2014) Non-viral oligonucleotide antimiR-138 delivery to mesenchymal stem cell sheets and the effect on osteogenesis. Biomaterials, 35, 7734–7749. [DOI] [PubMed] [Google Scholar]
- [111].Yan J, Chang B, Hu X, Cao C, Zhao L and Zhang Y. (2018) Titanium implant functionalized with antimiR-138 delivered cell sheet for enhanced peri-implant bone formation and vascularization. Mater Sci Eng C Mater Biol Appl, 89, 52–64. [DOI] [PubMed] [Google Scholar]
- [112].Qadir AS, Um S, Lee H, Baek K, Seo BM, Lee G, Kim GS, Woo KM, Ryoo HM and Baek JH (2015) miR-124 negatively regulates osteogenic differentiation and in vivo bone formation of mesenchymal stem cells. J Cell Biochem, 116, 730–742. [DOI] [PubMed] [Google Scholar]
- [113].Peng B, Chen Y and Leong KW (2015) MicroRNA delivery for regenerative medicine. Advanced drug delivery reviews, 88, 108–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sun X, Guo Q, Wei W, Robertson S, Yuan Y and Luo X. (2019) Current Progress on MicroRNA-Based Gene Delivery in the Treatment of Osteoporosis and Osteoporotic Fracture. International J Endocrinol, 2019, 6782653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Wang Z. (2011) The guideline of the design and validation of MiRNA mimics. Methods in molecular biology (Clifton, N.J.), 676, 211–223. [DOI] [PubMed] [Google Scholar]
- [116].Wang Z. (2009) In Wang Z (ed.), MicroRNA Interference Technologies. Springer Berlin; Heidelberg, Berlin, Heidelberg, pp. 101–110. [Google Scholar]
- [117].Akkouch A, Eliason S, Sweat ME, Romero-Bustillos M, Zhu M, Qian F, Amendt BA and Hong L. (2019) Enhancement of MicroRNA-200c on Osteogenic Differentiation and Bone Regeneration by Targeting Sox2-Mediated Wnt Signaling and Klf4. Hum Gene Ther, 30, 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Akkouch A, Zhu M, Romero-Bustillos M, Eliason S, Qian F, Salem AK, Amendt BA and Hong L. (2019) MicroRNA-200c Attenuates Periodontitis by Modulating Proinflammatory and Osteoclastogenic Mediators. Stem Cells Dev, 28, 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Hong L, Sharp T, Khorsand B, Fischer C, Eliason S, Salem A, Akkouch A, Brogden K and Amendt BA (2016) MicroRNA-200c Represses IL-6, IL-8, and CCL-5 Expression and Enhances Osteogenic Differentiation. PLoS One, 11, e0160915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Cao H, Yu W, Li X, Wang J, Gao S, Holton NE, Eliason S, Sharp T and Amendt BA (2016) A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene therapy, 23, 527–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Lennox KA and Behlke MA (2010) A direct comparison of anti-microRNA oligonucleotide potency. Pharmaceutical Res, 27, 1788–1799. [DOI] [PubMed] [Google Scholar]
- [122].Geary RS, Watanabe TA, Truong L, Freier S, Lesnik EA, Sioufi NB, Sasmor H, Manoharan M and Levin AA (2001) Pharmacokinetic properties of 2’-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J Pharmacol Experimental Therapeutics, 296, 890–897. [PubMed] [Google Scholar]
- [123].Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M and Stoffel M. (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature, 438, 685–689. [DOI] [PubMed] [Google Scholar]
- [124].Cheng P, Chen C, He HB, Hu R, Zhou HD, Xie H, Zhu W, Dai RC, Wu XP, Liao EY et al. (2013) miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res, 28, 1180–1190. [DOI] [PubMed] [Google Scholar]
- [125].Xu R, Shen X, Si Y, Fu Y, Zhu W, Xiao T, Fu Z, Zhang P, Cheng J and Jiang H. (2018) MicroRNA-31a-5p from aging BMSCs links bone formation and resorption in the aged bone marrow microenvironment. Aging Cell, 17, e12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM and Slack FJ (2012) Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proceedings of the National Academy of Sciences, 109, E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF et al. (2009) MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proceedings of the National Academy of Sciences, 106, 3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Zhang X and Godbey WT (2006) Viral vectors for gene delivery in tissue engineering. Advanced drug delivery reviews, 58, 515–534. [DOI] [PubMed] [Google Scholar]
- [129].Wang D, Tai PWL and Gao GP (2019) Adeno-associated virus vector as a platform for gene therapy delivery. Nature Reviews Drug Dis, 18, 358–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Milone MC and O’Doherty U. (2018) Clinical use of lentiviral vectors. Leukemia, 32, 1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].James E and Shuyuan Y. (2005) Retrovirus Silencing and Vector Design: Relevance to Normal and Cancer Stem Cells? Current Gene Therapy, 5, 367–373. [DOI] [PubMed] [Google Scholar]
- [132].Monaghan M, Browne S, Schenke-Layland K and Pandit A. (2014) A Collagen-based Scaffold Delivering Exogenous MicroRNA-29B to Modulate Extracellular Matrix Remodeling. Mol Therapy, 22, 786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, Sergi LS, Benedicenti F, Ambrosi A, Di Serio C. et al. (2006) Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nature Biotechnology, 24, 687–696. [DOI] [PubMed] [Google Scholar]
- [134].Wang FS, Chuang PC, Lin CL, Chen MW, Ke HJ, Chang YH, Chen YS, Wu SL and Ko JY (2013) MicroRNA-29a protects against glucocorticoid-induced bone loss and fragility in rats by orchestrating bone acquisition and resorption. Arthritis and Rheumatism, 65, 1530–1540. [DOI] [PubMed] [Google Scholar]
- [135].Liu Z, Chang H, Hou Y, Wang Y, Zhou Z, Wang M, Huang Z and Yu B. (2018) Lentivirus mediated microRNA 26a overexpression in bone mesenchymal stem cells facilitates bone regeneration in bone defects of calvaria in mice. Mol Med Rep, 18, 5317–5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Zinn E and Vandenberghe LH (2014) Adeno-associated virus: fit to serve. Current Opinion in Virology, 8, 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Yang Y-S, Xie J, Wang D, Kim J-M, Tai PWL, Gravallese E, Gao G and Shim J-H (2019) Bone-targeting AAV-mediated silencing of Schnurri-3 prevents bone loss in osteoporosis. Nat Comm, 10, 2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Lo W-H, Hwang S-M, Chuang C-K, Chen C-Y and Hu Y-C (2009) Development of a Hybrid Baculoviral Vector for Sustained Transgene Expression. Molecular Therapy, 17, 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kost TA and Kemp CW (2016) Fundamentals of Baculovirus Expression and Applications. Advances Experimental Med and Biol, 896, 187–197. [DOI] [PubMed] [Google Scholar]
- [140].Li KC, Chang YH, Yeh CL and Hu YC (2016) Healing of osteoporotic bone defects by baculovirus-engineered bone marrow-derived MSCs expressing MicroRNA sponges. Biomaterials, 74, 155–166. [DOI] [PubMed] [Google Scholar]
- [141].Karvande A, Kushwaha P, Ahmad N, Adhikary S, Kothari P, Tripathi AK, Khedgikar V and Trivedi R. (2018) Glucose dependent miR-451a expression contributes to parathyroid hormone mediated osteoblast differentiation. Bone, 117, 98–115. [DOI] [PubMed] [Google Scholar]
- [142].Leng Q, Chen L and Lv Y. (2020) RNA-based scaffolds for bone regeneration: application and mechanisms of mRNA, miRNA and siRNA. Theranostics, 10, 3190–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Tsekoura EK, K.C RB and Uludag H. (2017) Biomaterials to Facilitate Delivery of RNA Agents in Bone Regeneration and Repair. ACS Biomaterials Science & Engineering, 3, 1195–1206. [DOI] [PubMed] [Google Scholar]
- [144].Zhang G, Guo B, Wu H, Tang T, Zhang B-T, Zheng L, He Y, Yang Z, Pan X, Chow H. et al. (2012) A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat Med 18, 307–314. [DOI] [PubMed] [Google Scholar]
- [145].Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G. et al. (2013) miR-214 targets ATF4 to inhibit bone formation. Nat Med, 19, 93–100. [DOI] [PubMed] [Google Scholar]
- [146].Wang H, Hu Z, Shi F, Dong J, Dang L, Wang Y, Sun Z, Zhou H, Zhang S, Cao X. et al. (2018) Osteoblast-targeted delivery of miR-33–5p attenuates osteopenia development induced by mechanical unloading in mice. Cell Death & Disease, 9, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Liu J, Dang L, Li D, Liang C, He X, Wu H, Qian A, Yang Z, Au DW, Chiang MW et al. (2015) A delivery system specifically approaching bone resorption surfaces to facilitate therapeutic modulation of microRNAs in osteoclasts. Biomaterials, 52, 148–160. [DOI] [PubMed] [Google Scholar]
- [148].Wang D, Miller SC, Shlyakhtenko LS, Portillo AM, Liu X-M, Papangkorn K, Kopečková P, Lyubchenko Y, Higuchi WI and Kopeček J. (2007) Osteotropic Peptide That Differentiates Functional Domains of the Skeleton. Bioconjugate Chemistry, 18, 1375–1378. [DOI] [PubMed] [Google Scholar]
- [149].Son S, Namgung R, Kim J, Singha K and Kim WJ (2012) Bioreducible Polymers for Gene Silencing and Delivery. Accounts of Chemical Research, 45, 1100–1112. [DOI] [PubMed] [Google Scholar]
- [150].Shi B, Zheng M, Tao W, Chung R, Jin D, Ghaffari D and Farokhzad OC (2017) Challenges in DNA Delivery and Recent Advances in Multifunctional Polymeric DNA Delivery Systems. Biomacromolecules, 18, 2231–2246. [DOI] [PubMed] [Google Scholar]
- [151].Pan T, Song W, Gao H, Li T, Cao X, Zhong S and Wang Y. (2016) miR-29b-Loaded Gold Nanoparticles Targeting to the Endoplasmic Reticulum for Synergistic Promotion of Osteogenic Differentiation. ACS Applied Materials & Interfaces, 8, 19217–19227. [DOI] [PubMed] [Google Scholar]
- [152].Schade A, Delyagina E, Scharfenberg D, Skorska A, Lux C, David R and Steinhoff G. (2013) Innovative strategy for microRNA delivery in human mesenchymal stem cells via magnetic nanoparticles. International J Molecular Sciences, 14, 10710–10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Raftery R, Brien FJ and Cryan S-A (2013) Chitosan for Gene Delivery and Orthopedic Tissue Engineering Applications. Molecules, 18, 5611–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Dang JM and Leong KW (2006) Natural polymers for gene delivery and tissue engineering. Advanced Drug Delivery Reviews, 58, 487–499. [DOI] [PubMed] [Google Scholar]
- [155].Katas H and Alpar HO (2006) Development and characterisation of chitosan nanoparticles for siRNA delivery. J Control Release, 115, 216–225. [DOI] [PubMed] [Google Scholar]
- [156].Di Martino A, Sittinger M and Risbud MV (2005) Chitosan: A versatile biopolymer for orthopaedic tissue-engineering. Biomaterials, 26, 5983–5990. [DOI] [PubMed] [Google Scholar]
- [157].LogithKumar R, KeshavNarayan A, Dhivya S, Chawla A, Saravanan S and Selvamurugan N. (2016) A review of chitosan and its derivatives in bone tissue engineering. Carbohydr. Polym, 151, 172–188. [DOI] [PubMed] [Google Scholar]
- [158].Chen X, Gu S, Chen B-F, Shen W-L, Yin Z, Xu G-W, Hu J-J, Zhu T, Li G, Wan C. et al. (2015) Nanoparticle delivery of stable miR-199a-5p agomir improves the osteogenesis of human mesenchymal stem cells via the HIF1a pathway. Biomaterials, 53, 239–250. [DOI] [PubMed] [Google Scholar]
- [159].Wu G, Feng C, Hui G, Wang Z, Tan J, Luo L, Xue P, Wang Q and Chen X. (2016) Improving the osteogenesis of rat mesenchymal stem cells by chitosan-based-microRNA nanoparticles. Carbohydrate polymers, 138, 49–58. [DOI] [PubMed] [Google Scholar]
- [160].Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G. et al. (2014) miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature, 512, 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [161].Inoue K, Deng Z, Chen Y, Giannopoulou E, Xu R, Gong S, Greenblatt MB, Mangala LS, Lopez-Berestein G, Kirsch DG et al. (2018) Bone protection by inhibition of microRNA-182. Nat Commun, 9, 4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Jordan M, Schallhorn A and Wurm FM (1996) Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res, 24, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Olton D, Li J, Wilson ME, Rogers T, Close J, Huang L, Kumta PN and Sfeir C. (2007) Nanostructured calcium phosphates (NanoCaPs) for non-viral gene delivery: influence of the synthesis parameters on transfection efficiency. Biomaterials, 28, 1267–1279. [DOI] [PubMed] [Google Scholar]
- [164].Lee D, Upadhye K and Kumta PN (2012) Nano-sized calcium phosphate (CaP) carriers for nonviral gene deilvery. Materials Science and Engineering: B, 177, 289–302. [Google Scholar]
- [165].Levingstone TJ, Herbaj S and Dunne NJ (2019) Calcium Phosphate Nanoparticles for Therapeutic Applications in Bone Regeneration. Nanomaterials (Basel, Switzerland), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Curtin CM, Cunniffe GM, Lyons FG, Bessho K, Dickson GR, Duffy GP and O’Brien FJ (2012) Innovative collagen nano-hydroxyapatite scaffolds offer a highly efficient non-viral gene delivery platform for stem cell-mediated bone formation. Adv Mater, 24, 749–754. [DOI] [PubMed] [Google Scholar]
- [167].Jung H, Kim SA, Yang YG, Yoo H, Lim SJ and Mok H. (2015) Long chain microRNA conjugates in calcium phosphate nanoparticles for efficient formulation and delivery. Archives of pharmacal research, 38, 705–715. [DOI] [PubMed] [Google Scholar]
- [168].Kesharwani P, Gajbhiye V and Jain NK (2012) A review of nanocarriers for the delivery of small interfering RNA. Biomaterials, 33, 7138–7150. [DOI] [PubMed] [Google Scholar]
- [169].Kelly DC, Raftery RM, Curtin CM, O’Driscoll CM and O’Brien FJ (2019) Scaffold-Based Delivery of Nucleic Acid Therapeutics for Enhanced Bone and Cartilage Repair. J Orthopaedic Res, 37, 1671–1680. [DOI] [PubMed] [Google Scholar]
- [170].Chen S, Li F, Zhuo RX and Cheng SX (2011) Efficient non-viral gene delivery mediated by nanostructured calcium carbonate in solution-based transfection and solid-phase transfection. Molecular bioSystems, 7, 2841–2847. [DOI] [PubMed] [Google Scholar]
- [171].Woldetsadik AD, Sharma SK, Khapli S, Jagannathan R and Magzoub M. (2017) Hierarchically Porous Calcium Carbonate Scaffolds for Bone Tissue Engineering. Acs Biomaterials Science & Engineering, 3, 2457–2469. [DOI] [PubMed] [Google Scholar]
- [172].Yu H-D, Zhang Z-Y, Win KY, Chan J, Teoh SH and Han M-Y (2010) Bioinspired fabrication of 3D hierarchical porous nanomicrostructures of calcium carbonate for bone regeneration. Chemical Communications, 46, 6578–6580. [DOI] [PubMed] [Google Scholar]
- [173].Slowing II, Vivero-Escoto JL, Wu C-W and Lin VSY (2008) Mesoporous silica nanoparticles as controlled release drug delivery and gene transfection carriers. Advanced Drug Delivery Reviews, 60, 1278–1288. [DOI] [PubMed] [Google Scholar]
- [174].Zhou Y, Quan G, Wu Q, Zhang X, Niu B, Wu B, Huang Y, Pan X and Wu C. (2018) Mesoporous silica nanoparticles for drug and gene delivery. Acta Pharmaceutica Sinica B, 8, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Xia T, Kovochich M, Liong M, Meng H, Kabehie S, George S, Zink JI and Nel AE (2009) Polyethyleneimine coating enhances the cellular uptake of mesoporous silica nanoparticles and allows safe delivery of siRNA and DNA constructs. ACS Nano, 3, 3273–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Liu H-J, Luan X, Feng H-Y, Dong X, Yang S-C, Chen Z-J, Cai Q-Y, Lu Q, Zhang Y, Sun P. et al. (2018) Integrated Combination Treatment Using a “Smart” Chemotherapy and MicroRNA Delivery System Improves Outcomes in an Orthotopic Colorectal Cancer Model. Advanced Functional Materials, 28, 1801118. [Google Scholar]
- [177].Vo TN, Kasper FK and Mikos AG (2012) Strategies for controlled delivery of growth factors and cells for bone regeneration. Advanced Drug Delivery Reviews, 64, 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Pelled G, Ben-Arav A, Hock C, Reynolds DG, Yazici C, Zilberman Y, Gazit Z, Awad H, Gazit D and Schwarz EM (2010) Direct gene therapy for bone regeneration: gene delivery, animal models, and outcome measures. Tissue engineering. Part B, Reviews, 16, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Zhao H and Chai Y. (2015) Stem Cells in Teeth and Craniofacial Bones. J Dent Res, 94, 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Huang GT, Gronthos S and Shi S. (2009) Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res, 88, 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Mao X, Liu Y, Chen C and Shi S. (2017) Mesenchymal Stem Cells and Their Role in Dental Medicine. Dental clinics of North America, 61, 161–172. [DOI] [PubMed] [Google Scholar]
- [182].Friedenstein AJ, Petrakova KV, Kurolesova AI and Frolova GP (1968) Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation, 6, 230–247. [PubMed] [Google Scholar]
- [183].Liu Y, Wang S and Shi S. (2012) The role of recipient T cells in mesenchymal stem cell-based tissue regeneration. International J of Biochem Cell Biol, 44, 2044–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Zhang Z, Gupte MJ and Ma PX (2013) Biomaterials and stem cells for tissue engineering. Expert Opinion Biological Therapy, 13, 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Zhang Z, Hu J and Ma PX (2012) Nanofiber-based delivery of bioactive agents and stem cells to bone sites. Advanced Drug Delivery Rev, 64, 1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Nguyen MK, Jeon O, Dang PN, Huynh CT, Varghai D, Riazi H, McMillan A, Herberg S and Alsberg E. (2018) RNA interfering molecule delivery from in situ forming biodegradable hydrogels for enhancement of bone formation in rat calvarial bone defects. Acta biomaterialia, 75, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Li Y, Fan L, Liu S, Liu W, Zhang H, Zhou T, Wu D, Yang P, Shen L, Chen J. et al. (2013) The promotion of bone regeneration through positive regulation of angiogenic–osteogenic coupling using microRNA-26a. Biomaterials, 34, 5048–5058. [DOI] [PubMed] [Google Scholar]
- [188].Wu G, Feng C, Quan J, Wang Z, Wei W, Zang S, Kang S, Hui G, Chen X and Wang Q. (2018) In situ controlled release of stromal cell-derived factor-1α and antimiR-138 for on-demand cranial bone regeneration. Carbohydrate polymers, 182, 215–224. [DOI] [PubMed] [Google Scholar]
- [189].Dahlin RL, Kasper FK and Mikos AG (2011) Polymeric nanofibers in tissue engineering. Tissue engineering. Part B, Reviews, 17, 349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Zamani M, Prabhakaran MP and Ramakrishna S. (2013) Advances in drug delivery via electrospun and electrosprayed nanomaterials. International J Nanomedicine, 8, 2997–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Low WC, Rujitanaroj P-O, Lee D-K, Messersmith PB, Stanton LW, Goh E and Chew SY (2013) Nanofibrous scaffold-mediated REST knockdown to enhance neuronal differentiation of stem cells. Biomaterials, 34, 3581–3590. [DOI] [PubMed] [Google Scholar]
- [192].Adler AF and Leong KW (2010) Emerging links between surface nanotechnology and endocytosis: Impact on nonviral gene delivery. Nano Today, 5, 553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Holzwarth JM and Ma PX (2011) Biomimetic nanofibrous scaffolds for bone tissue engineering. Biomaterials, 32, 9622–9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Matson JB, Zha RH and Stupp SI (2011) Peptide self-assembly for crafting functional biological materials. Current Opinion in Solid State and Materials Science, 15, 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Wang X, Ding B and Li B. (2013) Biomimetic electrospun nanofibrous structures for tissue engineering. Materials Today, 16, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Lee S, Jin G and Jang JH (2014) Electrospun nanofibers as versatile interfaces for efficient gene delivery. J Biological Engineering, 8, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Zhou F, Jia X, Yang Y, Yang Q, Gao C, Hu S, Zhao Y, Fan Y and Yuan X. (2016) Nanofiber-mediated microRNA-126 delivery to vascular endothelial cells for blood vessel regeneration. Acta biomaterialia, 43, 303–313. [DOI] [PubMed] [Google Scholar]
- [198].Blakeney BA, Tambralli A, Anderson JM, Andukuri A, Lim D-J, Dean DR and Jun H-W (2011) Cell infiltration and growth in a low density, uncompressed three-dimensional electrospun nanofibrous scaffold. Biomaterials, 32, 1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Miszuk JM, Xu T, Yao Q, Fang F, Childs JD, Hong Z, Tao J, Fong H and Sun H. (2018) Functionalization of PCL-3D Electrospun Nanofibrous Scaffolds for Improved BMP2-Induced Bone Formation. Appl Mater Today, 10, 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Xu T, Miszuk JM, Zhao Y, Sun H and Fong H. (2015) Electrospun polycaprolactone 3D nanofibrous scaffold with interconnected and hierarchically structured pores for bone tissue engineering. Adv Healthc Mater, 4, 2238–2246. [DOI] [PubMed] [Google Scholar]
- [201].Xu T, Yao Q, Miszuk JM, Sanyour HJ, Hong Z, Sun H and Fong H. (2018) Tailoring weight ratio of PCL/PLA in electrospun three-dimensional nanofibrous scaffolds and the effect on osteogenic differentiation of stem cells. Colloids and surfaces. B, Biointerfaces, 171, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Yao Q, Cosme JG, Xu T, Miszuk JM, Picciani PH, Fong H and Sun H. (2017) Three dimensional electrospun PCL/PLA blend nanofibrous scaffolds with significantly improved stem cells osteogenic differentiation and cranial bone formation. Biomaterials, 115, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Chen VJ and Ma PX (2004) Nano-fibrous poly(L-lactic acid) scaffolds with interconnected spherical macropores. Biomaterials, 25, 2065–2073. [DOI] [PubMed] [Google Scholar]
- [204].Chen VJ, Smith LA and Ma PX (2006) Bone regeneration on computer-designed nano-fibrous scaffolds. Biomaterials, 27, 3973–3979. [DOI] [PubMed] [Google Scholar]
- [205].Liu X, Won Y and Ma PX (2006) Porogen-induced surface modification of nano-fibrous poly(L-lactic acid) scaffolds for tissue engineering. Biomaterials, 27, 3980–3987. [DOI] [PubMed] [Google Scholar]
- [206].Wei G, Jin Q, Giannobile WV and Ma PX (2006) Nano-fibrous scaffold for controlled delivery of recombinant human PDGF-BB. J Control Release, 112, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Wei G, Jin Q, Giannobile WV and Ma PX (2007) The enhancement of osteogenesis by nano-fibrous scaffolds incorporating rhBMP-7 nanospheres. Biomaterials, 28, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Wei G and Ma PX (2006) Macroporous and nanofibrous polymer scaffolds and polymer/bone-like apatite composite scaffolds generated by sugar spheres. J Biomedical Materials Res. Part A, 78, 306–315. [DOI] [PubMed] [Google Scholar]
- [209].Feng K, Sun H, Bradley MA, Dupler EJ, Giannobile WV and Ma PX (2010) Novel antibacterial nanofibrous PLLA scaffolds. J Control Release, 146, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Hu J, Smith LA, Feng K, Liu X, Sun H and Ma PX (2010) Response of human embryonic stem cell-derived mesenchymal stem cells to osteogenic factors and architectures of materials during in vitro osteogenesis. Tissue Eng Part A, 16, 3507–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Liu Y, Hu J and Sun H. (2020) Mineralized nanofibrous scaffold promotes phenamil-induced osteoblastic differentiation while mitigating adipogenic differentiation. J Tissue Eng Regen Med, 14, 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Sun H, Feng K, Hu J, Soker S, Atala A and Ma PX (2010) Osteogenic differentiation of human amniotic fluid-derived stem cells induced by bone morphogenetic protein-7 and enhanced by nanofibrous scaffolds. Biomaterials, 31, 1133–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Yao Q, Fuglsby KE, Zheng X and Sun H. (2020) Nanoclay-functionalized 3D nanofibrous scaffolds promote bone regeneration. J Mater Chem B. [DOI] [PubMed] [Google Scholar]
- [214].Yao Q, Liu Y, Selvaratnam B, Koodali RT and Sun H. (2018) Mesoporous silicate nanoparticles/3D nanofibrous scaffold-mediated dual-drug delivery for bone tissue engineering. J Control Release, 279, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Yao Q, Liu Y, Tao J, Baumgarten KM and Sun H. (2016) Hypoxia-Mimicking Nanofibrous Scaffolds Promote Endogenous Bone Regeneration. ACS Appl Mater Interfaces, 8, 32450–32459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Yao Q, Sandhurst ES, Liu Y and Sun H. (2017) BBP-Functionalized Biomimetic Nanofibrous Scaffold Can Capture BMP2 and Promote Osteogenic Differentiation. J Mater Chem B, 5, 5196–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Liu Z, Chen X, Zhang Z, Zhang X, Saunders L, Zhou Y and Ma PX (2018) Nanofibrous Spongy Microspheres To Distinctly Release miRNA and Growth Factors To Enrich Regulatory T Cells and Rescue Periodontal Bone Loss. ACS Nano, 12, 9785–9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Caplan AI and Correa D. (2011) The MSC: an injury drugstore. Cell Stem Cell, 9, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].El Andaloussi S, Lakhal S, Mager I and Wood MJA (2013) Exosomes for targeted siRNA delivery across biological barriers. Advanced Drug Delivery Rev, 65, 391–397. [DOI] [PubMed] [Google Scholar]
- [220].Ha D, Yang NN and Nadithe V. (2016) Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharmaceutica Sinica B, 6, 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Yeo RWY, Lai RC, Zhang B, Tan SS, Yin YJ, Teh BJ and Lim SK (2013) Mesenchymal stem cell: An efficient mass producer of exosomes for drug delivery. Advanced Drug Delivery Rev, 65, 336–341. [DOI] [PubMed] [Google Scholar]
- [222].Zhou J and Rossi JJ (2014) Cell-type-specific, Aptamer-functionalized Agents for Targeted Disease Therapy. Molecular therapy. Nucleic acids, 3, e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Zhou J and Rossi J. (2017) Aptamers as targeted therapeutics: current potential and challenges. Nature reviews. Drug discovery, 16, 440. [DOI] [PubMed] [Google Scholar]
- [224].Luo ZW, Li FX, Liu YW, Rao SS, Yin H, Huang J, Chen CY, Hu Y, Zhang Y, Tan YJ et al. (2019) Aptamer-functionalized exosomes from bone marrow stromal cells target bone to promote bone regeneration. Nanoscale, 11, 20884–20892. [DOI] [PubMed] [Google Scholar]
- [225].Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, Xia ZY, Zhou HD, Cao X, Xie H. et al. (2015) MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest, 125, 1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]