Abstract
Alarming rate of resistance to the existing antibiotics exhibits the importance of developing new antibiotic molecules from relatively under explored sources as well as implementing alternative approaches like antibiotic adjuvants. Six previously undescribed fungal polyketides, kaneoheoic acids A-F (1–6) were isolated from a fungal strain Fusarium sp. FM701 which was collected from a muddy sample of Hawaiian beach. The structures of these six compounds were elucidated by spectroscopic interpretation, including HRESIMS and NMR, and electronic circular dichroism (ECD) analysis. All six compounds that were inactive when tested alone showed significant antibacterial activity against Staphylococcus aureus and Bacillus subtilis, in the range of 10–80 μg/mL when assayed in combination with either chloramphenicol (half of the MIC, 1 μg/mL), an FDA approved antibiotic or disulfiram (6 μg/mL), an established antibiotic adjuvant that augmented the activity of antibiotics.
Keywords: Hawaiian fungi, Fusarium sp., polyketides, Antibacterial activity, Disulfiram
1. Introduction
Due to rapid growth of population and growing resistance to existing anti-biotic drugs, there is a need for the development of new antibiotics from relatively new natural sources. One approach is to uncover new biologically active molecules from unique ecological niches, which have the potential to be developed further into drugs. Fungi, as a rich source, produce a huge number of biologically active secondary metabolites, including a wide variety of clinically significant drugs. For example, fungi play a vital role in the production of beta-lactam antibiotics like penicillin and cephalosporin, as well as the immunosuppressant cyclosporine and cholesterol lowering agents, compactin and lovastatin [Aly et al., 2011; Manzoni and Rollini, 2002]. Considering the biodiversity of the fungal kingdom and the fact that only a small fraction of fungi has ever been explored for the secondary metabolites with biological activity, it is obvious that a variety of biologically active compounds are still to be discovered. Located in the central Pacific Ocean, Hawaii has its own ecosystems with a unique biodiversity and fungi play a vital role in this system. In our continuing search for biologically active compounds from Hawaiian fungi [Zaman et al., 2020; Wang et al., 2019a, Wang et al., 2020; Li et al., 2019, 2018; Fei-Zhang et al., 2016; Huang et al., 2017], we isolated a fungus Fusarium sp. FM701 (Genbank accession # MW130722) from a muddy sample collected at the Kaneohe Bay, Oahu, Hawaii. Fusarium species is well-known to produce mycotoxins such as fumonisins, fusaric acid, trichothecenes, fusaproliferin, moniliformin, and enniatins, with unique structures including polyketides, alkaloids, and terpenoids [Fotso et al., 2002]. These types of compounds are interesting due to broad-spectrum of biological properties, for example, antifungal, antibacterial, insecticidal, and cytotoxic activities [Song et al., 2015]. Recently, a crude methanolic extract of Fusarium sp. FM701, which was not active by itself at the concentration of 80 μg/mL against gram-positive bacteria Staphylococcus aureus and Bacillus subtilis, showed promising antibacterial activity when tested together with antibiotic adjuvant disulfiram. Although disulfiram alone exhibited some antibacterial activity against S. aureus [Ejim et al., 2011], CA-MRSA CA-347 [Long, 2017], and HA-MRSA COL [Long, 2017] with MIC values of 32, 16 and 8 μg/mL, respectively, our results showed that when tested against S. aureus (ATCC® 12600™) and Bacillus subtilis (ATCC®6633™) disulfiram alone was not active at 24 μg/mL. Remarkably, disulfiram also has the ability to enhance the activity of FDA-approved antibiotics [Ejim et al., 2011], indicating that pathogenic bacteria are more susceptible to antibiotic agents in the presence of disulfiram. Our bioassay-guided fractionation using disulfiram as an adjuvant led to the identification of six previously undescribed polyketides (1–6). Herein, we report the isolation, structural elucidation by HR-ESIMS, NMR spectral interpretation, and ECD analysis, and the biological evaluation of compounds 1–6 from Fusarium sp. FM701.
2. Results and discussion
2.1. Structural elucidation of the previously undescribed compounds
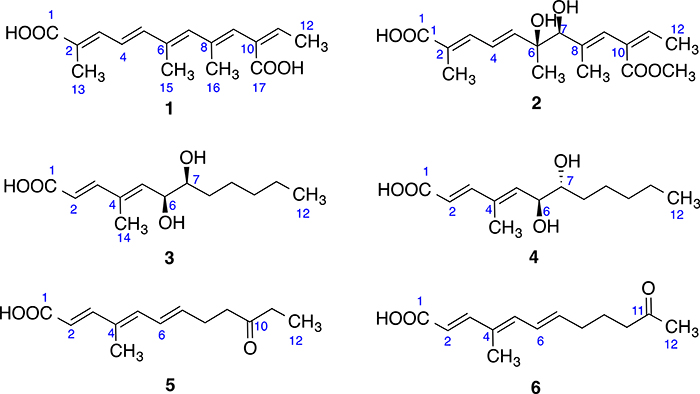
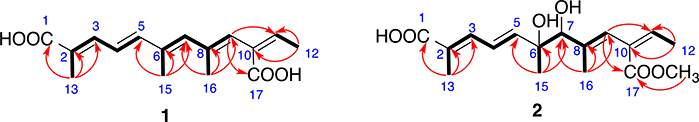
Compound 1 (Fig. 1) was obtained as whitish powder and its molecular formula was determined as C16H20O4 by HRESIMS, requiring 7 degrees of unsaturation. Comprehensive analysis of the 1H, 13C and HSQC NMR spectra indicated the presence of 16 carbons including four methyl groups (4 × CH3), six olefinic methines (6 × CHᆖC) and six non-protonated carbons including two carboxyl groups (2 × -COO-) (Table 1). 1H–1H COSY spectrum establishes three spin systems, –C(CH3) = CH–CHᆖCH-, –C(CH3) = CH–C(CH3) = CH-, and-CᆖCH–CH3 (Fig. 2). HMBC correlations (Fig. 2) from H3-15 to C5, C6 and C7 enabled us to connect the first two spin systems together as –C(CH3) = CH–CHᆖCH–C(CH3) = CH–C(CH3) = CH-. In the HMBC spectrum of 1, H-9 correlated to C-11 and C-17 (a carbonyl), indicating that these three spin systems formed a 2,6,8,10-tetrasubstituted straight chain with five double bonds at 2-, 4-, 6-, 8, and 10-positions [-C(CH3) = CH–CHᆖCH–C(CH3) = CH–C(CH3) = CH–C(COO)ᆖCH–CH3]. HMBC correlations from H3-13 to C-1 (a carbonyl), C-2 and C-3 established the flat structure of compound 1 as HOOC–C(CH3) = CH–CHᆖCH–C(CH3) = CH–C(CH3) = CH–C(COOH)ᆖCH–CH3. The conjugated double bonds at 2-, 4-, 6-, 8, and 10-positions were determined to be trans on the basis of NOESY correlations and comparison of chemical shifts with literature [Vesonder, 1996; Alécio et al., 1998]. Compound 1 is a derivative of 2,4, 5,8,10-dodecapentaenoic acid with a carboxyl group at 10-position and methyl groups at 2-, 6-, and 8-positions [Jaffe et al., 1966]. Hence, the structure of compound 1 was determined as shown, which was given a trivial name kaneoheoic acid A.
Fig. 1.
Chemical structures of compounds 1–6.
Table 1.
1H and 13C NMR spectroscopic data for compounds 1 and 2 in CD3OD.
| no. | 1 |
2 |
||
|---|---|---|---|---|
| δC | δH mult. J (Hz) | δC | δH multi. J (Hz) | |
| 1 | 171.3 | 171.1 | ||
| 2 | 126.9 | 126.6 | ||
| 3 | 137.8 | 7.28 d (10.6) | 137.0 | 7.23d (11.4) |
| 4 | 122.6 | 6.63 t dd (11.3, 15.2) | 138.8 | 6.70 dd (11.3, 15.2) |
| 5 | 144.0 | 6.66d (15.1) | 145.7 | 6.25 d (15.2) |
| 6 | 134.5 | 75.2 | ||
| 7 | 137.6 | 6.26 s | 81.7 | 4.06 s |
| 8 | 136.3 | 141.0 | ||
| 9 | 125.7 | 6.11 s | 120.7 | 5.99 s |
| 10 | 132.9 | 130.1 | ||
| 11 | 136.3 | 6.86 m | 138.8 | 6.94 m |
| 12 | 14.3 | 1.79 d (7.2) | 14.2 | 1.76 d (7.1) |
| 13 | 11.4 | 1.99 s | 11.2 | 1.95 s |
| 15 | 12.75 | 2.11 s | 23.5 | 1.36 s |
| 16 | 17.5 | 1.76 s | 14.1 | 1.59 s |
| 17 | 170.9 | 168.0 | ||
| 19 | 50.4 | 3.73 s | ||
Fig. 2.
Key COSY (bolds) and HMBC (red arrows) correlations of compounds 1 and 2.
The molecular formula of compound 2 (Fig. 1) was determined as C17H24O6 by HR-ESIMS analysis, with 6 degrees of unsaturation. 1H, 13C and HSQC spectra confirmed 17 carbons in 2 including five methyl (5 × CH3, including one O–CH3), one oxygenated methane (1 × CH–O), five olefinic carbons (5 × CHᆖC), and six non-protonated carbons including one oxygenated carbon and two carboxyl groups (2 × -COO-) (Table 1). The 1H NMR and HSQC spectra of 2 were similar to those of 1 except 6-, 7-, and 17-positions. Like 1, COSY spectrum of 2 showed three spin systems, –C(CH3) = CH–CHᆖCH-, –C(CH3)–CH–C(CH3) = CH-, and- CᆖCH–CH3 (Fig. 2). HMBC correlations (Fig. 2) from H3-15 to C-5, C-6 (oxygenated), and C-7 (oxygenated), and from the oxygenated methyl (O–CH3) to C-17 indicated that 2 was a derivative of 1 with a diol at 6-/7-positions and a methyl ester at 17-position. The double bonds at 2-, 4-, 8, and 10-positions were determined to be trans on the basis of NOESY correlations and comparison of chemical shifts with literature [Vesonder, 1996; Alécio et al., 1998].
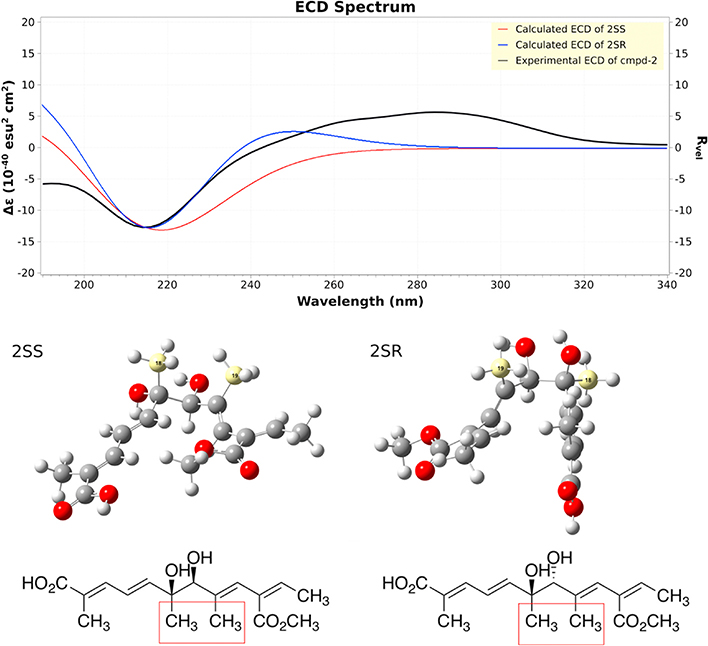
To determine the configuration of compound 2, ECD spectra were collected (Fig. 3). Compound 2 showed a strong negative Cotton effect at 210–220 nm. The calculated weighted ECD spectra of 2SS and 2SR showed a strong negative Cotton effect at 210–220 nm, coinciding with the experimental ECD spectrum (Fig. 3), which excluded 2RR and 2RS as the possible structure of 2. To verify the above analysis, we carried out NMR calculations of 2SS and 2SR with the conformers generated for ECD calculation. Results showed that the calculated NMR data of 2SS matched the experimental NMR data of 1 better than 2SR (Table S1), which enabled final assignment of the absolute configuration of 2 as shown. Compound 2 was given a trivial name kaneoheoic acid B.
Fig. 3.
Experimental and calculated ECD of compound 2.
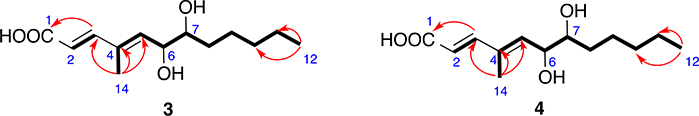
Compound 3 (Fig. 1) exhibited a prominent deprotonated molecule peak at m/z 241.1433 [M - H]− in the HR-ESIMS spectrum, suggesting a molecular formula of C13H22O4. 1H NMR and HSQC data (Table 2) defined two methyls (2 × CH3), four methylenes (4 × CH2), two oxygenated aliphatic methines (2 × CH–O), three olefinic methines (3 × CHᆖC), one carbonyl (1 × O–CᆖO), and one non-protonated carbon. 1H–1H COSY spectrum demonstrated two spin systems –CHᆖCH- and –C(CH3) = CH–CH(O)–CH(O)–CH2–CH2–CH2–CH2–CH3 (Fig. 4). HMBC correlations from H3-14 to C-3, C-4, and C-5 connected the two spin systems as –CHᆖCH–C(CH3)=CH–CH(O)–CH(O)–CH2–CH2–CH2–CH2–CH3. HMBC correlation from H-3 to C-1 (Fig. 4) established the flat structure as HOOC–CHᆖCH–C(CH3)=CH–CH(OH)–CH(OH)–CH2–CH2–CH2–CH2–CH3. The double bonds at 2- and 4-positions were also determined to be trans. Compound 3 was structurally similar to 2E,4E,6R-4,6-dimethyl-dodecadienoic acid [Yu et al., 2020], but has 1,2-diol moiety at 6- and 7-position instead of a methyl group at 6-position.
Table 2.
NMR spectroscopic data for compounds 3–6 in CD3OD.
| no. | 3 |
4 |
5 |
6 |
||||
|---|---|---|---|---|---|---|---|---|
| δC | δH multi. J (Hz) | δC | δH multi. J (Hz) | δC | δH multi. J (Hz) | δC | δH multi. J (Hz) | |
| 1 | 170.1 | 170.1 | 170.1 | 170.7 | ||||
| 2 | 120.7 | 5.96 d (15.6) | 120.7 | 5.96 d (15.6) | 117.3 | 5.86 d (15.6) | 117.3 | 5.86 d (15.5) |
| 3 | 146.4 | 7.25 d (15.5) | 147.8 | 7.27 d (15.5) | 148.9 | 7.30 d (15.5) | 148.3 | 7.30 d (15.4) |
| 4 | 135.9 | 134.7 | 131.4 | 132.3 | ||||
| 5 | 137.4 | 5.88 d (8.9) | 138.5 | 5.85 d (9.0) | 138.3 | 6.41 d (11.3) | 137.5 | 6.38 d (11.1) |
| 6 | 70.9 | 4.33 dd (5.1, 8.9) | 70.9 | 4.30 dd (5.9, 9.0) | 127.0 | 6.51 t (11.9) | 126.9 | 6.51 t (13.0) |
| 7 | 74.2 | 3.55 m | 74.3 | 3.48 m | 139.3 | 5.97 m | 137.7 | 5.97 m |
| 8 | 32.3 | 1.55 m | 32.3 | 1.38 m 1.50 m |
32.4 | 2.20 m | 27.0 | 2.46 m |
| 9 | 25.2 | 1.35 m 1.55 m |
25.2 | 1.36 m 1.51 m |
22.9 | 1.70 m | 40.9 | 2.62 t (7.2) |
| 10 | 31.7 | 1.33 m | 31.6 | 1.33 m | 42.4 | 2.53 t (7.3) | 212.3 | |
| 11 | 22.3 | 1.34 m | 22.2 | 1.34 m | 210 | 35.3 | 2.50 m | |
| 12 | 13.0 | 0.92 t (6.6) | 13.0 | 0.94 t (6.8) | 28.7 | 2.14 s | 6.77 | 1.05 t (7.3) |
| 14 | 11.8 | 1.88 s | 11.8 | 1.90 s | 11.4 | 1.90 s | 11.4 | 1.90 s |
Fig. 4.
Key COSY (bolds) and HMBC (red arrows) correlations of compounds 3 and 4.
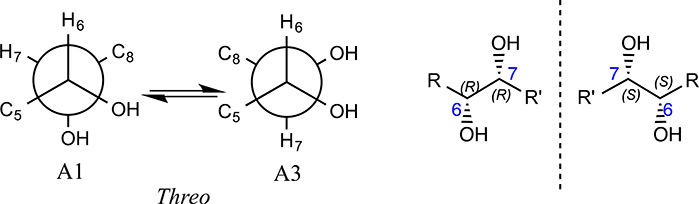
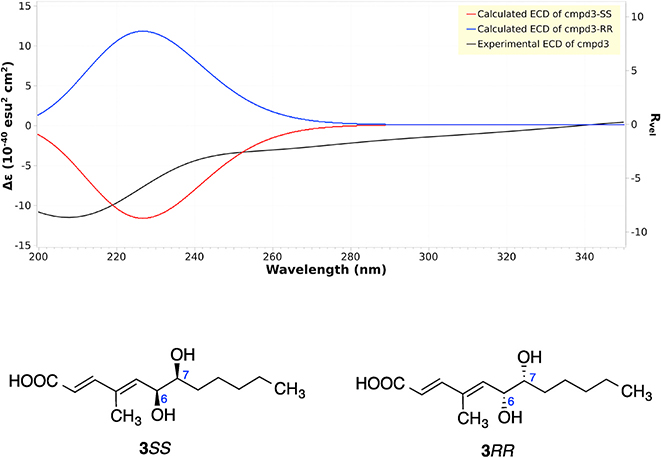
To determine the relative configurations of compound 3, we next carried out a J-based configuration analysis to determine the configuration of 6- and 7-positions. For the 1,2 methine system at C6–7, the 3JH-6,H-7 (4.9 Hz), 2JH-6,C-7 (−1.6Hz), 2JH-7,C-8 (−2.6Hz), and 3JH-7,C-5 (1.8 Hz) values indicated a Threo configuration for compound 3, which means that it could be 3RR or 3SS (Fig. 5). To determine the absolute configuration of compound 3, ECD spectra were collected, and ECD calculations were carried out. Compound 3 showed a strong negative Cotton effect at 210–215 nm, which was similar to the calculated weighted ECD spectra of 3SS (Fig. 6). Hence, the structure including the absolute configuration of compound 3 was determined as shown, and it was given a trivial name kaneoheoic acid C.
Fig. 5.
J-based configuration analysis of compound 3.
Fig. 6.
Experimental and calculated ECD of compound 3.
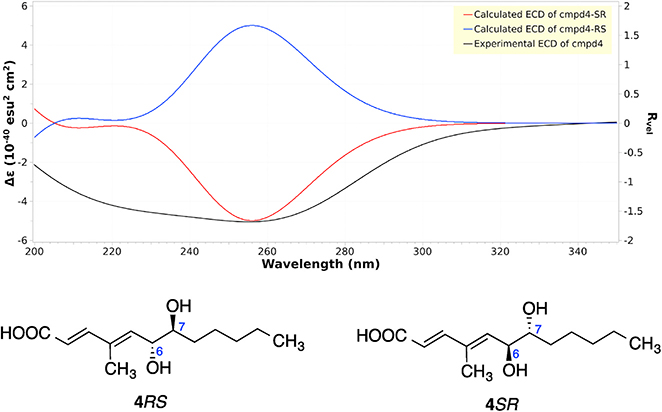
Compound 4 (Fig. 1) also exhibited a peak at m/z 241.1433 [M - H]− in the HR-ESIMS spectrum, however with a different retention time. The NMR spectra of 4 were almost the same as those of compound 3. So it is most likely a stereoisomer of compound 3, which means it is either 4SR or 4RS. ECD calculations were carried out to determine the absolute configuration of compound 4, which showed a strong negative Cotton effect at 240–260 nm, coinciding with the calculated weighted ECD spectra of 4SR (Fig. 7). Hence, the structure including the absolute configuration of compound 4 was determined as shown, and it was given a trivial name kaneoheoic acid D.
Fig. 7.
Experimental and calculated ECD of compound 4.
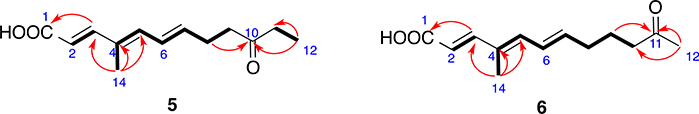
Compound 5 (Fig. 1) is a light brownish powder whose molecular formula was determined to be C13H18O3 from HR-ESIMS. 1H–1H COSY spectrum exhibited three spin systems, –CHᆖCH-, and –C(CH3)= CH–CHᆖCH–CH2–CH2-, and –CH2-CH3. The double bonds at 2-, 4- and 6-positions were determined to be trans. HMBC correlations (Fig. 8) from H3-14 to C-3, C-4, and C-5, from H3-12 to C-10 (a carbonyl), and C-11, from H2-8 to C-10, and from H-3 to C-1 (a carboxyl) connected the three spin systems together, and established the structure as shown. Compound 5 was given a trivial name kaneoheoic acid E.
Fig. 8.
Key COSY (bolds) and HMBC (red arrows) correlations of compounds 5 and 6.
Compound 6 (Fig. 1) was isolated as light brownish powder, and was determined to have the same molecular formula C13H18O3 as 5 on the basis of HR-ESIMS. However, 6 had a different retention time, suggesting that 6 was a stereoisomer of compound 5. The NMR spectra of 6 were very similar to those of 5. The only difference between compound 5 and compound 6 was the location of the ketone group, which was at 11-position in 6 instead of 10-position in 5. HMBC correlations from the methyl singlet at 12-position to C-11 (a ketone) and C-10 (Fig. 8) confirmed the location of the double bond. Hence, the structure of compound 6 was determined as shown, and it was given a trivial name kaneoheoic acid F. Compound 6 was an analog of 2E,4E,6E-6-hydroxy-4-methyl-dodecatrienoic acid [Lorenzen et al., 1996], but has a ketone at 11-position instead of a hydroxy group at 11-position.
2.2. Antibacterial and anti-proliferative activity of the kaneoheoic acids (1–6)
All these six previously undescribed compounds (1–6) are derivatives of dodecanoic acid (lauric acid) with different functional groups including double bound, hydroxy, methyl, ketone, carboxyl and carboxymethyl ester. It has been reported that lauric acid and its derivatives have antibacterial (Rouse et al., 2005), antifungal (Rihakova et al., 2001), antitumour (Kato et al., 1971), anti-inflammatory (Calder and Grimble, 2002), antimycobacterial (Saravanakumar et al., 2008) and antiviral (Villamor et al., 2007) activities. To test the biological activities of kaneoheoic acids A-F (1–6), we evaluated them in our antibacterial and anti-proliferative assays.
Compounds 1–6 were evaluated for their antibacterial activity against S. aureus, B. subtilis. and E. coli. Although none of them were active by themselves, even at 80 μg/mL, all the compounds showed significant antibacterial activity against S. aureus when given with 6 μg/ mL disulfiram. In the presence of disulfiram (6 μg/mL), compounds 4 and 5 (MIC 10 μg/mL each) were more potent than compounds 1 and 6 (MIC 20 μg/mL each), which were more active than compounds 2 and 3 (MIC 40 μg/mL each). Moreover compounds 2–6 also showed mild antibacterial activities against B. subtilis with MICs ranging from 40 to 80 μg/mL, when tested in the presence of chloramphenicol with half of its MIC value (1 μg/mL) (Table 3 and S2).
Table 3.
Activities of compounds 1–6 against S. aureus (ATCC® 12600™) and B. subtilis (ATCC®6633™) in the presence of disulfiram (6 μg/mL) or chloramphenicol (1 μg/mL).
| Compound | MIC [μg/mL] |
||||
|---|---|---|---|---|---|
| S. aureus |
B. subtilis |
||||
| Compound alone | Compound + Disulfiram [6 μg/mL] | Compound alone | Compound + Chloramphenicol [1 μg/mL] | ||
| 1 | NA | 20 | NA | NA | |
| 2 | NA | 40 | NA | 80 | |
| 3 | NA | 40 | NA | 40 | |
| 4 | NA | 10 | NA | 80 | |
| 5 | NA | 10 | NA | 80 | |
| 6 | NA | 20 | NA | 40 | |
NA→ Not Active.
To compare our findings with commercially available short, medium, and long chain fatty acids, we also tested five fatty acids such as butyric acid [CH3-(CH2)2–COOH], capric acid [CH3-(CH2)8–COOH], lauric acid [CH3-(CH2)10–COOH], myristic acid [CH3-(CH2)12–COOH] and linoleic acid [C18H32O2, CH3-(CH2)4-ZCH=CH–CH2-ZCH=CH-(CH2)7-COOH] for their antibacterial properties against S. aureus and B. subtilis in the presence of disulfiram or chloramphenicol. Although most of them showed very mild antibacterial activity with MIC values ranging from 63 μg/mL to 1.1 mg/mL when tested alone, in almost every case the MIC values decreased 2–4 folds when combined with either disulfiram or chloramphenicol (Table S3). These findings confirmed the synergistic potentials of the newly isolated fatty acids (1–6) from Fusarium sp. FM701. Although much less potent than compounds 1–6, lauric acid (a medium chain fatty acid) with twelve carbon atoms like compounds 1–6 was the most active among all the purchased fatty acids. Lauric acid and some other fatty acids have been investigated for their antibacterial activity extensively [Butt et al., 2016; Fisher et al., 2012; Rouse et al., 2005], but lauric acid in combination with disulfiram has not been studied before for its anti-S. aureus activity. The results strongly suggested that the lengths of the fatty acids and functional groups in the molecules (for examples, double bonds, hydroxy groups, and methyl groups) are important for the antibacterial activity.
Compounds 1–6 were further assayed for their anti-proliferative activity (Cao et al., 2010) against A2780 human ovarian cancer cells and HEK293 human embryonic kidney cells, but none was active at 40 μM (the highest concentration tested). The results indicated that non-toxic fatty acids like compounds 1–6 have potential to be antibiotics when used together with the adjuvant disulfiram.
3. Conclusion
In conclusion, six previously undescribed medium chain polyketide-derived fatty acids, kaneoheoic acids A-F (1–6) from Fusarium sp. FM701 are unique in structure with promising antibacterial potentials against gram-positive pathogenic bacteria. Compared to the other fatty acids, these previously undescribed fatty acids from Fusarium sp. FM701 showed significant antibacterial activities against S. aureus and B. subtilis when screened co-currently with either antibiotic adjuvant disulfiram or the FDA approved antibiotic chloramphenicol. The combination of FDA-approved non-antibiotic drugs (e.g., disulfiram) which acts as antibiotic adjuvant or enhancer with non-toxic fatty acids provides an opportunity to expand a previously untapped bioactive chemical space in the field of antibiotic drug discovery. The findings from the present study warrant future research on the mechanism of action behind these synergistic activities.
4. Experimental
4.1. General experimental procedure
Optical rotations, CD and FT-IR spectra were measured with a Rudolph research analytical autoPol automatic polarimeter, JASCO J-815 CD and Thermo scientific nicolet iS10 IR spectrometer, respectively. 1D and 2D NMR spectra were recorded on a Bruker AM-400 spectrometer. The 3.35 ppm and 49.3 ppm resonances of CD3OD were used as internal references for 1H and 13C NMR spectra, respectively. An Agilent 6530 accurate-Mass Q-TOF LC-MS spectrometer was used to record high-resolution mass spectra. Preparative HPLC was carried out on an Ultimate 3000 chromatographic system with a Phenomenex preparative column (Phenyl-Hexyl, 5 μm, 100 × 21.2 mm) and semi-preparative HPLC on an Ultimate 3000 chromatographic system with a Phenomenex semi-preparative column (C8, 5 μm, 250 × 10 mm), a Dionex Ultimate 3000 DAD detector and a Dionex Ultimate 3000 automated fraction collector; and all solvents were HPLC grade. Diaion HP-20 was used to run open column chromatography.
4.2. Strain isolation and fermentation
The strain FM701 was isolated from a muddy sample collected at the Kaneohe Bay, Oahu, Hawaii. The strain was sub-cultured on potato dextrose agar containing 15% marine sea water and maintained at −80 °C in 20% glycerol at Daniel K. Inouye College of Pharmacy, University of Hawaii at Hilo, HI, USA. The frozen strain from −80 °C freezer was activated on PDA plates at 28 °C for 5 days, then it was cut into small pieces and inoculated into 15 L autoclaved sterilized liquid medium [mannitol 20 g, glucose 10 g, monosodium glutamate 5 g, KH2PO4 (0.5 g), MgSO4·7H2O 0.3 g and yeast extract 3 g for 1 L distilled water; pH 6.5 prior sterilization] for fermentation at 24 °C for 28 days in static condition.
4.3. Molecular identification
DNA extraction: DNA was extracted according to the literature (Liu et al., 2000), with slight modifications. Mycelium was added to 500 μl of lysis buffer (400 mM Tris-HCl [pH 8.0], 60 mM EDTA, 150 mM NaCl, 1% sodium dodecyl sulfate) and incubated at 85°C for 20 minutes. After adding 150 μl of 3 M sodium acetate (pH 5.2), the tube was vortexed briefly and centrifuged (12,500 x g) for 1 minute. The supernatant was transferred to another tube and centrifuged again. After transferring the supernatant to a new tube, an equal volume of isopropanol was added and mixed by inversion. The tube was centrifuged for 2 minutes and the supernatant was discarded. The DNA pellet was washed twice with 300 μl of 70% ethanol. The DNA was air dried at room temperature for 45 minutes, then dissolved in 100 μl of 10 mM Tris-HCl (pH 8.0).
Sequencing of ITS region: The ITS region was amplified with the ITS1 and ITS4 primers. The PCR reaction included 1X High Fidelity PCR Buffer (Invitrogen), 2 mM MgSO4, 0.2 mM dNTP mix, 4% DMSO, 0.2 μM of each primer, 1 U Platinum Taq DNA Polymerase High Fidelity (Invitrogen), and 10 ng of genomic DNA. The PCR cycling conditions were 95°C for 3 minutes, followed by 35 cycles of 95°C for 30 seconds, 50°C for 30 seconds and 72°C for 1 minute, and a final extension of 72°C for 5 minutes. The PCR product was purified using Mag-Bind Total Pure NGS beads (Omega Bio-tek), then sequenced using a 3730xl DNA Analyzer (Applied Biosystems). The sequence was compared to the NCBI nucleotide collection (limited to sequences from type material) using the Basic Local Alignment Search Tool (BLAST), and was deposited in GenBank under the accession no. MW130722.
4.4. Extraction and isolation
The mycelia of FM107 were filtered and extracted with acetone under ultrasonic (2 L × 3 times), followed by removal of acetone under reduced pressure to afford an aqueous solution. After combining the aqueous mycelia extract and supernatant solution, it was subjected to HP-20 column eluted with MeOH–H2O (10, 50, 90 and 100%) to afford four fractions (Fr 1–4). Fraction 3 (12 g) was separated by prep-HPLC (Phenyl-Hexyl, 5 μm, 100 × 21.2 mm; 8 mL/min) eluted with 40–80% MeOH–H2O in 20 min to yield sub-fractions (SFr 3–1–20). Compound 1 (0.9 mg, tR 13 min) was separated from SFr 3–13 by using a semi-preparative HPLC (50% isocratic of MeOH–H2O with 0.1% formic acid for 20 min). SFr 3–10 was purified by using a semi-preparative HPLC (60% isocratic of MeOH–H2O with 0.1% formic acid for 20 min; 3 mL/min) to afford compounds 2 (1.2 mg, tR 12 min), 3 (0.8 mg, tR 17 min), and 4 (1.6 mg, tR 19 min) while compound 5 (1.8 mg, tR 16 min) and 6 (2 mg, tR 19 min) were isolated from SFr 3–8 and SFr 3–9 also by using a semi-preparative HPLC (75% isocratic of MeOH–H2O with 0.1% formic acid for 20 min; 3 mL/min).
4.5. Spectroscopic data of compounds 1–6
Kaneoheoic acid A (1):
White, amorphous powder; 13.4 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 340.09 (2.40) nm; IR νmax 3328, 2942, 2834, 1660, 1441, 1401, 1115, 1014 cm−1; 1H and 13C NMR data, Table 1; HRESIMS m/z 275.1284 [M - H]− (calcd for C16H19O4, 275.1283).
Kaneoheoic acid B (2):
White, amorphous powder; 17.3 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 265.44 (3.10) nm; IR νmax 3328, 2942, 2831, 1447, 1022 cm−1; 1H and 13C NMR data, Table 1; HRESIMS m/z 323.1499 [M - H]− (calcd for C17H23O6, 323.1495).
Kaneoheoic acid C (3):
Brownish powder; 18.7 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 255.13 (3.41) nm; IR νmax 3315, 2943, 2831, 2043, 1448, 1410, 1115, 1022 cm−1; 1H and 13C NMR data, Table 2; HRESIMS m/z 241.1433 [M - H]− (calcd for C13H21O4, 241.1440).
Kaneoheoic acid D (4):
Brownish powder; −14.8 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 265.64 (2.85) nm; IR νmax 3313, 2898, 2803, 2434, 1632, 1472, 1422, 1121, 1020 cm−1; 1H and 13C NMR data, Table 2; HRESIMS m/z 241.1433 [M - H]− (calcd for C13H21O4, 241.1440).
Kaneoheoic acid E (5):
Brownish powder; 17.3 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 305.70 (3.28) nm; IR νmax 3312, 2942, 2813, 2358, 1642, 1445, 1412, 1114, 1020 cm−1; 1H and 13C NMR data, Table 2; HRESIMS m/z 223.12137 [M + H]+ (calcd for C13H19O3, 223.12895).
Kaneoheoic acid F (6):
Brownish powder; 12.2 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 305.30 (3.14) nm; IR νmax 3302, 2948, 2824, 2434, 1632, 1445, 1416, 1115, 1017 cm−1; 1H and 13C NMR data, Table 2; HRESIMS m/z 223.12690 [M + H]+ (calcd for C13H19O3, 223.12895).
4.6. Computational section [Wang et al., 2018, 2019a, Wang et al., 2019b]
Mixed torsional/low-frequency mode conformational searches were carried out by means of the MacroModel v 11.5 software using the OPLS 2005 with an implicit solvent model for water, retaining the geometries within 5.02 kcal/mol of the global minimum. Geometry re-optimization and frequency calculation were performed with the B3LYP functional with the 6–31+G (d, p) basis set using the Gaussian 09 software. NMR shielding tensors were computed with the GIAO method in Gaussian 09 using the B3LYP functional with the 6–311+G (2d, p) basis set, with methanol as solvent of the integrated equation formalism polarized model (IEFPCM). The unscaled chemical shifts were scaled using regression analysis parameters created by the method of Tantillo et al. [Lodewyk et al., 2011]. and Boltzmann-weighted average shielding tensor data set were calculated thereafter. ECD calculations were performed with the APFD functional with the 6–311+G (2d, p) basis set with methanol as solvent. The Boltzmann-averaged spectrum was obtained by GaussView 5.0.
4.7. Antibacterial assay
Antibacterial assay was conducted by using the previously described method [Zaman et al., 2020]. Bacteria were grown on agar plates [Tryptic Soy Agar (TSA), Brain heart infusion agar (BHIA) or Luria–Bertani Agar(LBA)] for 1 day at 37 °C and then added to a liquid medium (TSB for S. aureus, BHIB for B. subtilis and LB for E. coli). After incubation at 37 °C for 20 h, the cultures were diluted with TSB, BHIB or LB media to obtain an OD600 value of approx. 0.1. One hundred microliter of fresh media with samples at the desired concentration of 80 μg/mL (dissolved in DMSO) was put in the first well and then a two-fold dilution continued to the lowest concentration. The bacterium-containing media (100 μL) were then added to each well of 96-well plates. Additionally, samples were tested in combination with the FDA approved antibiotic chloramphenicol at 1 μg/mL, and two antibiotic adjuvants disulfiram (for S. aureus and Bacillus subtilis) or Loperamide (for E. coli) at 6 μg/mL. DMSO (5%), chloramphenicol (1 μg/mL) and the antibiotic adjuvants (6 μg/mL) were used as negative controls in these sets of experiments. Chloramphenicol, which is active against S. aureus, Bacillus subtilis and E. coli at MIC values of 6.25 μg/ml, 10 μg/ml and 3.2 μg/ml, respectively, was employed as the a positive control.
Supplementary Material
Acknowledgement
This work was financially supported by start-up funding from Daniel K. Inouye College of Pharmacy (DKICP), Seed Grants from University of Hawaii at Hilo (UHH), and the Victoria S. and Bradley L. Geist Foundation (15ADVC-74420, 17CON-86295, and 20CON-102163) (to SC). Funding for this work was also supported by Hawaii IDeA Network for Biomedical Research Excellence III and IV (INBRE-III and INBRE-IV) project: NIGMS Grant 5P20GM103466. We would also like to express our gratitude to Mr. Justin Reinicke for his help with NMR and for his kind assistance with optical rotation and ECD data collection.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alécio AC, Bolzani V da Sliva, Young MCM, Kato MJ, Furlan M, 1998. Antifungal amide from leaves of Piper hispidum. J. Nat. Prod. 61, 637–639. [DOI] [PubMed] [Google Scholar]
- Aly H, Debbab A, Proksch P, 2011. Fungal endophytes: unique plant inhabitants with great promises. Fungal Divers. 50, 3–19. [DOI] [PubMed] [Google Scholar]
- Butt U, ElShaer A, Chaidemenou A, Alany RG, Snyder Lori AS, Alany RG, 2016. Fatty acid microemulsion for the treatment of neonatal conjunctivitis: quantification, characterisation and evaluation of antimicrobial activity. Drug Deliv. Transl. Res. 6 (6), 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder PC, Grimble RF, 2002. Polyunsaturated fatty acids, inflammation and immunity. Eur. J. Clin. Nutr. 56 (3), S14–S19. [DOI] [PubMed] [Google Scholar]
- Cao S, Brodie PJ, Callmander M, Randrianaivo R, Rakotobe E, Rasamison VE, Kingston DGI, 2010. Saponins and a lignan derivative of Terminalia tropophylla from the Madagascar Dry Forest. Phytochemistry 71, 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejim L, Farha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright GD, 2011. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 7, 348–350. [DOI] [PubMed] [Google Scholar]
- Fei-Zhang J, Li C, Cao S, 2016. Hawaii natural compounds are promising to reduce ovarian cancer deaths. Canc. Biol. Ther. 17, 709–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CL, Drake DR, Dawson DV, Blanchette DR, Brogden KA, Wertz PW, 2012. Antibacterial activity of sphingoid bases and fatty acids against gram-positive and gram-negative bacteria quick view other sources. Antimicrob. Agents Chemother. 56 (3), 1157–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotso J, Leslie JF, Smith JS, 2002. Production of beauvericin, moniliformin, fusaproliferin, and fumonisins B1, B2, and B3 by fifteen ex-type strains of Fusarium species. Appl. Environ. Microbiol. 68, 5195–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Li C, Sarotti AM, Turkson J, Cao S, 2017. Sphaerialactonam, a γ-lactam–isochromanone from the Hawaiian endophytic fungus Paraphaeosphaeria sp. FT462. Tetrahedron Lett. 58, 1330–1333. [Google Scholar]
- Jaffe HH, Miller Albert L., 1966. Tautomeric equilibriums. VIII. A direct spectroscopic method quick view other sources. Tetrahedron Lett. 14, 1489–1491. [Google Scholar]
- Kato A, Ando K, Tamura G, Arima K, 1971. Effects of some fatty acid esters on the viability and transplantability of Ehrlich ascites tumor cells. Canc. Res. 31, 501–504. [PubMed] [Google Scholar]
- Li C, Sarotti AM, Wu X, Yang B, Turkson J, Chen Y, Liu Q, Cao S, 2019. An unusual benzoisoquinoline-9-one derivative and other related compounds with antiproliferative activity from Hawaiian endophytic fungus Peyronellaea sp. FT431. Molecules 24, e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Hu Z, Liu Q, Wu X, Cao S, 2018. Two new tricycloalternarenes from Hawaiian endophytic fungus Didymella sp. FT433. Tetrahedron Lett. 59, 3381–3383. [Google Scholar]
- Liu Don, Coloe Sue, Baird Rob, Pedersen John, 2000. Rapid mini-preparation of fungal DNA for PCR. J. Clin. Microbiol. 38 (1), 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodewyk MW, Siebert MR, Tantillo DJ, 2011. Computational prediction of 1H and 13C chemical shifts: a useful tool for natural product, mechanistic, and synthetic organic chemistry. Chem. Rev. 112, 1839–1862. [DOI] [PubMed] [Google Scholar]
- Long TE, 2017. Repurposing thiram and disulfiram as antibacterial agents for multidrug-resistant Staphylococcus aureus infections. Antimicrob. Agents Chemother. 61 (9) e00898–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen K, Anke T, Sterner O, 1996. 11-Hydroxy-4-methyl-2,4,6-dodecatrienoic acid from fermentations of a mucor species quick view other sources. Phytochemistry 43 (4), 791–792. [DOI] [PubMed] [Google Scholar]
- Manzoni M, Rollini M, 2002. Biosynthesis and biotechnological production of statins by filamentous fungi and application of these cholesterol-lowering drugs. Appl. Microbiol. Biotechnol. 58, 555–564. [DOI] [PubMed] [Google Scholar]
- Rihakova Z, Plockova M, Filip V, Smidrkal J, 2001. Antifungal activity of lauric acid derivatives against Aspergillus Niger. Eur. Food. Re.s Technol. 213, 488–490. [Google Scholar]
- Rouse MS, Rotger M, Piper KE, Steckelberg JM, Scholz M, Andrews J, Patel R, 2005. In vitro and in vivo evaluations of the activities of lauric acid monoester formulations against Staphylococcus aureus. Antimicrob. Agents Chemother. 49, 3187–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanakumar DEM, Folb PI, Campbell BW, Smith P, 2008. Antimycobacterial activity of the red alga Polysiphonia virgata. Pharm. Biol. 46 (4), 254–260. [Google Scholar]
- Song Q, Nan Z, Gao K, Song H, Tian P, Zhang X, Li C, Xu W, Li X, 2015. Antifungal, phytotoxic, and cytotoxic activities of metabolites from epichloë bromicola, a fungus obtained from Elymus tangutorum grass. J. Agric. Food Chem. 63, 8787–8792. [DOI] [PubMed] [Google Scholar]
- Vesonder RF, 1996. (all trans)-2,4,6,8,10,12-Tetradecahexene-1,14-dial, a New Pigment from Conidiobolus paulus. J. Nat. Prod. 59, 441–442. [DOI] [PubMed] [Google Scholar]
- Villamor E, Koulinska IN, Furtado J, Baylin A, Aboud S, Manji K, Campos H, Fawzi WW, 2007. Long chain n-6 polyunsaturated fatty acids in breast milk decrease the risk of HIV transmission through breastfeeding. Am. J. Clin. Nutr. 86 (3), 682–689. [DOI] [PubMed] [Google Scholar]
- Wang Q, Hu Z, Luo X, Liu J, Li G, Cao S, Liu Q, 2019b. Clavukoellians A–F, highly rearranged nardosinane sesquiterpenoids with antiangiogenic activity from clavulariakoellikeri. J. Nat. Prod. 82, 1331–1337. [DOI] [PubMed] [Google Scholar]
- Wang C, Wu X, Bai H, Zaman KAU, Hou S, Saito J, Wongwiwatthananukit S, Kim KS, Cao S, 2020. Antibacterial and NF-κB inhibitory lumazine peptides, aspochalasin, γ-butyrolactone derivatives, and cyclic peptides from a Hawaiian Aspergillus flavipes. J. Nat. Prod. 83 (7), 2233–2240. [DOI] [PubMed] [Google Scholar]
- Wang F, Hu Z, Li C, Wu X, Cao S, 2019a. Circumdatin M, a new benzodiazepine alkaloid with a unique pyrimidone-4-pyrone moiety from a Hawaiian marine fungus Aspergillus sp. FM242. Tetrahedron Lett. 60, 1724–1726. [Google Scholar]
- Wang Q, Hu Z, Li X, Wang A, Wu H, Liu J, Cao S, 2018. Q. Liu, salviachinensines A–F, antiproliferative phenolic derivatives from the Chinese medicinal plant Salvia chinensis. J. Nat. Prod. 81, 2531–2538. [DOI] [PubMed] [Google Scholar]
- Yu H, Tian Y, Zong Y, Zhang J, Xu Tao, 2020. Total synthesis and structural reassignment of aranorosinol A, aranorosinol B, and EI-2128–1 quick view other sources. J. Org. Chem. 85 (6), 4335–4343. [DOI] [PubMed] [Google Scholar]
- Zaman KA, Wu X, Hu Z, Hou S, Saito J, Kondratyuk TP, Pezzuto JM, Cao S, 2020. NF-κB inhibitory and antibacterial helvolic and fumagillin derivatives from Aspergillus terreus. J. Nat. Prod. 83 (3), 730–737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.