Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the cause of the COVID-19 pandemic that has infected over a hundred million people globally. There have been more than two million deaths recorded worldwide, with no end in sight until a widespread vaccination will be achieved. Current research has centred on different aspects of the virus interaction with cell surface receptors, but more needs to be done to further understand its mechanism of action in order to develop a targeted therapy and a method to control the spread of the virus. Lipids play a crucial role throughout the viral life cycle, and viruses are known to exploit lipid signalling and synthesis to affect host cell lipidome. Emerging studies using untargeted metabolomic and lipidomic approaches are providing new insight into the host response to COVID-19 infection. Indeed, metabolomic and lipidomic approaches have identified numerous circulating lipids that directly correlate to the severity of the disease, making lipid metabolism a potential therapeutic target. Circulating lipids play a key function in the pathogenesis of the virus and exert an inflammatory response. A better knowledge of lipid metabolism in the host-pathogen interaction will provide valuable insights into viral pathogenesis and to the development of novel therapeutic targets.
Keywords: Lipid Metabolism, coronavirus, SARS-CoV-2, COVID-19, dyslipidaemia
Abbreviations: AA, arachidonic acid; ACE2, angiotensin-converting enzyme 2; AUC, area under curve; COVID-19, coronavirus disease 19; cPLA2 α, cytosolic phospholipase A2α; CTD, C-terminal domain; DAGs, diacylglycerols; DHA, C22:6 docosahexaenoic acid; DIC, disseminated intravascular coagulation; DMV, double-membrane vesicles; DPA, C22: 5 docosapentaenoic acid; DVT, deep vein thrombosis; EBOV, Ebola virus; EPA, C20:5 eicosapentaenoic acid; ER, endoplasmic reticulum; ERGIC, ER-Golgi intermediate compartment; GM3, monosialodihexosyl ganglioside; HDL, high density lipoproteins; IDL, intermediate density lipoproteins; IFNα, interferon α; ISTH, International Society on Thrombosis and Haemostasis; LC-MS, liquid chromatography; LDs, lipid droplets; LMs, lipid mediators; LPA, lysophosphatidic acid; LPE, lysophosphatidylethanolamine; LPLs, Lysophospholipids; LPC, lysophosphatidylcholine; MCS, membrane contact site; MERS-CoV, Middle East respiratory syndrome coronavirus; MVB, multivesicular body; NEFAs, non-esterified fatty acids; NET, neutrophil extracellular trap; NPC1, receptor Niemann-Pick C1; NTD, N-terminal domain; ORF, open-reading frame; PAs, phosphatidic acids; PAF, platelet-activating factor; PCs, phosphatidylcholines; PEs, phosphoethanolamines; PI3K, phosphoinositide 3-kinase; PLA2, phospholipase A2; plasmenyl-PC, plasmenyl phosphatidylcholine; PSTPIP2, proline-serine-threonine phosphatase-interacting protein 2; PtdIns, Phosphoinositides; PtdIns3P, monophosphate phosphatidylinositol 3-phosphate; PtdIns(3,5)P2, bisphosphate phosphatidylinositol 3,5-bisphosphate; PUFAs, poly-unsaturated fatty acids; QqQ, triple quadrupoles; RBD, receptor-binding domain; RO, replication organelles; RvE3, resolvin E3; SARS-CoV, Severe Acute Respiratory Syndrome; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; S protein, spike protein; S1P, sphingosine 1-phosphates; SMs, sphingomyelins; SPE, solid-phase extraction; SPME, solid-phase microextraction; SREBPs, sterol regulatory element-binding proteins; TAGs, triacylglycerols; TC, total cholesterol; TMPRSS2, type II transmembrane serine host cell protease; VLDL, very low-density lipoproteins; VTE, venous thromboembolism.
1. Introduction
The year 2020 will be certainly remembered as the year of the coronavirus disease 19 (COVID-19) pandemic. Towards the end of 2019, a novel coronavirus infectious disease generated by the severe acute syndrome coronavirus 2 (SARS-CoV-2) made its appearance in Wuhan, China. As it was the case with the other two recent coronavirus spreads, 2003 Severe Acute Respiratory Syndrome (SARS-CoV) and 2012 Middle East respiratory syndrome (MERS-CoV), and many other so-called “bird flu” worldwide, it is believed to be a zoonotic virus, originated in a live market from bats and then transferred to human hosts [1]. Within a few weeks, the first cases were detected in Europe, in particular in Italy, and in March 2020, the World Health Organization announced the beginning of a global pandemic [2]. As the 1918 Spanish flu spectre began to appear on the horizon, countries took different approaches to tackle the fast-rising spread of the infection. Some countries such as China, South Korea and Italy in Europe, followed by France and Spain, severely hit by an alarming increase of cases and mortality, chose to implement unprecedented strict measures to contain the spread. Others, like UK and USA, adopted softer strategies, ignoring the WHO guidelines or, as in the case of the Swedish government, chose to initially implement control strategies that could be perceived as a model to achieve herd-immunity [3,4]. These different decisions have somehow reflected on the ability of different countries to keep under control the disease. Nevertheless, despite all the efforts, more than two million deaths have been recorded worldwide (January 2021), the number of cases and deaths is steadily rising in some places such as USA, Brazil and India, and the pandemic is far from over [5]. The attention of the world scientists has been suddenly monopolized on COVID-19 research, in a frantic fight against time to develop a vaccine and, in parallel, to find pharmacological therapies able to treat the severe pneumonia symptoms caused by this virus. A huge number of drugs, natural compounds, and traditional Chinese remedies have been tested, and the guidelines issued by the Chinese National Health Commission revised many times. In the last version, antivirals such as ribavirin, interferon α (IFNα), chloroquine phosphate, lopinavir/ritonavir and arbidol have been proposed; in some cases, these have proven to be helpful in lowering the severity of the infection, but an effective antiviral specific to Covid-19 is yet to be found [6]. The extremely high infectiousness of this virus, especially in the presymptomatic phase, and a large number of asymptomatic patients, are thought to be the main booster for this pandemic [7,8] and the reason behind the difficulties in its containment. SARS-CoV-2 is a respiratory virus that spreads from person to person mainly through aerosol droplets released when talking, coughing or sneezing, but also by contact with contaminated surfaces. For this reason, although a huge debate arose on whether masks application could be helpful, useless or damaging, and this issue is still controversial, their use has been proven to play a role in protecting against the infection [9]. Once in the upper respiratory tract, SARS-CoV-2 binds to the receptor angiotensin-converting enzyme 2 (ACE2), a membrane-bound aminopeptidase involved in particular in the circulatory system functions and immune response, highly expressed in particular the ciliated nasal epithelial cells, heart and lungs [10], causing cold-like symptoms or no symptoms at all. After approximately 3 to 5 days, as the virus replicates in the lower respiratory tract, some patients develop pneumonia. While the majority of these patients recover spontaneously, a portion of them (about 20%) worsen in a few days presenting high fever, hypoxia, inflammation, blood clotting, lymphocytopenia and cardiovascular problems, which lead to mechanical ventilation for about a quarter of them, and to death for 50 to 80% of these patients [7]. Concomitant morbidities, age, and enhanced ACE2 expression can all negatively affect the prognosis. Moreover, the immune system response to the virus infiltration seems also to play a role in accelerating the breakdown of the epithelial-endothelial barrier in the lungs, causing further deterioration [7]. In order to completely clarify the mechanisms of action of SARS-CoV-2 and, therefore, be able to produce a specific effective treatment, it is important to unravel the role played by lipids in the development of this infection. The essential role of lipids in biology, as cell membrane components, and in cellular communication, membrane trafficking, energy storage and heat insulation is well known. They are also known to be crucial in enabling viruses to cross the host cell membrane and replicate, and their normal metabolism in the host cell is altered as a consequence of viral infections [11]. Studies on COVID-19 patients’ plasma have found that the levels of lipid modification correspond to the gravity of the infection, and several research projects have identified COVID-19-associated lipid biomarkers [12]. Moreover, lipid-lowering therapies such as statins and lipoprotein apheresis have been proposed as a therapeutic strategy to reduce clinical complications in patients severely affected by COVID-19 [13]. Increasing bioactive lipid levels has also been suggested as a method to reduce ACE2 receptors number and affinity to SARS-CoV-2 [14]. Therefore, targeting membrane sphingolipids and interfering with the virus lipid metabolism could represent a path to follow towards the development of COVID-19 treatments.
2. Coronaviruses within virus classification
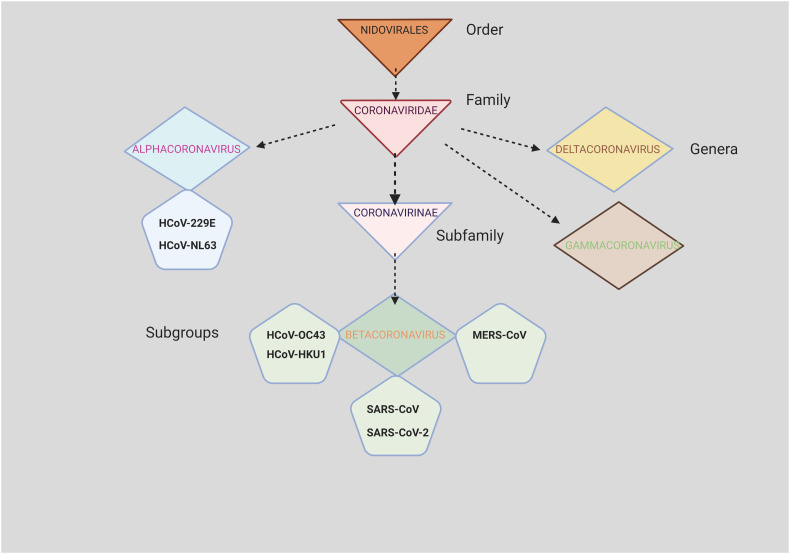
Viruses are small, simple living organisms that survive only by invading host cells of animals, bacteria or plants and replicate within these cells, causing diseases during the process. They rely on the host cell for both their energy supply and metabolism. Viruses are divided according to their genetic characteristics [15] into two broad groups: DNA viruses and RNA viruses, and they can be “naked” or surrounded by an external layer composed of lipids, proteins and sometimes glycoproteins, called “envelope”. DNA viruses can be double-stranded enveloped (Hepadnaviruses, Herpesviruses, Poxviruses), and unenveloped (Adenoviruses, Papillomaviruses, Polyomaviruses) or single-stranded (Parvoviruses). RNA viruses represent the majority amongst all viruses. They can be classified based on the sense that their amino acid sequence is translated by the host cell. Viral RNAs that are positioned in the same sense as mRNAs and are immediately translated into protein are called “positive-sense”, and can be unenveloped (Caliciviruses, Picomaviruses) or enveloped (Coronaviruses, Flaviviruses, Togaviruses); owing to this conformation, the host cell is able to transcribe and synthesize the viral proteins quicker. On the contrary, “negative sense” RNAs have to be reversed by an enzyme called RNA replicase for translation, and they are all enveloped (Arenaviruses, Bunyaviruses, Filoviruses, Orthomyxoviruses, Paramixoviruses and Rhabdoviruses). Some viruses can be “ambisense”, positive or negative, and have a double protein shell protecting the genetic material called “double capsid” (Reoviruses), while others (Retrovirus) are RNA “positive-sense”, but they use a DNA intermediate for replication. From the point of view of the type of structure, RNA viruses can be classified as icosahedral, equilateral triangles forming a spherical shape (including both naked or enveloped viruses), or helical, filaments forming a tube with a cavity, (enveloped viruses only). Coronaviruses belong to the order of Nidovirales and to the Coronaviridae family (Figure 1 ); they are RNA “positive-sense” enveloped viruses, with a helical shape and a viral genome length between 26-32 kilobases [16]. This family of viruses comprises two subfamilies: the Coronavirinae and the Torovirinae subfamily, capable of infecting various types of animals. The Torovirinae subfamily includes the Torovirus (equine and bovine) and the Bafinivirus (white bream virus). The Coronavirinae subfamily is divided into: i) Alphacoronavirus (including alphacoronavirus 1 and porcine epidemic diarrhoea virus), ii) Betacoronavirus (including severe acute respiratory syndrome-related coronavirus and Betacoronavirus 1), and iii) Gammacoronavirus (including Avian coronavirus and Beluga whale coronavirus). The SARS-CoV-2, belongs to the Betacoronavirus group, together with the viruses responsible for the previous SARS-CoV and MERS-CoV pandemics.
Figure 1.
Family tree of the seven coronaviruses that infect humans.
3. Structure of severe acute syndrome coronavirus 2 (SARS-CoV-2)
Even though viruses display different morphologies, ranging from a helical to Icosahedral, prolate, head or tail, complex or asymmetrical shape, these non-living particles depend on host organisms to survive and share a common basic structure. Three main elements concur to form a virus structure: 1) the nucleic acid, containing single or double-stranded DNA or RNA and enzymes for genetic transcription, which is between 5 to 200 kb in size; 2) a capsid surrounding this genetic material, a viral protective layer of proteins called “capsomeres” which gives the distinctive shape to the virus, and; 3) a glycoprotein envelope formed by a bilayer of lipids and proteins, essential for attaching to the host cell membrane and penetrating inside. Coronaviruses display on the envelope, as a distinctive feature, spike proteins that give the resemblance of a crown. SARS-CoV-2 particles (virions) are 125 nm wide spherical entities, with a characteristic crown on the surface, and a positive-sense single-stranded RNA enveloped by a nucleocapsid, forming a symmetrical helical shape containing a large genome of 27-34 kb. Four structural proteins characterize this virus: the transmembrane spike protein (S protein) (180-200 kDa), the envelope protein (8-12 kDa) involved in the virion production and expulsion from the host cell, the membrane protein (25-30 kDa) responsible for the virion shape, all located in the envelope, and the nucleocapsid protein located inside the capsid and binding the RNA genome. Sixteen non-structural proteins are formed after the invasion of the host cell via open-reading frame (ORF) translation. The S protein has a homotrimeric conformation containing the receptor-binding domain S1 subunit, comprising, in turn, an N-terminal domain (NTD) and a C-terminal domain (CTD), and the S2 subunit contributing to membrane fusion [17]. This protein allows SARS-CoV-2 virions to attach to host cell membrane receptors and, subsequently invade those cells. SARS-CoV-2 S1 subdomain CTD binds the ACE2 receptor on the surface of the host cell [18]. Unlike SARS-CoV, SARS-CoV-2 contains a polybasic furin cleavage site at the interface of the two subunits S1 and S2 of the spike protein [19]. Strikingly, this site is found in other pathogens such as Ebola, virulent bird flu strains, and HIV-1. When this site is cleaved by furin, the spike protein can interact with and activate an additional cell surface receptor, neuropilin 1 (NRP1). NRP1, when expressed together with ACE2, and type II transmembrane serine host cell protease (TMPRSS2), greatly potentiates viral entry and infection [20,21].
Two different entry mechanisms are possible at this point; an early pathway in which virions blend at the plasma membrane, or a late pathway in which virions amalgamate at the endosomal membrane, according to proteases accessibility [22]. The early entry is mediated by TMPRSS2, which promotes viral fusion and nucleocapsid access into the cytoplasm by cleaving the S protein at the S1/S2 and S2’ sites. For the late entry pathway, the virus is endocytosed, and the viral S protein is cleaved in the presence of cysteine protease cathepsin L before the genome being liberated into the host cell. In vivo studies have shown the importance of lipid rafts in facilitating coronaviruses entry in the host cells. In particular, cholesterol seems to play an essential role and, in its absence, both viral entry and fusion have been shown to be compromised [23]. Once in the cytoplasm, the virus exploits the host cell structures for genome replication; in the endoplasmic reticulum (ER) S protein constituents are produced, joined in the ER-Golgi intermediate compartment (ERGIC), the S protein is pre-cleaved and then released by exocytosis into the extracellular space.
4. Mechanisms of virus-host cell interaction
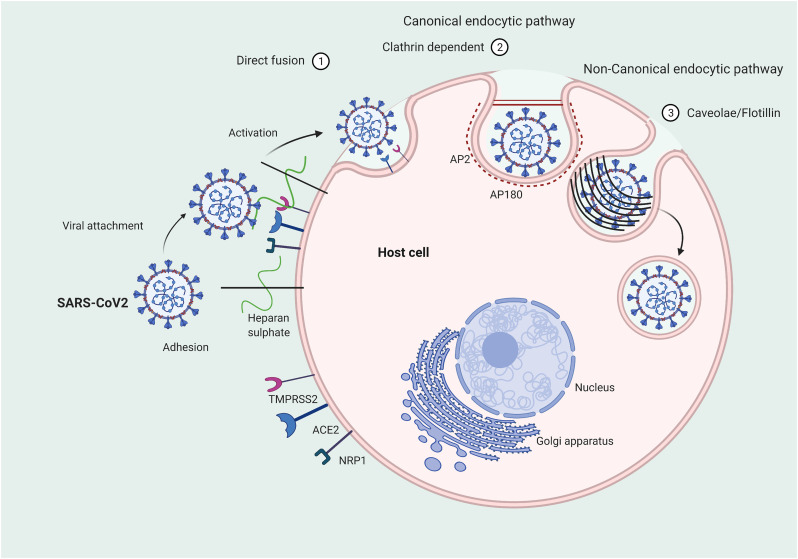
Lipids play a key role in the initial stages of the interaction of viruses with host membranes [24]. The very first step of this complex process is the virus adhesion to the cell membrane [25]. This involves unspecific binding with negatively charged sugars of glycoproteins and glycolipids that represents a first dock before the engagement with a specific receptor. Heparan sulphate proteoglycans, expressed by the majority of eukaryotic cells, have been shown to play a role in the adhesion of many viruses to the cell external membrane, including SARS-CoV-2 (Figure 2 ). Indeed, a recent study demonstrated that SARS-CoV-2 spike protein interacts with both cellular heparan sulphate and ACE2 through its receptor-binding domain (RBD) domain [26]. In the latter study, the authors proposed a model in which viral attachment and infection involve heparan sulphate-dependent enhancement binding to ACE2 and consequently, inhibition of viral adhesion by exogenous heparin is suggested as a novel therapeutic approach [26]. A new insight into the SARS-CoV-2 pathogenicity is emerging from the recently published study by Toelzer et al., demonstrating a direct binding between fatty acids and SARS-CoV-2 proteins [27]. The authors used the cryo-electron microscopy technique to examine the spike glycoprotein structure of SARS-CoV-2. The analysis revealed the presence of three binding pockets in the receptor-binding domains (RBDs) firmly binding linoleic acid (LA). Further experiments confirmed the existence of these LA-binding sites in SARS-CoV and MERS-CoV. In in vitro experiments, this LA binding was found to cause a locked conformation of the spike protein resulting in a decreased association between this protein and ACE2 host receptor. Subsequently, human epithelial cells infected with SARS-CoV-2 were treated with different concentrations of Remdesivir, an anti-viral drug that has shown some effectiveness in COVID-19 clinical trials, in combination with LA. The addition of the latter resulted in a synergistic effect permitting a remarkable reduction of Remdesivir’s dosage in order to block virus’ replication. This finding could be a strategy to overcome the significant side effects observed in SARS-CoV-2 patients treated with Remdesivir [28]. Moreover, as fatty acid-binding pockets were used to developed anti-viral drugs successfully employed in clinical trials against rhinovirus [[29], [30], [31], [32]], similarly, the discovery of LA-binding pockets could be exploited for designing specific inhibitors able to obstruct virus-host interactions by blocking the spike protein in the closed conformation.
Figure 2.
Mechanisms of SARS-CoV-2 interaction with host cell and entry. Three different possible endocytic pathways are shown: 1) direct fusion; 2) clathrin-dependent pathway; 3) caveolae- or flotillin-dependent pathway.
The subsequent step involves the virus entry into cell cytosol using either a direct fusion mechanism or the exploitation of the endocytic pathways of the host cells. In both cases, this is a critical energy-consuming stage involving viral membrane fusion. While enveloped virus entry through direct fusion with the plasma membrane appears to be the most convenient strategy, more recent evidence suggests that virus entry mostly occurs via endocytosis and that the use of the endosomal route provides multiple advantages [33].
Viruses have been shown to use different canonical and non-canonical endocytic pathways [34]. The canonical pathways involve the formation of clathrin-coated vesicles, whereas the non-canonical pathways require either caveolae, flotillin-1, glycophosphatidylinositol-anchored proteins or micropinocytosis.
It has been found that SARS-CoV enters cells via an endocytosis pathway dependent on pH and receptor engagement but clathrin- and caveolae-independent [35]. In the latter study, it has been shown that cholesterol- and sphingolipid-rich lipid raft microdomains in the plasma membrane are involved in SARS-CoV entry [35]. However, further research is needed to confirm this mechanism of entry.
Phosphoinositides (PtdIns) play critical roles in membrane dynamics, trafficking, and cellular signalling. In particular, the monophosphate phosphatidylinositol 3-phosphate (PtdIns3P) and the bisphosphate phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2] have been mostly linked with endosomes and endosomal functions [36]. Recent studies suggest that Ebola virus (EBOV) necessitates PtdIns(3,5)P2 production in cells to support efficient transport to the receptor Niemann-Pick C1 (NPC1) that localizes in late endosomes and lysosomes [37]. Inhibition of PIKFYVE, the enzyme that phosphorylates PtdIns3P to produce PtdIns(3,5)P2, impeded colocalization of EBOV with NPC1 and induced virus concentration in intracellular vesicles with characteristics of early endosomes. Interestingly, other viruses such as Zaire ebolavirus and SARS-CoV-2 have been indicated to use analogous trafficking pathways for efficient entry. More importantly, PIKFYVE inhibitor Apilimod strongly impairs the infection of Zaire ebolavirus and SARS-CoV-2 suggesting the potential for targeting PIKFYVE in developing antiviral agents against SARS-CoV-2 [38].
Furthermore, inhibition of class III phosphoinositide 3-kinase (PI3K), the enzyme that produces PtdIns3P from PtdIns, has been shown to potently inhibit SARS-CoV-2 infection of human lung model ex vivo [39]. Similarly, class I PI3K inhibition has been shown to affect SARS-CoV-2 replication [40]. In addition, Type III phosphatidylinositol-4-kinase enzymes, that produce phosphatidylinositol-4-phosphate, have been utilized in replication organelles by different viruses [41].
However, strategies aiming at the inhibition of PI-kinases should be taken with caution considering the potential side-effect that can be caused. In particular, PIKFYVE inhibition has been shown to generate large vacuoles in cells and lead to apoptosis in vitro [42] and in vivo [43].
5. Replication organelles and virus-induced double-membrane vesicles
Cell Membranes perform essential functions of containment, signalling and molecule exchange that regulate all biological processes. Membrane lipid bilayers are asymmetrically modified by factors both internal and external, using the energy released by changing the membrane lipid composition, and from proteins composing the membrane or present in the cytoplasm. They can be curved inward (positively), in the process of endocytosis, or outward (negatively), generating extracellular vesicles [44]. Every positive-sense RNA virus, and coronaviruses among them, manipulate host cell membranes to build structures, termed viral replication organelles (RO), able to grant RNA synthesis and viral replication [45]. These structures, in addition to hijacking host factors, metabolites, lipids and proteins relevant to replication, may have a role in protecting the viral genetic material from being exposed to the host immune system [46]. Even though among distinct viruses there are great differences in the composition, structure and development of RO, they can be grouped into two main morphological categories; spherule formations and tubular and/or vesicular formations [47]. Spherule formations are membrane invaginations within host cell organelles, such as the ER, mitochondria and endolysosomes, not entirely separated from the cytosol to facilitate access to the required molecules and outdoor transport of vRNA. The second category includes host membrane protrusions forming tubular or vesicular organelles, amongst which, structures referred to as double-membrane vesicles (DMV) have attracted particular interest. Nidoviruses and coronaviruses in particular, have been observed via electron tomography to produce a wide range of double-membrane formations, displaying a regular membrane interspace and various shapes such as zippered ER, convoluted membranes, open double-membrane and double-membrane vesicles [48]. Studies on noroviruses DMVs have revealed membrane connections, called membrane contact sites (MCSs), in between vesicles, between DMVs and other structures of viral origins, and in particular between DMVs and ER, suggesting a fundamental role of the latter in DMV development [46,47]. Viruses take control of these points of contact, which allow lipid trafficking between cytosol and RO, by interfering with cellular pathways or engaging MCS proteins, to ensure RO development and activity. Biogenesis of nidovirus DMV differs from other positive-sense RNA viruses. For some viruses, such as picornaviruses, a single membrane protrudes forming vesicles or tubules and subsequently the formation of paired membrane, two tightly placed lipid bilayers, is observed, forming cisternae that curve eventually giving shape to complete DMVs. In nidoviruses, including coronaviruses, membrane pairing occurs first in the ER, forming cisternae that will afterward curve and go through either one fission, generating a connected DMV, or two fissions to produce a disconnected floating DMV [46]. SARS-CoV non-structural proteins nsp3, nsp4 and nsp6 have been discovered to be essential for the biogenesis of double-membrane RO and viral RNA synthesis [48]. However, the complete understanding of the role of viral proteins in this process requires further studies. Another important element in viral biogenesis is the part played by host factors in supporting the virus life cycle. Regular cellular pathways in host cells are hijacked and exploited by viruses to survive and spread. For example, membrane proteins ER-associated reticulons and F-BAR domain-containing proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2) are thought to be instrumental in viral DMV establishment [49]. The autophagy and secretory pathways are also involved in this process as GBF1, a secretory protein, has been found to be correlated to DMV number in coronaviruses [50]. As essential membrane components and key factors regulating membrane curvature and DMV generation, lipids are also indispensable to coronaviruses development. Lysophospholipids (LPLs) levels have been found to be positively linked to coronaviruses DMV biosynthesis in in vitro studies [45].
Additionally, in the same study, an enzyme involved in lipid metabolism, cytosolic phospholipase A2α (cPLA2 α), has been shown to affect coronavirus replication and DMV development. Lipidomic studies on human coronavirus-infected cells, using an ultra-high-performance liquid chromatography-mass spectrometry, revealed a significant modification in the host cell lipid composition upon infection, remarkably linked to coronavirus spreading [51]. Although the formation and function of DMV are beginning to be understood, there is still not enough evidence in regards to the intrinsic benefits of using such formations for viral RNA synthesis. Another open question is whether this process would still be possible without these structures, given that a few rare viruses, such as a mutant CVB3 enterovirus, are able to replicate without RO [52].
It has been hypothesized that RO might shield the virus genetic material from the immune system, and some studies support this theory [[53], [54], [55], [56], [57]]. Nevertheless, the variety in RO morphology, ranging from closed spherules to semi-open or open structures, makes this postulation rather questionable. For semi-open DMVs, viral replication could occur safely inside by the inner membrane, while essential changes of metabolites would still be possible. On the other hand, completely exposed RO open structures, as in the case of picornaviruses, would not offer any protection. As for nidovirus, which has DMV close formations [46,58], material exchanges with the cytosol could take place via alternative routes, such as membrane pores, even though the exact location of RNA synthesis involving the inner membrane or the outer, or both, is not certain [46].
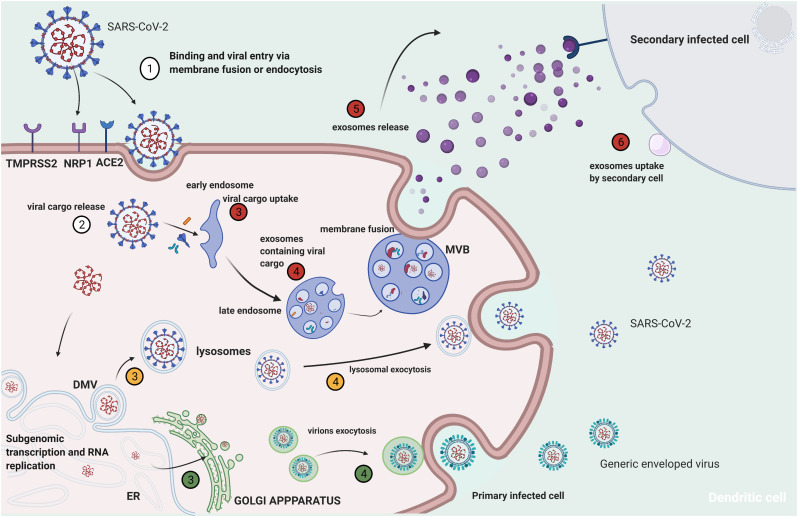
The release of the complete virions from the host cell is a little investigated aspect of SARS-CoV-2 replication cycle (Figure 3 ). Recent work identified a novel mechanism of SARS-CoV-2 egress involving lysosomal exocytosis and not the biosynthetic secretory pathway generally utilized by enveloped viruses [59]. However, other investigators demonstrated the existence of two distinct egress pathways, using the cryo-imaging approach, involving tunnels via exocytosis release or through membrane rupture [60]. Therefore, further investigations are required to clarify this important step of SARS-CoV-2 cell infection cycle.
Figure 3.
Potential mechanisms of SARS-CoV-2 secondary cell infection. Red: exosome-mediated viral cargo release; yellow: lysosome-mediated virus release; green: virus release by alternative secretory pathway.
6. Lipid metabolic remodelling of the host cell
After endocytosis, viruses establish their ideal niche, beginning the first step of viral gene expression and then proceed to replication. One of the major targets in this stage is the manipulation of the metabolic state of the infected cell at different levels. Therefore, other than physical interaction with cellular membranes, viruses exploit and modulate lipid signalling and metabolism to benefit their replication. Indeed, lipids constitute not only the backbone of membranes but function as signalling molecules and energy sources. Therefore, reprogramming of lipid metabolism is a key feature of virus-host interaction [41], similarly to the way cancer cells induce lipid reprogramming [61]. It should be noted that reprogramming of the host lipid metabolism could also be attributed to the host response to infection.
Several studies found that viruses are able to enhance the expression and activity of critical enzymes in lipid biosynthesis. Among these key enzymes, there is a family of membrane-bound transcription factors in the endoplasmic reticulum, sterol regulatory element-binding proteins (SREBPs), which are critical in lipid regulation [62]. SREBPS are transcription factors that bind to promoters of genes involved in lipid biosynthesis and, consequently, they regulate cellular lipid metabolism and homeostasis. SREBPs are reported to be highly upregulated in Middle East respiratory syndrome coronavirus (MERS-CoV) and influenza A virus infections [63]. Lipid biosynthesis pathways regulated by SREBP include necessary steps for virus replication such as the downstream viral protein palmitoylation and double-membrane vesicles formation. Therefore, SREBP represents a potential target for the development of broad-spectrum antiviral strategies. Another key enzyme in lipid biosynthesis is the fatty acid synthase that has been found upregulated in viral infection mediated by many viruses such as hepatitis C virus, Dengue virus, West Nile Virus and influenza virus.
Lipid droplets (LDs), are lipid-rich cellular organelles that regulate lipid homeostasis and metabolism [64]. They serve as a store for cholesterol esters and triacylglycerols (TAGs) utilized for membrane formation and maintenance but also supply essential lipids to produce signalling molecules and metabolic energy. LDs also have a more dynamic role since proteins present in the lipid droplet coat regulate lipid homeostasis and, therefore, LDs play a very important role in the regulation of intracellular lipid storage and lipid metabolism [65]. Single-strand RNA viruses, including coronaviruses, target LDs during their replication phase [66,67]. Given the role of LDs in viral replication, it has been proposed to use drugs interfering with LDs as therapeutics able to counteract SARS-CoV-2 infection [66,68]. Recent work from Dias et al. demonstrated that the virus replication and the heightening inflammatory mediator production in SARS-CoV-2 infection are favoured by the participation of LDs. In addition, they showed that the pharmacological inhibition of DGAT-1, which is a key enzyme for LD formation, reduced SARS-CoV2 replication, inflammatory mediator production and cell death, opening new therapeutic strategies [69].
7. Lipid rafts
Pathogens, and viruses among them, are obliged to find survival stratagems in order to avoid host defence mechanisms, successfully invade cells, replicate and spread. One of those strategies they rely on involves exploiting lipid microdomains, designated “lipid rafts” located primarily on the cell plasma membrane, but also reported in other internal organelles. Several diverse types of viruses, including coronaviruses, take advantage of these signal transduction platforms in order to penetrate inside cells during the fusion and internalization phase, for sending signals and transporting viral proteins, and also in the course of viral assembly before exocytosis [70]. Lipid rafts are specialized dynamic nanoscale regions, 10 to 200 nm in size, and consisting of sphingolipids, glycosphingolipids, cholesterol and GPI-linked proteins. They perform many tasks including cell signalling, membrane trafficking, cell polarity regulation, polarized secretion, epithelial cells’ transcellular transport, endocytosis and autophagy [70,71]. In addition to their analogous lipoid composition, lipid rafts are also non-ionic detergent-insoluble, are isolated by sucrose density gradient, and are quick to change structure upon stimulation [[72], [73], [74]]. Their protein conformation determines whether lipid rafts are categorized as “planar”, composed mainly of flotillin proteins, or “caveolae”, enriched in caveolin proteins [75], both mediating lipid raft-associated signal transduction. The hypothesis of lipid rafts playing an essential role in pathogenic microbe invasion of host cells has been confirmed by numerous studies. Lipid raft disrupting or cholesterol-reducing/removing compounds, such as statins and methyl-β-cyclodextrin, were able to block pathological infections [[76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86]]. Recently, various roles of lipid rafts in the coronavirus life cycle have been indicated. In the first phase of viral infection, coronaviruses bind to their particular receptors, ACE2 in the case of SARS-CoV and SARS-CoV-2, which are found inside lipid rafts or caveolae, producing structural transformations in the viral molecules and starting a signalling cascade activating the virus internalization. Viral particles are then incorporated into early and late endosomes where acidity induces the fusion between the viral envelope and the endosome membrane. The next phase includes the translation of viral genomic RNA and sub-genomic RNA synthesis. Nucleocapsid proteins are produced in the cytoplasm while the ER is the translation site for spike proteins, matrix and envelope. The autophagy apparatus is also exploited by coronaviruses to reinforce virus replication. Finally, by an exocytosis process, the newly synthesized virions are enclosed in smooth-wall vesicles or possibly multivesicular bodies and moved to the cell surface [87,88]. Given the prominent contribution of lipid rafts to virus survival and propagation, their therapeutic targeting could be a successful strategy to counteract infections [89] and, in particular, a valid tool in the fight against COVID-19 pandemic.
8. Autophagy and coronaviruses
Autophagy is a denomination derived from the Greek word meaning “self-eating”; it refers to a complex mechanism used routinely by cells to preserve homeostasis, which is enhanced in challenging conditions such as lack of nutrients or oxygen, stress, DNA damage, reactive oxygen species, or pathogenic infections [90,91]. During this process, cellular parts, organelles and proteins are packed in autophagosomes, double-membrane vesicles that transport their content to the lysosome; autophagosomes fuse with the lysosome originating autolysosomes, where the cargo is processed and broken down by specific enzymes. ATG5 and ATG7 genes are autophagy regulators, and LC3 and p62 are the main proteins used as markers for autophagy. In fact, levels of LC3-II, lipidated LC3, are directly correlated to the number of autophagosomes in cells [92] thus showing the presence of autophagy; conversely, as p62 is degraded during this process, increased levels of this protein reveal autophagy inhibition and vice versa [93]. Strict cooperation has been shown to exist between lipid rafts and autophagy, as caveolin-1, a major component of plasma membrane lipid rafts, interferes with the expression of ATG12-ATG5, a conjugation system essential for autophagosome development [94]. In addition, autophagy seems to be affected by the same molecules involved in the chemical and structural configuration of lipid rafts [95].
Although autophagy has been shown to have a role in counteracting infections, transporting viral particles to lysosomes and boosting hosts’ immune response, certain viruses are known to be able to neutralize lysosomal degradation and to utilize autophagy to survive and replicate [96]. Coyne et al. have found that cells that take up exosomes carrying microRNAs from placenta cells are very resistant to virus-induced autophagy [97]. Also, some virus replications can be limited by inducing autophagy, as in the case of Sindbis virus affecting the central nervous system in mice [98]. On the other hand other viruses, such as herpesviruses, have found means of protecting themselves from autophagy degradation by expressing proteins that impede autophagosomes development [99]. Many positive-sense RNA viruses, enteroviruses in particular, induce autophagy and exploit this mechanism to avail themselves of membranes necessary for the construction of their replication areas [100]. Nidovirales seem to have the ability to both employ autophagy-derived materials for their synthesis and to exploit autophagy mechanisms to decrease viral degradation [101]. Conflicting results have been published regarding MERS-CoV, which has been described to induce autophagy by some authors [102] and to inhibit the same process by others [103]. Similarly, while it is known that SARS-CoV utilizes the autophagy pathway for replication [104,105], SARS-CoV-2 was recently discovered to decrease autophagy through AMPK and mTORC1 downregulation, with the advantage of limiting viral product degradation and profiting by the increase of double-membrane vesicles [106]. Some known autophagy promoters such as spermidine, niclosamide and nitazoxanide have been shown to have an effect against SARS-CoV-2 in in vitro studies [16,107] and, as a consequence of these findings, the autophagy promoter rapamycin has been used in the clinical treatment of this infection, achieving some results (NCT04482712).
9. Exosomes
Exosomes are microvesicles released by all mammalian cells, which act as signalling organelles during normal physiological and pathological conditions. Important cellular homeostasis functions of exosomes include: cell repair and restoration, stimulation of the immune system, maturation of the nervous system, and maintenance of foetal development [108]. On the other hand, exosomes play a pro-cancer role by promoting the pre-metastatic niche, metastasis formation and inducing chemoresistance. Exosomes can also initiate and promote neurodegenerative diseases, participate in cardiovascular disorders and contribute to viral infection spread by carrying pathogenic factors (Figure 3) [108].
Among the variety of vesicles secreted by eukaryotic cells, exosomes are the smallest, between 30 and 150 nm in size. They carry a distinct RNA, lipid and protein cargo depending on their cells of origin, together with some common proteins such as membrane transport and fusion proteins (GTPases, Annexins, Flotillin), heat shock proteins (Hsc70, Hsp 90), tetraspanins (CD63, CD81, CD82), lipid-related proteins and phospholipases, as well as proteins involved in multivesicular body biogenesis (Alix, TSG101). Mass spectrometry studies have found more than 4400 proteins in connection to exosomes [109,110]. Exosome progenitors are early endosomes. These intracellular organelles collect endocytosed cargo and intraluminal vesicles and become late endosomes or MVBs once increased in size. These MVBs, found in the perinuclear cytoplasm, could either function as a storage unit for the cell, fuse with lysosomes for cargo degradation or, upon adequate stimulation, fuse with the cell plasma membrane to release the newly born exosomes into the extracellular space by exocytosis. Lipid metabolism, and especially PI3K, play an important part in MVBs biogenesis. Some proteins, such as Hrs and the ESCRT sorting complexes are also essential in the regulation of vesicle formation. Once in the extracellular space, exosomes are ready to be recognized and uploaded by recipient cells, mainly via endocytosis, fusion or antigen-recognition mechanism. Due to the presence of opsonins such as lactoadherin on the surface, exosomes are phagocytosed from the body fluids by immune cells in the liver and spleen. Specific cell adhesion transmembrane proteins make it possible for exosomes to attach on the surface of recipient cells. For instance, adhesion occurs when ICAM-1, CD44, CD49d, CD54 and CD11a proteins on the T-cell surface are exposed to CD81 and CD9 on exosomes. In the antigen recognition mechanism, the major histocompatibility complex on exosomes is recognized and bound by T-cells.
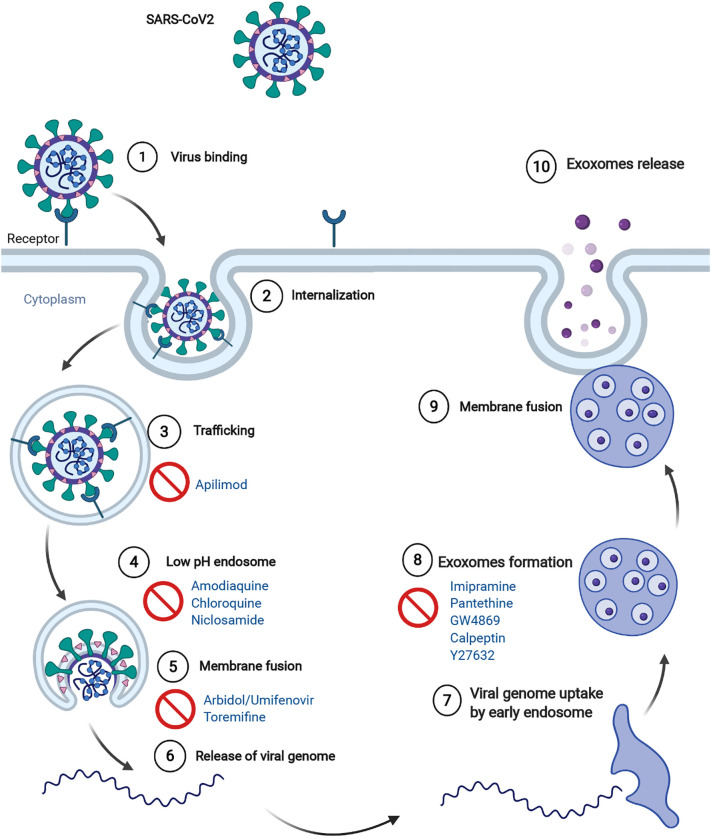
Virus and exosome biogenesis share evident similarities, and during infections viruses utilize the exosomal pathway to both enter cells and invade other tissues. Coronaviruses are similar in size to exosomes (about 125 nm), they both carry proteins and RNAs, and lipidomic studies on exosome membrane have found a remarkable resemblance with the composition of the virus membrane [111]. Viruses have been shown to modulate the production and molecular cargo of exosomes [112,113]. Indeed, microRNAs, proteins and even intact virions can be packed into exosomes [113,114]. Upon viral infection, host cells are activated to produce exosomes loaded with viral pro-inflammatory and cytotoxic molecules that are sent to nearby tissues where they induce inflammation, intravascular coagulation, and trigger the endothelium causing sepsis and tissue damage. In a recent in vitro study, lung epithelial cells incorporated with SARS-CoV-2 genes were found to produce exosomes containing viral RNAs, and caused inflammation and viral genes uptake in human-induced pluripotent stem cell-derived cardiomyocytes they were put in contact with [115]. Moreover, our recent studies have confirmed the presence of SARS-CoV-2 RNA in circulating exosomes isolated from COVID-19 patients [116]. Some studies suggested the possibility of SARS-CoV-2 using integrin as an additional receptor as its spike protein has been found to be able to bind integrins [117]. Similarly, tetraspanins, transmembrane glycoproteins abundant in exosomes and involved in their endocytosis mechanism, are known to be linked to certain viruses, including coronaviruses, and to be involved in several steps of their life cycle [118,119]. Recent data support the importance of exosome trafficking in viral infections. In particular, the investigation of the interactome between 26 of the 29 SARS-CoV-2 proteins and human proteins revealed that 332 human proteins have a high confidence score of interaction [120]. Of those proteins, about 40% were found to be involved in vesicular trafficking pathways, including exosomes trafficking and lipid modification [119]. Strategies targeting exosome biogenesis and release, as a mechanism for SARS-CoV-2 spread, have been recently proposed [121,122]. Many compounds have been shown to inhibit the release of exosomes such as calpeptin, Y27632, pantethine, imipramine, GW4869 (Figure 4 ) [123]. Further studies are required to validate the effects of these drugs – singly and in combination – in preclinical models of SARS-CoV-2.
Figure 4.
Steps in viral pathways inhibited by drugs acting on lipid metabolism or exosomes release.
10. Defining the changing lipidome to study SARS-CoV-2 infection
The lipidomic study of biological biofluids obtained from patients is particularly challenging compared to the analysis performed at cellular levels [149]. Therefore, before discussing the lipidomic studies performed in humans, we will briefly give an overview of all issues and obstacles that investigators face in this field, especially considering the complications related to handling infected materials. This is an outline of lipidomics approaches to study SARS-CoV-2 infection and COVID-19 pathogenesis.
10.1. Plasma and serum samples
Blood is a biological fluid that can be easily collected with minimal health risk. Its composition reflects human physiology in health and disease but also metabolic aspects. Plasma and serum, which are rich in lipids, are widely used for lipid biomarker discovery [[124], [125], [126], [127], [128]]. Plasma and serum are two distinct matrices; plasma is the liquid that remains when clotting is arrested with the introduction of an anticoagulant, while serum is obtained after blood has clotted. Thus, their lipid profiles differ even when obtained from the same blood draw [129,130].
For lipidomic research, the use of plasma is recommended. Plasma is the closest matrix to blood plasma in vivo, and it is better for robustness, for comparing and interpreting physiological conditions. The coagulation process involved in serum preparation leads to the generation or degradation of molecules, including lipids. This process affects the abundances of LPLs, sphingosine 1-phosphate (S1P), prostaglandins, leukotrienes, resolvins, oxylipins and other lipid species [131]. In clinical practice, the serum is largely used and therefore, may be more appropriate for diagnostic applications. Plasma should be obtained from whole blood collected immediately in tubes including dried or liquid anticoagulant to minimize clotting: K2EDTA, heparin and citrate are the most commonly used. The centrifugation condition of whole blood is another factor that can alter the plasma lipidome, especially in relation to the presence of residual platelets in plasma or their activation. In multicentre studies, the conditions should be standardized prior to the sample collection. Plasma samples should be stored at -80°C or lower temperatures. The containers utilized for sample collection should be airtight to avoid sublimation, which can also impair sample concentrations [132]. Preparing aliquots immediately after plasma/serum isolation will contribute to limit freeze-thaw cycles that may affect the abundance of specific lipid classes.
10.2. Cell samples
Sample stability is very important for lipidomic analysis [133]. Samples should be immediately processed or frozen because enzymatic and chemical processes can metabolize lipids. This is particularly important when oxidation products like oxidized phospholipids, oxylipins, and lysolipids are analysed. Samples should be stored at −80 °C, and the storage time should be as short as possible.
Sample homogenization is another important step for the analysis of cells. This procedure ensures a more efficient lipid extraction by rupturing the plasma membrane and giving access to intracellular domains. Cells can be disrupted by using cold sonication, a pebble mill with beads or a nitrogen cavitation bomb [133,135].
10.3. Viral inactivation
Sample manipulation of SARS-CoV-2 infected material must be performed using appropriate biosafety level containment laboratories until pathogen inactivation. For example, Burnum-Johnson et al. observed the total deactivation of both bacterial and viral pathogens with exposed lipid envelopes, including MERS-CoV, proposing a procedure for concomitant pathogen inactivation and extraction of samples for multi-omics profiling [136]. For lipidomic analysis, the sample could be considered safe after the addition of an alcoholic solution, which is usually employed for protein precipitations and lipid extraction.
11. Dysregulation of lipids in COVID-19 disease
The role of lipids in COVID-19 has been investigated especially on bio-fluids from infected patients. Several studies reported the involvement of lipids and lipid metabolism (Table 1 ) [[137], [138], [139]].
Table 1.
latest published or in pre-print articles that report lipids alterations in COVID-19 disease. The bio-fluid analysed, the number of samples, the platform used and the main altered lipid species are shown.
| Bio-fluid | N. of samples | Platform | Main altered lipids | Reference |
|---|---|---|---|---|
| Serum | 108 | High-Res MS | Glycerophospholipids | Shen et al. [137] |
| Serum | 50 | QQQ | GM3s, free fatty acids | Song et al. [138] |
| Plasma | 35 | QQQ | NEFAs, TAGs, PCs | Wu et al. [153] |
| Serum | 57 | QQQ | PUFAs and lipid mediators | Schwarz et al. [142] |
| Plasma | 161 | High-Res MS | PCs, LPCs, PEs, LPEs, NEFAs | Barberis et al. [145] |
| Serum | 49 | High-Res MS | Acylcarnitines, NEFAs | Thomas et al. [139] |
| Serum | 236 | NMR | Short chain fatty acids | Bruzzone et al. [147] |
| Plasma | 102 | High-Res MS and QQQ | TAGs, PCs | Overmyer et al. [146] |
Shen et al. conducted proteomic and metabolomic profiling of sera from 46 COVID-19 patients and 53 control individuals [137]. They identified differentially expressed factors that correlate with disease severity, highlighting an alteration of multiple immune and metabolic components in clinically severe patients. In particular, a strong down-regulation of over 100 lipids, comprising sphingolipids, glycerophospholipids and fatty acids, was identified. Biomembranes are composed of lipids such as sphingolipids and glycerophospholipids that have not only a structural role but regulate signal transduction and immune activation pathways as well as inflammatory responses [140]. The involvement of glycerophospholipids and fatty acids such as arachidonic acid has been reported in HCoV-229E-infected cells and MERS-CoV replication [51], while the down-regulation of choline and its derivatives and the up-regulation of phosphocholine, were associated to activated macrophage-mediated immunity [141]. The overall down-regulation of serum lipids in COVID-19 patients is linked to liver damage, which is supported by changes in bilirubin and bile acids.
Targeted and untargeted mass spectrometry was used to investigate the lipidome and metabolome of plasma obtained from a cohort of 76 subjects that included 26 healthy controls and 50 COVID-19 patients (mild, moderate, and severe) [138]. The authors noticed diminutions in major classes of plasma glycerophospholipids, comprising phosphatidic acids (PAs), PIs, and PCs and an increase of LPLs such as LPAs, LPIs, and LPCs. They also found that plasma lipidome of COVID-19 was similar to that of monosialodihexosyl ganglioside (GM3)-enriched exosomes, with increased levels of sphingomyelins (SMs) and GM3s, and decreased diacylglycerols (DAGs). Simultaneous elevation in non-esterified fatty acids (NEFAs) such as NEFA18:1 and NEFA 18:2 was shown in mild and moderate COVID-19, while TAGs were significantly decreased uniquely in mild cases. In addition, exosomes of COVID-19 patients with higher disease severity were progressively enhanced in GM3s, suggesting that GM3-rich exosomes may participate in pathological functions linked to COVID-19 pathogenesis.
Lipids alterations were also reported in a cohort of Chinese patients with COVID-19 [153]. General dyslipidaemia was observed using a targeted metabolomic and lipidomic analyses of plasma. Serial samples were gathered and analysed during the course of COVID-19 disease. The cohort included nine patients with fatal outcome, 11 patients diagnosed with severe symptoms, and 14 patients diagnosed as having mild symptoms. Most of the significantly changed lipids were upregulated and characterized by a positive correlation with the course of disease deterioration in fatal cases. DAGs, NEFAs, and TAGs, were found in greater amounts in the fatality group. Lipolysis of adipose tissue enhances as a consequence of infection, converting TAG to NEFA and DAG, but also enhancing the recycling of fatty acids back into TAGs [153]. Phosphatidylcholines (PCs) were constantly decreased during the course of COVID-19 fatal cases.
The reduction of specific immune regulatory lipid mediators (LMs) and the augmentation of pro-inflammatory species from moderate to severe COVID-19 disease was reported in a study that involved 19 healthy patients, 18 COVID-19 patients who did not need ICU admission (moderate) and 20 patients that needed ICU admission (severe) [142]. An increase of free poly-unsaturated fatty acids (PUFAs) and diminished amounts of PUFA-containing plasmalogens were reported [142]. The authors were able to identify an over-expression of the C20:4 series through the classification of lipids containing C20:4 arachidonic acid (AA), C20:5 eicosapentaenoic acid (EPA), C22:5n-3 docosapentaenoic acid (DPA) or C22:6 docosahexaenoic acid (DHA). PUFAs can be released from their related glycerolipids and afterwards converted to a variegate number of immune signalling molecules (eicosanoids and docosanoids) [143,144]. Through a correlation analysis performed on 67 eicosanoid and docosanoid species and 15 cytokines, Schwarz et al. showed that all LMs were directly correlated to the infection. They also found that patients with a moderate disease are distinguished by significantly greater amounts of the pro-resolving LM resolvin E3 (RvE3), an additional increase in free PUFAs levels, AA-, EPA-, DPA and DHA-derived mono-hydroxylated species and AA-derived dihydroxylated species were associated to severe patients. The authors thus suggested that a dysregulation of LMs might take part in disease progression.
The involvement of LMs was reported by Barberis et al [145]. The authors performed untargeted lipidomics and metabolomics analyses on plasma samples of 161 subjects, 103 of whom were infected with SARS-CoV-2. Out of the 103 COVID-19 patients enrolled, 19 were critical (admitted to ICU) and 84 non-critical. The results confirmed a strong alteration of lipids, in particular of acylcarnitines, diacylglycerols, fatty acids, glycerophosphoethanolamines (LPEs, PEs), glycerophosphocholines (LPCs, PCs), sphingomyelins, and triacylglycerols. PCs were significantly down-regulated (65.4%) as a consequence of the infection, while several free fatty acids were up-regulated (FA 18:1, FA 18:2, FA 22:6, FA 44:5, and FA 20:4). Moreover, higher lysophosphatidylethanolamine (LPE) and lysophophatidylcholine (LPC) levels were identified. In addition, they found that the abundance of FA 20:4, a well-known LMs, and of FA 18:1 was directly correlated to the severity of the disease.
Fatty acid metabolism was associated with SARS-CoV-2 infection by Thomas et al [139]. In their work, they performed a metabolomics and lipidomics approach on patients stratified by circulating levels of IL-6. Researchers analysed forty-nine subjects, 33 were COVID-19–positive subjects and 16 COVID-19–negative subjects. Short- and medium-chain acylcarnitines were significantly diminished in all patients with COVID-19. All NEFAs, apart from nonanoic acid, were augmented in all patients with COVID-19.
Overmyer et al. carried out RNA-Seq and high-resolution mass spectrometry on 128 blood samples collected from COVID-19 positive and negative patients with different disease gravities and outcomes. They observed enrichment of TAGs in COVID-19 patients, while plasmenyl–phosphatidylcholines (plasmenyl-PCs) were scarce in COVID-19 samples compared to non-COVID-19 samples [146].
Finally, lipids alterations were detected using NMR spectroscopy by Bruzzone et al. [147]. The authors analysed 263 symptomatic patients, and they found a pathogenic rearrangement of the lipoprotein particle size and composition. Patients were characterized by increased amount of ketone bodies like acetoacetic acid, 3-hydroxybutyric acid and acetone, but also of 2-hydroxybutyric acid, a reflection of hepatic glutathione synthesis and marker of oxidative stress. Authors suggested that serum accumulation of TAG and TAG-VLDL (VLDL, very low-density lipoproteins) in COVID-19 patients might be a consequence of the decreased hepatic ability to oxidize acetyl-CoA in the mitochondria. In fact, ketone bodies are mainly synthesized in the liver from fatty acid oxidation-derived acetyl-CoA.
To sum up, lipid alterations exhibited a correlation with the gravity of the disease, suggesting that the development of the infection influenced whole-body metabolism and specifically lipid metabolism. In addition, the balance between immune-regulating lipids, pro-inflammatory lipids and lipid mediators can modulate the immune response during COVID-19 disease.
12. Lipid metabolism as a central player in SARS-CoV-2 infection
Based on the literature on COVID-19 disease, it is evident that lipid metabolism has a key role in SARS-CoV-2 infection. This is supported not only by lipidomic results, but also by the strong involvement of apolipoproteins and steroid hormones. A decrease of apolipoproteins APOA1, APOA2, APOH, APOL1, APOD, and APOM was identified after the infection [137], while APOA1 underexpression was reported through the transition of COVID-19 patients from mild to severe disease [148]. Most of these apolipoproteins are associated with macrophage functions, which is closely related to lipid metabolism. The augmentation of steroid hormones, which can stimulate the activity of macrophages, was also shown [137,139].
In addition, Song et al. [138] discovered that plasma lipids are associated with pathologically relevant clinical indices: PUFA-PCs and plasmalogen-PCs, showed significant negative correlations with clinical parameters of systemic inflammation (IL-6, CRP, procalcitonin [PCT], ESR, and SF), suggesting that diminutions in plasma PUFA-PCs and plasmalogen-PCs are correlated with worsen systemic inflammation. Moreover, plasma GM3s were highly and negatively associated with T cell count and CD4+ T cell count, which progressively diminished as disease severity augmented, suggesting that higher levels of GM3s in plasma are correlated with diminutions in circulating CD4+ T cell counts in COVID-19.
Dysregulations of lipid metabolism resulted in exceedingly drastic changes in fatal COVID-19 cases than in survivors, as reported by Wu et al [153]. Interestingly, patients hospitalized with either severe or mild symptoms, showed abnormal lipid profiles long after major clinical signs had disappeared.
It has already been shown that, in severe COVID-19 inflammation, arachidonic acid is involved in the production of cytokines, causing a cytokine storm [150,151]. The same cytokines are able to induce the release of unsaturated fatty acids as a defence mechanism against invading microorganisms such as viruses. This highlights the important role of lipids, and in particular of glycerophospholipids and fatty acids, in inflammation and virus protection.
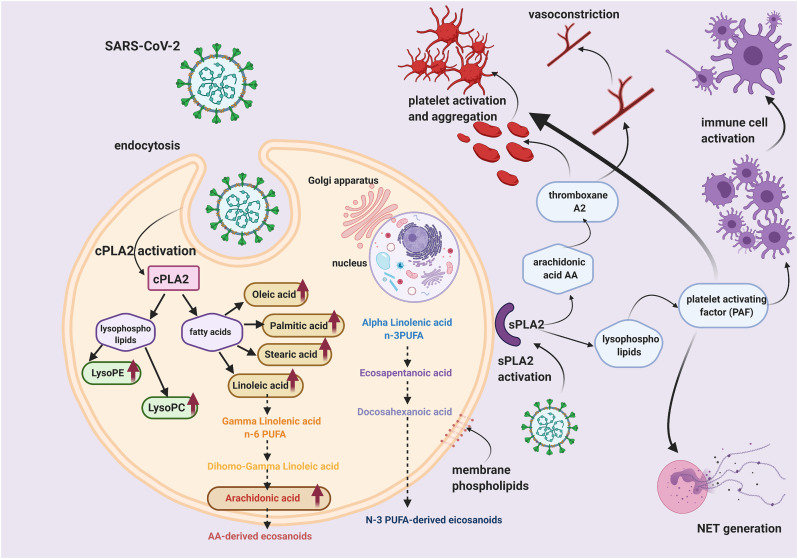
For example, elevated levels of long-chain PUFAs, in the absence of elevated long-chain acylcarnitines, which could mirror impaired mitochondrial fatty acid oxidation, might indicate that phospholipase A2 (PLA2) is involved in COVID-19. In fact, increased PLA2 activity is correlated with augmented synthesis of bioactive lipids by the metabolism of omega-6 PUFAs and lysophosphatidylcholine (Figure 5 ).
Figure 5.
Pathways activated by cytosolic phospholipase A2 (cPLA2) and secretory phospholipase A2 (sPLA2) potentially involved in virus entry and pathogenesis.
COVID-19 patients are also characterized by an increase of serum TAG, TAG-VLDL, TAG-IDL (IDL, intermediate density lipoproteins), TAG-LDL (LDL, low-density lipoproteins) and TAG-HDL (HDL, high density lipoproteins) [147]. At the same time, the total cholesterol (TC) was decreased. All these examinations imply a remodelling of the lipoprotein particle phenotype in COVID-19 patients, and thus of a great part of circulating lipids. Consequently, these observations denote an augmented cardiovascular risk for COVID-19 patients [147].
13. Lipids as biomarkers of SARS-CoV-2 infection
The discovery of potential biomarkers for the identification of COVID-19 disease is a challenge that could be overcome using mass spectrometry. Shen et al. investigated the potentiality of characterising the severity of COVID-19 patients based on the molecular signatures of proteins and metabolites [137]. They employed machine learning models on selected proteins (22) and metabolites (7), obtaining an area under the curve (AUC) of 0.957 in the training set. The classification of an independent cohort of patients allowed the correct classification of almost all severe patients. They also further validated the ability to predict severe patients by analysing a second test cohort using targeted mass spectrometry, confirming the performances.
A panel of plasma lipids and metabolites able to distinguish COVID-19 was also identified by Song et al [138], reaching an AUC of 0.975. Among these molecules, lysophophoslipids including lysophosphatidic acid (LPA) 18:1 and lysophosphatidylcholine 18:1, were increased in COVID-19. While non-polar lipids, including medium-chain TAG 48:1(18:0), long-chain TAG 60:3(18:1), and DAG 34:1(16:1/18:0) were reduced. Sphingolipids such as SM d18:1/18:1 and GM3 d18:1/25:0 were generally increased with the disease [138]. Wu et al. generated ROC curves of modulated lipids for discriminating between patients with COVID-19 and healthy controls, indicating glycerol 3-phosphate as the best biomarker, with an AUC value of 1.00 [153]. Finally, Barberis and co-authors reported that molecular species 14:0/22:6-PC, 16:1/22:6-PC, and 18:1/20:4-PE were promising potential biomarkers, with an area under the curve (AUC) values of 0.96, 0.97, and 0.94, respectively, whilst their integrated ROC reported an AUC of 0.97 [145]. The best biomarkers were arachidonic acid (FA 20:4), oleic acid (FA 18:1), 18:2/20:4-PE, and 16:1/18:2-PE, with AUC values of 0.99, 0.98 0.92, and 0.92, respectively. They investigated and validated the diagnostic performances of these biomarkers by analysing non-COVID-19 patients with similar symptoms and those with the COVID-19 infection, confirming the validity of potential biomarkers. Although some interesting potential biomarkers were identified, a multi-centric study has yet to be performed. Moreover, a wide range of confounding diseases, especially the ones associated with inflammation, should be considered.
14. Lipids and lipid metabolism as potential therapeutic targets of COVID-19 disease
As reported in previous sections, lipids not only are strongly altered in COVID-19, but are also entailed in the pathogenesis and progression of the infection. For example, the imbalance between lipid mediators and lipid enzymes that mediate inflammation or resolution play a central role in the development of the disease. Thus, lipids can be used as therapeutic pharmacologic agents, or lipid pathways can be targeted with drugs. Shen et al. suggested that COVID-19 patients might ameliorate from increased arachidonic acid because the exogenous supplement of arachidonic acid significantly impaired HCoV-229E and MERS-CoV replication [154] and because they found a decrease of glycerophospholipids. The authors also suggested the use of drugs inhibiting lipid syntheses such as statins, which had already been proposed to treat HCV [154] and COVID-19 [155]. On the other hands, Barberis et al. identified a down-regulation of glycerophospholipids and an up-regulation of LPLs, arachidonic acid and oleic acid, suggesting a strong involvement of PLA2 in the pathogenesis and progression of COVID-19 [145]. The involvement of PLA2 is also supported by Thomas et al. that found increased amounts of long-chain PUFAs, that are particularly enriched in the sn-2 position of phospholipids. Augmented PLA2 activity is correlated with greater production of bioactive lipids by the metabolism of omega-6 PUFAs and lysophosphatidylcholine. Thus, the inhibition of PLA2 activity or the augmenting of omega-3 PUFA levels that are transformed in bioactive lipids with anti-inflammatory activity might decrease disease severity. Indeed, for their anti inflammatory and antiviral properties, the use of omega-3 PUFA has been proposed, in combination with other anti-viral agents, for the treatments of COVID-19 patients. Interestingly, several clinical trials (NCT04647604, NCT04553705, NCT04495816, NCT04658433) are currently recruiting patients to validate the protective effect of an omega-3 PUFA enriched diet on patients severely affected by COVID-19 and to highlight the possible preventive role of a healthy diet in forthcoming outbreaks [156,157]. Finally, Wu et al. and Bruzzone et al. showed that many of the metabolites, lipid and lipoproteins alterations in COVID-19 are associated with hepatic functions, suggesting the targeting of liver activity as therapeutic strategy [147,153].
15. COVID-19-associated dyslipidaemia
Dyslipidaemia is a group of diverse abnormal blood lipid conditions outside of the normal range – generally describing hypercholesterolemia, elevated LDL cholesterol, reduced HDL cholesterol and hypertriglyceridemia – that promote atherosclerosis [158,159]. Dyslipidaemia can be separated into two broad categories, primary and secondary. Primary dyslipidaemia is an inherited disease that can have clinical indications earlier in life, particularly if the defect is monogenetic [160,161]. Secondary dyslipidaemia is multifactorial and is acquired through lifestyle and environmental factors. Prolonged inflammation and infection are two examples of non-genetic factors that can elicit abnormal lipid profile in the circulation. Patients with inflammatory diseases such as rheumatoid arthritis and systemic lupus erythematosus are noted for their substantial decrease in HDL cholesterol and apolipoprotein A-I levels [162,163], and low HDL and increased serum triglyceride levels, respectively. Similarly, the reduction of these lipoproteins is observed during infections, and is predictive of severity and mortality [[164], [165], [166]]. Thus, it is not surprising that COVID-19 patients showed worsen hypolipidaemia in association with the disease progression and severity of the symptoms [12,167,168]. Table 2 summarises several COVID-10 associated dyslipidaemia studies. Although cholesterol may be a predictive marker of poor outcome for COVID-19 infection, the exact molecular mechanism underpinning the functional role of lipids during SARS-CoV-2 infection is still not completely understood. Two large epidemiological studies recently described an increased risk of infection-related hospitalizations with low HDL-c levels [169,170], which support several previous findings that HDL-c has an essential part in host defence, including anti-thrombotic [[171], [172], [173]], anti-oxidant [174,175] and anti-inflammatory [171,176,177] properties. While there is not enough evidence to suggest that a reduced cholesterol level is the direct contributory factor in, or cause of, SARS-CoV-2 susceptibility, calls to examine cholesterol as a therapeutic target are gaining momentum [178,179]. Stemmed from retrospective analyses that showed COVID-19 patients on statins had better outcome [180,181], a yet to be peer-reviewed in vitro study by Moeller et al. suggests a decreased rate of SARS-CoV-2 infection under statin treatment, possibly due to reduced membrane cholesterol impacting ACE2-viral spike protein interactions [182].
Table 2.
Observational studies on COVID-19-associated dyslipidaemia
| Studies | Lipid features in COVID-19 patients | References |
|---|---|---|
| A retrospective study of 597 patients from Union Hospital of Tongji Medical College, Wuhan | ↓ LDL-c, HDL-c, TC and TAG in severe vs mild disease | Wei et al. [167] |
| A retrospective study of 21 patients at Zhongnan Hospital of Wuhan University in Wuhan | ↓ LDL-c, HDL-c, and TC in severe vs mild disease LDL returned to pre-infection level upon recovery |
Fan et al. [168] |
| A retrospective study of 71 patients at Wenzhou Central Hospital, Wuhan | ↓ LDL-c, HDL-c, and TC in COVID-19 patients vs healthy | Hu et al. [222] |
| A retrospective study of 114 patients at Wenzhou Central Hospital, Wuhan | Significant ↓ LDL-c, HDL-c, TAG and TC in COVID-19 patients vs healthy Significant ↓ HDL-c in severe vs COVID cases |
Hu et al. [12] |
| A retrospective analysis of 228 COVID patients at Public Health Treatment Center of Changsha, China | ↓ LDL-c, HDL-c in with disease severity | Wang et al. [223] |
| A clinical data collected from 70 patients at Yeungnam University Medical Center, Republic of Korea, as part of a sterol regulatory element binding protein-2 (SREBP-2) study | ↓ LDL-c, HDL-c in severe vs mild | Wonhwa et al. [224] |
HDL-c = high-density lipoprotein cholesterol, LDL-c = low-density lipoprotein cholesterol, TC = total cholesterol, TAG =triacylglycerol.
16. The role of lipids in COVID-19-associated coagulopathy
It is gradually becoming clear that SARS-CoV-2 causes a range of haematological issues. Whilst it is not unexpected that the elderly and patients with pre-existing health issues have elevated susceptibility to severe and life-threatening thrombotic complications during the infection, reports of young and healthy individuals experiencing varying degrees of blood clotting symptoms, from deep vein thrombosis (DVT) to strokes, are increasing in numbers. COVID-19-induced hematologic abnormalities are diverse. Studies have indicated that lymphopenia (low blood lymphocytes), neutrophilia (high count of circulating neutrophils) and thrombocytopenia (low platelet count), information readily obtained from routine blood analysis, are predictive of COVID-19 severity and mortality [[183], [184], [185], [186]]. Similarly, coagulation tests for increased plasma D-dimers, fibrinogen degradation products and prothrombin time (time required for blood plasma to clot) are associated with morbidity and death [[187], [188], [189], [190]]. The combination of elevated prothrombin time, increased levels of D-dimer and thrombocytopenia are features of disseminated intravascular coagulation (DIC), a disorder of blood clots blocking small vessels throughout the body, resulting in a variety of symptoms including fever, shortness of breath, and bleeding. A study by Tang et al. showed a striking 71.4% of the COVID-19 non-survivors met the criteria for DIC according to the International Society on Thrombosis and Haemostasis (ISTH), compared to 0.6% of survivors [188]. DIC can be triggered by infection, injury or certain types of cancer, through the extrinsic coagulation pathway when blood is exposed to tissue factor, commonly, on injured endothelium. Although there are some similar laboratory parameters, COVID-19-associated coagulopathy is quite different from DIC [191]. COVID-19 patients often suffer thrombotic complications rather than DIC-associated bleeding issues. Al-Samkeri et al. reported 9.5% overall thrombotic complication rate (approximately half of these individuals had confirmed venous thromboembolism (VTE), while 4.8% of patients showed overall bleeding event [192]. Thus, anticoagulants are increasingly being recommended by expert groups to treat hospitalized COVID-19 and critically ill patients [193,194]. The haemostatic system is complex and highly regulated interactions between the vessel wall, platelets, and plasma proteins, all of which lipids are acknowledged as essential components [[195], [196], [197], [198]]. For example, oxidized LDL and VLDL can act as a surface to support prothrombinase complex activity and, other reactions within the coagulation cascade [199,200]. However, one group of lipid-associated proteins is receiving increased attention during COVID-19 pandemic. The activity of enzymes of the family PLA2 is well known to be significantly elevated during inflammation and infection, including by SARS-CoV [[201], [202], [203]]. During viral infection, especially with positive-sense RNA viruses, the PLA2 activity has been demonstrated to reflect the remodelling of the host cellular membrane structure to accommodate viral replication [45,204,205]. PLA2 catalyses the cleavage of membrane glycerophospholipids, producing LPLs and fatty acids (Figure 5). There are more than 30 PLA2s or related enzymes in mammalian cells, which are subdivided into three major families, the cytosolic cPLA2 and the extracellular or secretory sPLA2 (Figure 5) [206]. PLA2-generated products such as AA and LPLs are crucial in the biosynthesis of potent cellular mediators: eicosanoids and platelet-activating factor [207,208]. In the context of coagulation and thrombosis, the products of PLA2 are highly relevant. The phospholipid platelet-activating factor (PAF) is an agonist of platelet activation and aggregation [209,210]. PAF can also stimulate neutrophils to release neutrophil extracellular trap (NET), a mesh of proteins and DNA that is capable of initiating the coagulation cascade, platelet aggregation and thrombosis [[211], [212], [213], [214]]. Similarly, AA and its eicosanoid products, such as thromboxane A2, have platelet-activating effects [215,216]. Indeed, increased platelet activity, hypercoagulability, and NET markers have been detected in COVID-19 patients with thrombotic complications [[217], [218], [219], [220]]. It remains to be investigated whether targeting virus-induced lipid dysfunction by inhibiting PLA2 activation is a useful strategy to ameliorate inflammation and clotting issues.
17. Conclusion and future perspectives
SARS-CoV-2 is the cause of the COVID-19 pandemic that has infected about a hundred million people globally (as of January 21st, 2021). In the last few months, an unprecedented scientific effort has been directed at studying COVID-19, particularly in examining its peculiar capacity to attack many different types of human cells and what drives its unusually high mortality rate [221]. An effective research approach is urgently needed to further understand the mechanism of action of this virus in order to develop a targeted therapy and a method to control its spreading. Different therapeutical approaches have been tried, using antiviral such as Remdesivir, corticosteroids or monoclonal antibodies, with marginal success in some cases, but a specific cure targeting this virus and impeding the occurrence of health complications for patients is yet to be found. In particular, it is currently impossible to anticipate the prognosis for infected patients and so the few treatments available start usually at a late stage when patients are already in critical conditions. Thus, the possibility of early treatment is currently hindered. Viruses are known to exploit the host resource and machinery, particularly lipid metabolism pathways, to invade cells and replicate. Therefore, therapeutic approaches targeting conserved cellular mechanism that virus manipulates to enter host cells and replicate are particularly attractive. Strategies to specifically target lipid metabolism have been tried, such as the use of cholesterol depleting agents or autophagy inducers. One way of responding in a short time frame and avoiding severe adverse effects is to look [149] to drugs that have received approval to treat other diseases. Unfortunately, given the frequency of viral epidemics that we have witnessed in the last decade (SARS, MERS, HIV/AIDS, H1N1, Ebola virus etc.), it is likely that we will see the appearance of a new pandemic in the near future. Consequently, a therapeutical approach to COVID-19 that can be adapted and exploited in the event of future outbreaks is urgently needed.
Vaccine or antibodies, if successfully developed, will be specific for SARS-CoV-2 and are unlikely to be useful in protecting against other viruses [222]. Therefore, a more general strategy to block virus spread, such as drugs targeting key steps in lipid metabolism, has the potential to be extended to counteract other viral infections.
Declaration of Competing Interest
The authors have no competing interests to declare.
Acknowledgements
This work was partially supported by the AGING Project – Department of Excellence – DIMET, Università del Piemonte Orientale”, MIUR ITALY to MM. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. Figures were created with BioRender.com.
References
- 1.Morens D.M., Daszak P., Taubenberger J.K. Escaping Pandora's Box - Another Novel Coronavirus. N Engl J Med. 2020;382:1293–1295. doi: 10.1056/NEJMp2002106. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . 11 March 2020. WHO Director-General's opening remarks at the media briefing on COVID-19; p. 2020. [Google Scholar]
- 3.Coronavirus: three things all governments and their science advisers must do nowNature. 2020;579:319–320. doi: 10.1038/d41586-020-00772-4. [DOI] [PubMed] [Google Scholar]
- 4.Vogel G. 2020. ‘It’s been so, so surreal.’ Critics of Sweden’s lax pandemic policies face fierce backlash. [Google Scholar]
- 5.J.H.C.R.C. (CRC) 2020. COVID-19 data in motion. [Google Scholar]
- 6.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 7.Matheson N.J., Lehner P.J. How does SARS-CoV-2 cause COVID-19? Science. 2020;369:510–511. doi: 10.1126/science.abc6156. [DOI] [PubMed] [Google Scholar]
- 8.Yu X., Yang R. COVID-19 transmission through asymptomatic carriers is a challenge to containment. Influenza Other Respir Viruses. 2020;14:474–475. doi: 10.1111/irv.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenhalgh T., Schmid M.B., Czypionka T., Bassler D., Gruer L. Face masks for the public during the covid-19 crisis. Bmj. 2020;369:m1435. doi: 10.1136/bmj.m1435. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. 2020;17:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abu-Farha M., Thanaraj T.A., Qaddoumi M.G., Hashem A., Abubaker J., Al-Mulla F. The Role of Lipid Metabolism in COVID-19 Virus Infection and as a Drug Target. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21103544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X., Chen D., Wu L., He G., Ye W. Declined serum high density lipoprotein cholesterol is associated with the severity of COVID-19 infection. Clin Chim Acta. 2020;510:105–110. doi: 10.1016/j.cca.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morawietz H., Julius U., Bornstein S.R. Cardiovascular diseases, lipid-lowering therapies and European registries in the COVID-19 pandemic. Cardiovasc Res. 2020;116:e122–e125. doi: 10.1093/cvr/cvaa176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das U.N. Response to: Bioactive Lipids and Coronavirus (COVID-19)-further Discussion. Arch Med Res. 2020;51:445–449. doi: 10.1016/j.arcmed.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin E.V., Dolja V., Krupovic M., Varsani A., Wolf Y.I., Yutin N., et al. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol Mol Biol Rev. 2020;84 doi: 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C.W., Peng T.T., Hsu H.Y., Lee Y.Z., Wu S.H., Lin W.H., et al. 2020. Repurposing old drugs as antiviral agents for coronaviruses. Biomed J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Antón-Plágaro C., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020 Nov 13;370(6518):861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020 Nov 13;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sturley S.L., Rajakumar T., Hammond N., Higaki K., Márka Z., Márka S., et al. Potential COVID-19 therapeutics from a rare disease: weaponizing lipid dysregulation to combat viral infectivity. J Lipid Res. 2020;61:972–982. doi: 10.1194/jlr.R120000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radenkovic D., Chawla S., Pirro M., Sahebkar A., Banach M. Cholesterol in Relation to COVID-19: Should We Care about It? J Clin Med. 2020;9 doi: 10.3390/jcm9061909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altan-Bonnet N. Lipid Tales of Viral Replication and Transmission. Trends Cell Biol. 2017;27:201–213. doi: 10.1016/j.tcb.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazzon M., Marsh M. Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Res. 2019;8 doi: 10.12688/f1000research.19694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020 Nov 12;183(4):1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toelzer C., Gupta K., Yadav S.K.N., Borucu U., Davidson A.D., Kavanagh Williamson M., et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370:725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olalla J. Remdesivir for the Treatment of Covid-19 - Preliminary Report. N Engl J Med. 2020;383:993–994. doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- 29.Badger J., Minor I., Kremer M.J., Oliveira M.A., Smith T.J., Griffith J.P., et al. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc Natl Acad Sci U S A. 1988;85:3304–3308. doi: 10.1073/pnas.85.10.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveira M.A., Zhao R., Lee W.-M., Kremer M.J., Minor I., Rueckert R.R., et al. The structure of human rhinovirus 16. Structure. 1993;1:51–68. doi: 10.1016/0969-2126(93)90008-5. [DOI] [PubMed] [Google Scholar]
- 31.Casanova V., Sousa F.H., Stevens C., Barlow P.G. Antiviral therapeutic approaches for human rhinovirus infections. Future Virol. 2018;13:505–518. doi: 10.2217/fvl-2018-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K.K.W. To, Yip C.C.Y., Yuen K.Y. Rhinovirus - From bench to bedside. J Formos Med Assoc. 2017;116:496–504. doi: 10.1016/j.jfma.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Lorizate M., Kräusslich H.G. Role of lipids in virus replication. Cold Spring Harb Perspect Biol. 2011;3:a004820. doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glebov O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. Febs j. 2020 Sep;287(17):3664–3671. doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., et al. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maffucci T., Falasca M. Analysis, regulationand roles of endosomal phosphoinositides. Methods Enzymol. 2014;535:75–91. doi: 10.1016/B978-0-12-397925-4.00005-5. [DOI] [PubMed] [Google Scholar]
- 37.Qiu S., Leung A., Bo Y., Kozak R.A., Anand S.P., Warkentin C., et al. Ebola virus requires phosphatidylinositol (3,5) bisphosphate production for efficient viral entry. Virology. 2018;513:17–28. doi: 10.1016/j.virol.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 38.Kang Y.L., Chou Y.Y., Rothlauf P.W., Liu Z., Soh T.K., Cureton D., et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuen C.K., Wong W.M., Mak L.F., Wang X., Chu H., Yuen K.Y., et al. Suppression of SARS-CoV-2 infection in ex-vivo human lung tissues by targeting class III phosphoinositide 3-kinase. J Med Virol. 2020 Oct 7 doi: 10.1002/jmv.26583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klann K., Bojkova D., Tascher G., Ciesek S., Münch C., Cinatl J. Growth Factor Receptor Signaling Inhibition Prevents SARS-CoV-2 Replication. Mol Cell. 2020;80:164–174. doi: 10.1016/j.molcel.2020.08.006. e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ketter E., Randall G. Virus Impact on Lipids and Membranes. Annu Rev Virol. 2019;6:319–340. doi: 10.1146/annurev-virology-092818-015748. [DOI] [PubMed] [Google Scholar]
- 42.Martin S., Harper C.B., May L.M., Coulson E.J., Meunier F.A., Osborne S.L. Inhibition of PIKfyve by YM-201636 dysregulates autophagy and leads to apoptosis-independent neuronal cell death. PLoS One. 2013;8:e60152. doi: 10.1371/journal.pone.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Min S.H., Suzuki A., Stalker T.J., Zhao L., Wang Y., McKennan C., et al. Loss of PIKfyve in platelets causes a lysosomal disease leading to inflammation and thrombosis in mice. Nat Commun. 2014;5:4691. doi: 10.1038/ncomms5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jarsch I.K., Daste F., Gallop J.L. Membrane curvature in cell biology: An integration of molecular mechanisms. J Cell Biol. 2016;214:375–387. doi: 10.1083/jcb.201604003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller C., Hardt M., Schwudke D., Neuman B.W., Pleschka S., Ziebuhr J. Inhibition of Cytosolic Phospholipase A(2)α Impairs an Early Step of Coronavirus Replication in Cell Culture. J Virol. 2018;92 doi: 10.1128/JVI.01463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolff G., Melia C.E., Snijder E.J., Bárcena M. Double-Membrane Vesicles as Platforms for Viral Replication. Trends Microbiol. 2020 Dec;28(12):1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagy P.D., Strating J.R., van Kuppeveld F.J. Building Viral Replication Organelles: Close Encounters of the Membrane Types. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005912. e1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.V'Kovski P., Al-Mulla H., Thiel V., Neuman B.W. New insights on the role of paired membrane structures in coronavirus replication. Virus Res. 2015;202:33–40. doi: 10.1016/j.virusres.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chao T.C., Su W.C., Huang J.Y., Chen Y.C., Jeng K.S., Wang H.D., et al. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), a host membrane-deforming protein, is critical for membranous web formation in hepatitis C virus replication. J Virol. 2012;86:1739–1749. doi: 10.1128/JVI.06001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verheije M.H., Raaben M., Mari M., Te Lintelo E.G., Reggiori F., van Kuppeveld F.J., et al. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000088. e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan B., Chu H., Yang D., Sze K.H., Lai P.M., Yuan S., et al. Characterization of the Lipidomic Profile of Human Coronavirus-Infected Cells: Implications for Lipid Metabolism Remodeling upon Coronavirus Replication. Viruses. 2019;11 doi: 10.3390/v11010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neufeldt C.J., Joyce M.A., Van Buuren N., Levin A., Kirkegaard K., Gale M., Jr., et al. The Hepatitis C Virus-Induced Membranous Web and Associated Nuclear Transport Machinery Limit Access of Pattern Recognition Receptors to Viral Replication Sites. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005428. e1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinkert D., Bartenschlager R., Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paul D., Hoppe S., Saher G., Krijnse-Locker J., Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol. 2013;87:10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du X., Zhang Y., Zou J., Yuan Z., Yi Z. Replicase-mediated shielding of the poliovirus replicative double-stranded RNA to avoid recognition by MDA5. J Gen Virol. 2018;99:1199–1209. doi: 10.1099/jgv.0.001111. [DOI] [PubMed] [Google Scholar]
- 57.van Hemert M.J., van den Worm S.H., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000054. e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snijder E.J., Limpens R., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., et al. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000715. e3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghosh S., Dellibovi-Ragheb T.A., Kerviel A., Pak E., Qiu Q., Fisher M., et al. β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell, 183. 2020;1520-1535:e1514. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendonça L., Howe A., Gilchrist J.B., Sun D., Knight M.L., Zanetti-Domingues L.C., et al. SARS-CoV-2 Assembly and Egress Pathway Revealed by Correlative Multi-modal Multi-scale Cryo-imaging. bioRxiv. 2020 Nov 5 doi: 10.1038/s41467-021-24887-y. 2020.2011.2005.370239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tutuncuoglu B., Cakir M., Batra J., Bouhaddou M., Eckhardt M., Gordon D.E., et al. The Landscape of Human Cancer Proteins Targeted by SARS-CoV-2. Cancer Discov. 2020;10:916–921. doi: 10.1158/2159-8290.CD-20-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimano H., Sato R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat Rev Endocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 63.Yuan S., Chu H., Chan J.F., Ye Z.W., Wen L., Yan B., et al. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat Commun. 2019;10:120. doi: 10.1038/s41467-018-08015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyers A., Weiskittel T.M., Dalhaimer P. Lipid Droplets: Formation to Breakdown. Lipids. 2017;52:465–475. doi: 10.1007/s11745-017-4263-0. [DOI] [PubMed] [Google Scholar]
- 65.Roberts M.A., Olzmann J.A. Protein Quality Control and Lipid Droplet Metabolism. Annual Review of Cell and Developmental Biology. 2020;36:115–139. doi: 10.1146/annurev-cellbio-031320-101827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pagliari F., Marafioti M.G., Genard G., Candeloro P., Viglietto G., Seco J., et al. Is There a Role for Lipid Droplets in SARS-CoV-2 Infection? Frontiers in Molecular Biosciences. 2020:7. doi: 10.3389/fmolb.2020.578964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bosch M., Sánchez-Álvarez M., Fajardo A., Kapetanovic R., Steiner B., Dutra F., et al. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science. 2020;370 doi: 10.1126/science.aay8085. [DOI] [PubMed] [Google Scholar]
- 68.Moriel-Carretero M. The hypothetical role of phosphatidic acid in subverting ER membranes during SARS-CoV infection. Traffic. 2020;21:545–551. doi: 10.1111/tra.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dias S.S.G., Soares C., Ferreira A.C., Sacramento C.Q., Fintelman-Rodrigues N., Temerozo J.R., et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009127. e1009127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosenberger C.M., Brumell J.H., Finlay B.B. Microbial pathogenesis: lipid rafts as pathogen portals. Curr Biol. 2000;10:R823–R825. doi: 10.1016/s0960-9822(00)00788-0. [DOI] [PubMed] [Google Scholar]
- 71.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 72.Brown D.A., Rose J.K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 73.Sargiacomo M., Sudol M., Tang Z., Lisanti M.P. Signal transducing molecules and glycosyl-phosphatidylinositol-linked proteins form a caveolin-rich insoluble complex in MDCK cells. J Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simons K., Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 75.McGuinn K.P., Mahoney M.G. Lipid rafts and detergent-resistant membranes in epithelial keratinocytes. Methods Mol Biol. 2014;1195:133–144. doi: 10.1007/7651_2014_71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nomura R., Kiyota A., Suzaki E., Kataoka K., Ohe Y., Miyamoto K., et al. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J Virol. 2004;78:8701–8708. doi: 10.1128/JVI.78.16.8701-8708.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi K.S., Aizaki H., Lai M.M. Murine coronavirus requires lipid rafts for virus entry and cell-cell fusion but not for virus release. J Virol. 2005;79:9862–9871. doi: 10.1128/JVI.79.15.9862-9871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Riethmüller J., Riehle A., Grassmé H., Gulbins E. Membrane rafts in host-pathogen interactions. Biochim Biophys Acta. 2006;1758:2139–2147. doi: 10.1016/j.bbamem.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 79.Lu Y., Liu D.X., Tam J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem Biophys Res Commun. 2008;369:344–349. doi: 10.1016/j.bbrc.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baglivo M., Baronio M., Natalini G., Beccari T., Chiurazzi P., Fulcheri E., et al. Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: a possible strategy for reducing SARS-COV-2 infectivity? Acta Biomed. 2020;91:161–164. doi: 10.23750/abm.v91i1.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105960. 105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodrigues-Diez R.R., Tejera-Muñoz A., Marquez-Exposito L., Rayego-Mateos S., Santos Sanchez L., Marchant V., et al. Statins: Could an old friend help in the fight against COVID-19? Br J Pharmacol. 2020 Nov;177(21):4873–4886. doi: 10.1111/bph.15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dou X., Li Y., Han J., Zarlenga D.S., Zhu W., Ren X., et al. Cholesterol of lipid rafts is a key determinant for entry and post-entry control of porcine rotavirus infection. BMC Vet Res. 2018;14:45. doi: 10.1186/s12917-018-1366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wudiri G.A., Nicola A.V. 2017. Cellular Cholesterol Facilitates the Postentry Replication Cycle of Herpes Simplex Virus 1, J Virol, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee C.J., Lin H.R., Liao C.L., Lin Y.L. Cholesterol effectively blocks entry of flavivirus. J Virol. 2008;82:6470–6480. doi: 10.1128/JVI.00117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Españo E., Nam J.H., Song E.J., Song D., Lee C.K., Kim J.K. Lipophilic statins inhibit Zika virus production in Vero cells. Sci Rep. 2019;9:11461. doi: 10.1038/s41598-019-47956-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fecchi K., Anticoli S., Peruzzu D., Iessi E., Gagliardi M.C., Matarrese P., et al. Coronavirus Interplay With Lipid Rafts and Autophagy Unveils Promising Therapeutic Targets. Front Microbiol. 2020;11:1821. doi: 10.3389/fmicb.2020.01821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sviridov D., Mukhamedova N., Miller Y.I. Lipid rafts as a therapeutic target. J Lipid Res. 2020;61:687–695. doi: 10.1194/jlr.TR120000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kroemer G., Mariño G., Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Runwal G., Stamatakou E., Siddiqi F.H., Puri C., Zhu Y., Rubinsztein D.C. LC3-positive structures are prominent in autophagy-deficient cells. Sci Rep. 2019;9:10147. doi: 10.1038/s41598-019-46657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bjørkøy G., Lamark T., Pankiv S., Øvervatn A., Brech A., Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 94.Mao J., Lin E., He L., Yu J., Tan P., Zhou Y. Autophagy and Viral Infection. Adv Exp Med Biol. 2019;1209:55–78. doi: 10.1007/978-981-15-0606-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khandia R., Dadar M., Munjal A., Dhama K., Karthik K., Tiwari R., et al. A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells. 2019;8 doi: 10.3390/cells8070674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pan C., Jin L., Wang X., Li Y., Chun J., Boese A.C., et al. Inositol-triphosphate 3-kinase B confers cisplatin resistance by regulating NOX4-dependent redox balance. J Clin Invest. 2019;129:2431–2445. doi: 10.1172/JCI124550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Delorme-Axford E., Donker R.B., Mouillet J.F., Chu T., Bayer A., Ouyang Y., et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U S A. 2013;110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Orvedahl A., MacPherson S., Sumpter R., Jr., Tallóczy Z., Zou Z., Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deretic V., Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jackson W.T., Giddings T.H., Jr., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cong Y., Verlhac P., Reggiori F. The Interaction between Nidovirales and Autophagy Components. Viruses. 2017;9 doi: 10.3390/v9070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kindrachuk J., Ork B., Hart B.J., Mazur S., Holbrook M.R., Frieman M.B., et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob Agents Chemother. 2015;59:1088–1099. doi: 10.1128/AAC.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gassen N.C., Niemeyer D., Muth D., Corman V.M., Martinelli S., Gassen A., et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat Commun. 2019;10:5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu C.H., Yeh S.H., Tsay Y.G., Shieh Y.H., Kao C.L., Chen Y.S., et al. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J Biol Chem. 2009;284:5229–5239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zúñiga S., Cruz J.L., Sola I., Mateos-Gómez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gassen N.C., Papies J., Bajaj T., Dethloff F., Emanuel J., Weckmann K., et al. Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. bioRxiv. 2020 https://www.biorxiv.org/content/10.1101/2020.04.15.997254v1 2020.2004.2015.997254. [Google Scholar]
- 107.Xu J., Shi P.Y., Li H., Zhou J. Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential. ACS Infect Dis. 2020;6:909–915. doi: 10.1021/acsinfecdis.0c00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De Toro J., Herschlik L., Waldner C., Mongini C. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front Immunol. 2015;6:203. doi: 10.3389/fimmu.2015.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Welton J.L., Khanna S., Giles P.J., Brennan P., Brewis I.A., Staffurth J., et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wubbolts R., Leckie R.S., Veenhuizen P.T., Schwarzmann G., Möbius W., Hoernschemeyer J., et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 111.Skotland T., Sandvig K., Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 112.McNamara R.P., Dittmer D.P. Extracellular vesicles in virus infection and pathogenesis. Curr Opin Virol. 2020;44:129–138. doi: 10.1016/j.coviro.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raab-Traub N., Dittmer D.P. Viral effects on the content and function of extracellular vesicles. Nat Rev Microbiol. 2017;15:559–572. doi: 10.1038/nrmicro.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Altan-Bonnet N., Perales C., Domingo E. Extracellular vesicles: Vehicles of en bloc viral transmission. Virus Res. 2019;265:143–149. doi: 10.1016/j.virusres.2019.03.023. [DOI] [PubMed] [Google Scholar]
- 115.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Marrero M. Correa, et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182 doi: 10.1016/j.cell.2020.06.034. 685-712.e619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barberis E., Vanella V.V., Falasca M., Caneparo V., Cappellano G., Raineri D., et al. Circulating Exosomes Are Strongly Involved in SARS-CoV-2 Infection. Front. Mol. Biosci. 2021;8:29. doi: 10.3389/fmolb.2021.632290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sigrist C.J., Bridge A., Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 2020;177:104759. doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Earnest J.T., Hantak M.P., Park J.E., Gallagher T. Coronavirus and influenza virus proteolytic priming takes place in tetraspanin-enriched membrane microdomains. J Virol. 2015;89:6093–6104. doi: 10.1128/JVI.00543-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martin F., Roth D.M., Jans D.A., Pouton C.W., Partridge L.J., Monk P.N., et al. Tetraspanins in viral infections: a fundamental role in viral biology? J Virol. 2005;79:10839–10851. doi: 10.1128/JVI.79.17.10839-10851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Urciuoli E., Peruzzi B. Inhibiting Extracellular Vesicle Trafficking as Antiviral Approach to Corona Virus Disease 2019 Infection. Front Pharmacol. 2020;11:580505. doi: 10.3389/fphar.2020.580505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Patil M., Singh S., Henderson J., Krishnamurthy P. Mechanisms of COVID-19-induced cardiovascular disease: Is sepsis or exosome the missing link? J Cell Physiol. 2020 Oct 20 doi: 10.1002/jcp.30109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Catalano M., O'Driscoll L. Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J Extracell Vesicles. 2020;9:1703244. doi: 10.1080/20013078.2019.1703244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Quehenberger O., Dennis E.A. The human plasma lipidome. N Engl J Med. 2011;365:1812–1823. doi: 10.1056/NEJMra1104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Harkewicz R., Dennis E.A. Applications of mass spectrometry to lipids and membranes. Annu Rev Biochem. 2011;80:301–325. doi: 10.1146/annurev-biochem-060409-092612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang K., Han X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem Sci. 2016;41:954–969. doi: 10.1016/j.tibs.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gorden D.L., Myers D.S., Ivanova P.T., Fahy E., Maurya M.R., Gupta S., et al. Jr., D.W. Russell, S. Subramaniam, H.A. Brown, Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J Lipid Res. 2015;56:722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Loomba R., Quehenberger O., Armando A., Dennis E.A. Polyunsaturated fatty acid metabolites as novel lipidomic biomarkers for noninvasive diagnosis of nonalcoholic steatohepatitis. J Lipid Res. 2015;56:185–192. doi: 10.1194/jlr.P055640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu Z., Kastenmüller G., He Y., Belcredi P., Möller G., Prehn C., et al. Differences between human plasma and serum metabolite profiles. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu X., Hoene M., Wang X., Yin P., Häring H.U., Xu G., et al. Serum or plasma, what is the difference? Investigations to facilitate the sample material selection decision making process for metabolomics studies and beyond. Anal Chim Acta. 2018;1037:293–300. doi: 10.1016/j.aca.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 131.Ono Y., Kurano M., Ohkawa R., Yokota H., Igarashi K., Aoki J., et al. Sphingosine 1-phosphate release from platelets during clot formation: close correlation between platelet count and serum sphingosine 1-phosphate concentration. Lipids Health Dis. 2013;12:20. doi: 10.1186/1476-511X-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kugler K.G., Hackl W.O., Mueller L.A., Fiegl H., Graber A., Pfeiffer R.M. The Impact of Sample Storage Time on Estimates of Association in Biomarker Discovery Studies. J Clin Bioinforma. 2011;1:9. doi: 10.1186/2043-9113-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ulmer C.Z., Koelmel J.P., Jones C.M., Garrett T.J., Aristizabal-Henao J.J., Vesper H.W., et al. A Review of Efforts to Improve Lipid Stability during Sample Preparation and Standardization Efforts to Ensure Accuracy in the Reporting of Lipid Measurements. Lipids. 2021 Jan;56(1):3–16. doi: 10.1002/lipd.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jurowski K., Kochan K., Walczak J., Barańska M., Piekoszewski W., Buszewski B. Analytical Techniques in Lipidomics: State of the Art. Crit Rev Anal Chem. 2017;47:418–437. doi: 10.1080/10408347.2017.1310613. [DOI] [PubMed] [Google Scholar]
- 135.Vasconcelos B., Teixeira J.C., Dragone G., Teixeira J.A. Optimization of lipid extraction from the oleaginous yeasts Rhodotorula glutinis and Lipomyces kononenkoae. AMB Express. 2018;8:126. doi: 10.1186/s13568-018-0658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Burnum-Johnson K.E., Kyle J.E., Eisfeld A.J., Casey C.P., Stratton K.G., Gonzalez J.F., et al. MPLEx: a method for simultaneous pathogen inactivation and extraction of samples for multi-omics profiling. Analyst. 2017;142:442–448. doi: 10.1039/c6an02486f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell, 182. 2020;e15:59–72. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Song J.W., Lam S.M., Fan X., Cao W.J., Wang S.Y., Tian H., et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab, 32. 2020;e185:188–202. doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Thomas T., Stefanoni D., Reisz J.A., Nemkov T., Bertolone L., Francis R.O., et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hannun Y.A., Obeid L.M. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sanchez-Lopez E., Zhong Z., Stubelius A., Sweeney S.R., Booshehri L.M., Antonucci L., et al. Choline Uptake and Metabolism Modulate Macrophage IL-1β and IL-18 Production. Cell Metab. 2019;e1357:1350–1362. doi: 10.1016/j.cmet.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schwarz B., Sharma L., Roberts L., Peng X., Bermejo S., Leighton I., Casanovas-Massana A., Minasyan M., Farhadian S., Ko A.I., Yale IMPACT Team, Dela Cruz C.S., Bosio C.M. Severe SARS-CoV-2 infection in humans is defined by a shift in the serum lipidome resulting in dysregulation of eicosanoid immune mediators. J Immunol. 2021 Jan 15;206(2):329–334. doi: 10.4049/jimmunol.2001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dennis E.A., Norris P.C. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Norris P.C., Gosselin D., Reichart D., Glass C.K., Dennis E.A. Phospholipase A2 regulates eicosanoid class switching during inflammasome activation. Proc Natl Acad Sci U S A. 2014;111:12746–12751. doi: 10.1073/pnas.1404372111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Barberis E., Timo S., Amede E., Vanella V., Puricelli C., Cappellano G., et al. Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21228623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Overmyer K.A., Shishkova E., Miller I.J., Balnis J., Bernstein M.N., Peters-Clarke T.M., et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2020 Jan 20;12(1):23–40.e7. doi: 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bruzzone C., Bizkarguenaga M., Gil-Redondo R., Diercks T., Arana E., García de Vicuña A., et al. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience. 2020;23:101645. doi: 10.1016/j.isci.2020.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nie S., Zhao X., Zhao K., Zhang Z., Zhang Z., Zhang Z. Metabolic disturbances and inflammatory dysfunction predict severity of coronavirus disease 2019 (COVID-19): a retrospective study. medRxiv. 2020 2020.2003.2024.20042283. [Google Scholar]
- 149.Wu Z., Shon J.C., Liu K.H. Mass Spectrometry-based Lipidomics and Its Application to Biomedical Research. J Lifestyle Med. 2014;4:17–33. doi: 10.15280/jlm.2014.4.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Das U.N. Can Bioactive Lipids Inactivate Coronavirus (COVID-19)? Arch Med Res. 2020;51:282–286. doi: 10.1016/j.arcmed.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wu, et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. National Science Review. 2020;7:1157–1168. doi: 10.1093/nsr/nwaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Heaton N.S., Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19:368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fedson D.S., Opal S.M., Rordam O.M. Hiding in Plain Sight: an Approach to Treating Patients with Severe COVID-19 Infection. mBio. 2020;11 doi: 10.1128/mBio.00398-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Weill P., Plissonneau C., Legrand P., Rioux V., Thibault R. May omega-3 fatty acid dietary supplementation help reduce severe complications in Covid-19 patients? Biochimie. 2020;179:275–280. doi: 10.1016/j.biochi.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hathaway D., Pandav K., Patel M., Riva-Moscoso A., Singh B.M., Patel A., et al. Omega 3 Fatty Acids and COVID-19: A Comprehensive Review. Infect Chemother. 2020;52:478–495. doi: 10.3947/ic.2020.52.4.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stern M.P., Haffner S.M. Dyslipidemia in type II diabetes. Implications for therapeutic intervention. Diabetes Care. 1991;14:1144–1159. doi: 10.2337/diacare.14.12.1144. [DOI] [PubMed] [Google Scholar]
- 159.Jornayvaz F.R., Samuel V.T., Shulman G.I. The role of muscle insulin resistance in the pathogenesis of atherogenic dyslipidemia and nonalcoholic fatty liver disease associated with the metabolic syndrome. Annu Rev Nutr. 2010;30:273–290. doi: 10.1146/annurev.nutr.012809.104726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Patni N., Ahmad Z., Wilson D.P. In: Endotext, MDText.com, Inc. Copyright © 2000-2020. Feingold K.R., Anawalt B., Boyce A., Chrousos G., De Herder W.W., Dungan K., …Wilson D.P., editors. MDText.com, Inc.; South Dartmouth (MA): 2000. Genetics and Dyslipidemia. [Google Scholar]
- 161.Hegele R.A. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10:109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 162.Choi H.K., Seeger J.D. Lipid profiles among US elderly with untreated rheumatoid arthritis--the Third National Health and Nutrition Examination Survey. J Rheumatol. 2005;32:2311–2316. [PubMed] [Google Scholar]
- 163.Lazarevic M.B., Vitic J., Mladenovic V., Myones B.L., Skosey J.L., Swedler W.I. Dyslipoproteinemia in the course of active rheumatoid arthritis. Semin Arthritis Rheum. 1992;22:172–178. doi: 10.1016/0049-0172(92)90017-8. [DOI] [PubMed] [Google Scholar]
- 164.Barlage S., Gnewuch C., Liebisch G., Wolf Z., Audebert F.X., Glück T., et al. Changes in HDL-associated apolipoproteins relate to mortality in human sepsis and correlate to monocyte and platelet activation. Intensive Care Med. 2009;35:1877–1885. doi: 10.1007/s00134-009-1609-y. [DOI] [PubMed] [Google Scholar]
- 165.Gruber M., Christ-Crain M., Stolz D., Keller U., Müller C., Bingisser R., et al. Prognostic impact of plasma lipids in patients with lower respiratory tract infections - an observational study. Swiss Med Wkly. 2009;139:166–172. doi: 10.4414/smw.2009.12494. [DOI] [PubMed] [Google Scholar]
- 166.Cirstea M., Walley K.R., Russell J.A., Brunham L.R., Genga K.R., Boyd J.H. Decreased high-density lipoprotein cholesterol level is an early prognostic marker for organ dysfunction and death in patients with suspected sepsis. J Crit Care. 2017;38:289–294. doi: 10.1016/j.jcrc.2016.11.041. [DOI] [PubMed] [Google Scholar]
- 167.Wei X., Zeng W., Su J., Wan H., Yu X., Cao X., et al. Hypolipidemia is associated with the severity of COVID-19. J Clin Lipidol. 2020;14:297–304. doi: 10.1016/j.jacl.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Fan J., Wang H., Ye G., Cao X., Xu X., Tan W., et al. Letter to the Editor: Low-density lipoprotein is a potential predictor of poor prognosis in patients with coronavirus disease 2019. Metabolism. 2020;107:154243. doi: 10.1016/j.metabol.2020.154243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Trinder M., Walley K.R., Boyd J.H., Brunham L.R. Causal Inference for Genetically Determined Levels of High-Density Lipoprotein Cholesterol and Risk of Infectious Disease. Arterioscler Thromb Vasc Biol. 2020;40:267–278. doi: 10.1161/ATVBAHA.119.313381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Madsen C.M., Varbo A., Tybjærg-Hansen A., Frikke-Schmidt R., Nordestgaard B.G. U-shaped relationship of HDL and risk of infectious disease: two prospective population-based cohort studies. European heart journal. 2018;39:1181–1190. doi: 10.1093/eurheartj/ehx665. [DOI] [PubMed] [Google Scholar]
- 171.Birjmohun R.S., van Leuven S.I., Levels J.H., van 't Veer C., Kuivenhoven J.A., Meijers J.C., et al. High-density lipoprotein attenuates inflammation and coagulation response on endotoxin challenge in humans. Arterioscler Thromb Vasc Biol. 2007;27:1153–1158. doi: 10.1161/ATVBAHA.106.136325. [DOI] [PubMed] [Google Scholar]
- 172.van der Stoep M., Korporaal S.J., Van Eck M. High-density lipoprotein as a modulator of platelet and coagulation responses. Cardiovasc Res. 2014;103:362–371. doi: 10.1093/cvr/cvu137. [DOI] [PubMed] [Google Scholar]
- 173.Calkin A.C., Drew B.G., Ono A., Duffy S.J., Gordon M.V., Schoenwaelder S.M., et al. Reconstituted high-density lipoprotein attenuates platelet function in individuals with type 2 diabetes mellitus by promoting cholesterol efflux. Circulation. 2009;120:2095–2104. doi: 10.1161/CIRCULATIONAHA.109.870709. [DOI] [PubMed] [Google Scholar]
- 174.Shih D.M., Gu L., Xia Y.R., Navab M., Li W.F., Hama S., et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;394:284–287. doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- 175.Kopprasch S., Pietzsch J., Graessler J. The protective effects of HDL and its constituents against neutrophil respiratory burst activation by hypochlorite-oxidized LDL. Mol Cell Biochem. 2004;258:121–127. doi: 10.1023/b:mcbi.0000012842.19059.c5. [DOI] [PubMed] [Google Scholar]
- 176.Pajkrt D., Doran J.E., Koster F., Lerch P.G., Arnet B., van der Poll T., et al. Antiinflammatory effects of reconstituted high-density lipoprotein during human endotoxemia. J Exp Med. 1996;184:1601–1608. doi: 10.1084/jem.184.5.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.De Nardo D., Labzin L.I., Kono H., Seki R., Schmidt S.V., Beyer M., et al. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat Immunol. 2014;15:152–160. doi: 10.1038/ni.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Castiglione V., Chiriacò M., Emdin M., Taddei S., Vergaro G. Statin therapy in COVID-19 infection. Eur Heart J Cardiovasc Pharmacother. 2020;6:258–259. doi: 10.1093/ehjcvp/pvaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schmidt N.M., Wing P.A.C., McKeating J.A., Maini M.K. Cholesterol-modifying drugs in COVID-19. Oxf Open Immunol. 2020;1 doi: 10.1093/oxfimm/iqaa001. iqaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang X.J., Qin J.J., Cheng X., Shen L., Zhao Y.C., Yuan Y., et al. In-Hospital Use of Statins Is Associated with a Reduced Risk of Mortality among Individuals with COVID-19. Cell metabolism, 32. 2020;e174:176–187. doi: 10.1016/j.cmet.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mehra M.R., Desai S.S., Kuy S., Henry T.D., Patel A.N. Cardiovascular Disease, Drug Therapy, and Mortality in Covid-19. N Engl J Med. 2020;382:e102. doi: 10.1056/NEJMoa2007621. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 182.Moeller R., Zapatero-Belinchon F.J., Lasswitz L., Kirui J., Brogden G., Gunesch A.P., et al. Effect of statins on SARS-CoV-2 infection. medRxiv. 2020 2020.2007.2013.20152272. [Google Scholar]
- 183.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.Q., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Violetis O.A., Chasouraki A.M., Giannou A.M., Baraboutis I.G. COVID-19 Infection and Haematological Involvement: a Review of Epidemiology, Pathophysiology and Prognosis of Full Blood Count Findings. SN Compr Clin Med. 2020:1–5. doi: 10.1007/s42399-020-00380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liang W.H., Guan W.J., Li C.C., Li Y.M., Liang H.R., Zhao Y., et al. Clinical characteristics and outcomes of hospitalised patients with COVID-19 treated in Hubei (epicentre) and outside Hubei (non-epicentre): a nationwide analysis of China. Eur Respir J. 2020:55. doi: 10.1183/13993003.00562-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liao D., Zhou F., Luo L., Xu M., Wang H., Xia J., et al. Haematological characteristics and risk factors in the classification and prognosis evaluation of COVID-19: a retrospective cohort study. Lancet Haematol. 2020;7:e671–e678. doi: 10.1016/S2352-3026(20)30217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Arachchillage D.R.J., Laffan M. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18:1233–1234. doi: 10.1111/jth.14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang L., Yan X., Fan Q., Liu H., Liu X., Liu Z., et al. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J Thromb Haemost. 2020;18:1324–1329. doi: 10.1111/jth.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fogarty H., Townsend L., Ni Cheallaigh C., Bergin C., Martin-Loeches I., Browne P., et al. More on COVID-19 coagulopathy in Caucasian patients. Br J Haematol. 2020;189:1060–1061. doi: 10.1111/bjh.16791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Aggarwal M. J. Dass, M. Mahapatra, Hemostatic Abnormalities in COVID-19: An Update. Indian J Hematol Blood Transfus. 2020:1–11. doi: 10.1007/s12288-020-01328-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Al-Samkari H., Karp Leaf R.S., Dzik W.H., Carlson J.C.T., Fogerty A.E., Waheed A., et al. COVID-19 and coagulation: bleeding and thrombotic manifestations of SARS-CoV-2 infection. Blood. 2020;136:489–500. doi: 10.1182/blood.2020006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hardy M., Lecompte T., Douxfils J., Lessire S., Dogné J.M., Chatelain B., et al. Management of the thrombotic risk associated with COVID-19: guidance for the hemostasis laboratory. Thromb J. 2020;18:17. doi: 10.1186/s12959-020-00230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Thachil J., Tang N., Gando S., Falanga A., Cattaneo M., Levi M., et al. ISTH interim guidance on recognition and management of coagulopathy in COVID-19. J Thromb Haemost. 2020;18:1023–1026. doi: 10.1111/jth.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Seegers W.H., Cole E.R., Aoki N., Oliveira A. Fundamental function of ac-globulin and lipid in blood clotting. Nature. 1963;200:1014–1015. doi: 10.1038/2001014a0. [DOI] [PubMed] [Google Scholar]
- 196.Billimoria J.D., Irani V.J., Maclagan N.F. Blood lipid fractionation and blood clotting in ischaemic heart disease. J Atheroscler Res. 1965;5:102–111. doi: 10.1016/s0368-1319(65)80013-8. [DOI] [PubMed] [Google Scholar]
- 197.Deguchi H., Elias D.J., Griffin J.H. Minor Plasma Lipids Modulate Clotting Factor Activities and May Affect Thrombosis Risk. Res Pract Thromb Haemost. 2017;1:93–102. doi: 10.1002/rth2.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lauder S.N., Allen-Redpath K., Slatter D.A., Aldrovandi M., O'Connor A., Farewell D., et al. Networks of enzymatically oxidized membrane lipids support calcium-dependent coagulation factor binding to maintain hemostasis. Sci Signal. 2017:10. doi: 10.1126/scisignal.aan2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Moyer M.P., Tracy R.P., Tracy P.B., Van't Veer C., Sparks C.E., Mann K.G. Plasma lipoproteins support prothrombinase and other procoagulant enzymatic complexes. Arterioscler Thromb Vasc Biol. 1998;18:458–465. doi: 10.1161/01.atv.18.3.458. [DOI] [PubMed] [Google Scholar]
- 200.Rota S., McWilliam N.A., Baglin T.P., Byrne C.D. Atherogenic lipoproteins support assembly of the prothrombinase complex and thrombin generation: modulation by oxidation and vitamin E. Blood. 1998;91:508–515. [PubMed] [Google Scholar]
- 201.Hurley B.P., McCormick B.A. Multiple roles of phospholipase A2 during lung infection and inflammation. Infect Immun. 2008;76:2259–2272. doi: 10.1128/IAI.00059-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bhowmick R., Clark S., Bonventre J.V., Leong J.M., McCormick B.A., Cytosolic Phospholipase A. 2α Promotes Pulmonary Inflammation and Systemic Disease during Streptococcus pneumoniae Infection. Infection and immunity. 2017;85 doi: 10.1128/IAI.00280-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Vijay R., Hua X., Meyerholz D.K., Miki Y., Yamamoto K., Gelb M., et al. Critical role of phospholipase A2 group IID in age-related susceptibility to severe acute respiratory syndrome-CoV infection. J Exp Med. 2015;212:1851–1868. doi: 10.1084/jem.20150632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Xu S., Pei R., Guo M., Han Q., Lai J., Wang Y., et al. Cytosolic phospholipase A2 gamma is involved in hepatitis C virus replication and assembly. J Virol. 2012;86:13025–13037. doi: 10.1128/JVI.01785-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liebscher S., Ambrose R.L., Aktepe T.E., Mikulasova A., Prier J.E., Gillespie L.K., et al. Phospholipase A2 activity during the replication cycle of the flavivirus West Nile virus. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Murakami M., Taketomi Y., Miki Y., Sato H., Hirabayashi T., Yamamoto K. Recent progress in phospholipase A₂ research: from cells to animals to humans. Prog Lipid Res. 2011;50:152–192. doi: 10.1016/j.plipres.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 207.Tatulian S.A. Toward understanding interfacial activation of secretory phospholipase A2 (PLA2): membrane surface properties and membrane-induced structural changes in the enzyme contribute synergistically to PLA2 activation. Biophys J. 2001;80:789–800. doi: 10.1016/S0006-3495(01)76058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Niessen H.W., Krijnen P.A., Visser C.A., Meijer C.J., Erik Hack C. Type II secretory phospholipase A2 in cardiovascular disease: a mediator in atherosclerosis and ischemic damage to cardiomyocytes? Cardiovasc Res. 2003;60:68–77. doi: 10.1016/s0008-6363(03)00324-9. [DOI] [PubMed] [Google Scholar]
- 209.Chignard M., Le Couedic J.P., Tence M., Vargaftig B.B., Benveniste J. The role of platelet-activating factor in platelet aggregation. Nature. 1979;279:799–800. doi: 10.1038/279799a0. [DOI] [PubMed] [Google Scholar]
- 210.Uchiyama S., Yamazaki M., Maruyama S. Role of platelet-activating factor in aggregation of leukocytes and platelets in cerebral ischemia. Lipids. 1991;26:1247–1249. doi: 10.1007/BF02536541. [DOI] [PubMed] [Google Scholar]
- 211.Abdol Razak N., Elaskalani O., Metharom P. Pancreatic Cancer-Induced Neutrophil Extracellular Traps: A Potential Contributor to Cancer-Associated Thrombosis. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18030487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Elaskalani O., Abdol Razak N.B., Metharom P. Neutrophil extracellular traps induce aggregation of washed human platelets independently of extracellular DNA and histones. Cell communication and signaling : CCS. 2018;16:24. doi: 10.1186/s12964-018-0235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yost C.C., Cody M.J., Harris E.S., Thornton N.L., McInturff A.M., Martinez M.L., et al. Impaired neutrophil extracellular trap (NET) formation: a novel innate immune deficiency of human neonates. Blood. 2009;113:6419–6427. doi: 10.1182/blood-2008-07-171629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Fuchs T.A., Brill A., Duerschmied D., Schatzberg D., Monestier M., Myers D.D., Jr., et al. Extracellular DNA traps promote thrombosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15880–15885. doi: 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vargaftig B.B., Zirinis P. Platelet aggregation induced by arachidonic acid is accompanied by release of potential inflammatory mediators distinct from PGE2 and PGF2. Nat New Biol. 1973;244:114–116. doi: 10.1038/newbio244114a0. [DOI] [PubMed] [Google Scholar]
- 216.Rao G.H., White J.G. Role of arachidonic acid metabolism in human platelet activation and irreversible aggregation. Am J Hematol. 1985;19:339–347. doi: 10.1002/ajh.2830190404. [DOI] [PubMed] [Google Scholar]
- 217.Manne B.K., Denorme F., Middleton E.A., Portier I., Rowley J.W., Stubben C., et al. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136:1317–1329. doi: 10.1182/blood.2020007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hottz E.D., Azevedo-Quintanilha I.G., Palhinha L., Teixeira L., Barreto E.A., Pão C.R.R., et al. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136:1330–1341. doi: 10.1182/blood.2020007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Middleton E.A., He X.Y., Denorme F., Campbell R.A., Ng D., Salvatore S.P., et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Leppkes M., Knopf J., Naschberger E., Lindemann A., Singh J., Herrmann I., et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58:102925. doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mahase E. Covid-19: Experts advise cautious optimism for neutralising antibodies after early results. Bmj. 2020;371:m3937. doi: 10.1136/bmj.m3937. [DOI] [PubMed] [Google Scholar]
- 223.Hu X., Chen D., Wu L., He G., Ye W. Low Serum Cholesterol Level Among Patients with COVID-19 Infection in Wenzhou, China. SSRN Electronic Journal. 2020 https://ssrn.com/abstract=3544826 Available at SSRN: [Google Scholar]
- 224.Wang S.H., Hung H.C., Tsai C.C., Huang M.C., Ho K.Y., Wu Y.M., et al. Plasma polyunsaturated fatty acids and periodontal recovery in Taiwanese with periodontitis: a significant relationship. Arch Oral Biol. 2014;59:800–807. doi: 10.1016/j.archoralbio.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 225.Lee W., Ahn J.H., Park H.H., Kim H.N., Kim H., Yoo Y., et al. COVID-19-activated SREBP2 disturbs cholesterol biosynthesis and leads to cytokine storm. Signal Transduct Target Ther. 2020;5:186. doi: 10.1038/s41392-020-00292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]