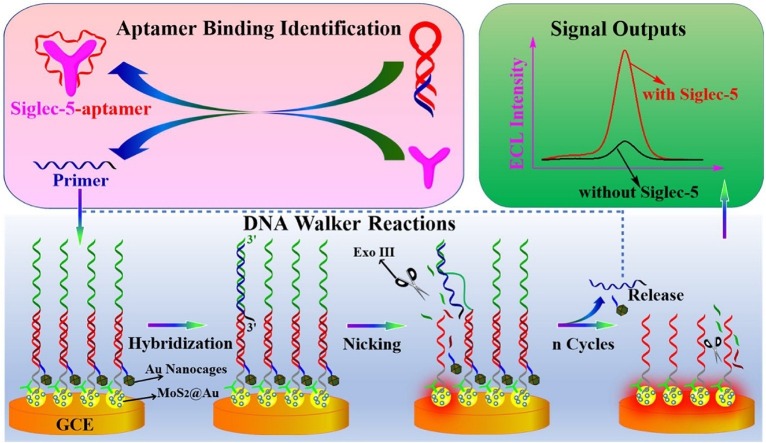
Graphical abstract
Keywords: Spike glycoprotein, MoS2 QDs, Siglec-5, Electrochemiluminescence, DNA walker, Aptasensor
Abstract
Lectins are highly specific binding proteins for glycoproteins which widely exist in living organisms, playing a vital role in exploring the biological evolution process, such as cellular proliferation, differentiation, carcinogenesis and apoptosis. Therefore, the content monitoring of lectin becomes particularly significant and urgent in the bioanalytical application. In this work, we fabricated an aptasensor, majorly capitalizing the eminent affinity between sialic acid-binding immunoglobulin (Ig)-like lectin 5 (Siglec-5) and nucleic acids aptamer (K19), with nontoxic MoS2@Au nanocomposites as electrochemiluminescence (ECL) emitters based on exonuclease III (Exo III)-powered DNA walker for the bioassays of Siglec-5. The DNA track was constructed on the emitters’ surface, providing a reliable platform for the DNA walker’s autonomous move. In the assay, the primer DNA in the DNA duplex was replaced by Siglec-5 due to the aptamer interactions and repeatedly released to participate in the movement of the DNA walker, further triggering cascade signal amplification. Finally, our aptasensor indicates significant potential for assays of Siglec-5 with a detection limit of 8.9 pM.
1. Introduction
The raging of 2019 novel coronavirus (2019-nCoV) with a rapid transmission spread in the worldwide area, had caused severe effects on travel, life and work of hundreds of millions to an unprecedented level in the past year [1,2]. However, no valid vaccines or therapeutic drugs were widely applied for the treatment of this highly contagious coronaviruses by now [3]. The three types of proteins encoded by the 2019-nCoV genome, including non-structural proteins, structural proteins, and accessory proteins are envisaged as potential targets for the development of human coronavirus antiviral agents [4]. The study of spike glycoprotein, an important structural protein that plays a crucial role in the interaction between viruses and cellular receptors, is beneficial for understanding the infection process of virus entry into cells and for obtaining new interventions [5]. Lectins are highly specific binding proteins for various viral surface glycoproteins, and the evaluation of lectin efficacy and the delivery route provides a potential short-term strategy for the development of coronavirus drugs at the molecular scale [6]. Sialic acid-binding immunoglobulin (Ig)-like lectins (Siglecs) as a member of the lectin family, the significance and quantitative analysis of which also need further evaluation and accurate implementation.
Siglecs are cell-surface receptors belonging to the immunoglobulin superfamily and each cell surface receptor consists of 2–17 extracellular Ig domains that contain the sialic acid-binding sites [[7], [8], [9]]. These type-I transmembrane proteins are known to us for recognizing negatively charged sialic acids linked to the terminal of non-reducing end of most glycans on scaffolds of proteoglycans, glycosphingolipids, and glycoproteins [10]. They are predominantly expressed in hematopoietic cells and participates in the modulation of physiological cellular processes by a host of glycan–protein interactions [10].
In terms of sequence homology, the Siglecs family can be split into two groups. The Siglecs known as CD33-related contain Siglec-3 (CD33), Siglec-5 (CD170), Siglec-11 and Siglec-14, which show high sequence homology (∼50−99 % sequence identity) in the Siglecs subfamily [11]. The other category includes Siglec-1, Siglec-2 (CD22), Siglec-4 and Siglec-15 with low sequence homology (25–50 % sequence identity) [12,13]. As one of the most common Siglecs, Siglec-5 has serval-linked conformations (α2−3, α2−6, α2−8) to recognize specific sialic acids and is selectively expressed in human monocytes, neutrophils, basophils and dendritic cells [14]. Interestingly, this type of Siglec consists of two intracellular “immunoreceptor tyrosine-based inhibitory motifs” (ITIM) that prevent kinase-mediated activation responses and are utilized as inhibitory receptors for leukocytes [10,15]. For these advantages, Siglec-5 has been regarded as the disease-specific biomarker and possible target protein of scores of immune-related diseases such as primary Sjogren's syndrome (pSS) and acute myeloid leukemia (AML) [16]. To improve the survival rates of these diseases, novel methods with high sensitivity and selectivity to discover Siglec-5 for the assays and targeted therapy are needed.
Aptamer-based technology as a distinct strategy in support of individualized medicine has exhibited extraordinary potential, partly bringing about the identification and validation of molecular signatures [[17], [18], [19]]. However, the discoveries of currently available aptamers of the biomarkers and valid analytical platforms are becoming imperative and valuable in bioanalysis and therapeutics. Systematic evolution of ligands by exponential enrichment (SELEX) as a molecular tool to screen nucleic acids from a library of random single-stranded nucleic acid sequences, with specific affinity to target proteins which is commensurate with individual antibodies, provides an effective means for biomarker investigations [[20], [21], [22]]. Based on SELEX selection technology, aptamer K19 with equilibrium dissociation constants (Kd) of 12.37 nM for NB4 AML cells shows high affinity for Siglec-5 which is regarded as the biomarker of AML and target protein of the K19 [23]. Additionally, the construct of the aptamer-based platform is envisioned to realize a quantitative approach for biomarker discovery and further leukemic therapy.
The aptamer-based analysis platforms are typically fabricated based on fluorescence (FL), electrochemistry (EC), and electrochemiluminescence (ECL), which are widely used in the areas of bioanalysis and clinical diagnosis [[24], [25], [26]]. ECL as a unique analytical technique is so charming for its characteristics of the low-cost device, simple operation and high sensitivity, avoiding the shortcoming of fluorescence using external background light and combining the advantage of electrochemistry with high sensitivity [27,28]. Besides, electrochemiluminescence-resonance energy transfer (ECL-RET) means the energy transfer between the adjacent emitters and the receptors, which was widely utilized in the design of biosensors and provided an efficient method for the detection of tumor markers [29,30]. In the last few years, ECL emitters ranged from traditional Ru(bpy)3 2+ to newly emerged materials such as Cd-based quantum dots (QDs), metal complexes, organic nanocrystals, and a series of high quantum efficiency materials [27,[31], [32], [33], [34]]. MoS2 QDs as classical graphene-analogous transition metal two-dimensional layered sulfide with a size-tunable bandgap, possess unique electronic characteristics, large specific area and heavy-metal-free properties, which have great applicable potential in multiphoton bioimaging, electrocatalytic activity as well as in ECL behavior [35,36]. Therefore, MoS2 QDs as biocompatible ECL emitters have the advantages of lower toxic metal ions and excellent characteristics compared to traditional Cd-based QDs [37]. However, limited by the intrinsic conductivity and the limited surface modification groups, the application of MoS2 QDs on the electrode surface for biosensors is limited [38]. In order to improve such conditions, compounding with gold nanoparticles provides an efficient approach which supplies a huge area for MoS2 QDs to attach, and facilitates the immobilization of nucleotide through the formation of Au-S bond, further promoting the construction of the sensor [39].
In the past decades, nucleic acid nanotechnology has supplied enormous possibilities for the operation of biochemical engineering [40]. In particular, dynamic nucleic acids nanotechnology is applied in artificial intelligence areas including smart biosensors, intelligent computing, nimble disease diagnosis and drug delivery [[41], [42], [43]]. One type of dynamic aspects is the DNA walker, and the foremost property of which is the performance in autonomous and highly directional mechanical movement in linear, planar and three-dimensional tracks through dynamic interactions [44]. DNA walker has been widely used in the fabrication of biosensors for the assay of target DNA, miRNA and bifunctional proteins. For example, Chai et al. utilized a bipedal DNA walker-based electrochemical strategy for the assays of specific sequences [45]. Peng et al. realized the analysis of miRNA using a bi-directional DNA machine and enzyme-free strategy [46]. Jiang et al. fabricated DNA nanomachine-based ECL biosensor for the detection of Mucin 1 [47]. The application of a DNA walker in the ultra-micro bioanalysis supplies reliable methods to ameliorate the bioanalytical platforms and improve diagnostic efficiency.
Herein, an electrochemiluminescence aptasensor with high sensitivity, rapid operation and simple device, using MoS2@Au nanocomposites as efficient emitter and Exo III-powered DNA walker for isothermal signal amplification, was fabricated for sensitive detection of Siglec-5. This strategy was based on ECL-RET, of which MoS2@Au nanocomposites as ECL emitter and the gold nanocage served as a quencher, for the reason that the absorbance of Au nanocages was overlapped with the ECL spectra of MoS2@Au nanocomposites, which significantly improved the quenching efficiency of the quenchers. Compared to gold nanoparticles or nanoshells, the absorbance spectra of Au nanocages had more overlapping parts with the ECL spectra of MoS2@Au nanocomposites [48,49]. In the presence of target Siglec-5, the multifunctional double-stranded DNA (dsDNA) probe formed by aptamer and primer DNA was disintegrated, and the primer DNA was released to further hybridize with the Au nanocages-labeled stator DNA (S2) probe already partially hybridized with staple DNA (S1). With the help of unidirectional digestion of Exo III, movement of single foot DNA walker on the MoS2@Au nanocomposites was conducted. The DNA nanomachine supplied an efficient way to transduce and amplify recognition signals. Most importantly, due to the excellent ECL emission of MoS2@Au nanocomposites, high-performance quenching platform and Exo III-powered signal amplification strategy using stochastic DNA walker, we had successfully fabricated an aptasensor for detecting Siglec-5 with high sensitivity and excellent selectivity.
2. Materials and method
2.1. Reagents and chemicals
Regents and chemicals, unless otherwise stated, were got from Sangon Biotech Co. Ltd. (Shanghai, China) and the use followed the instructions. Molybdenum disulfide powders and gold colloid solution with a diameter of 50 nm were obtained from XFNANO Co. Ltd. (Nanjing, China). 20-nm-sized Au nanocages were purchased from Nanoeast Biotech Co. Ltd. (Nanjing, China). Siglec-5 was from Sino Biological Inc. (Beijing, China). Related oligonucleotides used in our experiments came from Genscript Biotechnology Co., Ltd (Nanjing, China) and all sequences were summarized in Table S1. The water used in this strategy was purified with resistance 5 MΩ using a Milli-Q purification system (Branstead).
2.2. Instruments and measurements
Scanning electron microscopy (SEM) images and energy-dispersive X-ray (EDX) were from JSM-7800 F (Japan). ECL tests were carried out using ECL-6B equipped with a photomultiplier tube (PMT) with a voltage of 525 V offered from the State Key Laboratory of Analytical Chemistry for Life Science, Nanjing University. Besides, the potential sweep range for the ECL assay was 0–1.3 V and the assay condition was in 0.1 M phosphate buffered saline (PBS) containing 20 mM TEA. ECL measurements and a series of electrochemical measurements containing impedance spectra, cyclic voltammetry (CV) used a three-electrode system including a working electrode (glassy carbon electrode), a counter electrode (platinum), and a reference electrode (Ag/AgCl). The CV curve and impedance spectra (100 kHz ∼ 0.1 Hz) were obtained in PBS (0.1 M) with [Fe(CN)6]3−/4− (5 mM). The ECL emission spectra were got in 0.1 M PBS in the addition of 20 mM TEA and the concentration of Siglec-5 was 2 nM. UV–vis data were collected from Spectra Max M5e (Molecular Devices Co. Ltd).
2.3. Preparation of MoS2@Au
Firstly, molybdenum disulfide powders were pretreated to get a well-dispersed MoS2 QDs solution according to already published literatures [50,51]. In short, 300 mg molybdenum disulfide was firstly dissolved in 30 mL N, N-dimethylformamide (DMF). Subsequently, continuous sonication was performed for 6 h, and the obtained mixture was refluxed at 140 ℃ for 8 h. After that, the sonication of the resultant solution was conducted for 2.5 h and followed by centrifugation at 5000 rpm to get a pale yellow supernatant. Next, the supernatant was evaporated under vacuum to remove redundant DMF, and then deionized water was added to get MoS2 QDs solution.
Then, 2 mL MoS2 QDs aqueous solution was added to 4 mL 50-nm-sized gold colloid solution and slowly heated to 50 ℃ under stirring for 8 h. Besides, centrifugation at 10,000 rpm was performed to obtain MoS2@Au nanocomposites. Finally, the MoS2@Au nanocomposites were stored in the dark at 4 ℃ for the following utilization.
2.4. Preparation of stator DNA-Au nanocages composites
The synthesis of DNA-Au nanocages composites was referring to previous literature with slight modifications [52]. In short, 1 mL newly purchased 20-nm-sized nanocages solution (2 nM) with metallic luster and polyhedral structure (the SEM characterization was in Fig. S1) containing 200 nM mPEG-SH with a molecular weight of 5 kDa was mixed with 10 μL (1 wt%) Tween 20. After shaking the mixture for 3 min, 10 μL (100 μM) deprotected thiol-terminated stator DNA was put together and NaCl aqueous solution was added with a final concentration of 500 mM. Left at room temperature for 1 h, the mixture was centrifugated at 10,000 rpm. After being repeatedly washed three times, the precipitate was redispersed in 1 mL PBS (0.1 M) and put in the refrigeration at 4 ℃ for further modification.
2.5. Fabrication of ECL biosensor
Initially, a glassy carbon electrode with a diameter of 4 mm was constantly polished using 0.05 μm alumina, followed by sonication in deionized water to get a mirror electrode. After that, 10 μL prepared MoS2@Au solution was dropped onto the spotless GCE and dried for 1 h to form MoS2@Au modified electrode (abbreviation as MoS2@Au/GCE) by electrostatic adsorption. Next, the modified electrode was dipped into 50 μL PBS containing 1 μM thiol-terminated staple DNA probe which was pre-activated by 0.05 mM TCEP for 1 h, and incubated overnight to fabricate staple DNA probe modified MoS2@Au electrode (staple DNA/MoS2@Au/GCE) through the formation of Au-S covalent bond. Followed by immersed in 1.0 mM MCH for 45 min at room temperature to passivate the free position of the electrode, the biosensor architecture (MCH/staple DNA/MoS2@Au/GCE) had been obtained. And then, the modified electrode was dipped into the solution of stator DNA-Au nanocages (50 μL) to incubate for 3 h. Besides, the PBS was used to flush away unbound composites after each modification. Therefore, the aptasensor (stator DNA-Au nanocages/MCH/staple DNA/MoS2@Au/GCE) was finally successfully fabricated.
2.6. Detection of Siglec-5
Initially, the aptamer DNA (0.1 mM) and primer DNA (0.1 mM) mixture solution (pH 7.1) containing 10 mM Tris-HCl, 50 mM Mg2+ was prepared. Afterward, the solution was heated to 95 ℃ to untangle each DNA strand and slowly cooled to room temperature within 15 min to form the DNA duplex. The initial conformations of the aptamer DNA and hybridized aptamer DNA with primer DNA were showed in Fig. S2. Before use, the DNA duplex solution was annealed and stored in a -20 ℃ refrigerator. Subsequently, the DNA duplexes were diluted to 10 μM by binding buffer (140 mM NaCl, 12 mM NaHPO4, 3.0 mm KCl, 3 mM KH2PO4, pH 7.4) solution which contained series concentrations of Siglec-5. Subsequently, the mixture (50 μL) was gradually diluted into 500 μL by Tris-HCl buffer (10 mM, pH 7.1) to prepare assay solution in two steps, for each dilution 30 min incubation time at room temperature was required. Then, the aptasensor was incubated with 50 μL assay solution for 45 min. Then, Exo III with a final concentration of 10 U mL−1 was added to the assay solution to incubate for 50 min. For every modification of the electrode, PBS (100 mM, pH 7.1) was used to rinse out the non-specifically substances. As for endogenous assessment in cell nuclear extract samples, the process of cell culture and the preparation of nuclear extract were referred to our previous work [53]. ECL signals were obtained through PBS containing 20 mM triethylamine with the voltage scanning range from 0–1.3 V.
3. Results and discussion
3.1. The working principle
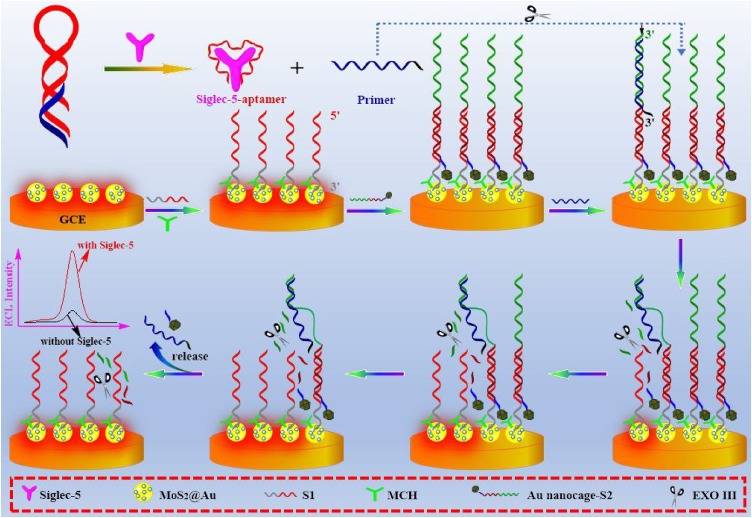
The fabrication of aptasensor and ECL-RET mechanism of sensing process of Siglec-5 was shown in Scheme 1 . Initially, ECL emitter MoS2@Au nanocomposites dripped onto GCE, staple DNA (S1) modified on top of MoS2@Au nanocomposites, and Au nanocage-labeled stator DNA were used together to construct a platform for DNA walker tracks. The MoS2@Au nanocomposites were presented as emitters indicating a “signal on” state, while stator DNA carrying a quencher (Au nanocage) at the 5′ termini suppressed emitting light and identified as a “signal off” state. Meanwhile, we designed primer DNA strands containing partial complementary sequences of K19 aptamer, which could hybridize with K19 aptamer to some extent. With the selective interaction between Siglec-5 and K19 aptamer, the pre-synthesized DNA duplexes of primer DNA and K19 aptamer split up, resulting in the release of primer DNA, here served as a single-foot DNA walker which was complementary to the sequences of stator DNA on the surface of architectural ECL platform. After binding to the Au nanocage-S2, the primer DNA moved autonomously after stator DNA sequences gradually digested by Exo III using exonuclease activity. The Exo III exclusively cleaved nucleotides from blunt or concave 3′ termini of dsDNA but expressed no activity on ssDNA or convex 3′ termini (more than four bases) of dsDNA [54]. Therefore, the sequence of stator DNA paired with staple DNA was merely digested, yielding a 6-nt oligo carrying Au nanocage, but the consumption of mononucleotides of primer DNA was blocked by extended sequences (black extensions) in 3′ termini. The released primer DNA then hybridized with another strand of stator DNA through toehold-mediated branch migration and triggered a new digestion activity, which tended to form automatically cyclical motions reaching cognate clarity as isothermal amplification reactions. As the walking process proceeded, the quencher was gradually removed from the stator DNA and the “signal on” state reappeared.
Scheme 1.
Schematics representation of electrochemiluminescence-based aptasensor for the assays of Siglec-5 using Exo III-driven DNA walkers.
3.2. Characterization of MoS2@Au nanocomposites
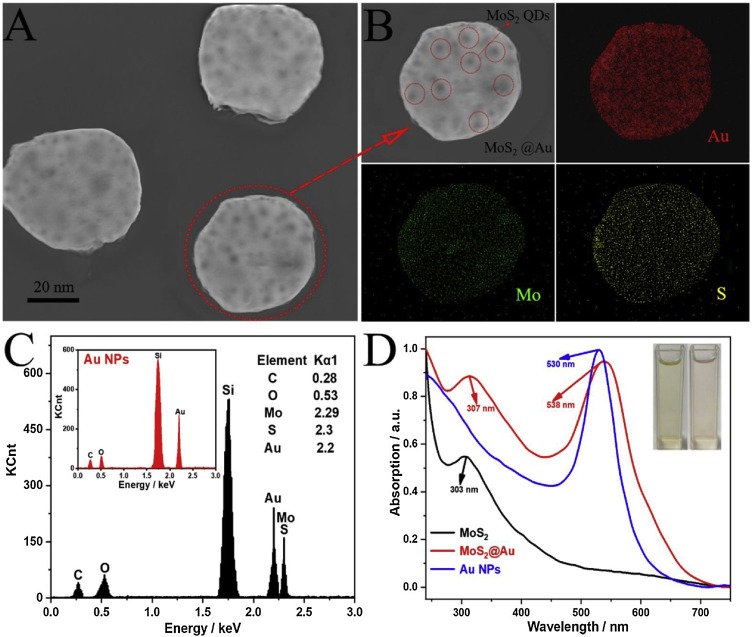
TEM was used to characterize the morphology and size of MoS2 as depicted in Fig. S3, illustrating that the as-prepared MoS2 had a uniform lateral diameter of around 6 nm, which was suitable for further modification with Au nanoparticles. Scanning electron microscope and energy-dispersive X-ray elemental mapping were used to characterize the formation of MoS2@Au nanocomposites. Fig. 1 A was typically morphological characterization of MoS2@Au nanocomposites, which showed that the 50-nm-sized gold nanoparticles (bright region) with clear MoS2 QDs (dark substance) on the surface, and the regular spherical gold nanoparticles were slightly distorted for the penetration of MoS2 QDs into the gold nanoparticles during the modification process. Further insight on the formation of MoS2@Au nanocomposites was verified by EDX element mapping. As shown in Fig. 1B, Mo and S elements which represented the main elements of MoS2 QDs were evenly distributed on the gold nanoparticles. The SEM image and the mapping pictures confirmed the successful decoration of MoS2 QDs on the gold nanoparticles. To further describe its elemental compositions, energy dispersive spectroscopy (EDS) was used to confirm the result as shown in Fig. 1C. The MoS2@Au nanocomposites contained obvious characteristic peaks of S (2.3 keV), Mo (2.29 keV) in addition to Au element (2.2 keV) peak of Au nanoparticles (inset of Fig. 1C).
Fig. 1.
SEM characterization of (A) MoS2@Au nanocomposites and its EDX elemental mapping (B). EDS spectra of MoS2@Au nanocomposites (C) and Au nanoparticles (inset of Fig. 1C). UV–vis absorption spectra (D) of MoS2 QDs (black line), MoS2@Au nanocomposites (red line) and 50-nm-sized Au nanoparticles (blue line). The insets of Fig. 1D were photographs of MoS2 QDs (left) and MoS2@Au nanoparticles (right) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Moreover, UV–vis spectra were also shown to characterize the optical properties of MoS2@Au nanocomposites. The peaks at 303 and 530 nm were earmarked for the excitonic traits of MoS2 QDs and Au nanoparticles. Besides, the MoS2@Au nanocomposites possessed two absorbance peaks respectively at 307 nm and 538 nm, with 4 nm and 8 nm red-shifted in comparison to the excitonic features of MoS2 QDs and Au nanoparticles, which was attributed to the energy transfer of photons.
3.3. PAGE characterization and hydrolysis investigation for the assay
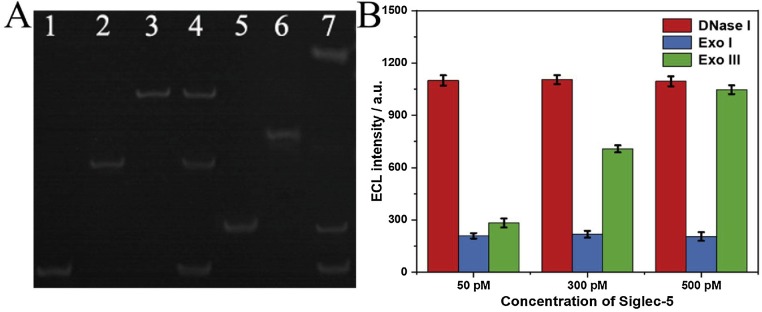
For the verification of the assay process of Siglec-5, we employed polyacrylamide gel electrophoresis (PAGE) to characterize the bioanalytical process, and the PAGE image was depicted in Fig. 2 A. The ssDNA contained primer DNA (lane 1), aptamer DNA (lane 2) and staple DNA (lane 5) presented single bright stripes. The dsDNA included primer DNA with aptamer DNA (lane 3) and staple DNA with stator DNA (lane 6), where slower speeds indicated the partial hybridizations of both were successfully achieved. Additionally, the single bright bands denoted their purities were relatively high. After preparations of the dsDNA, we verified the high affinity between aptamer DNA with Siglec-5 (lane 4), and the process of Exo III-powered DNA walker (lane 7) respectively. Three strips in lane 4 corresponded to the primer, aptamer and its duplex DNA probe, respectively. Therefore, we could infer that Siglec-5 competed with primer DNA for the hybridization with K19 aptamer, and thus some primer DNA probe was released. The band representing aptamer DNA indicated that the binding of aptamer and Siglec-5 was undermined during the PAGE process. The three bands in lane 7 contained two strips with the same migration speeds as the primer and the staple DNA as well as a newly emerged strip, revealing that with Exo III the sequences of stator DNA were digested and primer DNA probe released with high concentration. Also, the dark stripe with the lowest migration speed should be speculated as the residual hybridization strands of staple, stator and primer DNA probes during the walker process.
Fig. 2.
PAGE characterization (A) for the assay procedure. Lane 1, 2 and 5 were the primer, aptamer and staple DNA probe with the concentration of 1 μM, respectively. Lane 3 was 1 μM dsDNA (primer + aptamer). Lane 4 showed dsDNA +1.3 μM Siglec-5. Lane 6 was 1 μM hybridized strand (staple DNA + stator DNA). Lane 7 revealed the reaction of the hybridized strand (1 μM) + primer DNA (1 μM) + Exo III (20 U mL−1). Hydrolysis investigation (B) of enzyme system (DNase I, Exo I, and Exo III) for DNA walker of this aptasensor in detecting different concentrations (50, 300 and 500 pM) of Siglec-5.
For further systematically investigating the hydrolysis effects of different enzyme systems, for instance, the effects of DNase I, Exo I, and Exo III on nucleotides in this DNA walker were also explored (Fig. 2B). DNase I as an endonuclease could digest ssDNA and dsDNA randomly. Exo I and Exo III exclusively digested ssDNA and dsDNA in 3′ to 5′ direction, severally. The hydrolysis reactions were monitored according to the ECL response of the three enzymatic reactions under significant changes of Siglec-5 concentrations, showing that the ECL intensity enhanced along with the separation of ECL emitter and quencher (Au nanocages). As for DNase I, hydrolysis degree was the highest in the three types of nucleases, while did not change with the target protein concentration. Exo I showed a generally similar result that hydrolysis reaction had nothing to do with the target protein and the separation of emitter and quencher was blocked due to its cleaving characteristics. The hydrolysis efficiency of Exo III was significantly influenced by the target concentration due to its high participatory for DNA walker, which indicated the importance of Exo III for the assay of Siglec-5 of this aptasensor.
3.4. ECL and electrochemical activity of emitters
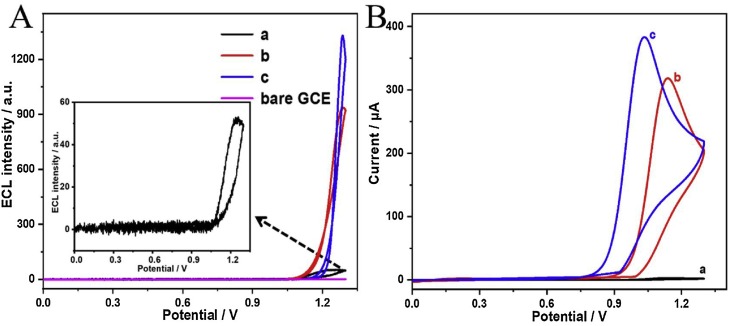
ECL and electrochemical behaviors of MoS2 QDs and MoS2 QDs modified Au nanoparticles were investigated to certificate the emitting mechanism of the emitter. As shown in Fig. 3 A, a feeble ECL signal of MoS2 QDs with approximately 50 a.u. was observed (curve a) in PBS. Meanwhile, its corresponding CV testing as depicted in Fig. 3B, indicated that the maximum current was slight (curve a). However, when TEA was added to the MoS2-based ECL system, the ECL intensity and maximum current were extremely raised, with ECL intensity increased to 910 a.u. (curve b in Fig. 3A), about 18.7 folds compared to the ECL system without co-reactant TEA, and maximum current dramatically enhanced to 325 μA (curve b in Fig. 3B). Besides, in the working buffer containing TEA, MoS2@Au nanocomposites exhibited more obvious enhancement compared to MoS2 QDs in ECL and electrochemical aspects, the ECL intensity of which increased to about 1311 a.u. (curve c in Fig. 3A), and the maximum current was heightened to 381 μA (curve c in Fig. 3B). We inferred that the Au nanoparticles here acted as the catalyst to improve the efficiency of charge transfer and reinforced ECL emission. One possible ECL mechanism of MoS2 QDs could be illustrated as below. In brief, TEA lost an electron to be oxidized to TEA• with strong reduction and participate to the ECL process of reducing MoS2 QDs for the generation of the electronically excited state of MoS2*. At last, MoS2* released energy (light) and back to ground state.
| TEA – e– → TEA•+ → TEA• + H+ | (1) |
| (2) |
| MoS2* → MoS2 + hν | (3) |
Fig. 3.
ECL-potential curve (A) and CV curve (B) in PBS of (a) MoS2 QDs, (b) MoS2 QDs with 20 mM TEA, (c) MoS2@Au nanoparticles with 20 mM TEA.
3.5. Stepwise modifications and feasibility of aptasensor
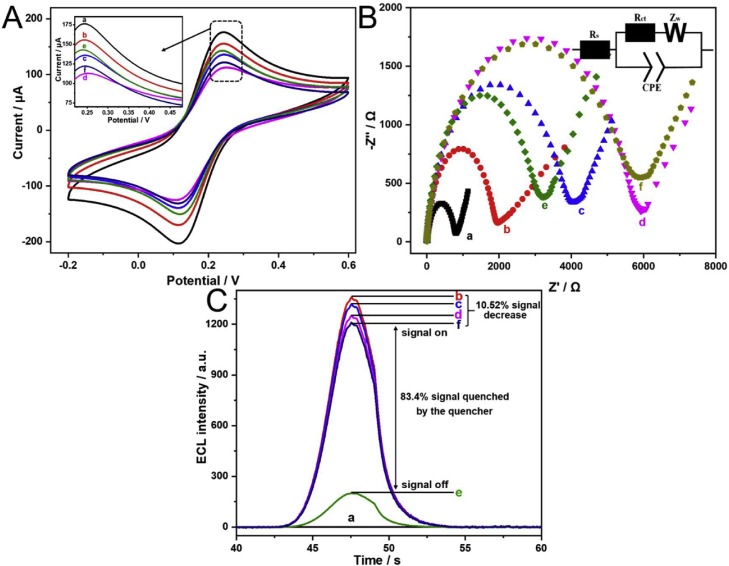
We applied CV, Nyquist plot of impedance spectra and ECL emission for the characterization of the step-by-step modifications of aptasensor on the electrode. As depicted in Fig. 4 A, the CV curve of the bare electrode with symmetrical redox peaks (curve a), indicating it was spotless. With MoS2@Au nanocomposites (curve b), staple DNA (c), MCH (curve d) successively modified onto the electrode, the redox peak decreased obviously, demonstrating that the impedance of electron transfer was increased with the adsorption of non-specific substances. Besides, we had characterized the size change of AuNPs before and after ssDNA linkage by DLS as shown in Fig. S4. The size change indicated that the modification of thiol-terminated staple DNA on the Au nanoparticle was successfully achieved. However, with the immobilization of stator DNA-Au nanocages (curve e), an increase of redox peak was noticed due to the superb conductivity of Au nanocages. As the DNA walker proceed (curve f), the redox current decreased due to the release of Au nanocages from the modified electrode. Besides, corresponding changes of electrode resistance also reflected the actual assembly process of the aptasensor. In Fig. 4B, the revised Randles circuit was used to simulate the electrochemical activities during the electrode modifications. The impedance contains real Z’ and imaginary Z’’, which respectively come from the resistance and capacitance of the working system. The solution resistance (Rs) of the system reflects the actual impedance between the GCE and the reference electrode. The resistance of charge transfer (Rct) accounts for interfacial resistance associated with the dielectric and insulation properties of modified substances on the working electrode, which can be incarnated by the high-frequency region of the Nyquist diagram. Constant phase element (CPE) represents a non-linear capacitor that illustrates the unevenness of the surface layer. Warburg (W) embodied the molecular motion of the electrode surface film caused by ion conduction, which explains the diffusion-limited electrochemical process. In this stepwise modification, Rct was driven up from a to d, which denoted the impedance between the working electrode, and the reference electrode was gradually increasing. With the modification of stator DNA-Au nanocages (curve e), Rct decreased slightly for that nanocages enhanced the efficiency of electron transfer and reduced the interfacial resistance. After the process of DNA walker (curve f), the Rct raised again for the reason that the nanocages were set free.
Fig. 4.
Gradual assembly of ECL aptasensor for Siglec-5 detection. (A) CV, (B) Nyquist plot (Z’ vs -Z’’) of impedance spectra and (C) ECL response for the modifications of the electrode. (a) GCE alone, (b) MoS2@Au nanocomposites/GCE, (c) staple DNA/MoS2@Au nanocomposites/GCE, (d) MCH/staple DNA/MoS2@Au nanocomposites/GCE, (e) stator DNA-Au nanocages/MCH/staple DNA/MoS2@Au nanocomposites/GCE, (f) stator DNA-Au nanocages/MCH/staple DNA/MoS2@Au nanocomposites/GCE after DNA walker process.
Besides, ECL response (Fig. 4C) was used to illustrate the feasibility of the aptasensor further. When MoS2@Au nanocomposites (curve b) adsorbed on the electrode, ECL signal with high intensity was recorded. As staple DNA (c), MCH (curve d) gradually assembled on the electrode, the ECL signals were slightly decreased. Stator DNA-Au nanocages (curve e) with high quenching efficiency quenched the ECL signal to a certain extent. However, the DNA walker (curve f) recovered the signal, which was owing to the overlap of absorbance of Stator DNA-Au nanocages and ECL spectra of MoS2@Au nanocomposites seen in Fig. S5. These electrochemical and ECL activities of the electrode meant the successful fabrication of the aptasensor and the feasibility of the DNA walker.
3.6. Optimization and assay performance of the aptasensor
To get the best sensing conditions, univariate analyses were performed on the concentration of primer DNA, the reaction time of Au nanocages-S2 with staple DNA, and the digestion time of Exo III to optimize the experimental parameters. As depicted in Fig. S6A, as the concentration of primer DNA increased, the ECL signal also increased between 1 μM–100 μM. Then, the ECL intensity no longer increased with the increasing concentration of primer DNA. So, 100 μM was selected as the optimal concentration for the fabrication of the ECL biosensor. Meanwhile, the reaction time between Au nanocages-S2 and staple probe was also optimized. Fig. S6B showed that the lowest ECL signal was achieved at 90 min. Therefore, we chose an incubation time of more than 90 min. Fig. S6C showed the ECL signal increased as the digestion time of Exo III increased from 20 min to 40 min, and then tended to a constant value after 50 min. Based on this, we determined the incubation time of Exo III. Furthermore, we observed that the ECL intensity increased with increasing digestion time at lower Siglec-5 concentration and did not show an inflection point, indicating that ECL intensity was affected by Siglec-5 concentration, thus validating the feasibility of our method.
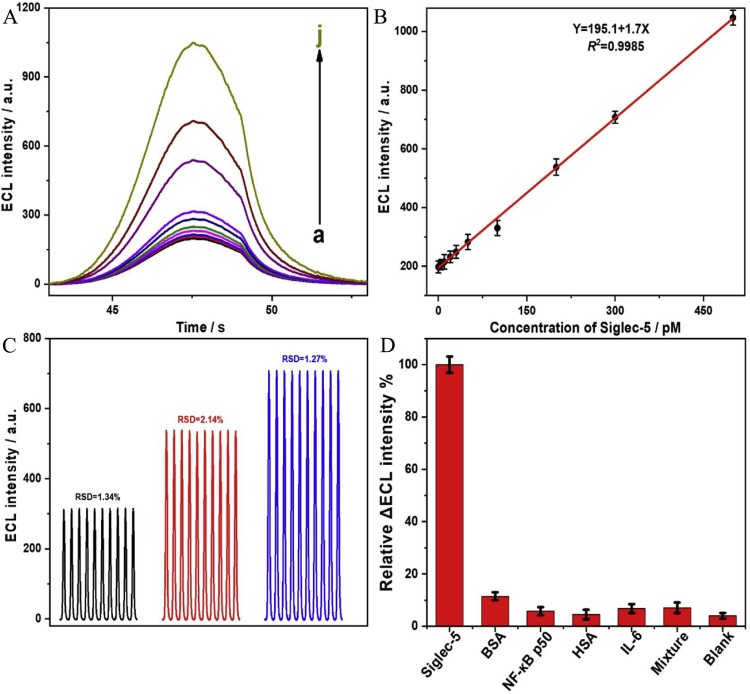
In order to verify the assay behaviors of the aptasensor, various concentrations of Siglec-5 were detected by the ECL devices. Fig. 5 A shows that the ECL intensity of the “signal on” state had enhanced with the increasing concentrations of target proteins from 10 pM to 500 pM. Besides, as depicted in Fig. 5B, the clear linear relationship between 10 pM to 500 pM was made, of which the ECL intensity (Y) and the Siglec-5 concentrations (X) conformed to the linear equation Y = 195.1 + 1.7X (R 2 = 0.9985) and limit of detection (LOD) was calculated as 8.9 pM (3σ), which was more sensitive compared with the method (with LOD of 0.27 nM) proposed by Lin et al. utilizing luminescent iridium(III) with G-quadruplex DNA, indicating that the aptasensor was with good sensitivity and suitable for Siglec-5 analysis [16]. For further exploring the detection performance, the recovery rate was also shown in Table 1 . The recovery rates of data 1 presented a narrow range (96.1 %–103.68 %) when detecting spiked Siglec-5 in Tris-HCl solution, expressing that this method possessed good feasibility. The endogenous Siglec-5 was measured to test the practical value of this method. Specifically, Siglec-5 in 10-fold diluted cell nuclear extracts was detected for the assessment of endogenous influences of other components in the actual detecting environment. As shown in Table 1 (data 2), the recovery was in the range of 95.1 %–104.13 %, indicating that the effect of endogenous components in the nuclear extract could be negligible. The intensity of ECL in the presence of 10-fold diluted serum with different concentrations of Siglec-5 was compared with the standard curve as depicted in Fig. S7. Moreover, both curves showed the same trend, indicating that serum interference did not affect the actual detection of Siglec-5.
Fig. 5.
(A) ECL emission spectra of the aptasensor in the presence of various concentrations of Siglec-5 (0, 5, 10, 20, 30, 50, 100, 200, 300 and 500 pM, from a to j). (B) Relationship between the ECL intensity and concentrations of Siglec-5 from 0 to 300 pM. The assays were carried with five duplicated measurements. (C) Reproducibility of the aptasensor with three different concentrations (100, 200, and 300 pM) under 10 continuous measurements. (D) Relatively increased ECL intensity of the aptasensor with 100 pM Siglec-5 or 10-fold amount of other nonspecific proteins.
Table 1.
Analytical performance of the aptasensor for detecting spiked Siglec-5 in Tris-HCl buffer (1) and 10-fold diluted nuclear extracts (2).
| Siglec-5 |
|||||
|---|---|---|---|---|---|
| Sample | Spiked (pM) | Detected 1 (pM) | Recovery 1 (%) | Detected 2 (pM) | Recovery 2 (%) |
| 1 | 0 | 0 | N/A | 53.47 | N/A |
| 2 | 10 | 9.81 | 98.10 | 62.98 | 95.10 |
| 3 | 30 | 28.84 | 96.10 | 82.44 | 96.57 |
| 4 | 50 | 51.84 | 103.7 | 107.2 | 101. 4 |
| 5 | 100 | 102.6 | 102.6 | 157.6 | 104.1 |
3.7. Calculation of Siglec-5 concentration in single A549 cell
The estimation of the content of Siglec-5 in A549 cell () was according to the following equation:
Where represents the molar amount of Siglec-5 in each A549 cell, represents the average volume of each A549 cell (2.31 pL), the value of which refers to the supplementary information of the literature [55]. represents the detected concentration in the diluted cell nuclear extracts without extra addition of Siglec-5 (53.47 pM), presents the detected volume of the testing sample (2 mL), and represents the total number of the A549 cells (5 × 107). Therefore, the calculated average concentration of Siglec-5 in single A594 cell was 0.926 nM.
3.8. Reproducibility and selectivity of this method
In addition, to verify the application prospects of the proposed aptasensor, we explored the reproducibility of the ECL signal by performing 10 consecutive scans of the cyclic potential at three target concentrations (100, 200 and 300 pM) as shown in Fig. 5C. The lower relative standard deviations (RSD) denoted that its repeatability was within the acceptable range (<5%). Since selectivity was of importance for aptasensor, further researches on it were conducted. Extra nonspecific proteins, for example, bovine serum albumin (BSA), NF-κB p50, human serum albumin (HSA), interleukin-6 (IL-6) and their mixture were selected to compare with Siglec-5 in the sensing assays (Fig. 5D). Besides, we selected a variety of other types of lectins, including galectin-3, Siglec-8, and snailagglutinin for investigating the resistance of the biosensor system to these interfering lectin proteins to highlight the specificity of the system for the specifically Siglec-5 (Fig. S8). These results revealed an unignorable disparity that the ECL signal enhancement for target protein was higher than that for 10-fold excess of other proteins, indicating the aptasensor with excellent selectivity for Siglec-5.
4. Conclusions
Related lectins with binding properties to spike glycoproteins are potentially valuable for the developing antiviral agents against 2019-nCoV. Thus, rapid identification and quantitative assays of lectins are necessary for the early discovery and analysis of coronaviruses. In our work, an ECL aptasensor using MoS2@Au nanocomposites as emitters and Exo III-propelled DNA walker as a signal amplification strategy was constructed for quantitative bioanalysis of Siglec-5. Employing the high binding affinity between Siglec-5 with K19 aptamer, we ingeniously transformed the assays of target protein into the assays of primer DNA, which further participated in the motion of the DNA walker for signal amplification. Besides, we constructed a notable DNA walking track on MoS2@Au nanocomposites and caused a “signal off” state, which with the DNA walker process gone ahead, the state reversed to “signal on”. Finally, our ECL system exhibited strong anti-interference ability with a superior selectivity for ultrasensitive assay of Siglec-5, performing a detection limit as low as 8.9 pM and providing a feasible analysis method for Siglec-5 in a single A549 cell, which possessed huge potential in evaluating the role of lectins at a cellular level and developing anti-coronavirus agents in 2019 novel coronaviruses epidemic.
CRediT authorship contribution statement
Zhenqiang Fan: Data curation, Formal analysis, Writing - review & editing. Bo Yao: Software, Validation. Yuedi Ding: Formal analysis. Minhao Xie: Validation. Jianfeng Zhao: Funding acquisition, Supervision. Kai Zhang: Conceptualization, Supervision. Wei Huang: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21705061), and China-Sweden Joint Mobility Project (No. 51811530018), Ministry of Education and Synergetic Innovation Center for Organic Electronics and Information Displays for financial support, the Jiangsu Provincial Key Medical Discipline (Laboratory) (ZDXKA2016017), and the Innovation Capacity Development Plan of Jiangsu Province (BM2018023).
Biographies
Zhenqiang Fan works on electrochemical based biosensors in Jiangsu Institute of Nuclear Medicine.
Yao Bo is a master student at Nanjing Tech University, China. His main research interests are biosensors. Yuedi Ding: Testing.
Dr. Yuedi Ding’s field and main subject of current research interests are the application of biosensors in single-cell assay.
Prof. Dr. Minhao Xie received his pH.D. degree at Jiangnan University, China. He currently works as a professor at Jiangsu Institute of Nuclear Medicine. His research interest is biosensor.
Dr. Jianfeng Zhao is an Associate Professor in the Institute of Advanced Materials of Nanjing Tech University. He received a pH.D. degree from Nanjing University of Posts and Telecommunications in 2011. His main research interests are micro/nano materials and chemical sensors.
Dr. Kai Zhang currently works as an associate professor at Jiangsu Institute of Nuclear Medicine. His current research interests include nanostructured functional materials and their application for chemical and biosensors
Prof. Dr. Wei Huang is in the Institute of Flexible Electronics of Northwestern Polytechnical University. He received his PhD degrees from Peking University in 1992. He is a member of the Chinese Academy of Sciences and also a foreign member of the Russian Academy of Sciences, as well ASA member of the Asian Pacific Academy of Materials. His current research interests include flexible electronics, organic electronics, bioelectronics, nanomaterials and nanotechnology.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2021.129592.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.York A. Novel coronavirus takes flight from bats? Nat. Rev. Microbiol. 2020;18:191. doi: 10.1038/s41579-020-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng X., Xu X., Li Y., Cheng L., Zhou X., Ren B. Transmission routes of 2019-nCoV and controls in dental practice. Int. J. Oral. Sci. 2020;12:9. doi: 10.1038/s41368-020-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G., Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug. Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 6.Chiodelli P., Rezzola S., Urbinati C., Federici Signori F., Monti E., Ronca R., Presta M., Rusnati M. Contribution of vascular endothelial growth factor receptor-2 sialylation to the process of angiogenesis. Oncogene. 2017;36:6531–6541. doi: 10.1038/onc.2017.243. [DOI] [PubMed] [Google Scholar]
- 7.Chang L., Chen Y.-J., Fan C.-Y., Tang C.-J., Chen Y.-H., Low P.-Y., Ventura A., Lin C.-C., Chen Y.-J., Angata T. Identification of Siglec ligands using a proximity labeling method. J. Proteome Res. 2017;16:3929–3941. doi: 10.1021/acs.jproteome.7b00625. [DOI] [PubMed] [Google Scholar]
- 8.Nycholat C.M., Duan S., Knuplez E., Worth C., Elich M., Yao A., O’Sullivan J., McBride R., Wei Y., Fernandes S.M., Zhu Z., Schnaar R.L., Bochner B.S., Paulson J.C. A sulfonamide sialoside analogue for targeting Siglec-8 and -F on immune cells. J. Am. Chem. Soc. 2019;141:14032–14037. doi: 10.1021/jacs.9b05769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blixt O., Han S., Liao L., Zeng Y., Hoffmann J., Futakawa S., Paulson J.C. Sialoside analogue arrays for rapid identification of high affinity Siglec ligands. J. Am. Chem. Soc. 2008;130:6680–6681. doi: 10.1021/ja801052g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cecioni S., Imberty A., Vidal S. Glycomimetics versus multivalent glycoconjugates for the design of high affinity lectin ligands. Chem. Rev. 2015;115:525–561. doi: 10.1021/cr500303t. [DOI] [PubMed] [Google Scholar]
- 11.Von Gunten S., Bochner B.S. Basic and clinical immunology of Siglecs. Ann. N. Y. Acad. Sci. 2008;1143:61–82. doi: 10.1196/annals.1443.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angata T., Varki A. Chemical diversity in the sialic acids and related α-keto acids: an evolutionary perspective. Chem. Rev. 2002;102:439–470. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 13.O’Reilly M.K., Collins B.E., Han S., Liao L., Rillahan C., Kitov P.I., Bundle D.R., Paulson J.C. Bifunctional CD22 ligands use multimeric immunoglobulins as protein scaffolds in assembly of immune complexes on B cells. J. Am. Chem. Soc. 2008;130:7736–7745. doi: 10.1021/ja802008q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pepin M., Mezouar S., Pegon J., Muczynski V., Adam F., Bianchini E.P., Bazaa A., Proulle V., Rupin A., Paysant J., Panicot-Dubois L., Christophe O.D., Dubois C., Lenting P.J., Denis C.V. Soluble Siglec-5 associates to PSGL-1 and displays anti-inflammatory activity. Sci. Rep. 2016;6 doi: 10.1038/srep37953. 37953-37953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tourdot B.E., Brenner M.K., Keough K.C., Holyst T., Newman P.J., Newman D.K. Immunoreceptor tyrosine-based inhibitory motif (ITIM)-mediated inhibitory signaling is regulated by sequential phosphorylation mediated by distinct nonreceptor tyrosine kinases: a case study involving PECAM-1. Biochemistry. 2013;52:2597–2608. doi: 10.1021/bi301461t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin S., Lu L., Kang T.-S., Mergny J.-L., Leung C.-H., Ma D.-L. Interaction of an Iridium(III) complex with G-Quadruplex DNA and its application in luminescent switch-on detection of Siglec-5. Anal. Chem. 2016;88:10290–10295. doi: 10.1021/acs.analchem.6b03128. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L., Wan S., Jiang Y., Wang Y., Fu T., Liu Q., Cao Z., Qiu L., Tan W. Molecular elucidation of disease biomarkers at the interface of chemistry and biology. J. Am. Chem. Soc. 2017;139:2532–2540. doi: 10.1021/jacs.6b10646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lao Y.-H., Phua K.K.L., Leong K.W. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano. 2015;9:2235–2254. doi: 10.1021/nn507494p. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., Feng Y., Duan S., Su L., Zhang J., He F. Mycobacterium tuberculosis strain H37Rv electrochemical sensor mediated by aptamer and AuNPs–DNA. ACS Sensors. 2019;4:849–855. doi: 10.1021/acssensors.8b01230. [DOI] [PubMed] [Google Scholar]
- 20.Berezovski M., Musheev M., Drabovich A., Krylov S.N. Non-SELEX selection of aptamers. J. Am. Chem. Soc. 2006;128:1410–1411. doi: 10.1021/ja056943j. [DOI] [PubMed] [Google Scholar]
- 21.Mosing R.K., Mendonsa S.D., Bowser M.T. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal. Chem. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 22.Tan Y., Guo Q., Xie Q., Wang K., Yuan B., Zhou Y., Liu J., Huang J., He X., Yang X., He C., Zhao X. Single-walled carbon nanotubes (SWCNTs)-assisted cell-systematic evolution of ligands by exponential enrichment (Cell-SELEX) for improving screening efficiency. Anal. Chem. 2014;86:9466–9472. doi: 10.1021/ac502166b. [DOI] [PubMed] [Google Scholar]
- 23.Yang M., Jiang G., Li W., Qiu K., Zhang M., Carter C.M., Al-Quran S.Z., Li Y. Developing aptamer probes for acute myelogenous leukemia detection and surface protein biomarker discovery. J. Hematol. Oncol. 2014;7:5. doi: 10.1186/1756-8722-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W., Hu G.-B., Yao L.-Y., Yang Y., Liang W.-B., Yuan R., Xiao D.-R. Matrix coordination-induced electrochemiluminescence enhancement of tetraphenylethylene-based hafnium metal–organic framework: An electrochemiluminescence chromophore for ultrasensitive electrochemiluminescence sensor construction. Anal. Chem. 2020;92:3380–3387. doi: 10.1021/acs.analchem.9b05444. [DOI] [PubMed] [Google Scholar]
- 25.Qi W., Lai J., Gao W., Li S., Hanif S., Xu G. Wireless electrochemiluminescence with disposable minidevice. Anal. Chem. 2014;86:8927–8931. doi: 10.1021/ac501833a. [DOI] [PubMed] [Google Scholar]
- 26.Han Z., Shu J., Liang X., Cui H. Label-free ratiometric electrochemiluminescence aptasensor based on nanographene oxide wrapped titanium dioxide nanoparticles with potential-resolved electrochemiluminescence. Anal. Chem. 2019;91:12260–12267. doi: 10.1021/acs.analchem.9b02318. [DOI] [PubMed] [Google Scholar]
- 27.Sun J., Zhou F., Hu H., Li N., Xia M., Wang L., Wang X., Wang G. Photocontrolled thermosensitive electrochemiluminescence hydrogel for isocarbophos detection. Anal. Chem. 2020;92:6136–6143. doi: 10.1021/acs.analchem.0c00719. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Jiang Z.W., Xiao S.Y., Huang C.Z., Li Y.F. Terbium(III) organic gels: Novel antenna effect-induced enhanced electrochemiluminescence emitters. Anal. Chem. 2018;90:12191–12197. doi: 10.1021/acs.analchem.8b03383. [DOI] [PubMed] [Google Scholar]
- 29.Wu M.-S., Shi H.-W., He L.-J., Xu J.-J., Chen H.-Y. Microchip device with 64-site electrode array for multiplexed immunoassay of cell surface antigens based on electrochemiluminescence resonance energy transfer. Anal. Chem. 2012;84:4207–4213. doi: 10.1021/ac300551e. [DOI] [PubMed] [Google Scholar]
- 30.Huo X.-L., Zhang N., Yang H., Xu J.-J., Chen H.-Y. Electrochemiluminescence resonance energy transfer system for dual-wavelength ratiometric mirna detection. Anal. Chem. 2018;90:13723–13728. doi: 10.1021/acs.analchem.8b04141. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Y., Huang Y., Lei J., Zhang L., Ju H. Design and biosensing of Mg2+-dependent dnazyme-triggered ratiometric electrochemiluminescence. Anal. Chem. 2014;86:5158–5163. doi: 10.1021/ac500965p. [DOI] [PubMed] [Google Scholar]
- 32.Wu Y., Han Z., Wei L., Sun H., Wang T., Chen J., Zhang R., Lu X. Depolymerization-induced electrochemiluminescence of insoluble porphyrin in aqueous phase. Anal. Chem. 2020;92:5464–5472. doi: 10.1021/acs.analchem.0c00208. [DOI] [PubMed] [Google Scholar]
- 33.Dini D. Electrochemiluminescence from organic emitters. Chem. Mater. 2005;17:1933–1945. [Google Scholar]
- 34.Zhang C., Wang Z., Liu Y., Yang J., Zhang X., Li Y., Pan L., Ke Y., Yan H. Nicking-assisted reactant recycle to implement entropy-driven DNA circuit. J. Am. Chem. Soc. 2019;141:17189–17197. doi: 10.1021/jacs.9b07521. [DOI] [PubMed] [Google Scholar]
- 35.Dong H., Tang S., Hao Y., Yu H., Dai W., Zhao G., Cao Y., Lu H., Zhang X., Ju H. Fluorescent MoS2 quantum dots: ultrasonic preparation, up-conversion and down-conversion bioimaging, and photodynamic therapy. ACS Appl. Mater. Interfaces. 2016;8:3107–3114. doi: 10.1021/acsami.5b10459. [DOI] [PubMed] [Google Scholar]
- 36.Lu S.-M., Li Y.-J., Zhang J.-F., Wang Y., Ying Y.-L., Long Y.-T. Monitoring hydrogen evolution reaction catalyzed by MoS2 quantum dots on a single nanoparticle electrode. Anal. Chem. 2019;91:10361–10365. doi: 10.1021/acs.analchem.9b02364. [DOI] [PubMed] [Google Scholar]
- 37.Zhao M., Chen A.-Y., Huang D., Chai Y.-Q., Zhuo Y., Yuan R. MoS2 quantum dots as new electrochemiluminescence emitters for ultrasensitive bioanalysis of lipopolysaccharide. Anal. Chem. 2017;89:8335–8342. doi: 10.1021/acs.analchem.7b01558. [DOI] [PubMed] [Google Scholar]
- 38.Gopalakrishnan D., Damien D., Shaijumon M.M. MoS2 quantum dot-interspersed exfoliated MoS2 nanosheets. ACS Nano. 2014;8:5297–5303. doi: 10.1021/nn501479e. [DOI] [PubMed] [Google Scholar]
- 39.Ye J., Zhu L., Yan M., Zhu Q., Lu Q., Huang J., Cui H., Yang X. Dual-wavelength ratiometric electrochemiluminescence immunosensor for cardiac Troponin I detection. Anal. Chem. 2019;91:1524–1531. doi: 10.1021/acs.analchem.8b04640. [DOI] [PubMed] [Google Scholar]
- 40.Panigaj M., Johnson M.B., Ke W., McMillan J., Goncharova E.A., Chandler M., Afonin K.A. Aptamers as modular components of therapeutic nucleic acid nanotechnology. ACS Nano. 2019;13:12301–12321. doi: 10.1021/acsnano.9b06522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X., Liu Y., Yung B., Xiong Y., Chen X. Nanotechnology-enhanced no-wash biosensors for in vitro diagnostics of cancer. ACS Nano. 2017;11:5238–5292. doi: 10.1021/acsnano.7b02618. [DOI] [PubMed] [Google Scholar]
- 42.Song X., Reif J. Nucleic acid databases and molecular-scale computing. ACS Nano. 2019;13:6256–6268. doi: 10.1021/acsnano.9b02562. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y., Chen F., Li Q., Wang L., Fan C. Isothermal amplification of nucleic acids. Chem. Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 44.Qu X., Zhu D., Yao G., Su S., Chao J., Liu H., Zuo X., Wang L., Shi J., Wang L. An exonuclease III-powered, on-particle stochastic DNA walker. Angew. Chem. Int. Ed. 2017;56:1855–1858. doi: 10.1002/anie.201611777. [DOI] [PubMed] [Google Scholar]
- 45.Chai H., Miao P. Bipedal DNA walker based electrochemical genosensing strategy. Anal. Chem. 2019;91:4953–4957. doi: 10.1021/acs.analchem.9b01118. [DOI] [PubMed] [Google Scholar]
- 46.Peng L., Zhang P., Chai Y., Yuan R. Bi-directional DNA walking machine and its application in an enzyme-free electrochemiluminescence biosensor for sensitive detection of micrornas. Anal. Chem. 2017;89:5036–5042. doi: 10.1021/acs.analchem.7b00418. [DOI] [PubMed] [Google Scholar]
- 47.Jiang X., Wang H., Wang H., Zhuo Y., Yuan R., Chai Y. Electrochemiluminescence biosensor based on 3-D DNA nanomachine signal probe powered by protein-aptamer binding complex for ultrasensitive mucin 1 detection. Anal. Chem. 2017;89:4280–4286. doi: 10.1021/acs.analchem.7b00347. [DOI] [PubMed] [Google Scholar]
- 48.Zhang K., Fan Z., Li H., Zhao J., Xie M. Determination of the concentration of transcription factor by using exonuclease III-aided amplification and gold nanoparticle mediated fluorescence intensity: A new method for gene transcription related enzyme detection. Anal. Chim. Acta. 2020;1104:132–139. doi: 10.1016/j.aca.2019.12.076. [DOI] [PubMed] [Google Scholar]
- 49.Li H., Ma X., Dong J., Qian W. Development of methodology based on the formation process of gold nanoshells for detecting hydrogen peroxide scavenging activity. Anal. Chem. 2009;81:8916–8922. doi: 10.1021/ac901534b. [DOI] [PubMed] [Google Scholar]
- 50.Dai W., Dong H., Fugetsu B., Cao Y., Lu H., Ma X., Zhang X. Tunable fabrication of molybdenum disulfide quantum dots for intracellular microRNA detection and multiphoton bioimaging. Small. 2015;11:4158–4164. doi: 10.1002/smll.201500208. [DOI] [PubMed] [Google Scholar]
- 51.Xu S., Li D., Wu P. One-pot, facile, and versatile synthesis of monolayer MoS2/ws2 quantum dots as bioimaging probes and efficient electrocatalysts for hydrogen evolution reaction. Adv. Funct. Mater. 2015;25:1127–1136. [Google Scholar]
- 52.Lu H.-J., Pan J.-B., Wang Y.-Z., Ji S.-Y., Zhao W., Luo X.-L., Xu J.-J., Chen H.-Y. Electrochemiluminescence energy resonance transfer system between RuSi nanoparticles and hollow Au nanocages for nucleic acid detection. Anal. Chem. 2018;90:10434–10441. doi: 10.1021/acs.analchem.8b02347. [DOI] [PubMed] [Google Scholar]
- 53.Fan Z., Lin Z., Wang Z., Wang J., Xie M., Zhao J., Zhang K., Huang W. Dual-wavelength electrochemiluminescence ratiometric biosensor for NF-κB p50 detection with dimethylthiodiaminoterephthalate fluorophore and self-assembled DNA tetrahedron nanostructures probe. ACS Appl. Mater. Interfaces. 2020;12:11409–11418. doi: 10.1021/acsami.0c01243. [DOI] [PubMed] [Google Scholar]
- 54.Shevelev I.V., Hübscher U. The 3′–5′ exonucleases. Nat. Rev. Mol. Cell Bio. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 55.Zhang K., Yang X.-J., Zhao W., Xu M.-C., Xu J.-J., Chen H.-Y. Regulation and imaging of gene expression via an RNA interference antagonistic biomimetic probe. Chem. Sci. 2017;8:4973–4977. doi: 10.1039/c7sc00909g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.