Summary:
Mutations in leucine-rich repeat kinase 2 (LRRK2) are the most frequent cause of familial Parkinson’s disease. LRRK2 is a multi-domain protein containing a kinase and GTPase. Using correlative light and electron microscopy, in situ cryo-electron tomography and subtomogram analysis, we reveal a 14-Å structure of LRRK2 bearing a pathogenic mutation that oligomerizes as a right-handed double-helix around microtubules, which are left-handed. Using integrative modeling, we determine the architecture of LRRK2, showing that the GTPase and kinase are in close proximity, with the GTPase closer to the microtubule surface, while the kinase is exposed to the cytoplasm. We identify two oligomerization interfaces mediated by non-catalytic domains. Mutation of one of these abolishes LRRK2 microtubule-association. Our work demonstrates the power of cryo-electron tomography to generate models of previously unsolved structures, in their cellular environment.
Graphical Abstract

In brief
Cryo-electron tomography combined with integrative modeling yields the structure of LRKK2 in a cellular context and shows how filaments of the protein embrace microtubules.
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disease. About 10% of patients show a clear family history, and mutations in the leucine-rich repeat kinase 2 (LRRK2) are the most frequent cause of familial PD (Roosen and Cookson, 2016; Thomas and Beal, 2007). LRRK2 is a 286 kDa protein with an N-terminal half containing repeating motifs and a C-terminal half composed of kinase and GTPase domains surrounded by protein-protein interaction domains (Kalia and Lang, 2015). Most of the pathogenic mutations are found in the two catalytic domains, but all result in hyperactivation of the kinase in cells (Steger et al., 2016). Hyperactivation of LRRK2’s kinase is also reported in non-familial PD cases (Di Maio et al., 2018), suggesting that suppression of kinase activity might be beneficial for a wide range of PD patients. However, how LRRK2 kinase activity is regulated on a molecular and cellular level and how abnormal LRRK2 kinase activity leads to PD remains unclear. LRRK2 is involved in several cellular processes, including neurite outgrowth and synaptic morphogenesis, membrane trafficking, autophagy, and protein synthesis (Kalia and Lang, 2015), and it has been shown to phosphorylate a subgroup of Rab GTPases, modulating their membrane localization and function (Steger et al., 2016).
In cells, wild-type LRRK2, as observed by immunofluorescence microscopy, is predominantly cytosolic (Eguchi et al., 2018). Exogenously expressed wild type LRRK2, visualized via epitope or fluorescent protein tags, is also mostly cytosolic while some of the common PD mutations can also be seen forming filamentous structures surrounding microtubules. Filament formation is enhanced in N1437H, R1441G/C, Y1699C and I2020T LRRK2 mutants and has been observed in multiple cell types (Blanca Ramirez et al., 2017; Kett et al., 2012; Schmidt et al., 2019). While it remains to be determined how enhanced microtubule association by the LRRK2 mutants may contribute to their pathogenicity, pharmacological inhibition of LRRK2’s kinase causes rapid and reversible recruitment of both wild-type and other mutant LRRK2 proteins to microtubules (Blanca Ramirez et al., 2017; Deng et al., 2011; Schmidt et al., 2019).
A major barrier to understanding the mechanism of LRRK2 is the lack of structural information on the protein, which to date only includes structures of domains, either single or in tandem. Many of these are from a distantly related bacterial homolog (Deyaert et al., 2019; Gotthardt et al., 2008; Guaitoli et al., 2016; Schmidt et al., 2019), two are single domains from the human protein (Deng et al., 2008; Zhang et al., 2019), and two are electron microscopy (EM) structures, whose resolution was insufficient to reveal domains of LRRK2 (Guaitoli et al., 2016; Sejwal et al., 2017).
Given the paucity of structural information about LRRK2 and its importance as a target for PD treatment, we set out to determine the structure of human LRRK2 using in situ cryo-electron tomography (Cryo-ET). Achieving high-resolution in situ using cryo-ET is greatly helped by helical structures, because they combine symmetry with all of the views required to generate a fully sampled structure. Given the helical nature of microtubules, we reasoned that microtubule-associated LRRK2 would be an ideal sample. Here we report a 14 Å structure of human LRRK2(I2020T) bound to microtubules as ordered filaments, determining the overall domain structure of LRRK2 in cells and the architecture of the microtubule-associated filaments.
Results and Discussion
Visualization of microtubule-associated LRRK2 by cryo-correlative light and electron microscopy
To determine the structure of microtubule-associated LRRK2, we used cryo-correlative light and electron microscopy (cryo-CLEM) to image LRRK2 filaments in situ (Figure 1). To increase the population of LRRK2-decorated microtubules we could image, we used several approaches. First, we chose to express LRRK2 bearing the pathogenic I2020T mutation, because it has previously been shown to spontaneously undergo filament formation and decorate cellular microtubules by immuno-electron microscopy (Kett et al., 2012). Second, we stabilized microtubules with Taxol, which was previously shown to increase the percentage of cells showing LRRK2 filamentous structures, but was not necessary for their formation (Kett et al., 2012). We began by growing human HEK293T cells expressing amino-terminally YFP-tagged LRRK2 with the I2020T mutation, hereafter LRRK2(I2020T), on EM grids. After vitrification, cells containing filamentous LRRK2(I2020T) were identified by cryo-fluorescence microscopy. Cryo-focused ion beam (cryo-FIB) milling of these cells was then used to prepare lamellae for cryo-ET (Figure 1A–C). The LRRK2(I2020T)-decorated microtubules were identified by CLEM as regions where microtubule bundles in transmission electron microscopy (TEM) images overlapped with the YFP signal of LRRK2(I2020T) in the fluorescence images (Figure 1D–F). Subsequently, cryo-ET was performed in these regions (Figure 1G–H).
Figure 1. Cryo-correlative light and electron microscopy of mutant LRRK2-decorated microtubule bundles.
(A) Fluorescence micrograph (FM) of a grid with HEK-293T cells expressing YFP-tagged LRRK2(I2020T), shown in green.
(B) Overlay of the fluorescence signal (green) corresponding to LRRK2(I2020T) and cryo-SEM image (greyscale) of the area marked in (A). A cell containing a large LRRK2(I2020T) aggregate, which have been previously reported (Blanca Ramirez et al., 2017; Kett et al., 2012), can be seen on the left while a cell containing LRRK2(I2020T) filaments is shown in the square.
(C) Overlay of the FM image and an SEM image after FIB-milling of the area shown in B.
(D) Overlay of the FM image (green) and cryo-TEM image of the area shown in B. (E and F) Enlarged TEM image of the area marked in D, with (E) and without (F) the overlaid FM image. The asterisks indicate mitochondria, which appear fragmented.
(G) Slice of a tomogram showing a LRRK2-decorated microtubule bundle.
(H) Annotated undecorated (green) and decorated (green with LRRK2 in gray) microtubules and membranes (purple) in the tomogram shown in H.
Scale bars: 500 µm (A), 20 µm (B), 5 µm (C), 2 µm (D), 1 µm (E, F), 100 nm (G).
We first characterized the microtubules in our tomograms. Microtubules are formed from heterodimers of alpha- and beta-tubulin and polymerize into polar left-handed helical arrays, which in human cells typically consist of 13 protofilaments (Chaaban and Brouhard, 2017). We annotated each microtubule individually by computationally determining its coordinates (Figure S1A). Within bundles, most of the decorated microtubules were aligned parallel to each other (Figure 2B–E), with a center-to-center distance of 70 ± 18 nm (Figure 2E). This regular distribution of angles and distances suggests that pathogenic LRRK2 mediates microtubule bundling. Most microtubules were either undecorated or fully decorated, and these two populations co-existed within the same cellular regions (Figure 2A). Protofilament number and polarity of each microtubule was determined by subtomogram analysis (Figure S1B). Microtubules within a given bundle largely had the same polarity (Figure 2B,C). Unexpectedly, we observed populations of non-conventional 11- and 12-protofilament microtubules (5% and 56%, respectively), in addition to the 13-protofilament microtubules (39%) that are typically found in human cells (Figure 2D) (Chaaban and Brouhard, 2017). To determine if bundling and microtubule protofilament number was specific to LRRK2(I2020T) expression, we examined microtubule populations in cells treated with Taxol, but without LRRK2(I2020T) expression. Under these conditions we did not observe any decorated microtubules or bundles and found that microtubules consisted primarily of 13-protofilament (92%) microtubules (Figure 2F–I).
Figure 2. LRRK2(I2020T)-decorated microtubules have various protofilament numbers and are found in bundles.
(A) Slice of a tomogram of HEK cells expressing LRRK2(I2020T) showing undecorated (white arrow) and LRRK2-decorated (green arrows) microtubules. (B) Slice of a tomogram showing bundles of LRRK2(I2020T)-decorated microtubules.
(C) Segmented microtubules from the tomogram in B. Individual microtubules found in bundles often have consistent polarity. The yellow arrows at the end of the microtubules point towards their plus end.
(D) Distribution of protofilament number in microtubules in filamentous LRRK2-enriched areas of LRRK2(I2020T)-expressing cells (green bars) and of decorated and undecorated microtubules within each class (black and white bars, respectively).
(E) Relative angle and distance between microtubule segments reveal parallel bundles with a regular center-to-center distance between microtubules of 70 ± 18 nm (peak ~67 nm).
(F) Slice of a tomogram showing microtubules in HEK-293T cells treated with 5 µM Taxol.
(G) Annotated microtubules (in green) of the tomogram shown in F.
(H) Distribution of protofilament number in microtubules in cryo-ET data of Taxol-treated cells from two biological duplicates. No microtubules are decorated in the absence of LRRK2(I2020T).
(I) Relative angle and distance between microtubules from Taxol-treated cellular tomograms. The average distance is 47 ± 32 nm (peak ~31 nm), showing no bundling.
Scale bars: 100 nm (A), 200 nm (B,F).
LRRK2 forms a right-handed double helix around microtubules
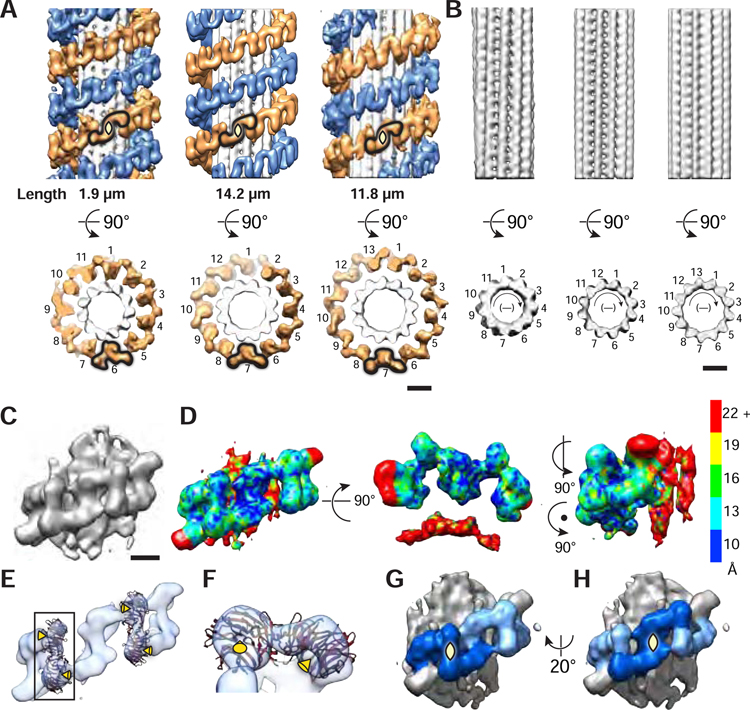
After classification of microtubules according to protofilament numbers, we used subtomogram analysis to resolve the helical arrangement of LRRK2(I2020T) bound to microtubules (Figure 3A and Figure S1C,D). We found that LRRK2(I2020T) forms a right-handed double helix around 11-, 12- and 13-protofilament microtubules (Figure 3A). Thus, there is a symmetry mismatch between the right-handed LRRK2(I2020T) and the left-handed microtubule helices (Figure 3A,B; helical parameters in Table S1). Along the LRRK2(I2020T) helix, we identified repeating units (protomers) with a C2 symmetry axis perpendicular to the filament axis. The number of LRRK2(I2020T) protomers per turn matched the microtubule protofilament number, with 11, 12 and 13 LRRK2(I2020T) protomers per turn around 11-, 12- and 13-protofilament microtubules, respectively (Figure 3A). LRRK2(I2020T) showed a clear preference for 11- and 12-protofilament microtubules (Figure 2D). The interaction between LRRK2 bound to 11-protofilament microtubules also appeared better resolved than LRRK2 bound to 12- and 13-protofilament microtubules, despite its smaller dataset (Figure 3A). The physiological significance of the preferential LRRK2 decoration and possible stabilization of microtubules with non-conventional protofilament numbers remains to be determined.
Figure 3. LRRK2 forms double-stranded right-handed helices around microtubules with different protofilament numbers.
(A) Structures of LRRK2 bound to microtubules composed of 11, 12, and 13 protofilaments. The S-shaped protomer is highlighted in each reconstruction by a black outline, and its 2-fold symmetry axis indicated with a yellow diamond. The total length of microtubules used for averaging (in µm) is shown below each microtubule type. The end- on views show only the golden strand of the double helix for clarity and highlight that the number of protomers matches the number of protofilaments in each microtubule type. A protomer is highlighted by a black outline in this view as well.
(B) Structure of the microtubules shown in A. Particles were reprocessed using an alignment mask that excluded LRRK2. The polarity of microtubules in B is indicated with (−), meaning that the plus end of the microtubule is pointing into the page, and the minus end towards the reader.
(C) LRRK2 protomer density determined by subtomogram analysis (Figure S1F–I,K,L).
(D) Local resolution of the map in C determined by ResMap.
(E) The same subtomogram average shown in C is displayed at a higher threshold to emphasize the hole in the WD40’s β-propeller (indicated by yellow arrowheads) and to highlight the fit of the known crystal structure of a LRRK2 WD40 dimer into the density.
(F) Close up of a WD40 dimer showing the holes at the center of the beta propellers, with yellow arrowheads pointing towards them as in (E).
(G and H) Location of two different 2-fold symmetry axis in the LRRK2 filament (both perpendicular to the filament’s axis and highlighted with yellow diamonds). The first one (G) lies at the WD40:WD40 interface while the second (H) is at the center of the protomer identified in A.
Scale bars: A,B: 10 nm, C,D,E,G,H: 5nm.
Subtomogram analysis combined with integrative modeling reveals the architecture of LRRK2
To determine the three-dimensional architecture of LRRK2 in microtubule-associated filaments, we performed further subtomogram analysis by extracting individual protomer subtomograms from 12-protofilament microtubule-bound LRRK2, as this was the most abundant species in our dataset (Figure 3A, Figure S1F–N). We obtained a map at a resolution of 14 Å (Figure 2C and Figure S1K), with higher local resolution in parts of the LRRK2 protomer (Figure 3D). To date, the only eukaryotic in situ structures of comparable resolution are of ribosomes (Delarue et al., 2018; Schaffer et al., 2019b) and proteasomes (Guo et al., 2018b), whose structures had previously been determined by other methods.
A salient feature in our map was adjacent donut-shaped densities (Figure 3C,F). We could unambiguously assign the identity of these densities as the LRRK2 WD40 domains by docking their recently reported X-ray structure, which was solved as a dimer (Zhang et al., 2019) (Figures 3E,F). The LRRK2 filament density shows two two-fold axes of symmetry perpendicular to the microtubule axis, suggesting two sites of homotypic interaction (Figure 3G,H), one of which forms a Z-shaped protomer dimerizing through the WD40:WD40 interface (Figure 3G). The second homotypic interaction results in a S-shaped dimer and most likely involves a COR:COR interface (Figure 3H; see below). Thus, the repeating unit of the LRRK2 helix is a protomer representing a LRRK2 dimer, which can be defined by either a WD40:WD40 or a COR:COR interface (Figure 3G,H). We did not observe the characteristic extended arch-shaped densities for the N-terminal Armadillo (ARM) and Ankyrin (ANK) or Leucine-rich repeat (LRR) domains in the best resolved area of the protomer close to the microtubule. This is consistent with the previous observation that the four C-terminal domains, ROC, COR, kinase and WD40 domains (Figure 4A), are sufficient for microtubule association in cells (Kett et al., 2012).
Figure 4. The in situ architecture of LRRK2 bound to microtubules.
(A) Schematic representation of the domain organization of LRRK2. ARM: Armadillo repeat domain; ANK: Ankyrin repeat domain; LRR: Leucine-rich repeat domain; ROC: Ras of complex (GTPase); CORN/CORC: C-terminal of ROC; KIN: Kinase. Familial PD mutations are indicated.
(B) Subtomogram average of the microtubule-bound LRRK2 filament (Figure 3C) is shown with the protomer used for model building displayed at higher threshold (blue) within the density of the filament (semi-transparent lighter blue).
(C) Molecular model of microtubule-bound LRRK2, shown in the same two orientations used in B. Domains are colored for one protomer (a LRRK2 dimer) with neighboring LRRK2 monomers shown in light grey. A cartoon representation of the domain architecture of the protomer (left-side view) is shown next to it.
(D) Two views of the molecular model for a LRRK2 monomer seen along the axis of the LRRK2 filament (and the microtubule). The second monomer in the protomer is shown as an outline. The ROC and KIN domains are labeled to highlight their proximity and their location next to, and away from the microtubule, respectively.
(E) Molecular model of the protomer viewed from the microtubule surface. ROC and CORC domains are labeled to highlight that dimerization is mediated by the CORC domain.
(F) A low-threshold isosurface of a cryo-ET map of LRRK2 bound to a 12-protofilament microtubule highlights a connection between adjacent strands in the LRRK2 double helix (colored in blue).
(G) Subtomogram average obtained using alignment mask B (Figure S1J,M,N,P), highlighting hook-like structures (in blue) protruding from the central protomer.
(H) The same molecular model for the LRRK2 protomer shown in C with a model for the LRR repeats, which was docked into the blue densities shown in G.
(I) Schematic representation of the LRRK2 filament using the same cartoon of a LRRK2 dimer introduced in C. The filament is viewed in a direction perpendicular to its axis (and that of the microtubule). The blue lines connecting adjacent strands represent the LRR domains.
(J) Close-up of the middle map in Figure 3A showing a stretch of LRRK2 filament corresponding to the schematic representation in I.
(K, L) The two possible LRRK2 dimers—COR C-mediated in blue (K) and WD40-mediated in purple (L)—are shown in solid colors inside the semi-transparent density for the filament, and their location within the cartoon in I is highlighted by contours of the same color. Scale bar: 5 nm (B–E, G, H), 20 nm (F).
See also Figures S2–S6.
Next, we used an integrative modeling approach (Lasker et al., 2009) (Figure S2) to build a model of LRRK2 into the S-shaped protomer map (Figure 4). We combined available crystallographic and homology models of the ROC, COR (split into CORN and CORC), kinase and WD40 domains, using the WD40 domain as an anchor point. Since inhibitors that capture the kinase domain in an ATP-bound active-like state (Scott et al., 2017; Williamson et al., 2017) induce filament formation (Blanca Ramirez et al., 2017; Deng et al., 2011) and all filament-forming LRRK2 mutants (N1437H, R1441G/C, Y1699C and I2020T) show increased kinase activity in cells (Blanca Ramirez et al., 2017; Kett et al., 2012; Steger et al., 2016), our structure most likely represents an active-like kinase conformation. Thus, we used a humanized kinase-active model derived from an amoeba Roco4 protein (PDB: 4F0F) (Gilsbach et al., 2012) for our integrative modeling. Systematic sampling of combinations of domain placements within the cryo-EM density map, subject to polypeptide connectivity and steric-clash restraints, yielded a single possible architecture of LRRK2(I2020T) in terms of the relative spatial arrangement of its domains, represented by 1,167 models which were further categorized into 53 clusters with slight variations in the centroid and orientation of each domain (Figure S2, S6). One of these models of human LRRK2(I2020T), displayed inside the subtomogram average of the protomer, is shown in Figure 4C–E.
Our LRRK2 model recapitulates the monomeric ROC-COR configuration observed in the X-ray structure of the homologous bacterial protein (PDB: 6HLU) (Deyaert et al., 2019), even though the ROC and COR domains were modelled as separate units during integrative modeling (Figure S3 B vs. C). Comparing the architecture of the monomeric bacterial ROC-COR (PDB: 6HLU) and our LRRK2 monomer models indicated that among the 53 clusters, one cluster particularly resembles the bacterial ROC-COR conformation with an average backbone RMSD of ~10 Å, reflecting slight changes in orientation (Figure S3 F,G). However, the dimerization interfaces of LRRK2 differ significantly from the interfaces seen in two crystal structures of the bacterial LRRK2 homologs (Deyaert et al., 2019; Gotthardt et al., 2008) (Figure S3 D vs. E), and from those in an integrative model of LRRK2 in solution based on an EM map using negative stain (Guaitoli et al., 2016).
In our structure, the ROC (GTPase) domain is closest to the microtubule surface (Figure 4C–E), consistent with observations that the ROC domain of LRRK2 is sufficient for interaction with tubulin heterodimers (Gandhi et al., 2008). GTP binding and hydrolysis by the ROC domain have been proposed to be involved in dimerization (Deyaert et al., 2019), filament formation around microtubules (Blanca Ramirez et al., 2017), and regulation of kinase activity (Biosa et al., 2013). In our model, the ROC and kinase domains are in close spatial proximity (cf. sequence proximity), hinting at direct crosstalk between the catalytic (GTPase and kinase) domains of LRRK2 (Figure 4D). However, higher resolution structures are needed to pinpoint these contacts and map the pathogenic mutations.
Our model shows the kinase domain distal to the microtubule and exposed to the cytosol (Figure 4C). Although we expected that the microtubule-bound form of LRRK2 would contain the kinase in its active-like conformation, and thus used that state in our integrative modeling, we wanted to evaluate whether the conformation of the kinase could influence the arrangements of the other domains. To test this, we replaced the kinase-active domain with a humanized version of the kinase-inactive conformation of the same protein (PDB: 4F0G) (Gilsbach et al., 2012) and found that significantly less models satisfied all restraints at every step of integrative modeling. In order to directly compare both conformations of the kinase domain, we selected models present in the the final ensemble for both conformations and evaluated the percentage of steric backbone clashes. We measured an average of 50% more clashes in the kinase-inactive domain across all models. In order to resolve these clashes, the overlapping domains would need to be moved outside of the cryo-ET density (Figure S3H), suggesting both that our structure reflects an active-like kinase conformation, and that a switch between the active and inactive conformations of the kinase can result in large conformational changes of LRRK2.
The N-terminal domains are likely involved in connections between neighboring strands of LRRK2
Outside the filament that we modeled, our tomograms showed densities connecting the two strands of the LRRK2 helices that decorate microtubules, suggesting inter-helix contacts (Figure 4F). We generated an 18 Å resolution map encompassing a larger volume around the protomer that revealed two hook-like structures stemming from the protomer (Figure 4G, Figure S1J,M,N,P). Given their shape and adjacency to the ROC domain, these densities likely represent the LRR domains (Figure 4H). Thus, it is likely there is a third homotypic interaction, mediated by the LRR domains, that connects the two strands of the double helix, contributing to its regular spacing along the microtubule (Figure 4F). In our density maps, the N-terminal ARM and ANK domains were not resolved, presumably due to flexibility. However, we observed additional density in the periphery of the LRRK2-decorated microtubules that is absent from undecorated microtubules (Figure S1E). The density approximately spans the distance between decorated microtubules within a bundle, suggesting that the ARM and ANK domains form connections between adjacent microtubules.
The WD40:WD40 protomer interface is necessary for the formation of LRRK2 filaments
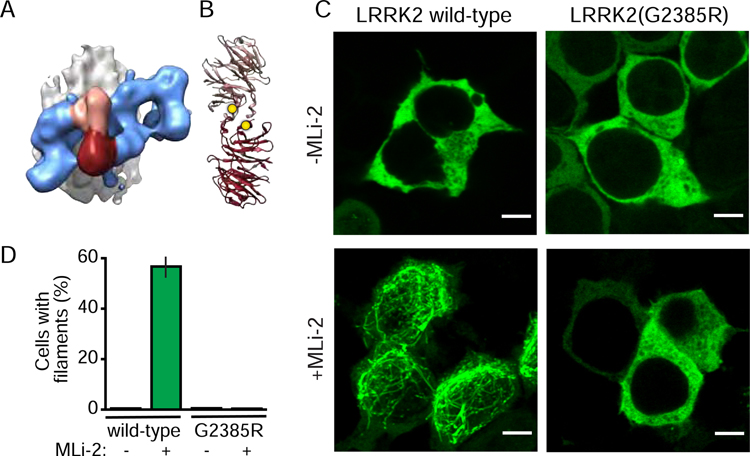
Lastly, to test our model we set out to determine if disrupting one of the two dimer interfaces we observed affected the formation of microtubule-associated LRRK2 filaments in cells. Since the precise COR:COR interface could not be determined due to the limited resolution of the EM density, we focused on the LRRK2 WD40:WD40 interface. The G2385R mutation located in the WD40 domain is a risk factor for PD (Fung et al., 2006; Tan, 2006) and has been shown to disrupt dimerization of isolated WD40 domains in vitro (Zhang et al., 2019). G2385 is located near the WD40:WD40 interface (Fig 4A, B). We hypothesized that the G2385R mutation might impair filament formation in cells. To test this, we expressed Halo-tagged wild-type or G2385R LRRK2 in human HEK-293T cells in the presence or absence of the LRRK2-specific kinase inhibitor MLi-2 (Fell et al., 2015), which has been shown to induce LRRK2 filament formation (Blanca Ramirez et al., 2017; Schmidt et al., 2019). We found that MLi-2 induced filament formation in cells expressing wild-type LRRK2, but not in cells expressing G2385R LRRK2 (Figure 5C, D) in agreement with previous studies using another LRRK2 kinase inhibitor, LRRK2-IN-1 (Rudenko et al., 2012). These data support our model that the WD40:WD40 protomer interface is necessary for the formation of LRRK2 filaments.
Figure 5. The WD40:WD40 interface is necessary for LRRK2 filament formation.
(A) The subtomogram average of the microtubule-bound LRRK2(I2020T) filaments is shown with the microtubule in grey, the LRRK2(I2020T) filament in blue, and the WD40-WD40 dimer connecting two protomers in pink/maroon.
(B) The crystal structure of the WD40 dimer with monomers colored as in A, and the G2385 residues at the interface shown as yellow spheres.
(C) HEK-293T cells transiently expressing Halo-tagged LRRK2 were labeled using a TMR-ligand. Representative images are shown for cells expressing wild-type (left panels) and G2385R (right panels) LRRK2, incubated in the absence (top) and presence (bottom) of 100 nM MLi-2, a LRRK2-specific kinase inhibitor, for 2 hours. Scale bar: 5 nm.
(D) Percentage of transfected cells showing LRRK2 filament formation, defined by bright green rod-like filamentous structures. Data represent three independent experiments. The total number of transfected cells in the analysis was 1705 for wild-type/-MLi-2, 1374 for wild-type/+MLi-2, 1464 for G2385R/-MLi-2, and 1472 for G2385R/+MLi-2.
Conclusions
We have shown that recent advances in cryo-ET and subtomogram analysis can be used to generate models of proteins in situ through an integrative modeling approach, making it possible to generate structural hypotheses in a cellular context. In order to achieve this, there must be orthogonal means to ensure that the structures being analyzed correspond to the right targets. In few selected cases, e.g., ribosomes and associated proteins, the molecular complexes can be directly recognized by comparing them to known structures (Albert et al., 2020; Guo et al., 2018a; Kovtun et al., 2018). However, in most cases, proteins cannot yet be unequivocally identified directly from cryo-ET alone. For those cases, cryo-CLEM is key to identifying the location of the molecules under study (Bharat and Kukulski, 2019; Dahlberg et al., 2020; Hoffman et al., 2020; Moser et al., 2019) To date, using cryo-CLEM and cryo-FIB milling results in significantly lower throughput than cryo-ET alone, limiting the number of structures that can be studied in situ to highly abundant or overexpressed macromolecules. Technological advances in sample preparation and hardware (Buckley et al., 2020; Fäßler et al., 2020; Gorelick et al., 2019; Kuba et al., 2020; Schaffer et al., 2019a; Toro-Nahuelpan et al., 2020; Wu et al., 2020; Zachs et al., 2020), and data processing and analysis (Bepler et al., 2020; Eisenstein et al., 2019; Himes and Zhang, 2018; Martinez-Sanchez et al., 2020; Rickgauer et al., 2017; Song et al., 2020; Tegunov et al., 2020; Xu et al., 2019) will expand this reach in the future (Böhning and Bharat, 2020).
In this work, we determined the structure of a large portion of the full-length human LRRK2 bound to microtubules. The structure includes its catalytic kinase and GTPase domains and all regions of LRRK2 where PD mutations are found. Because all filament-forming LRRK2 mutants show increased kinase activity in cells (Blanca Ramirez et al., 2017; Kett et al., 2012; Steger et al., 2016), and because kinase inhibitors that stabilize an active-like conformation induce LRRK2 filament formation (Blanca Ramirez et al., 2017; Schmidt et al., 2019), we present a model of a human kinase-active LRRK2 structure found in cells. Our structure revealed the domain organization of LRRK2 and showed that the two non-catalytic domains in its C-terminal half (COR and WD40) are involved in the formation of microtubule-associated filaments. The structure showed that LRRK2’s kinase and GTPase are in close spatial proximity, providing a structural explanation for the reported functional cross-talk between these domains (Ito et al., 2007; Terheyden et al., 2016; West et al., 2007), reviewed in (Terheyden et al., 2016). The structure also showed that LRRK2’s kinase faces the cytosol and is thus capable of interacting with potential substrates. Some of the major challenges that will be addressed in future work include imaging endogenously-expressed LRRK2, directly visualizing it while interacting with microtubules and their associated proteins, as well as with other LRRK2 partners, in situ. Our work has highlighted how recent advances in cryo-CLEM, cryo-ET and integrative modeling make it possible to tackle these major cell biological questions.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Elizabeth Villa (evilla@ucsd.edu).
Materials Availability
No new unique reagents were generated in this work.
Data and Code Availability
Cryo-ET structures and representative tomograms have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-20825, EMD-20826, EMD-20827 and EMD-20828. The corresponding tilt series were deposited in the Electron Microscopy Public Image Archive (EMPIAR) with the accession code EMPIAR-10377 and EMPIAR-10378. The LRRK2 models are deposited in the Protein Data Bank with accession code PDB-6XR4.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
HEK-293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing GlutaMAX-I [Thermo Fisher Scientific, (TFS)], 10% HyClone bovine calf serum (GE Healthcare) and 100 U/mL HyClone penicillin-streptomycin (GE Healthcare) at 37°C and 5% CO2.
Plasmids
The plasmid encoding full-length LRRK2(I2020T) fused with YFP at the N-terminus was used as described previously (Kett et al., 2012). The plasmid containing full-length LRRK2 with an N-terminal Halo-tag was cloned using the Gibson Assembly method. The LRRK2 gene was cloned by using the primer sets: 5’-GCGATAACATGGCTAGTGGCAGC-3’ and 5’-GGGGTTATGCTAGTTACTCAACAGATGTTCGTCTC-3’ with pENTR221-LRRK2 (Addgene #39529) as the template. The N-terminal Halo-tag was cloned by using the primer sets: 5’-GAGTAACTAGCATAACCCCTTGGC-3’ and 5’-CACTAGCCATGTTATCGCTCTGAAAGTACAGATC-3’ with the pHTN HaloTag® CMV-neo Vector (Promega) as a template. The two segments were assembled using the NEBuilder® HiFi DNA Assembly Kit following their suggested protocol. The G2358R mutation was made by using the NEB Q5-site directed mutagenesis kit, and the primers were designed based on the online tool NEBBaseChanger. The constructs were sequenced and expressed in HEK-293T cells to confirm the expression of full-length LRRK2(I2020T).
METHOD DETAILS
TEM grid preparation
For expression of LRRK2(I2020T), the HEK-293T cells were transfected with a plasmid encoding full-length LRRK2(I2020T) fused with YFP at the N-terminus using a Lipofectamine 3000 transfection kit (TFS) according to the manufacturer’s protocol. After 2 days, 5 µM paclitaxel (aka Taxol, from Cell Signaling technology) was added. After 16 to 24 hours of Taxol treatment, the cells were detached, and counted with a hemocytometer. Quantifoil 200 mesh holey carbon R4/1 or R2/1 copper grids (Quantifoil Micro Tools) were glow-discharged for 60 s at 0.2 mbar with 20 mA using a PELCO easiGlow glow discharge system (Ted Pella). Just before deposition of cells on TEM grids, 1.5 µl of poly-lysine solution (Sigma) was applied and ~2000–6000 cells were deposited onto a grid by pipetting 3–10 µL of detached cells onto the grid. Blotting and plunging was performed in a humidity-controlled room (~35 % relative humidity) using a custom-made plunger (Max Planck Institute of Biochemistry). Grids were blotted manually using #1 Whatman filter paper on the side of the grid opposite to the Quantifoil carbon foil used as substrate for cell growth, and plunged into a 50/50 mixture of liquid ethane and propane (Airgas) cooled to liquid nitrogen temperature. The grids were clipped onto Autogrids (Thermo Fisher Scientific, hereinafter TFS) and samples were kept at liquid nitrogen temperature throughout the experiments.
Cryo-fluorescence microscopy
For cryo-fluorescence microscopy, grids were observed with a CorrSight microscope (TFS) using EC Plan-Neofluar 5x/0.16NA, EC Plan-Neofluar 20x/0.5NA, and EC Plan-Neofluar 40x/0.9NA air objectives (Carl Zeiss Microscopy), an Oligochrome light-source, which emits in 4 different channels (405/488/561/640 nm) (TFS) and a 1344×1024 px ORCA-Flash 4.0 camera (Hamamatsu). Data acquisition and processing was performed using MAPS 2.1 (TFS). After acquisition of a grid map at 5X magnification, regions of interest were imaged at 20x or 40x magnification, to identify cells with regions containing LRRK2 filamentous structures.
Cryo-focused ion beam milling
Micromachining of frozen hydrated cells was performed in a Scios DualBeam FIB/SEM microscope equipped with a prototype cryo stage (TFS), as detailed previously (Wagner et al., 2020). The sample chamber of the microscope was kept at a pressure below ~1×10 −6 mbar. A low-magnification SEM image encompassing the entire grid using an acceleration voltage of 5 kV, a beam current of 25 pA, and a dwell time of 200 ns was correlated with the grid map from the cryo-light microscope using MAPS 2.1 (TFS) to identify regions of interest on TEM grids. A platinum layer (typically ~2 µm) was deposited on the sample to improve its conductivity and to reduce streaking on the lamella caused by local variations in mass density that result in variations in lamella thickness, known as curtaining (Mahamid et al., 2016; Wagner et al., 2020). An integrated gas injection system (GIS) was used to deposit the precursor compound trimethyl(methylcyclopentadienyl)platinum(IV) (Hayles et al., 2007; Schaffer et al., 2017). To this end, the grid was placed 11.5 mm away from the GIS with a nominal stage tilt of 7°, and the GIS was opened for 13 seconds. Cells were targeted for FIB-milling if they satisfied the following conditions: (1) They showed a filamentous LRRK2 phenotype; (2) they were centered within a grid square, and (3) they were located on a grid square that was at most five squares from the center of the grid (the central ~1mm2 area of the grid). The stage was positioned at a nominal tilt of 11–18°, corresponding to a milling angle of 4–11°. FIB milling was performed in three steps with decreasing ion beam currents and a fixed acceleration voltage of 30 kV (Wagner et al., 2020). For a rough milling step, an ion beam current of 0.3 nA was used. The current was reduced to 0.1 nA for the intermediate step and to 30 pA for fine milling. The target lamella thickness was ~100 nm. During the milling process, lamella thickness was estimated utilizing thickness-dependent charging effects observed in the SEM images at different acceleration voltages (Schaffer et al., 2017). At the end of the session, the grids were transferred out of the FIB/SEM chamber under vacuum and stored in liquid nitrogen.
Cryo-electron tomography
Tomographic tilt series were recorded in a Tecnai G2 Polara (TFS) equipped with a field emission gun operated at 300 kV, a GIF Quantum 968 post-column energy filter (Gatan) and a K2 Summit 4k×4k pixel direct electron detector (Gatan). The FIB-milled grids were loaded into the TEM using modified Polara cartridges, which accommodate Autogrids securely (Rigort et al., 2012). The milling slot of each grid was aligned perpendicular to the tilt axis of the microscope (Wagner et al., 2020). Tilt series were acquired at a target defocus of 5 µm and a pixel size of 2.2 or 3.5 Å using the SerialEM software (Mastronarde, 2005) in low-dose mode (Table S2). The dose-symmetric tomography acquisition scheme (Hagen et al., 2017) was modified to account for the pre-tilt of the lamella, and to optimize the dose using a SerialEM algorithm that calculates the exposure according to a target average count per image, and does not take the image if the dose per tilt required would exceed a threshold value. The K2 detector was operated in counting and dose fractionation modes, with 0.075 to 0.1 s frames. The target tilt range was set to 120° (±60° starting at the estimated pre-tilt of the lamellae), with increments of 2 or 3°, and a target total electron dose was of 180 e/Å2 or 120 e/Å2 respectively (Table S2).
Tomogram reconstruction and annotation
Alignment of the tilt series and tomographic reconstructions were performed using Etomo, part of the IMOD package (Kremer et al., 1996). Since no fiducial markers were present on the lamellas, tilt-series alignment was performed using patch tracking. Contrast transfer function (CTF) correction was performed in IMOD (Kremer et al., 1996) using defocus values estimated by CTFFIND4 (Rohou and Grigorieff, 2015). Motion correction and dose weighting was performed on each image of the tilt series using MotionCor2 (Zheng et al., 2017); motion-corrected and dose-weighted images were replaced in the aligned tilt series. Tomograms were reconstructed using weighted back-projection. Tomograms were 4x-binned (without CTF correction) and used for segmentation of microtubules using the filament tracing function in Amira (TFS) with the following parameters: cylinder length: 600 Å, angular sampling: 5, mask cylinder radius: 140 Å, outer cylinder radius: 125 Å, inner cylinder radius: 75 Å, and missing wedge according to the individual tilt series. After filament tracing (Figure S1A), the resulting center coordinates were re-sampled equidistantly using a script written in MATLAB (Jasnin et al., 2013). For each point along the filaments, initial Euler angles were assigned for subtomogram analysis. Since microtubules are polar, within a filament all angles were assigned such as the direction along the filament axis was the same for all particles. The Euler angle orthogonal to the axis of the filament, i.e., the angle around the microtubule, was randomized to minimize the effect of the missing wedge (see details below).
Subtomogram analysis
To determine the protofilament number of single microtubules, 4x binned tomograms were used for extraction of subtomograms with a box size of 38 nm along the microtubule at 4 nm spacing using Dynamo (Castano-Diez et al., 2012), and an initial average for each microtubule was created using the pre-assigned Euler angles described above. Each filament was separately aligned and averaged using Dynamo, using the initial average as a reference. Three iterations of translational and orientational alignment of the first two Euler angles, followed by five iterations of translational alignment and orientational alignment of the third Euler angle around the axis of the microtubule, with the latter restricted to 18–36 degrees, were performed. Particle shifts were limited to 10–20 nm. As this would allow for the particles to shift by 4 nm and result in duplicate particles, we tested barring particle shifts along the helical axis of the filament, and obtained the same overall geometries. However, we used the shifts as described, as they resulted in better resolved features for the analysis. Since this analysis was performed exclusively to obtain the polarity and geometry of the microtubule and LRRK2 helices as described below and we did not actively exclude potential particle duplication that could cause resolution overestimation, we do not report the resolution of the resulting structures. Particles were low-pass filtered to 22–30 Å for alignment. 2D projections along the microtubule axis were calculated in MATLAB, and the 2D projections as well as the 3D averages were visually inspected in MATLAB and IMOD to determine protofilament number and plus/minus end polarity for each microtubule. Assignment of microtubule polarity can be achieved by inspecting a cross section of the averages (Figure S1B) from the plus end (‘anti-clockwise slew’ Figure S1B, left column) or from the minus end (‘clockwise slew’ Figure S1B, right column) respectively (Bouchet-Marquis et al., 2007). Microtubules with unclear or ambiguous protofilament number or polarity were discarded. Then, new particles with a box length of ~70 nm were extracted from 4x binned tomograms, and for each microtubule class (11-, 12- or 13-protofilament microtubules) all particles representing the respective class were submitted to alignment and averaging in Dynamo, using an initial average as a reference. In order to separate decorated microtubules from non-decorated microtubules, multi-reference alignment in Dynamo was conducted using two templates: (1) a LRRK2-decorated microtubule template (the average from the previous step) and (2) a non-decorated microtubule template (the average from the previous step with the density of LRRK2 masked out) using a hollow cylindrical classification mask corresponding to the LRRK2 density (Figure S1D). Alignment parameters were used as described above. In order to further improve the structure of LRRK2 bound to microtubules, particles sorted to class 1 (decorated microtubules) were further aligned by using a hollow cylindrical alignment mask that included LRRK2 but excluded the microtubule. For obtaining microtubule averages (Figure 3B), all particles (decorated and undecorated) with the same microtubule protofilament number and polarity were aligned by using a cylindrical alignment mask containing only the microtubule region and excluding the LRRK2 helix. The resulting averages (Figure 3A) were used to estimate the helical parameters of the LRRK2 double-stranded helix and microtubules as detailed below (Table S1). These averages showed that while the LRRK2 and microtubule helices have different polarities, the number of subunits per turn (11, 12, or 13) is the same for both helices (Figure 3A). Because imposing helical symmetry made the LRRK2 helix averages worse (data not shown), we extracted new, smaller subtomograms that would encompass the repeating unit of the LRRK2 helix and the adjacent microtubule region. Since the largest sample corresponded to the 12-protofilament (and 12-LRRK2 repeating units per turn) class, we considered only these helices for further analysis. Coordinates and initial Euler angles of particles along the LRRK2 helical path (Table S1) were used to extract particles from unbinned, dose-weighted and non-CTF-corrected tomograms, and used to create an initial average using orientations (Figure S1F). 3D refinement of extracted particles was performed in RELION (Bharat and Scheres, 2016; Bharat et al., 2015) to create a template for subsequent classification (Figure S1G). The resulting density map was used as a reference for 3D classification into three classes, employing a soft mask covering the LRRK2 protomer and a regularization parameter of 2–4 (Figure S1H). Class averages were inspected in UCSF Chimera (Pettersen et al., 2004), and classes with high levels of noise were excluded. A density corresponding to the ring-shaped WD40 domain was clearly resolved in one class (class 1 in Figure S1H). Particles belonging to this class were subjected to gold-standard refinement using two different masks (masks A and B; Figure S1I,J) and post-processing without B-factor sharpening in RELION (Figure S1K,M). Gold-standard Fourier Shell Correlation (FSC) was calculated (Figure S1L,N) for resolutions of 14.1 Å and 17.6 Å. Local-resolution maps (Figure 3D and S1O,P) were calculated using ResMap (Kucukelbir et al., 2014). Maps and structures were visualized using UCSF Chimera (Pettersen et al., 2004) and VMD (Humphrey et al., 1996).
Measuring distances/angles between microtubules
To calculate the angles and distances between filaments (microtubules), the distances between points sampled every 4 nm along each filament, and all points on all other filaments within the tomogram, were measured. The resulting distance matrix was then reduced to point pairs with distances of 100 nm or less. For each point along a filament, we calculated (1) a tangent vector along the filament at that point, considering the nearest points along the same filament, (2) the distance to the nearest point in each neighboring filament, and (3) the angle between the vectors at these two points, where an angle of zero corresponds to parallel filaments. A 2-D histogram of the distance between filaments vs angle between filaments was calculated in MATLAB.
Determining helical parameters
Helical parameters of non-symmetrized microtubules and LRRK2 helices were obtained using an autocorrelation function as implemented in Dynamo. To obtain the helical parameters for the microtubule and LRRK2 helices, averages were obtained by masking out the volume corresponding to the other helix, i.e., microtubule helical parameters were obtained using a map that excluded the LRRK2, and vice versa.
Integrative modeling of LRRK2 bound to microtubules
LRRK2 is a 286kDa protein formed of seven domains: armadillo (ARM), Ankyrin (ANK) and Leucin-rich repeat (LRR) domains, a Ras of complex proteins (ROC), and C-terminal of ROC domain (COR), kinase (KIN) and a WD40 domains (Figure 4A). To determine the architecture of LRRK2(I2020T) bound to microtubules, we used an integrative approach implemented in the open source Integrative Modeling Platform (IMP) package (Russel et al., 2012). The integrative modeling process consists of four stages: (1) gathering data, (2) choosing how to represent the system and translating the information into spatial restraints, (3) determining an ensemble of structures that satisfy these restraints and, (4) validating the model. This approach has been applied to determine the structure of numerous biological complexes including the 26S proteasome (Forster et al., 2010), the mediator complex (Robinson et al., 2015) as well as the nuclear pore complex (Kim et al., 2018). However, this integrative modeling approach has not, to our knowledge, been applied to solve a previously undetermined structure using in situ cryo-ET.
Given the resolution of the cryo-ET map, the modeling was limited to the location and orientation of the domains within the density, treated as rigid bodies. The clear identification of the characteristic donut shape of the WD40 in the cryo-ET density map provided the starting point for the LRRK2 domains allocation and allowed us to assess that the assignment of the LRRK2 is possible up to the last four domains (WD40, KIN, COR, ROC) while the density related to the LRR, ANK and ARM is visible only at higher threshold, presumably due to their intrinsic flexibility.
Stage 1: Gathering Information.
Three different types of data were used for structure determination:
Cryo-ET map of LRRK2(I2020T) protomer: a 14 Å in situ cryo-ET map was determined as detailed above (EMDB-20825). The protomer corresponds to a dimer LRRK2 bound to the microtubules (Fig 3C).
- Atomic models of the domains composing LRRK2:
- X-ray crystallography structure of the human LRRK2 WD40 dimer (Zhang et al., 2019) (PDB: 6DLP)
- Homology model of the kinase domain of human LRRK2 (Uniprot ID Q5S007, residue 1883–2135). We generated models for both the active and inactive conformations. The models were generated by homology modeling with SWISS-MODEL (Benkert et al., 2011; Bienert et al., 2017; Guex et al., 2009; Waterhouse et al., 2018). The crystal structure of the Roco4 Kinase domain bound to AppCp from D. discoideum was used as a template for the kinase-active (open) model (PDB: 4F0F, homology 43%), and the the crystal structure Roco4 kinase domain from D. discoideum (PDB: 4F0G, homology 43%) was used as a template for the kinase-inactive (closed) model (Gilsbach et al., 2012). Alignment was generated by ClustalOmega (Sievers et al., 2011) (Figure S4). Our initial integrative modeling used an active conformation of the kinase based on the fact that the LRRK2-specific Type-1 inhibitors MLi-2 and LRRK2-IN-1, which bind to the active form, increase filament formation (Blanca Ramirez et al., 2017; Schmidt et al., 2019).
- A model of the monomeric ROCCOR domain of human LRRK2 protein (Uniprot ID Q5S007, residue 1332–1838 was obtained by homology modeling with SWISS-MODEL (Benkert et al., 2011; Bienert et al., 2017; Guex et al., 2009; Waterhouse et al., 2018). The structure of C. tepidum Roco protein (Gotthardt et al., 2008) (PDB: 3DPU) was used as template and aligned to the human LRRK2 sequence of domains ROC and COR by using ClustalOmega (Sievers et al., 2011) (Figure S4). Normal model analysis (NMA) with Bio3D package in R (Grant et al., 2006) was performed on the ROCCOR template revealing a hinge in the COR domain (L1669-I1689) which corresponds to a missing loop area in the template PDB structure. Therefore, we split the ROCCOR domain into three separate rigid bodies namely, CORC, CORN, and ROC (Gotthardt et al., 2008). (PDB: 3DPU, homology 37%, 37% and 38% respectively; Figure S4 and S5). We did not directly use human ROC domain structure (Deng et al., 2008) (PDB: 2ZEJ) due to the potential domain swapping described earlier (Gotthardt et al., 2008). However, we replaced the coordinates of the region not involved in this interface (residues 1425–1441 and 1474–1457) in the final models of the ensemble.
Since all the domains are part of a single polypeptide chain of LRRK2, the amino acid stretches between the domains were considered as linkers connecting the N- and C-termini of consecutive domains. A worm-like chain model (Flory, (1953)) was used to model the connecting linkers, with an average end-to-end distance of , where N is the number of amino acids and l=3.1Å is the length of one amino acid measured as the typical average distance between alpha carbons of adjacent amino acids (Figure S5).
Stage 2: System Representation and Translation of Data into Spatial Restraints.
Each domain was represented as a rigid body at atomic resolution. Five types of spatial restraints were used during monomer configuration and dimer refinement in Stage 3.
Cryo-ET density restraint: (a) an IMP scoring term that considers the percentage of atoms that are included in the map, used both in monomer configuration and refinement. (b) Cross correlation of model to map implemented in MDFF (Trabuco et al., 2008), used to select models in monomer configuration.
Location of WD40: based on the characteristic donut shapes in the density and a rigid-body fit of the crystal structure of the human LRRK2 WD40 dimer to the density, the WD40 could be unequivocally located in the cryo-ET density map (see below) and thus its location was constrained to that region in the map (Figure S5), used in monomer configuration.
Chain connectivity restraints: distance restraints between the C-term and N-term of consecutive domains as described above (Figure S5), used both in monomer configuration and dimer refinement.
Excluded volume restraints: Overlap between domains is assessed as an excluded volume restraint (steric clashes of alpha carbons of less than 10% and 5%), used both in monomer configuration and dimer refinement respectively.
Dimer symmetry restraints: (a) Overlap between monomers was assessed using excluded volume (steric clashes of less that 10% of alpha carbon atoms between LRRK2 monomers), and used in the final stage of monomer configuration. (b) Symmetry restraints implemented in IMP were used during dimer refinement.
Stage 3: Ensemble Sampling:
To create an ensemble of LRRK dimers that incorporates the data available and satisfies the restraints, we applied a sampling protocol consisting of two steps: monomer configuration and dimer refinement.
A. Monomer configuration.
To determine the monomer configuration we used the MultiFit module of IMP (Lasker et al., 2009). First, rigid-body fitting of each of the individual five domains to a dimer protomer map were sampled at 5° resolution using colores (Chacon and Wriggers, 2002). The best 10,000 fits for WD40, ROC, CORN and CORC and 5,000 fits for KIN were considered. Additionally, the crystal structure of the human LRRK2 WD40 dimer (PDB: 6DLP) (Zhang et al., 2019) was rigid body fitted using the same parameters. Rigid-body fitting the WD40 dimer (PDB: 6DLP) into the cryo-ET density using the same parameters as above resulted in an unequivocal best fit that matched the WD40 monomers into the donut shaped regions of the map, revealing the same orientation between the monomers as in the crystal structure (Figure 3E). Thus, the WD40 fits for the monomer were filtered to be placed within the donut-shaped density to satisfy constraint ii. From these fits, we selected a single position and orientation that matched that of the dimer in the dimer crystal structure (Zhang et al., 2019). Second, we looked at all 5000 fits of the active kinase and selected those that satisfied the distance linker of 9 Å (restraint iii). Only six fits satisfied this restraint, and they had an RMSD of 4.7Å between them. Considering the resolution of our map, they were equivalent, and thus we used a single kinase fit for the subsequent steps to reduce computational time. Next, the fits for ROC, CORN and CORC were filtered to include only those fitted to the same monomer within the protomer map, and not overlapping with the placed WD40 and kinase, yielding 1188, 1446 and 1148 models respectively. MultiFit was then used to sample all possible combinations of the ROC, CORN and CORC domain fits and scored for spatial restraints i(a), iii and iv. A total of three million models were initially scored. From that, we filtered the solutions based on respecting the N-C distance (restraint iii) and having not major compenetration between domains (restraint iv; corresponding to an EV score of <100,000). This yielded 786,211 models.
Third, the ensemble was clustered based on an RMSD of 8 Å, leading to 256 clusters, that could be further classified into classes of different architectures, i.e., swaps between ROC, CORN and CORC domains. Next, the models were filtered to allow only clashes of up to 10% of the alpha carbons between domains as measured in VMD. From here, only two clusters remained, one with 31,140 models, and the other with 394 models. These two clusters differed in the location of the CORC and ROC domains (Figure S2).
Fourth, dimers were built from each monomer by applying a symmetry transformation determined from the cryo-ET map. All models in the second cluster had major clashes at the dimer interface. Thus, only one ensemble of 31,140 models remained corresponding to the same domain architecture. The ensemble was filtered to satisfy restraints i, iii, iv, and v(a), resulting in 1,167 dimer models.
B. Dimer refinement.
We generated an ensemble of LRRK2 dimers as detailed above. We then clustered the ensemble of 1,167 dimers into 53 clusters based on an RMSD of 8 Å. From each cluster, we refined a dimer using a simulated-annealing Monte-Carlo (MC) protocol implemented in IMP. During the refinement, local perturbations in positions and orientations of the ten rigid bodies were sampled and assessed using a scoring function including terms i-v. Minima from the refinement protocol were gathered and used in the refined ensemble assessment (stage 4).
Stage 4: Assessment of Data and Structure
Our extensive sampling rendered a single architecture of LRR2 dimers. In order to validate this structure, we first looked at the overall conformation and its congruency with other available data. We found that our model agrees with the following independent observations:
The LRR domain fits into the density and is in the proximity of the ROC domain (Figure 4H). The LRR model corresponds to residues 129–444 of the LRR in a crystal structure of a homologous Roco protein (PDB: 6HLU), and was fitted using the rigid-body fitting function of Chimera (Pettersen et al., 2004).
The ROC domain is proximal to the microtubules, consistent with observations that the ROC domain of LRRK2 is sufficient for interaction with tubulin heterodimers (Gandhi et al., 2008).
The dimerization of the WD40 observed in a recent crystal structure (Zhang et al., 2019) matches the architecture in our model, and is consistent with our data showing no filament formation (Figure 5). Although we used the crystal structure to assign the overall region occupied by the WD40, a preliminary global alignment using MultiFit (data not shown) confirmed the best conformation of the WD40 to fit that of the dimer crystal structure.
The configuration of ROCCOR of our LRRK2 models obtained by integrative modeling is similar to the locations of those domains within a monomer in the crystal structure of the bacterial homolog (PDB:6HLU) (Deyaert et al., 2019) even though the humanized ROC and COR domains (further separated as CORN and CORC) were modelled as separate units during integrative modeling.
The monomer ensemble consisting of 1,167 models was analyzed to assess the uncertainty on the positions and orientations of each domain (Figure S6). The analysis on the variability showed a narrow conformational space visited by COR, CORC and a broader conformational space visited by ROC (Figure S6). During refinement, the spread of the conformational variability was further constricted without major rearrangements. Thus, the ensemble reflects the variability of the structure given the available experimental data.
The average RMSD distribution between the ROCCOR of the C. tepidum Roco protein and the 53 clusters of LRRK2 models were calculated. Conformational space analysis on the lowest RMSD LRRK2 models (cluster 20) showed that the CORC and CORN domains were found in the same orientation as in the bacterial ROCCOR structure, and again found that the ROC domain explored a vaster conformational space. The conformational variability of the ROC domain observed in our data was previously described in the X-ray structure (PDB: 3DPU) where the ROC domain was solved only for one of the two monomers. Altogether, we determined the architecture of LRRK2, with certainty in the location of the domains, and varying uncertainty in their orientation, as expected from the resolution of our data. The molecular models represented throughout the manuscript depict one of the models in cluster 20. For each of the clusters, we selected a snapshot of the refinement and deposited the alpha-carbon coordinates to the Protein Data Bank (53 models in PDB 6XR4). The snapshots were selected so that the scoring function and the RMSD to the bacterial ROCCOR structure improved. However, due to the uncertainty of our data, the variability of the ensemble should be considered (Figure S6). The full monomer ensemble and dimer refinement runs are available upon request, as depositing these entire data is currently not possible in the Protein Data Bank.
We also evaluated the effect of the functional state of the kinase in our LRRK2 models. Our workflow first considered an active (closed) kinase conformation based on previous experiments that link the filament formation to the ATP-bound active-like state of the kinase (Scott et al., 2017; Williamson et al., 2017), and the fact that LRRK2(I2020T) is known to cause hyper-kinase activity in cells (Blanca Ramirez et al., 2017; Kett et al., 2012; Steger et al., 2016). In order to verify our assumption, we investigated the effect of using an inactive kinase domain model. First, we built a humanized inactive (open) kinase model using PDB ID 4F0G (open/inactive Roco4 Kinase domain from D. discoideum) using Swissmodel as described above. Next, we replaced the inactive kinase model in of 786K models generated in the second step of monomer refinement, as this is equivalent to running the full integrative modeling exercise with significantly lower computational cost. The replacement of the active by the inactive model was done by aligning it to the C-terminal part of the active kinase model we used previously, given that the linker between the WD40 and the C-terminal of the kinase is the most stringent restraint. The same analysis, namely clustering and excluding models based on domain clashes within the monomer followed by building dimers and excluding based on clashes between monomers, was performed using the kinase-inactive model. At every step, the subset of models that conformed to the restraints was between 50–67% the size of that for the kinase active conformation. The final ensemble had 786 models with the same overall architecture (location of the domains). In order to have a direct comparison between the ensembles with active-like and inactive-like kinase conformations, we selected the models that were present in both ensembles and calculated the clashes between the kinase and the ROCCOR domains by measuring the percentage of backbone atoms that were within 5 Å using VMD. The number of clashes in the inactive conformations was 17% higher than the active conformation in all models. Resolving the clashes would require the structure of the ROCCOR domain to move outside of the EM density (Figure S3H). Altogether, our analysis suggests that our cryo-ET map of LRRK2(I2020T) bound to microtubules represents a kinase-active state of LRRK2.
Confocal fluorescence microscopy and statistical analysis
For transient protein expression of N-terminally Halo-tagged full-length wild-type and G2385R LRRK2, HEK-293T cells were plated onto 6-well dishes containing poly-D-lysine-coated glass coverslips or onto poly-D-lysine-coated glass-bottom culture dishes (MatTek Corporation, Ashland, MA, USA, Cat. No. P35GC-0-14-C) and allowed to grow to 40–50% confluency. Cells were transfected using the Lipofectamine 3000 transfection kit (Thermo Fisher Scientific) according to the manufacturer’s protocol using 1 µg of tagged Halo-LRRK2 DNAs. After 48 hours, cells were treated with the LRRK2 inhibitor MLi-2 (100 nM) for 2 hours and then labelled with the TMR ligand (3 µM) (Promega). After 15 minutes, cells were washed with fresh media and then fixed with pre-warmed (37°C) 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 minutes at room temperature. Cells were then washed in PBS and mounted with an in-house made antifade agent gelvatol. Confocal imaging was performed with an inverted Olympus Fluoview 1000 laser scanning confocal microscope using a 561 nm laser line and a 60X oil immersion objective lens with numerical aperture 1.42. Z-stack images were acquired with a step size of 0.3 microns and processed using the Fiji software package (Schindelin et al., 2012). Cells expressing the different LRRK2 mutants were assessed for the presence of clear filamentous structures and quantified in three independent experiments.
QUANTIFICATION AND STATISTICAL ANALYSIS
The details of the quantification and all statistical analyses are included in the relevant sections of the METHOD DETAILS.
Supplementary Material
Figure S1. Subtomogram Analysis Workflow, Related to Figures 2 and 3
(A) Individual microtubules are automatically annotated using the filament-tracing function of Amira (TFS). The annotated microtubules (grey) are shown on a slice of a tomogram of LRRK2(I2020T)-decorated microtubules.
(B) Single microtubule averaging sampled at every 4 nm was performed using Dynamo to determine the protofilament number and polarity of individual microtubules using 4x-binned tomograms. Example cross-sections of subtomogram averages seen from the plus end (‘anti-clockwise slew’, left column) and from the minus end (‘clockwise slew’ right column). The total length of each class of microtubule unambiguously determined from LRRK2(I2020T) tomograms is shown.
(C) After sorting of microtubules according to protofilament number, larger subtomograms (70 nm) were re-extracted and averaged to obtain initial LRRK2/microtubule averages.
(D) Dynamo multi-reference alignment using two different templates: (1) LRRK2/microtubule and (2) microtubule only, using a hollow cylindrical classification mask corresponding to the LRRK2 density, resulting in averages for LRRK2-decorated 12-protofilament microtubule (class 1) and undecorated 12-protofilament microtubule (class 2). The same protocol was followed for 11-protofilament and 13-protofilament microtubules.
(E) Putative location of LRRK2’s N-terminal domains. Left: Cross-section view of the LRRK2/13-protofilament microtubule map. A weak outermost density is highlighted with a pair of blue dotted circles. Right: Cross-section view of an undecorated 13-protofilament microtubule at lower threshold. No outer density is observed in this cross-section. The dotted blue circles shown correspond to 50 nm and 67 nm in diameter, respectively, matching the peak of center-to-center distance between LRRK2-decorated microtubule bundles of ~67 nm (Figure 2E). Scale bar: 25 nm.
(F) Helical parameters of the LRRK2 helix on 12-protofilament microtubules were estimated and used for extraction of subtomograms containing the LRRK2 protomer from unbinned tomograms and used to create an initial average.
(G) 3D refinement of extracted particles was performed in RELION to create a template for subsequent classification.
(H) The refined average from (G) was used as a reference for 3D classification into three classes in RELION. Classes with high levels of noise (classes 2 and 3) were discarded.
(I,J) Particles belonging to the good class (class 1) were subjected to gold-standard refinement and post-processing in RELION using two different masks shown in grey (A and B).
(K, M) Final subtomogram averages from a LRRK2(I2020T) dimer bound to a 12-protofilament microtubule.
(L,N) FSC curves for the two maps shown in K and M, respectively.
(O,P) Local resolution of the maps shown in K and M, respectively, calculated using ResMap. Panel O is the same as Figure 3D, shown here again for clarity.
Figure S2. Integrative modeling of human LRRK2, Related to Figure 4 and STAR Methods
First, data generated by various experiments and computational methods are gathered. Second, the five C-terminal domains of LRRK2 are represented as rigid bodies and the data are translated into spatial restraints. Third, an ensemble of structures that satisfy the restraints are sampled using Multifit, an IMP module that allows for the simultaneous fitting of the rigid bodies while respecting the a priori restraints imposed. The ensemble is clustered into distinct domain arrangements and representative structures are further refined. Finally, the refined models are analyzed for accuracy and certainty (see STAR Methods for details).
Figure S3. Comparison of human LRRK2 with a bacterial Roco homolog protein, Related to Figure 4 and STAR Methods
(A) Domain organization of the human LRRK2 and Chlorobaculum tepidum Roco proteins.
(B) The ROCCOR domains of an integrative model (#909) of a monomeric human LRRK2 are shown in the same view as that in Figure 4D within the density corresponding to the protomer (dimer) used to build the model shown in white.
(C) The ROCCOR domains of the crystal structure of C. tepidum Roco monomer (PDB ID: 6HLU) were aligned to the ROCCOR region of human LRRK2 and are shown in the same orientation as in B. The LRR domain was omitted for clarity.
(D) The ROCCOR domains of the CORC-mediated dimer of human LRRK2 are shown inside the protomer’s density in the same views as in B (top) and Figure 4E (bottom). Domain names are indicated for both monomers and are distinguished by the presence or absence of an outline.
(E) The ROCCOR domains of the C. tepidum Roco dimer (PDB ID: 6HLU) are shown on the left in the same orientation as that in F above. The panel on the top shows the same structure viewed down its 2-fold symmetry axis (equivalent to that shown above for human LRRK2). Note that different rotations relate the two views of LRRK2 and the Roco protein, and that the dimer interface for the Roco protein involves both its ROC and CORC domains while the LRRK2 dimer is only mediated by its CORC domain.
(F) Distribution of RMSD values calculated between the ROCCOR of C. tepidum Roco protein and 53 clusters of LRRK2 models generated by integrative modeling. Cluster 20 has the closest structure, an arrow indicates the model used in this figure and Figure 4.
(G) Analysis of the variability in domain orientation for the cluster with the lowest RMSD against C. tepidum Roco (cluster 20). The three ROCCOR domains—COR C, CORN and ROC—are shown. The variability in domain orientation for all the models within the cluster is shown for each domain. The red arrow indicates the orientation of the domain as shown in B (model #909). Arrows matching the domain color indicate the orientations of other models in the cluster. Conformational variability of the ROC domain as observed in our models was previously described for an X-ray structure (PDB: 3DPU) where the ROC domain was solved only for one of the two monomers.
(H) Analysis of active vs inactive domains of microtubule-bound LRRK2. Left: Model of human LRRK2 with an active kinase model (same model as in Figure 4) shown inside the cryo-ET density. Right: Same model as in B modified to show the movement of the ROCCOR needed to accommodate the inactive-like kinase state. The ROCCOR was moved along with the N-terminus of the kinase to avoid clashes. The orange arrows show the movement of the ROCCOR domains, which places them outside of the density.
Scale bars: B,C,D,E: 2nm.
Figure S4. Sequence alignment used for homology modeling of LRRK2, Related to Figure 4 and STAR Methods
(A) Alignment of the LRRK2 kinase domain to the D. discoideum Roco4 kinase domain template structure PDB ID: 4F0F (bound to ADPCP).
(B) Alignment of the LRRK2 CORC (C-terminal end of ROCCOR) domain to the C. tepidum Roco ROCCOR template structure PDB ID: 3DPU.
(C) Alignment of the LRRK2 ROC and CORN to the C.tepidum Roco ROCCOR template structure PDB ID: 3DPU.
Figure S5. Experimental data, statistical inference and physical principles considered in the integrative modeling procedure, Related to Figure 4 and STAR Methods
(A) Comparative modeling templates for WD40, KIN, CORC, CORN, and ROC. The PDB template, the organism, the homology, and the relative modeled region are reported for each domain.
(B) Distance restraints and (C) excluded volumes pairs considered in the integrative modeling approach.
Figure S6. Ensemble Variability Analysis, Related to Figure 4 and STAR Methods
Analysis of the ensemble variability for WD40 (red), KIN (pink), CORC (orange), CORN (yellow) and ROC (green).
(A) Representative structure from the ensemble, with the position and orientation indicated by an arrow.
(B) Probability density function of the localization of the centroid of each domain within the density. A blue star indicates the potential location of the C-terminus of the LRR domain.
(C) Orientation variability analysis of each domain within the ensemble; each arrow indicates the orientation of the domain with respect to their center of mass colored accordingly to the domain analyzed. Scale bar: 5 nm.
Highlights:
Cryo-electron tomography and integrative modeling reveal LRRK2’s structure in situ
Parkinson’s disease-linked LRRK2 forms double-helical filaments around microtubules
Homotypic interactions between the WD40 and COR domains form the filaments
The GTPase and kinase domains face the microtubule and cytoplasm, respectively
Acknowledgements
We thank Daniel Castaño Díez and Paula Pérez Navarro for assistance with Dynamo; Guillaume Castillon, Alexis Rohou, Junru Hu, and members of the Villa lab for stimulating discussions and technical support. This work was supported by an NIH Director’s New Innovator Award 1DP2GM123494–01 (to E.V.), a Michael J. Fox Foundation grant #11425 (to E.V., S.T., and D.B.), an NIH NIGMS R01GM086197 and a Branfman Family Foundation award (to D.B). M.A. was supported by a postdoctoral fellowship from the Visible Molecular Cell Consortium at UC San Diego. T.L. is supported by a Taiwan MOE fellowship. We acknowledge the use of the UC San Diego cryo-Electron Microscopy Facility (partially supported by a gift from the Agouron Institute to UC San Diego) with special thanks to Tim Booth, the NIH National Center for Microscopy and Imaging Research (P41GM103412), and the San Diego Nanotechnology Infrastructure (SDNI) of UC San Diego, a member of the National Nanotechnology Coordinated Infrastructure, which is supported by the National Science Foundation (ECCS-1542148), with special thanks to Jeff Wu and Bernd Fruhberger.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
M. Audagnotto is an employee of AstraZeneca and has stock ownership and/or stock options or interests in the company.
References
- Albert S, Wietrzynski W, Lee C-W, Schaffer M, Beck F, Schuller JM, Salomé PA, Plitzko JM, Baumeister W, and Engel BD (2020). Direct visualization of degradation microcompartments at the ER membrane. Proceedings of the National Academy of Sciences 117, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P, Biasini M, and Schwede T (2011). Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bepler T, Kelley K, Noble AJ, and Berger B (2020). Topaz-Denoise: general deep denoising models for cryoEM and cryoET. bioRxiv, 838920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, and Kukulski W (2019). Cryo-Correlative Light and Electron Microscopy: Toward in situ Structural Biology. Correlative Imaging: Focusing on the Future, 137–153.
- Bharat TA, and Scheres SH (2016). Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc 11, 2054–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TAM, Russo CJ, Lowe J, Passmore LA, and Scheres SHW (2015). Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure 23, 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, and Schwede T (2017). The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res 45, D313–D319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biosa A, Trancikova A, Civiero L, Glauser L, Bubacco L, Greggio E, and Moore DJ (2013). GTPase activity regulates kinase activity and cellular phenotypes of Parkinson’s disease-associated LRRK2. Hum Mol Genet 22, 1140–1156. [DOI] [PubMed] [Google Scholar]
- Blanca Ramirez M, Lara Ordonez AJ, Fdez E, Madero-Perez J, Gonnelli A, Drouyer M, Chartier-Harlin MC, Taymans JM, Bubacco L, Greggio E, et al. (2017). GTP binding regulates cellular localization of Parkinson’s disease-associated LRRK2. Hum Mol Genet 26, 2747–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhning J, and Bharat TA (2020). Towards high-throughput in situ structural biology using electron cryotomography. Progress in Biophysics and Molecular Biology. [DOI] [PubMed]
- Bouchet-Marquis C, Zuber B, Glynn AM, Eltsov M, Grabenbauer M, Goldie KN, Thomas D, Frangakis AS, Dubochet J, and Chretien D (2007). Visualization of cell microtubules in their native state. Biol Cell 99, 45–53. [DOI] [PubMed] [Google Scholar]
- Buckley G, Gervinskas G, Taveneau C, Venugopal H, Whisstock JC, and de Marco A (2020). Automated cryo-lamella preparation for high-throughput in-situ structural biology. J Struct Biol 210, 107488. [DOI] [PubMed] [Google Scholar]
- Castano-Diez D, Kudryashev M, Arheit M, and Stahlberg H (2012). Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J Struct Biol 178, 139–151. [DOI] [PubMed] [Google Scholar]
- Chaaban S, and Brouhard GJ (2017). A microtubule bestiary: structural diversity in tubulin polymers. Mol Biol Cell 28, 2924–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon P, and Wriggers W (2002). Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol 317, 375–384. [DOI] [PubMed] [Google Scholar]
- Dahlberg PD, Saurabh S, Sartor AM, Wang J, Mitchell PG, Chiu W, Shapiro L, and Moerner WE (2020). Cryogenic single-molecule fluorescence annotations for electron tomography reveal in situ organization of key proteins in Caulobacter. Proceedings of the National Academy of Sciences 117, 13937–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Brittingham GP, Pfeffer S, Surovtsev IV, Pinglay S, Kennedy KJ, Schaffer M, Gutierrez JI, Sang D, Poterewicz G, et al. (2018). mTORC1 Controls Phase Separation and the Biophysical Properties of the Cytoplasm by Tuning Crowding. Cell 174, 338–349 e320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Lewis PA, Greggio E, Sluch E, Beilina A, and Cookson MR (2008). Structure of the ROC domain from the Parkinson’s disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc Natl Acad Sci U S A 105, 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee JD, Patricelli MP, Nomanbhoy TK, Alessi DR, et al. (2011). Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat Chem Biol 7, 203–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyaert E, Leemans M, Singh RK, Gallardo R, Steyaert J, Kortholt A, Lauer J, and Versees W (2019). Structure and nucleotide-induced conformational dynamics of the Chlorobium tepidum Roco protein. Biochem J 476, 51–66. [DOI] [PubMed] [Google Scholar]
- Di Maio R, Hoffman EK, Rocha EM, Keeney MT, Sanders LH, De Miranda BR, Zharikov A, Van Laar A, Stepan AF, Lanz TA, et al. (2018). LRRK2 activation in idiopathic Parkinson’s disease. Sci Transl Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi T, Kuwahara T, Sakurai M, Komori T, Fujimoto T, Ito G, Yoshimura SI, Harada A, Fukuda M, Koike M, et al. (2018). LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc Natl Acad Sci U S A 115, E9115–E9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein F, Danev R, and Pilhofer M (2019). Improved applicability and robustness of fast cryo-electron tomography data acquisition. Journal of structural biology 208, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fäßler F, Zens B, Hauschild R, and Schur FKM (2020). 3D printed cell culture grid holders for improved cellular specimen preparation in cryo-electron microscopy. bioRxiv, 2020.2006.2012.147678. [DOI] [PubMed]
- Fell MJ, Mirescu C, Basu K, Cheewatrakoolpong B, DeMong DE, Ellis JM, Hyde LA, Lin Y, Markgraf CG, Mei H, et al. (2015). MLi-2, a Potent, Selective, and Centrally Active Compound for Exploring the Therapeutic Potential and Safety of LRRK2 Kinase Inhibition. J Pharmacol Exp Ther 355, 397–409. [DOI] [PubMed] [Google Scholar]
- Flory PJ ((1953)). Principles of Polymer Chemistry. Cornell University Press. [Google Scholar]
- Forster F, Lasker K, Nickell S, Sali A, and Baumeister W (2010). Toward an integrated structural model of the 26S proteasome. Mol Cell Proteomics 9, 1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung HC, Chen CM, Hardy J, Singleton AB, and Wu YR (2006). A common genetic factor for Parkinson disease in ethnic Chinese population in Taiwan. BMC Neurol 6, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi PN, Wang X, Zhu X, Chen SG, and Wilson-Delfosse AL (2008). The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J Neurosci Res 86, 1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilsbach BK, Ho FY, Vetter IR, van Haastert PJ, Wittinghofer A, and Kortholt A (2012). Roco kinase structures give insights into the mechanism of Parkinson disease-related leucine-rich-repeat kinase 2 mutations. Proc Natl Acad Sci U S A 109, 10322–10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick S, Buckley G, Gervinskas G, Johnson TK, Handley A, Caggiano MP, Whisstock JC, Pocock R, and de Marco A (2019). PIE-scope, integrated cryo-correlative light and FIB/SEM microscopy. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt K, Weyand M, Kortholt A, Van Haastert PJ, and Wittinghofer A (2008). Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J 27, 2239–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, and Caves LS (2006). Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics 22, 2695–2696. [DOI] [PubMed] [Google Scholar]
- Guaitoli G, Raimondi F, Gilsbach BK, Gomez-Llorente Y, Deyaert E, Renzi F, Li X, Schaffner A, Jagtap PK, Boldt K, et al. (2016). Structural model of the dimeric Parkinson’s protein LRRK2 reveals a compact architecture involving distant interdomain contacts. Proc Natl Acad Sci U S A 113, E4357–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC, and Schwede T (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30 Suppl 1, S162–173. [DOI] [PubMed] [Google Scholar]
- Guo Q, Lehmer C, Martínez-Sánchez A, Rudack T, Beck F, Hartmann H, Pérez-Berlanga M, Frottin F, Hipp MS, and Hartl FU (2018a). In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell 172, 696–705. e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, Perez-Berlanga M, Frottin F, Hipp MS, Hartl FU, et al. (2018b). In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell 172, 696–705 e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen WJH, Wan W, and Briggs JAG (2017). Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J Struct Biol 197, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles MF, Stokes DJ, Phifer D, and Findlay KC (2007). A technique for improved focused ion beam milling of cryo-prepared life science specimens. J Microsc 226, 263–269. [DOI] [PubMed] [Google Scholar]
- Himes BA, and Zhang P (2018). emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging. Nature methods 15, 955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DP, Shtengel G, Xu CS, Campbell KR, Freeman M, Wang L, Milkie DE, Pasolli HA, Iyer N, Bogovic JA, et al. (2020). Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367, eaaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- Ito G, Okai T, Fujino G, Takeda K, Ichijo H, Katada T, and Iwatsubo T (2007). GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson’s disease. Biochemistry 46, 1380–1388. [DOI] [PubMed] [Google Scholar]
- Jasnin M, Asano S, Gouin E, Hegerl R, Plitzko JM, Villa E, Cossart P, and Baumeister W (2013). Three-dimensional architecture of actin filaments in Listeria monocytogenes comet tails. Proc Natl Acad Sci U S A 110, 20521–20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia LV, and Lang AE (2015). Parkinson’s disease. Lancet 386, 896–912. [DOI] [PubMed] [Google Scholar]
- Kett LR, Boassa D, Ho CC, Rideout HJ, Hu J, Terada M, Ellisman M, and Dauer WT (2012). LRRK2 Parkinson disease mutations enhance its microtubule association. Hum Mol Genet 21, 890–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, et al. (2018). Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O, Leneva N, Bykov YS, Ariotti N, Teasdale RD, Schaffer M, Engel BD, Owen DJ, Briggs JAG, and Collins BM (2018). Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature 561, 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Kuba J, Mitchels J, Hovorka M, Erdmann P, Berka L, Kirmse R, J, K.O., J, D.E.B., Goetze B, and Rigort A (2020). Advanced cryo-tomography workflow developments - correlative microscopy, milling automation and cryo-lift-out. J Microsc. [DOI] [PubMed]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Topf M, Sali A, and Wolfson HJ (2009). Inferential optimization for simultaneous fitting of multiple components into a CryoEM map of their assembly. J Mol Biol 388, 180–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Forster F, Hyman AA, Plitzko JM, and Baumeister W (2016). Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Kochovski Z, Laugks U, Meyer zum Alten Borgloh J, Chakraborty S, Pfeffer S, Baumeister W, and Lučić V (2020). Template-free detection and classification of membrane-bound complexes in cryo-electron tomograms. Nature Methods 17, 209–216. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Moser F, Pražák V, Mordhorst V, Andrade DM, Baker LA, Hagen C, Grünewald K, and Kaufmann R (2019). Cryo-SOFI enabling low-dose super-resolution correlative light and electron cryo-microscopy. Proceedings of the National Academy of Sciences 116, 4804–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rickgauer JP, Grigorieff N, and Denk W (2017). Single-protein detection in crowded molecular environments in cryo-EM images. Elife 6, e25648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigort A, Bauerlein FJ, Villa E, Eibauer M, Laugks T, Baumeister W, and Plitzko JM (2012). Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci U S A 109, 4449–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Pellarin R, Greenberg CH, Bushnell DA, Davis R, Burlingame AL, Sali A, and Kornberg RD (2015). Molecular architecture of the yeast Mediator complex. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosen DA, and Cookson MR (2016). LRRK2 at the interface of autophagosomes, endosomes and lysosomes. Mol Neurodegener 11, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko IN, Kaganovich A, Hauser DN, Beylina A, Chia R, Ding J, Maric D, Jaffe H, and Cookson MR (2012). The G2385R variant of leucine-rich repeat kinase 2 associated with Parkinson’s disease is a partial loss-of-function mutation. Biochem J 446, 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, and Sali A (2012). Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol 10, e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer M, Mahamid J, Engel BD, Laugks T, Baumeister W, and Plitzko JM (2017). Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J Struct Biol 197, 73–82. [DOI] [PubMed] [Google Scholar]
- Schaffer M, Pfeffer S, Mahamid J, Kleindiek S, Laugks T, Albert S, Engel BD, Rummel A, Smith AJ, Baumeister W, et al. (2019a). A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nature Methods 16, 757–762. [DOI] [PubMed] [Google Scholar]
- Schaffer M, Pfeffer S, Mahamid J, Kleindiek S, Laugks T, Albert S, Engel BD, Rummel A, Smith AJ, Baumeister W, et al. (2019b). A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat 16, 757–762. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SH, Knape MJ, Boassa D, Mumdey N, Kornev AP, Ellisman MH, Taylor SS, and Herberg FW (2019). The dynamic switch mechanism that leads to activation of LRRK2 is embedded in the DFGpsi motif in the kinase domain. Proc Natl Acad Sci U S A 116, 14979–14988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JD, DeMong DE, Greshock TJ, Basu K, Dai X, Harris J, Hruza A, Li SW, Lin SI, Liu H, et al. (2017). Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity. J Med Chem 60, 2983–2992. [DOI] [PubMed] [Google Scholar]
- Sejwal K, Chami M, Remigy H, Vancraenenbroeck R, Sibran W, Sutterlin R, Baumgartner P, McLeod R, Chartier-Harlin MC, Baekelandt V, et al. (2017). Cryo-EM analysis of homodimeric full-length LRRK2 and LRRK1 protein complexes. Sci Rep 7, 8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, Lopez R, McWilliam H, Remmert M, Soding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Shang Z, Fu X, Lou X, Grigorieff N, and Nicastro D (2020). In situ structure determination at nanometer resolution using TYGRESS. Nature Methods 17, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger M, Tonelli F, Ito G, Davies P, Trost M, Vetter M, Wachter S, Lorentzen E, Duddy G, Wilson S, et al. (2016). Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EK (2006). Identification of a common genetic risk variant (LRRK2 Gly2385Arg) in Parkinson’s disease. Ann Acad Med Singapore 35, 840–842. [PubMed] [Google Scholar]
- Tegunov D, Xue L, Dienemann C, Cramer P, and Mahamid J (2020). Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.7 Å inside cells. bioRxiv. [DOI] [PMC free article] [PubMed]
- Terheyden S, Nederveen-Schippers LM, and Kortholt A (2016). The unconventional G-protein cycle of LRRK2 and Roco proteins. Biochem Soc Trans 44, 1611–1616. [DOI] [PubMed] [Google Scholar]
- Thomas B, and Beal MF (2007). Parkinson’s disease. Hum Mol Genet 16 Spec No. 2, R183–194. [DOI] [PubMed] [Google Scholar]
- Toro-Nahuelpan M, Zagoriy I, Senger F, Blanchoin L, Théry M, and Mahamid J (2020). Tailoring cryo-electron microscopy grids by photo-micropatterning for in-cell structural studies. Nature Methods 17, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, and Schulten K (2008). Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner FR, Watanabe R, Schampers R, Singh D, Persoon H, Schaffer M, Fruhstorfer P, Plitzko J, and Villa E (2020). Preparing samples from whole cells using focused-ion-beam milling for cryo-electron tomography. Nat Protoc. [DOI] [PMC free article] [PubMed]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, et al. (2007). Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet 16, 223–232. [DOI] [PubMed] [Google Scholar]
- Williamson DS, Smith GP, Acheson-Dossang P, Bedford ST, Chell V, Chen IJ, Daechsel JCA, Daniels Z, David L, Dokurno P, et al. (2017). Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1). J Med Chem 60, 8945–8962. [DOI] [PubMed] [Google Scholar]
- Wu G-H, Mitchell PG, Galaz-Montoya JG, Hecksel CW, Sontag EM, Gangadharan V, Marshman J, Mankus D, Bisher ME, Lytton-Jean AKR, et al. (2020). Multi-Scale 3D Cryo-Correlative Microscopy for Vitrified Cells. bioRxiv, 2020.2005.2021.107771. [DOI] [PMC free article] [PubMed]
- Xu M, Singla J, Tocheva EI, Chang Y-W, Stevens RC, Jensen GJ, and Alber F (2019). De novo structural pattern mining in cellular electron cryotomograms. Structure 27, 679–691. e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachs T, Schertel A, Medeiros J, Weiss GL, Hugener J, Matos J, and Pilhofer M (2020). Fully automated, sequential focused ion beam milling for cryo-electron tomography. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Fan Y, Ru H, Wang L, Magupalli VG, Taylor SS, Alessi DR, and Wu H (2019). Crystal structure of the WD40 domain dimer of LRRK2. Proc Natl Acad Sci U S A 116, 1579–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Subtomogram Analysis Workflow, Related to Figures 2 and 3
(A) Individual microtubules are automatically annotated using the filament-tracing function of Amira (TFS). The annotated microtubules (grey) are shown on a slice of a tomogram of LRRK2(I2020T)-decorated microtubules.
(B) Single microtubule averaging sampled at every 4 nm was performed using Dynamo to determine the protofilament number and polarity of individual microtubules using 4x-binned tomograms. Example cross-sections of subtomogram averages seen from the plus end (‘anti-clockwise slew’, left column) and from the minus end (‘clockwise slew’ right column). The total length of each class of microtubule unambiguously determined from LRRK2(I2020T) tomograms is shown.
(C) After sorting of microtubules according to protofilament number, larger subtomograms (70 nm) were re-extracted and averaged to obtain initial LRRK2/microtubule averages.
(D) Dynamo multi-reference alignment using two different templates: (1) LRRK2/microtubule and (2) microtubule only, using a hollow cylindrical classification mask corresponding to the LRRK2 density, resulting in averages for LRRK2-decorated 12-protofilament microtubule (class 1) and undecorated 12-protofilament microtubule (class 2). The same protocol was followed for 11-protofilament and 13-protofilament microtubules.
(E) Putative location of LRRK2’s N-terminal domains. Left: Cross-section view of the LRRK2/13-protofilament microtubule map. A weak outermost density is highlighted with a pair of blue dotted circles. Right: Cross-section view of an undecorated 13-protofilament microtubule at lower threshold. No outer density is observed in this cross-section. The dotted blue circles shown correspond to 50 nm and 67 nm in diameter, respectively, matching the peak of center-to-center distance between LRRK2-decorated microtubule bundles of ~67 nm (Figure 2E). Scale bar: 25 nm.
(F) Helical parameters of the LRRK2 helix on 12-protofilament microtubules were estimated and used for extraction of subtomograms containing the LRRK2 protomer from unbinned tomograms and used to create an initial average.
(G) 3D refinement of extracted particles was performed in RELION to create a template for subsequent classification.
(H) The refined average from (G) was used as a reference for 3D classification into three classes in RELION. Classes with high levels of noise (classes 2 and 3) were discarded.
(I,J) Particles belonging to the good class (class 1) were subjected to gold-standard refinement and post-processing in RELION using two different masks shown in grey (A and B).
(K, M) Final subtomogram averages from a LRRK2(I2020T) dimer bound to a 12-protofilament microtubule.
(L,N) FSC curves for the two maps shown in K and M, respectively.
(O,P) Local resolution of the maps shown in K and M, respectively, calculated using ResMap. Panel O is the same as Figure 3D, shown here again for clarity.
Figure S2. Integrative modeling of human LRRK2, Related to Figure 4 and STAR Methods
First, data generated by various experiments and computational methods are gathered. Second, the five C-terminal domains of LRRK2 are represented as rigid bodies and the data are translated into spatial restraints. Third, an ensemble of structures that satisfy the restraints are sampled using Multifit, an IMP module that allows for the simultaneous fitting of the rigid bodies while respecting the a priori restraints imposed. The ensemble is clustered into distinct domain arrangements and representative structures are further refined. Finally, the refined models are analyzed for accuracy and certainty (see STAR Methods for details).
Figure S3. Comparison of human LRRK2 with a bacterial Roco homolog protein, Related to Figure 4 and STAR Methods
(A) Domain organization of the human LRRK2 and Chlorobaculum tepidum Roco proteins.
(B) The ROCCOR domains of an integrative model (#909) of a monomeric human LRRK2 are shown in the same view as that in Figure 4D within the density corresponding to the protomer (dimer) used to build the model shown in white.
(C) The ROCCOR domains of the crystal structure of C. tepidum Roco monomer (PDB ID: 6HLU) were aligned to the ROCCOR region of human LRRK2 and are shown in the same orientation as in B. The LRR domain was omitted for clarity.
(D) The ROCCOR domains of the CORC-mediated dimer of human LRRK2 are shown inside the protomer’s density in the same views as in B (top) and Figure 4E (bottom). Domain names are indicated for both monomers and are distinguished by the presence or absence of an outline.
(E) The ROCCOR domains of the C. tepidum Roco dimer (PDB ID: 6HLU) are shown on the left in the same orientation as that in F above. The panel on the top shows the same structure viewed down its 2-fold symmetry axis (equivalent to that shown above for human LRRK2). Note that different rotations relate the two views of LRRK2 and the Roco protein, and that the dimer interface for the Roco protein involves both its ROC and CORC domains while the LRRK2 dimer is only mediated by its CORC domain.
(F) Distribution of RMSD values calculated between the ROCCOR of C. tepidum Roco protein and 53 clusters of LRRK2 models generated by integrative modeling. Cluster 20 has the closest structure, an arrow indicates the model used in this figure and Figure 4.
(G) Analysis of the variability in domain orientation for the cluster with the lowest RMSD against C. tepidum Roco (cluster 20). The three ROCCOR domains—COR C, CORN and ROC—are shown. The variability in domain orientation for all the models within the cluster is shown for each domain. The red arrow indicates the orientation of the domain as shown in B (model #909). Arrows matching the domain color indicate the orientations of other models in the cluster. Conformational variability of the ROC domain as observed in our models was previously described for an X-ray structure (PDB: 3DPU) where the ROC domain was solved only for one of the two monomers.
(H) Analysis of active vs inactive domains of microtubule-bound LRRK2. Left: Model of human LRRK2 with an active kinase model (same model as in Figure 4) shown inside the cryo-ET density. Right: Same model as in B modified to show the movement of the ROCCOR needed to accommodate the inactive-like kinase state. The ROCCOR was moved along with the N-terminus of the kinase to avoid clashes. The orange arrows show the movement of the ROCCOR domains, which places them outside of the density.
Scale bars: B,C,D,E: 2nm.
Figure S4. Sequence alignment used for homology modeling of LRRK2, Related to Figure 4 and STAR Methods
(A) Alignment of the LRRK2 kinase domain to the D. discoideum Roco4 kinase domain template structure PDB ID: 4F0F (bound to ADPCP).
(B) Alignment of the LRRK2 CORC (C-terminal end of ROCCOR) domain to the C. tepidum Roco ROCCOR template structure PDB ID: 3DPU.
(C) Alignment of the LRRK2 ROC and CORN to the C.tepidum Roco ROCCOR template structure PDB ID: 3DPU.
Figure S5. Experimental data, statistical inference and physical principles considered in the integrative modeling procedure, Related to Figure 4 and STAR Methods
(A) Comparative modeling templates for WD40, KIN, CORC, CORN, and ROC. The PDB template, the organism, the homology, and the relative modeled region are reported for each domain.
(B) Distance restraints and (C) excluded volumes pairs considered in the integrative modeling approach.
Figure S6. Ensemble Variability Analysis, Related to Figure 4 and STAR Methods
Analysis of the ensemble variability for WD40 (red), KIN (pink), CORC (orange), CORN (yellow) and ROC (green).
(A) Representative structure from the ensemble, with the position and orientation indicated by an arrow.
(B) Probability density function of the localization of the centroid of each domain within the density. A blue star indicates the potential location of the C-terminus of the LRR domain.
(C) Orientation variability analysis of each domain within the ensemble; each arrow indicates the orientation of the domain with respect to their center of mass colored accordingly to the domain analyzed. Scale bar: 5 nm.
Data Availability Statement
Cryo-ET structures and representative tomograms have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-20825, EMD-20826, EMD-20827 and EMD-20828. The corresponding tilt series were deposited in the Electron Microscopy Public Image Archive (EMPIAR) with the accession code EMPIAR-10377 and EMPIAR-10378. The LRRK2 models are deposited in the Protein Data Bank with accession code PDB-6XR4.