Abstract
The evolution of SARS-CoV-2 could impair recognition of the virus by human antibody-mediated immunity. To facilitate prospective surveillance for such evolution, we map how convalescent plasma antibodies are impacted by all mutations to the spike’s receptor-binding domain (RBD), the main target of plasma neutralizing activity. Binding by polyclonal plasma antibodies is affected by mutations in three main epitopes in the RBD, but longitudinal samples reveal that the impact of these mutations on antibody binding varies substantially both among individuals and within the same individual over time. Despite this inter- and intra-person heterogeneity, the mutations that most reduce antibody binding usually occur at just a few sites in the RBD’s receptor-binding motif. The most important site is E484, where neutralization by some plasma is reduced >10-fold by several mutations, including one in the emerging 20H/501Y.V2 and 20J/501Y.V3 SARS-CoV-2 lineages. Going forward, these plasma escape maps can inform surveillance of SARS-CoV-2 evolution.
Keywords: SARS-CoV-2, spike, receptor-binding domain, RBD, polyclonal immunity, antibody escape, deep mutational scanning
Graphical abstract
Greaney et al. map all mutations to the SARS-CoV-2 spike receptor-binding domain that reduce binding of longitudinally sampled polyclonal convalescent plasma. Despite heterogeneity within and between individuals, they identify site E484 as an immunodominant site on the RBD. In some individuals, mutations to this site can decrease neutralization by >100-fold.
Introduction
Neutralizing antibodies against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike are associated with protection against infection in both humans (Addetia et al., 2020; Lumley et al., 2020) and animals (Alsoussi et al., 2020; Walls et al., 2020; Zost et al., 2020a). However, other human coronaviruses undergo antigenic evolution that erodes neutralizing antibody immunity (Eguia et al., 2020). This antigenic evolution is driven by positive selection for mutations in the viral spike, particularly in regions involved in receptor binding (Kistler and Bedford, 2021; Wong et al., 2017). To monitor for similar antigenic evolution of SARS-CoV-2, it is important to determine which viral mutations impact human polyclonal antibody immunity.
A multitude of recent studies have identified viral mutations that escape monoclonal antibodies targeting the SARS-CoV-2 spike (Baum et al., 2020; Greaney et al., 2021; Li et al., 2020; Liu et al., 2020b; Starr et al., 2021; Weisblum et al., 2020). However, it remains unclear how mutations that escape specific monoclonal antibodies will affect the polyclonal antibody response elicited by infection. Several recent studies have identified viral mutations that impact neutralization by polyclonal human plasma or sera. So far, these studies have relied on either selecting viral escape mutants with reduced neutralization sensitivity (Andreano et al., 2020; Weisblum et al., 2020) or characterizing the antigenic effects of specific mutations such as those observed in circulating viral isolates (Kemp et al., 2020b; Li et al., 2020; Liu et al., 2020b; Thomson et al., 2020; Wang et al., 2021; Wibmer et al., 2021). This work has shown that single mutations to the spike’s receptor-binding domain (RBD) or N-terminal domain (NTD) can appreciably reduce viral neutralization by polyclonal plasma, sometimes by as much as 10-fold. However, these studies characterize an incomplete subset of all possible mutations and thus do not completely describe the effects of viral mutations on recognition by polyclonal antibodies.
Here, we comprehensively map how all amino-acid mutations to the SARS-CoV-2 spike RBD affect binding by the antibodies in plasma collected from convalescent individuals ∼1 to ∼3 months post-symptom onset. We focus on the RBD because prior studies have reported that RBD-binding antibodies contribute the majority of the neutralizing activity of most human plasma (Piccoli et al., 2020; Steffen et al., 2020), a result we confirm. Our complete maps of how mutations impact plasma antibody binding identify three major epitopes in the RBD. However, antibody binding from different individuals is impacted differently by mutations in these epitopes, and sometimes the impacts of mutations also change over time for longitudinal samples from the same individual. Some mutations that reduce antibody binding also reduce viral neutralization by >10-fold. The site where mutations tend to have the largest effect on binding and neutralization is E484, which unfortunately is a site where mutations are present in several emerging SARS-CoV-2 lineages (Tegally et al., 2020; Voloch et al., 2020). However, some plasmas are more affected by mutations at other sites, while others are largely unaffected by any single mutation. Overall, these systematic maps of how mutations to the SARS-CoV-2 RBD affect recognition by human antibody immunity can inform surveillance of ongoing viral evolution.
Results
RBD-targeting antibodies dominate the neutralizing activity of most convalescent plasma
We characterized 35 plasma samples longitudinally collected from 17 different SARS-CoV-2-infected individuals between 15 and 152 days post-symptom onset (Figure S1A). Prior work has shown that these samples all have RBD-binding antibodies and neutralizing activity, with a median neutralization titer 50% (NT50) of ∼250 (range of 34 to >10,000) against lentiviral particles pseudotyped with the D614 variant of the SARS-CoV-2 spike. For most of the 17 individuals, both the RBD binding and the neutralizing activity decreased moderately from 1 to 4 months post-infection (Table S1) (Crawford et al., 2020a). We previously characterized binding of the IgG, IgA, and IgM antibody isotypes to RBD for all the plasma samples (Crawford et al., 2020a), and those data are re-plotted in Figure S1B.
Several recent studies have reported that RBD-binding antibodies contribute the majority of the neutralizing activity in most convalescent human plasma (Piccoli et al., 2020; Steffen et al., 2020). To confirm the importance of anti-RBD antibodies for the samples in our study, we used RBD-conjugated beads to deplete the plasma of RBD-binding antibodies and compared the neutralizing activity pre- and post-depletion. First, we validated that these depletions effectively removed RBD-directed antibodies but not other anti-spike antibodies. To do this, we created “synthetic” sera by combining non-neutralizing pre-pandemic sera with either an RBD-binding or NTD-binding monoclonal antibody. As expected, RBD antibody depletion completely eliminated binding of the anti-RBD synthetic sera to both RBD and spike but did not reduce the spike-binding activity of the anti-NTD synthetic sera (Figure S1C).
We then validated that the depletion removed all RBD-binding antibodies from the convalescent human plasma and examined how depletion of RBD-binding antibodies affected total plasma binding to the spike ectodomain (Figures 1A and S1D). To do this, we performed ELISAs for binding to the RBD and spike for each sample both pre- and post-depletion, using an anti-IgG secondary antibody. The depletion removed essentially all RBD-binding IgG antibodies but only modestly decreased the amount of IgG that bound to spike (Figures 1A and S1D). This result suggests that RBD-binding antibodies comprise a relatively modest proportion of all spike-binding IgG plasma antibodies in naturally infected individuals, consistent with studies reporting that less than half of spike-reactive B cells and monoclonal antibodies bind to RBD (Brouwer et al., 2020; Huang et al., 2020; Seydoux et al., 2020; Voss et al., 2020).
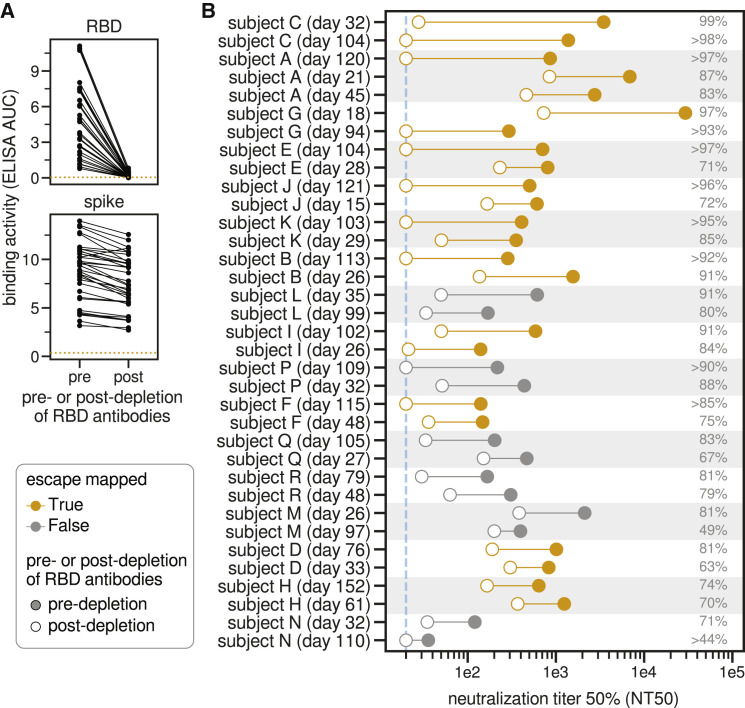
Figure 1.
RBD-binding antibodies are responsible for most of the neutralizing activity of human polyclonal plasma
(A) Change in binding of plasma to RBD and spike before and after depletion of RBD antibodies, measured by ELISA area under the curve (AUC). The dashed orange line is binding of pre-pandemic pooled sera collected in 2017 and 2018. Raw ELISA binding curves in Figure S1A.
(B) Neutralization titer 50% (NT50) of human plasma before and after depletion of RBD-binding antibodies. Legend is at left: filled and open circles are pre- and post-depletion samples, respectively, connected by a line. Orange indicates plasma for which we subsequently mapped mutations that reduce binding. The numbers at right indicate the percent of all neutralizing activity attributable to RBD-binding antibodies. Plasma are sorted in descending order of percent of neutralization due to RBD-binding antibodies, first by subject and then within subject. The dashed blue line is the limit of detection (NT50 of 20). Points on this line have an NT50 of 20, so the percent of neutralization due to RBD-binding antibodies may be an underestimate for these plasmas.
See Figure S1 and Table S1 for additional data including sample metadata, full ELISA and neutralization curves, and numerical values plotted here.
We next measured how depletion of RBD-binding antibodies affected neutralization of lentiviral particles pseudotyped with the now-predominant G614 variant of the SARS-CoV-2 spike and found that RBD-binding antibodies usually dominated the neutralizing activity (Figures 1B and S1C; Table S1). Specifically, the majority of the neutralizing activity was due to RBD-binding antibodies in nearly all samples (33 of 35 tested), and >90% of neutralizing activity was due to RBD-binding antibodies in over one-third of the samples (13 of 35 tested) (Figures 1B and S1E).
Notably, RBD-binding antibodies dominated the plasma neutralizing activity both at early (∼30 day) and late (∼100 day) time points post-symptom onset. For many individuals, the contribution of RBD-binding antibodies to plasma neutralizing activity increased over time, although this was not always the case (Figures 1B and S1F). For instance, the contribution of RBD-binding antibodies to neutralization increased over time for subjects E and J, but not subjects L or M. The strong contribution of RBD-binding antibodies to plasma neutralization demonstrates that mapping mutations that escape these antibodies is crucial for understanding the potential for SARS-CoV-2 antigenic evolution.
Complete mapping of RBD mutations that reduce binding by plasma collected ∼1 month post-symptom onset
To completely map RBD mutations that reduce binding by polyclonal plasma antibodies, we extended a deep-mutational scanning method previously developed to identify mutations that escape binding by monoclonal antibodies (Greaney et al., 2021). Briefly, we used libraries of yeast that each expressed a different RBD variant on their surface. The library covered nearly all possible single amino-acid mutations to the RBD (Starr et al., 2020). We incubated these yeast libraries with polyclonal human plasma and used fluorescence-activated cell sorting (FACS) with an IgG/IgA/IgM secondary antibody to enrich for yeast expressing RBD mutants that bound appreciably less plasma antibodies than unmutagenized RBD (Figures S2A–S2C). We then used deep sequencing to measure the frequency of each RBD mutation in the initial population and the antibody-escape FACS bin. We quantified the effect of each RBD mutation on plasma antibody binding as that mutation’s “escape fraction,” which is the fraction of all yeast cells expressing RBD with that mutation that fall into the FACS escape bin. These escape fractions range from 0 (no effect on plasma antibody binding) to 1 (all cells with this mutation are in the antibody-escape bin) (Figures S2A–S2C) (Greaney et al., 2021; Starr et al., 2020). All mapping experiments were performed in biological duplicate using independently constructed RBD mutant libraries; the replicates were highly correlated (Figures S2D and S2E), and we report the average measurements across the two libraries throughout.
We began by mapping mutations that reduced binding by plasma antibodies in samples collected from 11 individuals at approximately 30 days post-symptom onset (range 15–61 days). These samples had neutralizing titers against lentiviral particles pseudotyped with the G614 variant of the SARS-CoV-2 spike that ranged from 140 to 30,000, with the extent of neutralization attributable to RBD-targeting antibodies ranging from 63% to 99% (first time point for subjects shown in orange in Figure 1B). We quantified each RBD mutation’s escape fraction and visualized the results using “escape maps,” which are logo plots where the height of each letter is proportional to that mutation’s escape fraction (Figures 2A and S2A). Interactive versions of these escape maps are at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_HAARVI_sera. The total height of the letter stacks for each site represents the sum of the escape fractions for all mutations at that site, and so can range from 0 (no effect of any mutation) to 19 (all mutations at the site have an escape fraction of 1).
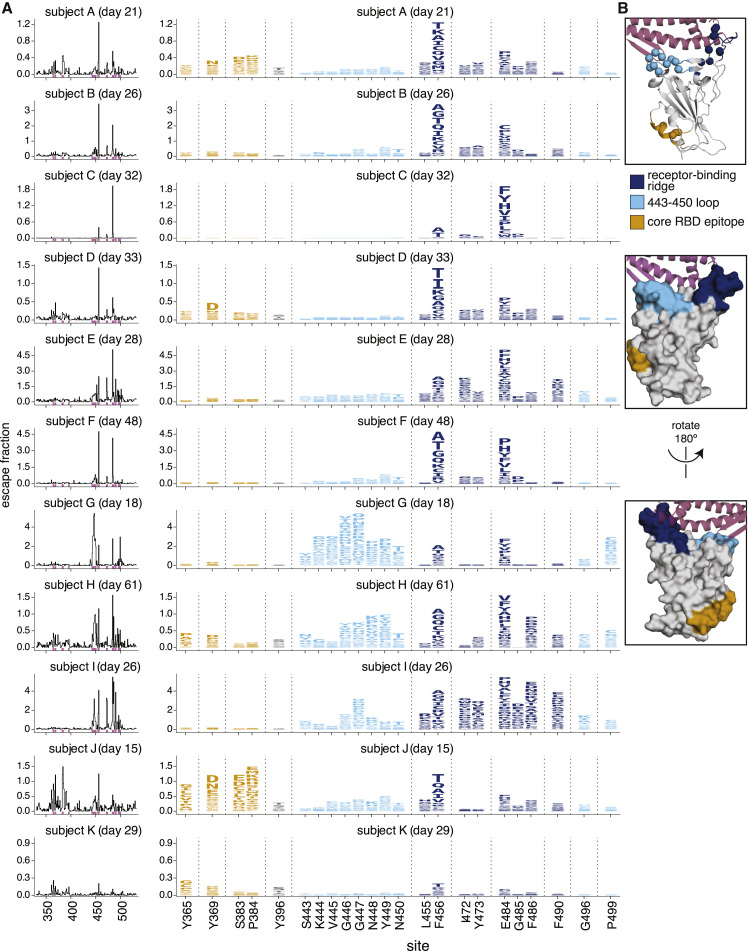
Figure 2.
Complete maps of RBD mutations that reduce binding by polyclonal plasma antibodies from 11 individuals
(A) The line plots at left indicate the total effect of all mutations at each site in the RBD on plasma antibody binding, with larger values indicating a greater reduction in antibody binding. The logo plots at right zoom in on individual mutations at key sites (indicated by purple highlighting on the x axis of the line plots). In these logo plots, the height of each letter is that mutation’s escape fraction, so larger letters indicate mutations that cause a greater reduction in antibody binding. Escape fractions are comparable across sites within a sample, but not necessarily between samples due to the use of sample-specific FACS gates—therefore, for each plasma, the y axis is scaled independently (see STAR methods). Sites in the logo plots are colored by RBD epitope.
(B) For coloring of the logo plots, we designated three RBD epitopes based on the structural locations where mutations had large effects on plasma antibody binding. The images show the structure of the RBD bound to ACE2 (PDB: 6M0J) (Lan et al., 2020) in several representations. The receptor-binding-ridge epitope is dark blue, the epitope containing the 443–450 loop is cyan, the core-RBD epitope is orange, the rest of the RBD is gray, and ACE2 is purple. For the cartoon rendering in the top structure, alpha carbons for sites of strong binding escape for any of the 11 plasma (i.e., all sites shown in the logo plots) are represented as spheres.
Interactive versions of these escape maps are available at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_HAARVI_sera/.
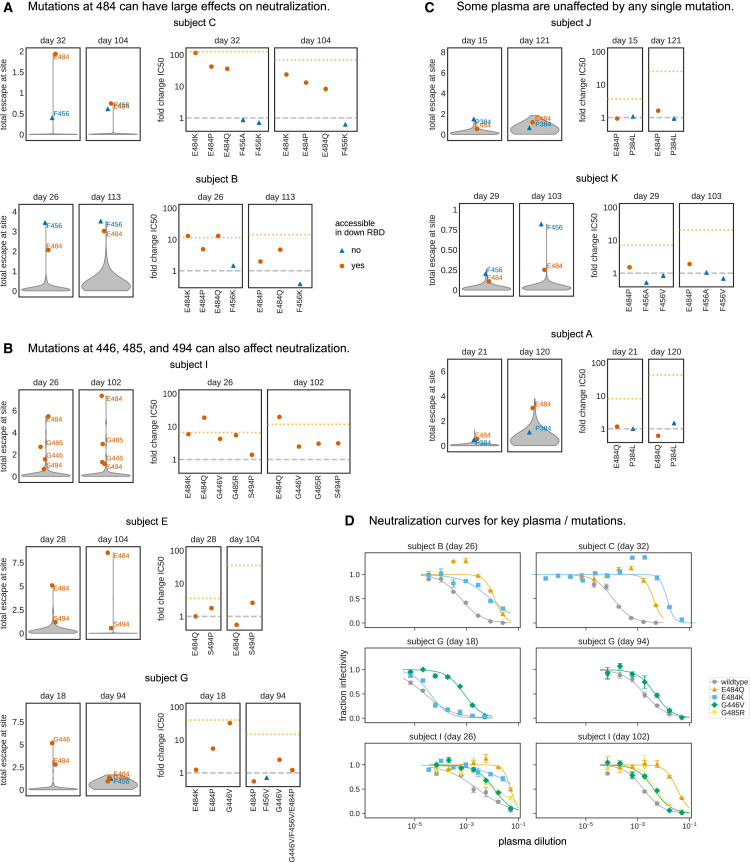
Although the effects of mutations on plasma antibody binding varied widely across individuals, the escape maps revealed several common patterns. Mutations that strongly reduced binding fell in one of three discrete regions of the RBD: the receptor-binding ridge within the receptor-binding motif (RBM), a loop in the RBM opposite the ridge (spanning sites 443–450, and the structurally adjacent sites at 494–501), or a surface patch in the core RBD (Figure 2B). The receptor-binding ridge and 443–450 loop are also targeted by many potently neutralizing antibodies, including the two antibodies in the REGN-COV2 cocktail (Greaney et al., 2021; Hansen et al., 2020; Starr et al., 2021). The core RBD epitope is targeted by monoclonal antibodies that tend to be less potently neutralizing but more broadly cross-reactive to SARS-like coronaviruses (Barnes et al., 2020a; Piccoli et al., 2020; Yuan et al., 2020b; Zost et al., 2020a). In particular, binding by all 11 samples was reduced by mutations at site F456, and binding by most samples (9 of 11) was reduced by mutations at site E484 (Figure 2A). Both of these sites are within the receptor-binding ridge epitope. Notably, E484 is a site at which mutations have recently been demonstrated to reduce neutralization by both monoclonal antibodies and human sera or plasma (Andreano et al., 2020; Greaney et al., 2021; Starr et al., 2021; Wang et al., 2021; Weisblum et al., 2020).
We grouped the samples into several classes based on which mutations most strongly reduced plasma antibody binding (Figure 3 and the interactive visualizations at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_HAARVI_sera/; sera were grouped manually based on qualitative examination of escape profiles). Binding by 6 of the 11 samples was reduced primarily by mutations in the receptor-binding ridge. For instance, binding by plasma antibodies from subject B (day 26) was most strongly affected by mutations at sites F456 and E484 (Figures 2 and 3A). Binding by three samples was strongly reduced by mutations across a broader swath of the RBM, including the 443–450 loop (Figure 3B). An example is subject G (day 18), which was strongly affected by mutations at sites 443–450 in addition to F456 and E484. Binding by two samples was most affected by mutations in the core RBD epitope (Figure 3C). The sites where mutations reduced binding by these core-RBD targeting plasma clustered around the lipid-binding pocket in the RBD, where binding of free fatty acids may contribute to locking spike into a “closed” conformation (Carrique et al., 2020; Toelzer et al., 2020). Notably, for the sample from subject K (day 29), no single RBD mutation had more than a small effect on plasma antibody binding (Figures 2 and 3D).
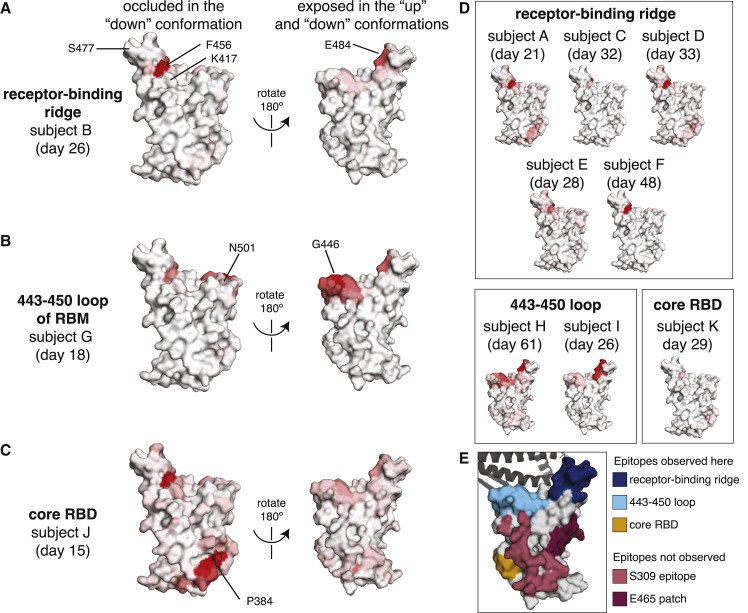
Figure 3.
Regions of the RBD where mutations strongly reduced binding by the antibodies in plasma collected from 11 individuals
The total effect of mutations at each site (sum of escape fractions) are projected onto the structure of the RBD (PDB: 6M0J), with white indicating no effect of mutations at that site and red indicating a large reduction in antibody binding. Two views of the RBD are shown: the surface of the RBD that is buried in the “down” conformation and the surface that is always exposed and accessible (Walls et al., 2020; Wrapp et al., 2020).
(A) For some individuals (typified by subject B), antibody binding is predominantly reduced by mutations in the receptor-binding ridge, particularly at sites F456 and E484.
(B) For some individuals (typified by subject G), antibody binding is strongly reduced by mutations in the 443–450 loop of the RBM in addition to the receptor-binding ridge.
(C) For a few individuals (typified by subject J), antibody binding is affected by mutations in the core RBD epitope around site P384.
(D) Samples from the other eight individuals fall in one of the three classes detailed in panels (A–C). For panels (A–D), the white-to-red coloring scale is set to span the same range as the y axis limits for that plasma in Figure 2.
(E) Mutations in two major surface regions (the S309 epitope and the sites near E465) do not strongly affect plasma antibody binding for any of the subjects. Shown is a surface representation of the RBD, with the three polyclonal plasma epitopes colored as in Figure 2. The S309 epitope and region near E465 (“E465 patch”) are shown in pink and maroon. ACE2 is shown in a dark gray cartoon representation.
Interactive versions of these structural visualizations are available at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_HAARVI_sera/.
There are some regions of the RBD where mutations did not strongly affect plasma antibody binding for any sample in our panel. These regions include the sites near the 343 glycan that are targeted by the SARS-CoV-1 cross-reactive antibody S309 (Pinto et al., 2020), and the region near residue E465 on the “lateral” side of the RBD, which to our knowledge is not an epitope for any known neutralizing antibodies (Figure 3E) (Barnes et al., 2020a; Greaney et al., 2021; Piccoli et al., 2020; Starr et al., 2020). Antibodies targeting these two regions may be rare, have low binding avidity in the context of polyclonal plasma, or be subdominant relative to other RBD epitopes (Barnes et al., 2020b; Piccoli et al., 2020; Weisblum et al., 2020).
How mutations affect plasma antibody binding can shift over time in the same individual
Next, we examined how the RBD mutations that affect plasma antibody binding change over time as an individual’s immune response matures. We speculated that such changes might occur because other studies have shown that anti-SARS-CoV-2 antibodies become more somatically hypermutated and less clonal in the months following recovery from infection (Gaebler et al., 2020; Nielsen et al., 2020; Rodda et al., 2021). Moreover, we reasoned that mapping mutations that affect plasma antibody binding several months after infection would be relevant for determining which viral mutations might alter the effectiveness of immunity if these individuals were re-exposed to a distinct SARS-CoV-2 variant in the future.
We performed escape mapping for samples collected at later time points (76–152 days post-symptom onset) from all 11 individuals for whom we had characterized plasma antibody binding at the ∼1-month time point. For some but not all individuals, there were substantive changes over time in how binding was affected by RBD mutations (Figures 4 , S3, and S4 and interactive visualizations at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_HAARVI_sera) (Hilton et al., 2020).
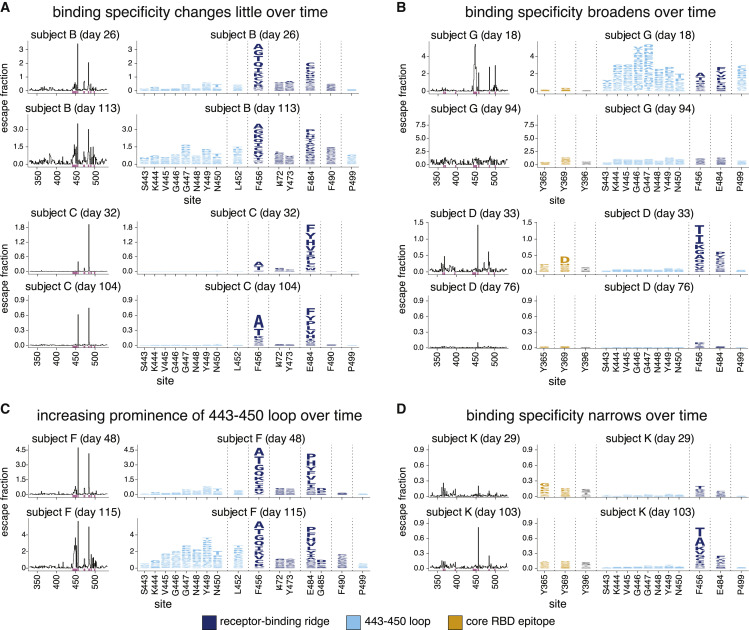
Figure 4.
The RBD mutations that affect plasma antibody binding change over time for some individuals
Escape maps, colored as in Figure 2, demonstrating temporal patterns: (A) no change over time, (B) broadening over time, (C) increasing prominence of one antigenic region, the 443–450 loop, or (D) narrowing over time. This figure shows the escape maps over time for 6 of the 11 individuals to illustrate representative trends; see Figure S3 for escape maps for all individuals at all time points. Figure S4 shows the effects of mutations at each site projected onto the RBD structure. Different sets of sites are shown in the logo plots in panels (A and C), and in panels (B and D). Sites highlighted in the logo plots are indicated in purple on the x axes of the associated line plots. The y axis limits were set as in Figure 2A (see STAR methods). Interactive versions of these visualizations are available at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_HAARVI_sera/.
Specifically, for over half of the 11 individuals, there was relatively little change in which RBD mutations affected plasma antibody binding (Figures 4A, S3, and S4). For two individuals, antibody binding became strikingly more broad and less affected by any single RBD mutation (Figures 4B, S3, and S4). For one individual, mutations in the 443–450 loop had a much larger effect on binding by plasma antibodies from the later time compared to the earlier time (Figure 4C, S3, and S4). Finally, for one individual, there was a strong narrowing of the response, with no single RBD mutation having a large effect on binding by plasma from the early time point, but mutations at F456 and to a lesser extent, E484, having large effects at the later time point (Figure 4D, S3, and S4). In summary, while the specificity of plasma antibody binding is often maintained over time, in some individuals, the specificity broadens to become relatively unaffected by any single RBD mutation, while in other individuals the specificity narrows so that single mutations have a greater impact.
However, there was no clear relationship between changes in the fine specificity of antibody binding and overall plasma neutralizing activity. For instance, subjects B and C maintained similar binding specificities over time (Figure 4A) even though the neutralization titers of both subjects’ plasma decreased (Figure 1B). Similarly, subject D showed major changes in binding specificity over time (Figure 4B), although this change in specificity was not accompanied by a substantial change in overall plasma neutralization titer.
For some plasma, RBD mutations that reduce antibody binding strongly reduce neutralization
To determine how mutations that reduced plasma antibody binding to the RBD affected viral neutralization, we characterized a subset of mutations in neutralization assays with spike-pseudotyped lentiviral particles. For these assays, we chose mutations that our mapping showed had substantial effects on plasma antibody binding by samples from multiple individuals, and prioritized mutations present in circulating isolates of SARS-CoV-2.
In many cases, single mutations that were mapped to strongly reduce plasma antibody binding also greatly reduced viral neutralization. The effect of mutations at site E484 were particularly striking (Figures 5A and 5B). For several plasmas, the neutralization titer dropped by over an order of magnitude against viruses pseudotyped with spikes with E484 mutated to K, Q, or P. For instance, these three mutations to E484 caused 35- to 115-fold decreases in the neutralization titer of the plasma collected from subject C (day 32) (Figures 5A and 5D). As another example, both E484K and E484Q reduced neutralization by the plasma from subject B (day 26) by 10-fold, the same reduction achieved by depleting the plasma of all RBD-binding antibodies (Figures 5A and 5D).
Figure 5.
Mutations mapped to reduce plasma antibody binding often reduce viral neutralization
(A–C) Violin plots at left show the distribution of how mutations at all sites in the RBD affect plasma binding in the mapping experiments. The plots at right then show the effects of tested mutations on neutralization (the fold-change in neutralization inhibitory concentration 50% [IC50]). For instance, the top row in (A) shows that mutations at E484 and F456 are mapped to reduce plasma antibody binding for subject C at both days 32 and 104, and that multiple different mutations at E484 but not F456 greatly reduced plasma neutralization (e.g., a >100-fold increase in IC50 for E484K for the day-32 plasma). Sites that are accessible in the down conformation of the RBD in the context of full spike are indicated by red circles (e.g., E484), and sites that are inaccessible in the RBD’s down conformation are indicated by blue triangles (e.g., F456). In the plots showing the fold-change in IC50s, the dashed gray line indicates a value of one (no change in neutralization), and the dotted orange line indicates the change in inhibitory concentration if all RBD-binding antibodies are removed (see Figure 1B).
(D) Full neutralization curves for a subset of plasma and viral mutants demonstrating how E484Q, E484K, G446V, and G485R substantially reduce viral neutralization for some plasma. Error bars are the standard error for n = 2 replicates.
For all neutralization curves used to determine changes in neutralization plotted in (A–C), see Figure S5. The y axis limits in the violin plots are set as the maximum of the y axis limit for all time points of a subject in the escape maps in Figures 2A and S3. Numerical IC50 values and fold-change IC50 relative to wildtype are listed at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/experimental_validations/results/mutant_neuts_results/mutants_foldchange_ic50.csv.
While mutations at E484 generally caused the largest drops in neutralization, other mutations mapped to decrease antibody binding for specific plasma also affected neutralization. A dramatic example was G446V, which caused a ∼30-fold decrease in the neutralization titer of subject G (day 18) (Figures 5B and 5D). Mutations G485R and S494P also caused lesser but still appreciable (∼3- to 5-fold) decreases in neutralization titer for a few plasmas (Figure 5B). However, no single mutation completely abrogated neutralization for any of the plasma samples (Figure S5C).
In general, there was good concordance between the mapping of how mutations affected plasma antibody binding and their impact on viral neutralization. This concordance can be seen in Figures 5A–5C, where the violin plots show the distributions of the effects of mutations on plasma antibody binding across all sites. The sites in the upper tails of these violin plots are ones where mutations had large effects on binding, and mutations to such sites usually reduced neutralization. The one major exception was site F456, where mutations often caused large reductions in binding to yeast-displayed RBD but never appreciably affected neutralization (Figures 2A and 5A–5C). This discrepancy is not because antibodies targeting this region are inherently non-neutralizing or unaffected by mutations at site 456, as F456A and F456K disrupt neutralization by two monoclonal antibodies with epitopes that include F456 (Figure S5B) (Greaney et al., 2021; Shi et al., 2020; Starr et al., 2021; Zost et al., 2020a). Rather, we hypothesize that the discrepancy is because we mapped how mutations affected binding using isolated RBD, but in the native viral context of full spike, RBD can be positioned in either a “down” or “up” conformation (Walls et al., 2020; Wrapp et al., 2020). All sites where mutations that reduced binding also affected neutralization are accessible in both conformations, but F456 is only accessible in the up conformation. Because the RBD is usually in the down conformation (Cai et al., 2020; Ke et al., 2020; Walls et al., 2020), we speculate that sites accessible only in the up conformation may be subdominant in the context of polyclonal plasma neutralization of full spike even if they are dominant when assaying binding to isolated yeast-displayed RBD.
The neutralization assays also validated one of the most notable findings from the mapping: that the antigenic effects of mutations varied markedly across samples from different individuals. For several samples, the maps of binding escape were relatively “flat” with no mutations having large effects, and for these samples, no tested mutations substantially affected neutralization (Figure 5C). Additionally, sometimes the effects of mutations changed over time for the same individual. Such a temporal change was especially notable for subject G: mapping of the day-18 sample showed a strong effect of mutations centered around G446, but by day 94, the escape map had flattened (Figure 4B). Concordant with the maps, G446V greatly decreased neutralization by subject G’s day-18 plasma, but had only a modest effect on the day-94 plasma, even when combined with mutations at several other key sites (Figures 5B and 5D). These facts highlight how the antigenic effects of mutations vary across people and time and suggest that some plasmas are more resistant than others to erosion by viral evolution.
RBD mutations that reduce plasma binding and neutralization in circulating SARS-CoV-2 isolates
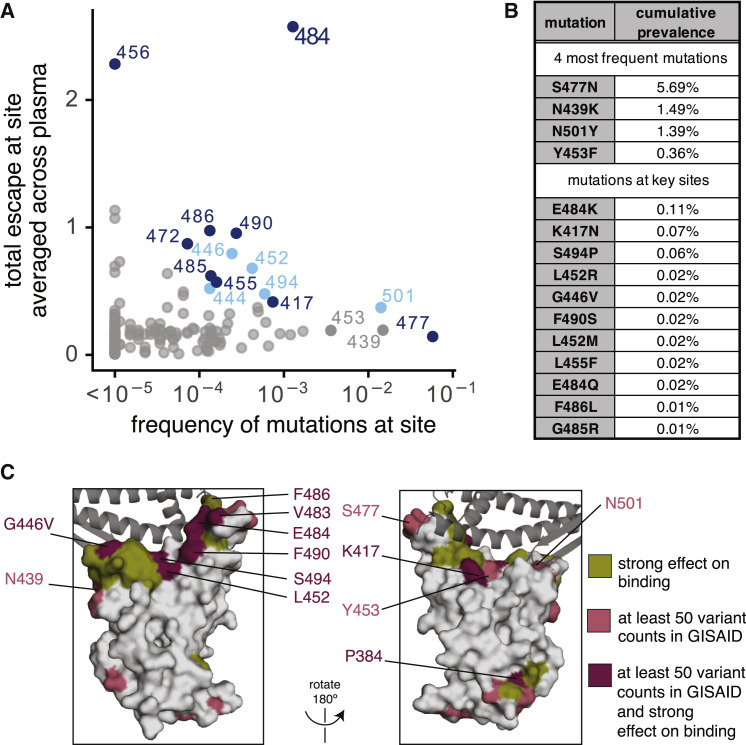
To determine the extent that mutations we mapped to affect plasma binding are present among circulating SARS-CoV-2 isolates, we determined the frequency of mutations at each RBD site among all SARS-CoV sequences in GISAID as of December 23, 2020 (Elbe and Buckland-Merrett, 2017). We then compared these frequencies to the effects of mutations at each site on plasma antibody binding, averaged across all samples (Figure 6 A).
Figure 6.
Frequencies of mutations that affect plasma antibody binding among circulating SARS-CoV-2 isolates
(A) Effects of mutations at each RBD site on plasma antibody binding versus frequency of mutations at each site among all SARS-CoV-2 sequences in GISAID as of December 23, 2020. Key sites (see STAR methods) are labeled and colored according to epitope region as in Figure 2.
(B) Cumulative prevalence for the four most frequent mutations and also any mutations at sites labeled in (A) with at least ten counts in GISAID.
(C) Surface representations of the RBD (PDB: 6M0J). Sites where mutations have a strong effect on binding, have circulating variation with >50 total counts in GISAID, or both, are colored in olive, pink, or maroon, respectively. See STAR methods for precise description of highlighted sites. ACE2 is shown as a dark gray cartoon.
The most concerning site of mutations is E484 (Figure 6A). E484 is the site where mutations tend to have the largest effect on plasma antibody binding to the RBD, and our neutralization assays (Figures 5A and 5D) and similar experiments by others (Andreano et al., 2020; Liu et al., 2020b; Weisblum et al., 2020) show that mutations to site E484 reduce the neutralization potency of some human plasmas by >10-fold, although other plasmas are unaffected by mutations at this site. Over 0.1% of all sequenced isolates have mutations at this site. Of note, E484K is present in the 20H/501Y.V2 viral lineage (also known as B.1.351, originally identified in South Africa) and the 20J/501Y.V3 viral lineage (also known as P.1, originally identified in Brazil) (Faria et al., 2021; Tegally et al., 2020; Voloch et al., 2020); another mutation at the same site (E484Q) has also been found in a smaller number of human isolates (Figure 6B). Consistent with the observation of E484K/Q mutation in naturally occurring SARS-CoV-2 isolates, these mutations have neutral to very mildly beneficial effects on RBD affinity for ACE2 (Starr et al., 2020). The 20H/501Y.V2 and 20J/501Y.V3 lineages have two other RBD mutations, N501Y and K417N or K417T, respectively, that co-occur with E484K (Faria et al., 2021; Tegally et al., 2020). K417N escapes neutralization by some monoclonal antibodies (Greaney et al., 2021; Starr et al., 2021), but mutations to site 417 only modestly affected binding by a few of the samples we assayed (the largest effects were for the last time point for subjects A and J; see Figure S3). N501Y increases affinity for ACE2 (Starr et al., 2020) and is also present in the 20I/501Y.V1 lineage (also known as B.1.1.7, originally identified in the UK) that may have increased transmissibility (Kemp et al., 2020a; Public Health England, 2020; Rambaut et al., 2020). Although other mutations at N501 have modest effects on binding by some monoclonal antibodies (Greaney et al., 2021; Starr et al., 2021), mutations at N501 do not strongly affect binding by any plasma we tested (Figures 2A and S3). However, several recent studies suggest that the combination of K417N + E484K + N501Y may cause a larger decrease in neutralization than any of these mutations alone (Cele et al., 2021; Wang et al., 2021; Wibmer et al., 2021).
Several other sites where we mapped mutations to affect plasma antibody binding for a few samples also have low-level variation (<0.1%) among circulating viruses (Figures 6A–6C). These include site G446, where the G446V mutation reduced neutralization by one sample by >10-fold (Figures 5B and 5D). Other key sites with circulating variation where mutations impact binding by some samples are indicated in Figure 6. Notably, site F456, where mutations consistently affect plasma antibody binding but not neutralization, has little variation among circulating viruses (Figure 6A)—perhaps in part because most mutations to site F456 decrease RBD affinity for ACE2 (Starr et al., 2020).
The four mutations at the highest frequency among sequenced viruses (S477N, N439K, N501Y, and Y453F; see Figure 6B) do not strongly affect plasma antibody binding to yeast-displayed RBD by any samples we tested. As mentioned above, N501Y increases affinity for ACE2, is present in the 20I/501Y.V1 (B.1.1.7) lineage (Kemp et al., 2020a), and is in the epitope defined by the 443–450 loop (Figures 2B and 3B)—but does not impact binding by any samples we tested, a result corroborated by live-virus neutralization assays (Muik et al., 2021; Rees-Spear et al., 2021). Y453F and N439K both also increase affinity for ACE2 (Starr et al., 2020; Thomson et al., 2020), and both escape some monoclonal antibodies (Baum et al., 2020; Starr et al., 2021; Thomson et al., 2020), but neither greatly impacts plasma antibody binding by the samples we tested. Finally, S477N also reduces neutralization by some monoclonal antibodies (Liu et al., 2020b) but did not greatly affect binding by the samples we tested.
In summary, our results suggest that E484 is the site of most concern for viral mutations that impact binding and neutralization by polyclonal plasma antibodies targeting the RBD. However, mutations at the other plasma antibody epitopes (e.g., the 443–450 loop, site 417, and residues around 484 such as 455, 485, 486, and 490) also have antigenic impacts.
Discussion
We comprehensively mapped how mutations to the SARS-CoV-2 RBD affected binding by the antibodies in convalescent human plasma. One major result is that plasma antibody binding is predominantly affected by mutations at just a few dominant epitopes in the RBD. In particular, E484 is the site in the RBD where mutations usually have the largest effect on binding and neutralization—possibly because E484 is often targeted by antibodies that utilize heavy-chain germline genes that are common among anti-SARS-CoV-2 RBD antibodies, IGHV3-53, and IGHV3-66 (Barnes et al., 2020a; Greaney et al., 2021; Robbiani et al., 2020; Weisblum et al., 2020; Yuan et al., 2020a; Zost et al., 2020b). Mutations at other structurally adjacent sites in the RBD’s receptor-binding ridge (e.g., L455, F456, G485, F486, and F490) can also have substantial antigenic effects. Another major epitope is centered on the loop formed by residues 443–450 in the RBD’s RBM, and mutations in this epitope sometimes strongly affect plasma antibody neutralization. A third epitope is in the core of the RBD distal from the RBM, although mutations here tend to have smaller effects on plasma antibody binding. Notably, RBD mutations reported by other studies to have large effects on plasma neutralization are also in the epitope centered around E484 or in the 443–450 loop (Andreano et al., 2020; Li et al., 2020; Wang et al., 2021; Weisblum et al., 2020; Wibmer et al., 2021).
While the major plasma epitopes are targeted by many characterized monoclonal antibodies (Barnes et al., 2020a; Baum et al., 2020; Greaney et al., 2021; Hansen et al., 2020; Starr et al., 2021), there are also sites where mutations that escape monoclonal antibodies have little effect on plasma antibody binding for any sample we tested. For instance, mutations in the S309 epitope footprint (Pinto et al., 2020) and at sites of escape from antibody C135 (e.g., R346 and N440) (Barnes et al., 2020a; Weisblum et al., 2020) had minimal effects on plasma antibody binding (Figures 3 and S3). This lack of concordance between the epitopes of plasma and monoclonal antibodies is consistent with other studies reporting that the specificities of potent monoclonal antibodies often do not recapitulate the plasma from which they were isolated (Barnes et al., 2020b; Weisblum et al., 2020). These antibodies may be rare in polyclonal plasma or the epitopes they target may be subdominant (Piccoli et al., 2020). However, subdominant epitopes may become more important as SARS-CoV-2 evolves: after mutations at immunodominant sites such as E484 partially erode plasma antibody neutralization, the remaining neutralization is presumably due to antibodies targeting previously subdominant epitopes.
Another key finding is that there is extensive person-to-person variation in how mutations affect plasma antibody binding and neutralization. For instance, the neutralizing activity of several samples was reduced by >10-fold by single mutations to site E484, but a few samples were essentially unaffected by E484 mutations. Similarly, mutations at sites in the 443–450 loop (e.g., G446V) caused a large drop in plasma antibody binding and neutralization for some samples but had little effect on others. This inter-individual heterogeneity is further compounded by the fact that the effects of mutations sometimes changed over time for samples longitudinally collected from the same individual. These temporal changes could be due to a disproportionate decay in one dominant antibody clonotype, or a relative increase in antibodies targeting other epitopes (Gaebler et al., 2020).
There are several limitations to our study. Most importantly, we only examined mutations to the RBD. While we and others (Piccoli et al., 2020; Steffen et al., 2020) have shown that RBD-binding antibodies contribute the majority of the plasma neutralizing activity of most convalescent human sera and plasma, antibodies also target other regions of the spike. For example, mutations and deletions in the NTD can affect plasma antibody neutralization (Andreano et al., 2020; Kemp et al., 2020b; Liu et al., 2020a; McCarthy et al., 2020; Voss et al., 2020). In addition, we only mapped samples from 11 individuals at two time points. Given the substantial inter- and intra-individual heterogeneity, mapping more samples may identify additional sites of importance. On a technical level, we assayed binding of antibodies to isolated RBD expressed by yeast, which implies several limitations. First, we are unable to map the effects of mutations that alter the spike’s overall conformation or affect antibodies spanning quaternary epitopes (Barnes et al., 2020a). Second, our mapping likely overestimates the contributions of antibodies that bind epitopes that are more accessible on isolated RBD than in the context of full spike (e.g., F456). Finally, the N-linked glycans on yeast-expressed proteins are more mannose-rich than those on mammalian-expressed proteins (Hamilton et al., 2003), which could affect measurements of how N-linked glycans affect antibody binding. However, the general consistency of our mapping with our pseudovirus neutralization assays and the plasma-escape mutations reported by others suggest that our study successfully defines the major RBD epitopes of convalescent human plasma antibodies.
The comprehensive nature of our mapping makes it possible to assess which circulating RBD mutations are likely to have the greatest impact on human immunity. In particular, our results predict that the emerging 20H/501Y.V2 and 20J/501Y.V3 viral lineages (originally identified in South Africa and Brazil, respectively) carrying the E484K mutation will have reduced susceptibility to neutralization by the polyclonal plasma antibodies of some individuals. In contrast, the N501Y mutation present in the 20I/501Y.V1 lineage (B.1.1.7) is unlikely to greatly affect neutralization by most human plasma, although it could contribute to increased viral titer or enhanced transmissibility (Kidd et al., 2020; Public Health England, 2020). The NTD deletions in this lineage, however, may have an antigenic effect (Andreano et al., 2020; Kemp et al., 2020b; McCarthy et al., 2020). Notably, very recent studies on viruses from these lineages largely confirm these predictions from our mapping (Cele et al., 2021; Rees-Spear et al., 2021; Wibmer et al., 2021). More generally, our mapping can be used to assess the likely antigenic impacts of additional viral mutations that emerge in the future.
Our mapping also reveals broader features of antibody immunity that are relevant to SARS-CoV-2 evolution. One reason that influenza virus undergoes such rapid antigenic evolution is that neutralizing human immunity often focuses on just a few residues in hemagglutinin, such that a single mutation can dramatically reduce neutralization (Lee et al., 2019). In contrast, antibody immunity to measles virus targets multiple co-dominant measles epitopes, meaning that no single mutation has a large effect on neutralization (Muñoz-Alía et al., 2020). Our results show that polyclonal antibody immunity to the SARS-CoV-2 RBD is sometimes focused as for influenza, but in other cases more broadly targets the RBD in a way that mitigates the effect of any single mutation. This heterogeneity in the antigenic impacts of RBD mutations implies that the immunity of different individuals will be impacted differently by viral evolution. It also suggests that an important area for future work is understanding how viral mutations impact vaccine-elicited immunity, and using this knowledge to design vaccines that are robust to viral antigenic evolution.
STAR★methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| COV2-2832 | Zost et al., 2020a | N/A |
| COV2-2165 | Zost et al., 2020a | N/A |
| COV2-2082 | Zost et al., 2020a | N/A |
| rREGN10987 | Starr et al., 2021 | NA |
| r4A8 | This paper | NA |
| FITC-conjugated chicken anti-cMyc antibody | Immunology Consultants Laboratory, Inc. | Cat# CMYC-45F |
| Alexa-647-conjugated Goat Anti-Human IgA+IgG+IgM | Jackson Immuno Research Laboratories | Cat# 109-605-064; RRID: AB_2337886 |
| DyLight-405-conjugated Goat Anti-Human IgA+IgG+IgM | Jackson Immuno Research Laboratories | Cat# 109-475-064; RRID: AB_2337792 |
| Goat anti-human IgG-Fc horseradish peroxidase (HRP) | Bethyl Labs | Cat# A80-104P; RRID: AB_67064 |
| Biological samples | ||
| Normal Human Serum, collected 2017-2018 and pooled from 75 donors | Gemini Biosciences | Cat# 100–110, lot H86W03J |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 S2Pecto | Dingens et al., 2020 | N/A |
| SARS-CoV-2 RBD | Dingens et al., 2020 | NA |
| SARS-CoV-2 (COVID-19) Spike protein RBD-coupled magnetic beads | AcroBiosystems | Cat# MBS-K002 |
| Critical commercial assays | ||
| Zymoprep Yeast Plasmid Miniprep II | Zymo Research | Cat# D2004 |
| TMB/E HRP substrate | Millipore Sigma | Cat# ES001 |
| Bright-Glo Luciferase Assay System | Promega | Cat# E2620 |
| HIV-1 p24 Antigen Capture Assay | Advanced Bioscience Laboratories | Cat# 5421 |
| Deposited data | ||
| PacBio CCSs linking variants to barcodes | Starr et al., 2020 | NCBI SRA: BioProject PRJNA639956, BioSample: SAMN15295683 |
| Illumina barcode sequencing | This paper | NCBI SRA: BioProject: PRJNA639956, BioSample: SAMN17185313 |
| ACE2-bound RBD crystal structure | Lan et al., 2020 | PDB: 6M0J |
| GISAID EpiCoV SARS-CoV-2 sequence isolates | GISAID | Full list of contributing labs and accessions: https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/data/gisaid_hcov-19_acknowledgement_table_2020_12_30.pdf |
| Experimental models: cell lines | ||
| Human: Embryonic Kidney (HEK293T) | ATCC | ATCC CRL-3216 |
| Human: Embryonic Kidney cells expressing human ACE2 (HEK293T-hACE2) | BEI Resources | BEI NR-52511 |
| Saccharomyces cerevisiae strain AWY101 | Wentz and Shusta, 2007 | AWY101 |
| Recombinant DNA | ||
| Plasmid: pETcon_SARS-CoV-2_RBD | Starr et al., 2020 | sequence at https://github.com/jbloomlab/SARS-CoV-2-RBD_DMS/tree/master/data/plasmid_maps/2649_pETcon-SARS-CoV-2-RBD-201aa.gb |
| Plasmid: HDM_Spikedelta21_D614G | Addgene | Addgene #158762 |
| Plasmid: HDM-Hgpm2 gag/pol lentiviral helper plasmid | BEI Resources | BEI NR-52517 |
| Software and algorithms | ||
| dms_variants, version 0.8.5 | GitHub | https://jbloomlab.github.io/dms_variants/ |
| dmslogo, version 0.6.2 | GitHub | https://jbloomlab.github.io/dmslogo/ |
| dms-view | Hilton et al., 2020 | https://dms-view.github.io/docs/ |
| neutcurve, version 0.5.2 | GitHub | https://jbloomlab.github.io/neutcurve/ |
| custom code | This paper | all analyses provided on github: https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera |
| Other | ||
| SARS-CoV-2 RBD mutant libraries | Starr et al., 2020 | N/A |
| Infinite M1000Pro plate reader | Tecan | N/A |
| BD FACSAria II | BD Biosciences | N/A |
Resource availability
Lead contact
Further information and requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact, Jesse Bloom (jbloom@fredhutch.org).
Materials availability
SARS-CoV-2 mutant libraries used in this study will be made available on request by the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
We provide data and code in the following ways:
The complete code for the full computational data analysis pipeline of the mapping experiments is available on GitHub at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera
A Markdown summary of the analysis workflow with renderings of all the code is on GitHub at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/results/summary/summary.md
The escape fraction measured for each mutation in Table S3 and also at https://raw.githubusercontent.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/main/results/supp_data/human_sera_raw_data.csv
The barcode counts for each RBD variant in each mapping condition are at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/results/counts/variant_counts.csv
All raw sequencing data are available on the NCBI Short Read Archive at BioProject: PRJNA639956 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA639956), BioSample: SAMN17185313 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN17185313).
The patient metadata and neutralization titers of convalescent plasma pre- and post-depletion of RBD-binding antibodies is available in Table S1 and online at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/experimental_validations/results/rbd_absorptions/TableS1.csv
Experimental model and subject details
SARS-CoV-2 convalescent human plasma
Plasma samples were previously described (Crawford et al., 2020a) and collected as part of a prospective longitudinal cohort study of individuals with SARS-CoV-2 infection in Seattle, WA February-July 2020. The plasma from 17 individuals were studied here (8/17 female; age range 23–76 years, mean 51.6 years, median 56 years). See Figure S1A for available clinical information and Table S1 for the sample metadata, which is also described in (Crawford et al., 2020a). That table also links the sample IDs used in (Crawford et al., 2020a) to the names used for the plasma in this paper. All plasma were heat-inactivated prior to use by treatment at 56 C for 60 min. Prior to use in each assay, plasma samples were centrifuged for 15 min at 2000 xg to pellet platelets. This study was approved by the University of Washington Human Subjects Institutional Review Board.
Cell lines
HEK293T (ATCC CRL-3216) cells were used to generate SARS-CoV-2 spike-pseudotyped lentiviral particles and 293T-ACE2 cells (BEI NR-52511) were used to titer the SARS-CoV-2 spike-pseudotyped lentiviral particles and to perform neutralization assays (see Method details).
Method details
RBD deep mutational scanning library
The yeast-display RBD mutant libraries are previously described (Greaney et al., 2021; Starr et al., 2020). Briefly, duplicate mutant libraries were constructed in the spike receptor binding domain (RBD) from SARS-CoV-2 (isolate Wuhan-Hu-1, GenBank accession number MN908947, residues N331-T531) and contain 3,804 of the 3,819 possible amino-acid mutations, with > 95% present as single mutants. Each RBD variant was linked to a unique 16-nucleotide barcode sequence to facilitate downstream sequencing. As previously described, libraries were sorted for RBD expression and ACE2 binding to eliminate RBD variants that are completely misfolded or non-functional (i.e., lacking modest ACE2 binding affinity) (Greaney et al., 2021).
FACS sorting of yeast libraries to select mutants with reduced binding by polyclonal plasma
Plasma mapping experiments were performed in biological duplicate using the independent mutant RBD libraries, as previously described for monoclonal antibodies (Greaney et al., 2021), with the following modifications: Mutant yeast libraries induced to express RBD were washed and incubated with plasma at a range of dilutions for 1 h at room temperature with gentle agitation. For each plasma, we chose a sub-saturating dilution such that the amount of fluorescent signal due to plasma antibody binding to RBD was approximately equal across plasma. The exact dilution used for each plasma is given in Table S2. After the plasma incubations, the libraries were secondarily labeled with 1:100 FITC-conjugated anti-MYC antibody (Immunology Consultants Lab, CYMC-45F) to label for RBD expression and 1:200 Alexa-647- or DyLight-405-conjugated goat anti-human-IgA+IgG+IgM (Jackson ImmunoResearch 109-605-064 or 109-475-064, respectively) to label for bound plasma antibodies. A flow cytometric selection gate was drawn to capture 3%–6% of the RBD mutants with the lowest amount of plasma binding for their degree of RBD expression (Figures S1A–S1C). We also measured what fraction of cells expressing unmutated RBD fell into this gate when stained with 1x and 0.1x the concentration of plasma. For each sample, approximately 10 million RBD+ cells (range 7.4e6 to 1.7e7 cells) were processed on the cytometer, with between 2e5 and 8e5 plasma-escaped cells collected per sample (see percentages in Table S2). Antibody-escaped cells were grown overnight in SD-CAA (6.7g/L Yeast Nitrogen Base, 5.0g/L Casamino acids, 1.065 g/L MES acid, and 2% w/v dextrose) to expand cells prior to plasmid extraction.
DNA extraction and Illumina sequencing
Plasmid samples were prepared from 30 OD units (1.6e8 cfu) of pre-selection yeast populations and approximately 5 OD units (∼3.2e7 cfu) of overnight cultures of plasma-escaped cells (Zymoprep Yeast Plasmid Miniprep II) as previously described (Greaney et al., 2021). The 16-nucleotide barcode sequences identifying each RBD variant were amplified by PCR and prepared for Illumina sequencing as described in (Starr et al., 2020). Barcodes were sequenced on an Illumina HiSeq 3500 with 50 bp single-end reads. To minimize noise from inadequate sequencing coverage, we ensured that each antibody-escape sample had at least 2.5x as many post-filtering sequencing counts as FACS-selected cells, and reference populations had at least 2.5e7 post-filtering sequencing counts.
Analysis of deep sequencing data to compute each mutation’s plasma escape fraction
Escape fractions were computed as described in (Greaney et al., 2021), with minor modifications as noted below. We used the dms_variants package (https://jbloomlab.github.io/dms_variants/, version 0.8.5) to process Illumina sequences into counts of each barcoded RBD variant in each pre-sort and antibody-escape population using the barcode/RBD look-up table from (Starr et al., 2021).
For each plasma selection, we computed the “escape fraction” for each barcoded variant using the deep sequencing counts for each variant in the original and plasma-escape populations and the total fraction of the library that escaped antibody binding via the formula provided in (Greaney et al., 2021). These escape fractions represent the estimated fraction of cells expressing that specific variant that fall in the plasma escape bin, such that a value of 0 means the variant is always bound by plasma and a value of 1 means that it always escapes plasma binding. We then applied a computational filter to remove variants with low sequencing counts or highly deleterious mutations that might cause antibody escape simply by leading to poor expression of properly folded RBD on the yeast cell surface (Greaney et al., 2021; Starr et al., 2020). Specifically, we removed variants that had (or contained mutations with) ACE2 binding scores < −2.35 or expression scores < −1, using the variant- and mutation-level deep mutational scanning scores from (Starr et al., 2020). Note that these filtering criteria are slightly more stringent than those used in (Greaney et al., 2021) but are identical to those used in (Starr et al., 2021).
We next deconvolved variant-level escape scores into escape fraction estimates for single mutations using global epistasis models (Otwinowski et al., 2018) implemented in the dms_variants package, as detailed at (https://jbloomlab.github.io/dms_variants/dms_variants.globalepistasis.html) and described in (Greaney et al., 2021). The reported scores throughout the paper are the average across the libraries; these scores are also in Table S3. Correlations in final single mutant escape scores are shown in Figures S2D and S2E.
For plotting and analyses that required identifying RBD sites of “strong escape” (e.g., choosing which sites to show in logo plots in Figures 2A, 2B, and S4 or label in Figure 4B), we considered a site to mediate strong escape if the total escape (sum of mutation-level escape fractions) for that site exceeded the median across sites by > 10-fold, and was at least 10% of the maximum for any site. We also included site K417, which did not meet this threshold but was of interest due to its frequency among circulating viruses.
Full documentation of the computational analysis is at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera.
Generation of pseudotyped lentiviral particles
We used spike-pseudotyped lentiviral particles that were generated essentially as described in (Crawford et al., 2020b), using a codon-optimized SARS-CoV-2 spike from Wuhan-Hu-1 that contains a 21-amino-acid deletion at the end of the cytoplasmic tail (Crawford et al., 2020a) and the D614G mutation that is now predominant in human SARS-CoV-2 (Korber et al., 2020). The plasmid encoding this spike, HDM_Spikedelta21_D614G, is available from Addgene (#158762), and the full sequence is at (https://www.addgene.org/158762). Point mutations were introduced into the RBD of this plasmid via site-directed mutagenesis. Therefore, all mutations tested in this paper are in the G614 background, and are compared to a “wildtype” spike with G614. The only exception is the C432D mutation, which was made in the D614 background and is included in Figure S5D to show a baseline titer for lentiviral particles pseudotyped with a non-functional spike variant.
To generate these spike-pseudotyped lentiviral particles (Crawford et al., 2020b), 6e5 HEK293T (ATCC CRL-3216) cells per well were seeded in 6-well plates in 2 mL D10 growth media (DMEM with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin). 24 h later, cells were transfected using BioT transfection reagent (Bioland Scientific, Paramount, CA, USA) with a Luciferase_IRES_ZsGreen backbone, Gag/Pol lentiviral helper plasmid (BEI NR-52517), and wildtype or mutant SARS-CoV-2 spike plasmids. Media was changed to fresh D10 at 24 h post-transfection. At 60 h post-transfection, viral supernatants were collected, filtered through a 0.45 μm SFCA low protein-binding filter, and stored at −80°C.
Titering and p24 ELISAs of pseudotyped lentiviral particles
Titers of spike-pseudotyped lentiviral particles were determined as described in (Crawford et al., 2020b) with the following modifications. Spike-pseudotyped lentiviral supernatants were diluted in D10 growth media starting with a 1:10 dilution followed by 7 serial 2-fold dilutions. 100 μL of each dilution was added to 1.25e4 293T-ACE2 cells (BEI NR-52511) grown overnight in 50 μL of D10 growth media in a 96-well black-walled poly-L-lysine coated plate (Greiner Bio-One, 655936). Relative luciferase units (RLU) were measured 60 h post-infection (Promega Bright-Glo, E2620) in the infection plates with a black back-sticker (Fisher Scientific, NC9425162) added to minimize background. Titers in RLU per mL were calculated for each dilution, and the median of each technical replicate normalized to p24 concentration (in pg/mL) is plotted in Figure S5D.
p24 ELISAs were conducted using the HIV-1 p24 Antigen Capture Assay following the manufacturer’s instructions (Advanced Bioscience Laboratories Cat. #5421). All lentiviral supernatants were diluted 1:100,000 and measured in technical duplicate. In Figure S5D, technical duplicate p24 concentrations (in pg/mL) were averaged and used to normalize RLU values for each lentiviral supernatant preparation.
Neutralization assays
293T-ACE2 cells (BEI NR-52511) were seeded at 1.25e4 cells per well in 50 μL D10 in poly-L-lysine coated, black-walled, 96-well plates (Greiner 655930). 24 h later, pseudotyped lentivirus supernatants were diluted to ∼200,000 RLU per well (determined by titering as described above and shown in Figure S5D and incubated with a range of dilutions of plasma for 1 h at 37°C. 100 μL of the virus-antibody mixture was then added to cells. The following dilutions were used for each virus: WT 1:50; P384L 1:20; F456K 1:10; E484Q 1:20; G485R 1:32; S494P 1:50; G446V/F456V/E484P 1:6; F456V 1:40; E484P 1:6; G446V 1:25; E484K 1:50; F456A 1:6.
At ?70 h post-infection, luciferase activity was measured using the Bright-Glo Luciferase Assay System (Promega, E2610). Fraction infectivity of each plasma antibody-containing well was calculated relative to a “no-plasma” well inoculated with the same initial viral supernatant (containing wildtype or mutant RBD) in the same row of the plate. We used the neutcurve package (https://jbloomlab.github.io/neutcurve version 0.5.2) to calculate the inhibitory concentration 50% (IC50) and the neutralization titer 50% (NT50), which is simply 1/IC50, of each plasma against each virus by fitting a Hill curve with the bottom fixed at 0 and the top fixed at 1. The full neutralization curves are in Figure S5.
Depletion of RBD-binding antibodies from polyclonal plasma
Magnetic beads conjugated to the SARS-CoV-2 RBD (AcroBiosystems, MBS-K002) were prepared according to the manufacturer’s protocol. Beads were resuspended in ultrapure water at 1 mg beads/mL and a magnet was used to wash the beads 3 times in PBS with 0.05% BSA. Beads were then resuspended in PBS with 0.05% BSA at 1 mg beads per mL. Beads (manufacturer-reported binding capacity of 10-40 μg/mL anti-RBD antibodies) were incubated with human plasma at a 3:1 ratio beads:plasma (150 μL beads + 50 μL plasma), rotating overnight at 4°C. A magnet (MagnaRack Magnetic Separation Rack, ThermoFisher CS15000) was used to separate antibodies that bind RBD from the supernatant, and the supernatant (the post-RBD antibody depletion sample) was removed. A mock depletion (pre-depletion sample) was performed by adding 150 μL of PBS + 0.05% BSA and incubating rotating overnight at 4°C. For the neutralization assays on these plasma depleted of RBD-binding antibodies shown in Figure S1E; the reported plasma dilution is corrected for the dilution incurred by the depletion process.
Measurement of plasma binding to RBD or spike by ELISA
The IgG ELISAs for spike protein and RBD were conducted as previously described (Dingens et al., 2020). Briefly, ELISA plates were coated with recombinant spike and RBD antigens described in (Dingens et al., 2020) at 2 μg/mL. Five 3-fold serial dilutions of plasma beginning at 1:100 were performed in phosphate-buffered saline with 0.1% Tween with 1% Carnation nonfat dry milk. Dilution series of the “synthetic” sera comprised of the anti-RBD antibody rREGN10987 (Hansen et al., 2020) or anti-NTD antibody r4A8 (Chi et al., 2020) and pooled pre-pandemic human plasma from 2017-2018 (Gemini Biosciences; nos. 100–110, lot H86W03J; pooled from 75 donors) were performed such that the anti-spike antibody was present at a highest concentration of 0.25 μg/mL. Both antibodies were recombinantly produced by Genscript. The rREGN10987 is that used in (Starr et al., 2021) and the variable domain heavy and light chain sequences for r4A8 were obtained from GenBank GI 1864383732 and 1864383733 (Chi et al., 2020) and produced on a human IgG1 and IgK background, respectively. Pre-pandemic plasma alone, without anti-RBD antibody depletion, was used as a negative control, averaged over 2 replicates. Secondary labeling was performed with goat anti-human IgG-Fc horseradish peroxidase (HRP) (1:3000, Bethyl Labs, A80-104P). Antibody binding was detected with TMB/E HRP substrate (Millipore Sigma, ES001) and 1 N HCl was used to stop the reaction. OD450 was read on a Tecan infinite M1000Pro plate reader. The area under the curve (AUC) was calculated as the area under the titration curve with the serial dilutions on a log-scale.
Analysis of RBD mutations among circulating SARS-CoV-2 isolates
All 283,908 spike sequences on GISAID as of Dec-23-2020 were downloaded and aligned via mafft (Katoh and Standley, 2013). Sequences from non-human origins and sequences containing gap characters or excessive mutations were removed, leaving 263,217 sequences. The code that performs this alignment and filtering is at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/results/summary/gisaid_rbd_mutations.md. The counts and frequencies of mutations at each RBD site were then computed using this filtered sequence set. We acknowledge all GISAID contributors for sharing sequencing data (https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/data/gisaid_hcov-19_acknowledgement_table_2020_12_30.pdf).
Sites and mutations highlighted in Figure 6 were chosen as follows. Sites in the RBM containing the 4 RBD mutations with the highest cumulative frequency (S477N, N439K, N501Y, and Y453F), the two sites with the highest total escape (F456 and E484), and sites that have > = 30 variant counts in GISAID and are sites of strong escape for any plasma, are labeled in Figure 6A. The labeled sites are colored according to epitope region as in Figure 2. Figure 6B highlights the 4 most frequent mutations and also any mutations at the other sites labeled in Figure 6A with at least 10 or more counts in GISAID. Figure 6C highlights sites where mutations have a strong effect on binding of at least 1 plasma or have circulating variation with > 50 counts in GISAID. Site K417 was also of interest due to the presence of the K417N/T mutations in recently identified lineages (Faria et al., 2021; Tegally et al., 2020), and thus is also highlighted in each panel in Figure 6.
Data visualization
The static logo plot visualizations of the escape maps in the paper figures were created using the dmslogo package (https://jbloomlab.github.io/dmslogo, version 0.6.2) and in all cases the height of each letter indicates the escape fraction for that amino-acid mutation calculated as described above. For each plasma, the y axis is scaled to be the greatest of (a) the maximum site-wise escape metric observed for that plasma, (b) 20x the median site-wise escape fraction observed across all sites for that plasma, or (c) an absolute value of 1.0 (to appropriately scale plasma that are not “noisy” but for which no mutation has a strong effect on plasma binding). Site C361 has been removed from the plots, because while mutations at this site reduce plasma binding, these mutations ablate a disulfide bond in the core RBD that is important for proper folding of the RBD and likely result in a grossly misfolded RBD and do not represent specific plasma escape mutations. Sites N501and K417 have been added to Figure S3 due to their frequencies among circulating viruses. The code that generates these logo plot visualizations is available at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/results/summary/escape_profiles.md.
In many of the visualizations (e.g., Figures 2, 4, 6A, S3, and S4), the RBD sites are categorized by epitope region (core-RBD epitope, receptor-binding ridge, or 443–450 loop) and colored accordingly. We define the core-RBD epitope as residues 365–372+382–386, the receptor-binding ridge epitope to be residues 417+455+456+471–490, and the 443–450 loop epitope to be residues 443–452+494–501. In Figure 4E, the S309 epitope is defined as RBD non-hydrogen atoms within 4Å of antibody in PDB: 6WPS, (Pinto et al., 2020).
For the static structural visualizations in the paper figures, the RBD surface (PDB: 6M0J, (Lan et al., 2020)) was colored by the site-wise escape metric at each site, with white indicating no escape and red scaled to be the same maximum used to scale the y axis in the logo plot escape maps, determined as described above. We created interactive structure-based visualizations of the escape maps using dms-view (Hilton et al., 2020) that are available at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_HAARVI_sera. The logo plots in these escape maps can be colored according to the deep mutational scanning measurements of how mutations affect ACE2 binding or RBD expression as described above.
Quantification and statistical analysis
Quantitative analyses were performed using custom code, available at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera. Plasma mapping experiments were performed in biological duplicate using the independent mutant RBD libraries.
To quantify the effects of mutations on binding by convalescent plasma antibodies, (see Method details section, “Analysis of deep sequencing data to compute each mutation’s plasma escape fraction”), we computed the “escape fraction” for each barcoded variant using the deep sequencing counts for each variant in the original and plasma-escape populations and the total fraction of the library that escaped antibody. To deconvolve the variant-level escape scores into escape fraction estimates for single mutations, we used global epistasis models (Otwinowski et al., 2018) implemented in the dms_variants package, as detailed at (https://jbloomlab.github.io/dms_variants/dms_variants.globalepistasis.html). The reported scores throughout the paper are the average across the biological duplicate libraries; these scores are also in Table S3. Correlations in final single mutant escape scores are shown in Figures S2D and S2E.
We did not determine statistical significance for the escape metrics as there is no established method for doing so. Rather, for plotting and analyses that required identifying RBD sites of “strong escape,” (e.g., choosing which sites to show in logo plots in Figures 2A, 2B, and S4 or label in Figure 4B), we considered a site to mediate strong escape if the total escape (sum of mutation-level escape fractions) for that site exceeded the median across sites by > 10-fold, and was at least 10% of the maximum for any site.
Additional resources
Interactive versions of the serum-escape logo plots can be found at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_HAARVI_sera.
Acknowledgments
We thank Adam Dingens for experimental assistance; Tulio de Oliveira for helpful advice; the Flow Cytometry and Genomics core facilities at the Fred Hutchinson Cancer Research Center for experimental support, especially Dolores Covarrubias and Andy Marty; Neil King, Alexandra Walls, David Veesler, and the UW Institute for Protein Design for purified RBD and spike proteins; and Seth Zost and James Crowe of Vanderbilt University for monoclonal antibodies. We also thank all research participants in the Hospitalized or Ambulatory Adults with Respiratory Viral Infections (HAARVI) study for their generous participation and all HAARVI study researchers and staff, especially Caitlin Wolf. This work was supported by the NIAID/NIH (R01AI141707 and R01AI127893 to J.D.B., T32AI083203 to A.J.G., and F30AI149928 to K.H.D.C.) and the Gates Foundation (INV-004949). The Scientific Computing Infrastructure at Fred Hutch is funded by ORIP grant S10OD028685. T.N.S. is a Washington Research Foundation Innovation Fellow at the University of Washington Institute for Protein Design and a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation (DRG-2381-19). J.D.B. is an Investigator of the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the US government or the other sponsors.
Author contributions
Conceptualization, A.J.G., H.Y.C., and J.D.B.; methodology, A.J.G., K.H.D.C., T.N.S., and J.D.B.; investigation, A.J.G., A.N.L., K.H.D.C., and K.D.M.; code, A.J.G., T.N.S., and J.D.B.; formal analysis, A.J.G. and J.D.B.; validation, A.J.G., A.N.L., and K.H.D.C.; resources, H.Y.C.; writing – original draft, A.J.G. and J.D.B.; writing – review and editing, all authors; supervision, H.Y.C. and J.D.B.
Declaration of interests
H.Y.C. is a consultant for Merck, Pfizer, Ellume, and Bill and Melinda Gates Foundation and has received support from Cepheid and Sanofi-Pasteur. The other authors declare no competing interests.
Published: February 8, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2021.02.003.
Supplemental information
This table contains the sample metadata from the second supplementary file of Crawford et al., (2020b) as well as the neutralization titers measured in the present study. The columns titled "NT50_pre" and "NT50_post" give the pre- and post-depletion neutralization measurements, respectively. The table is available online at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/experimental_validations/results/rbd_absorptions/TableS1.csv
The file gives the “escape fraction” for each mutation, as well as the total escape fraction at each site and the maximum escape fraction for any mutation at the site. The table is available online at https://raw.githubusercontent.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/main/results/supp_data/human_sera_raw_data.csv.
References
- Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.-L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrique L., Duyvesteyn H.M.E., Malinauskas T., Zhao Y., Ren J., Zhou D., Walter T.S., Radecke J., Huo J., Ruza R.R., et al. The SARS-CoV-2 Spike harbours a lipid binding pocket which modulates stability of the prefusion trimer. bioRxiv. 2020 doi: 10.1101/2020.08.13.249177. [DOI] [Google Scholar]
- Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Escape of SARS-CoV-2 501Y.V2 variants from neutralization by convalescent plasma. medRxiv. 2021 doi: 10.1101/2021.01.26.21250224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Dingens A.S., Eguia R., Wolf C.R., Wilcox N., Logue J.K., Shuey K., Casto A.M., Fiala B., Wrenn S., et al. Dynamics of neutralizing antibody titers in the months after SARS-CoV-2 infection. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12 doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingens A.S., Crawford K.H.D., Adler A., Steele S.L., Lacombe K., Eguia R., Amanat F., Walls A.C., Wolf C.R., Murphy M., et al. Serological identification of SARS-CoV-2 infections among children visiting a hospital during the initial Seattle outbreak. Nat. Commun. 2020;11:4378. doi: 10.1038/s41467-020-18178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguia R., Crawford K.H.D., Stevens-Ayers T., Kelnhofer-Millevolte L., Greninger A.L., Englund J.A., Boeckh M.J., Bloom J.D. A human coronavirus evolves antigenically to escape antibody immunity. bioRxiv. 2020 doi: 10.1101/2020.12.17.423313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Claro I.M., Candido D., Moyses Franco L.A., Andrade P.S., Coletti T.M., Silva C.A.M., Sales F.C., Manuli E.R., Aguiar R.S., et al. Virological; 2021. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Ladinsky M., Cho A., Jankovic M., Schaefer-Babajew D., et al. Evolution of Antibody Immunity to SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton S.R., Bobrowicz P., Bobrowicz B., Davidson R.C., Li H., Mitchell T., Nett J.H., Rausch S., Stadheim T.A., Wischnewski H., et al. Production of complex human glycoproteins in yeast. Science. 2003;301:1244–1246. doi: 10.1126/science.1088166. [DOI] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton S., Huddleston J., Black A., North K., Dingens A., Bedford T., Bloom J. dms-view: Interactive visualization tool for deep mutational scanning data. J. Open Source Softw. 2020;5:2353. doi: 10.21105/joss.02353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.-Y.A., Tan T., Chen T.-H., Huang C.-G., Harvey R., Hussain S., Chen C.-P., Harding A., Gilbert-Jaramillo J., Liu X., et al. Plasmablast-derived antibody response to acute SARS-CoV-2 infection in humans. bioRxiv. 2020 doi: 10.1101/2020.08.28.267526. [DOI] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp S., Harvey W., Datir R., Collier D., Ferreira I., Carabelii A., Robertson D.L., Gupta R.K. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion DH69/V70. bioRxiv. 2020 doi: 10.1101/2020.12.14.422555. [DOI] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R., Ferreira I.A., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. Neutralising antibodies drive Spike mediated SARS-CoV-2 evasion. medRxiv. 2020 doi: 10.1101/2020.12.05.20241927. [DOI] [Google Scholar]
- Kidd M., Richter A., Best A., Mirza J., Percival B., Mayhew M., Megram O., Ashford F., White T., Moles-Garcia E., et al. S-variant SARS-CoV-2 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-QPCR. medRxiv. 2020 doi: 10.1101/2020.12.24.20248834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler K.E., Bedford T. Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e. eLife. 2021 doi: 10.7554/eLife.64509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee J.M., Eguia R., Zost S.J., Choudhary S., Wilson P.C., Bedford T., Stevens-Ayers T., Boeckh M., Hurt A.C., Lakdawala S.S., et al. Mapping person-to-person variation in viral mutations that escape polyclonal serum targeting influenza hemagglutinin. eLife. 2019 doi: 10.7554/eLife.49324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Rothlauf P.W., Bloyet L.-M., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Ellebedy A.H., et al. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv. 2020 doi: 10.1101/2020.11.06.372037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley S.F., O’Donnell D., Stoesser N.E., Matthews P.C., Howarth A., Hatch S.B., Marsden B.D., Cox S., James T., Warren F., et al. Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Paul Duprex W. Natural deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. bioRxiv. 2020 doi: 10.1101/2020.11.19.389916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.-K., Saenger B., Swanson K.A., Muehl J., Chen W., Cai H., Sarkar R., Tuereci O., Dormitzer P.R., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. bioRxiv. 2021 doi: 10.1101/2021.01.18.426984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Alía M.Á., Nace R.A., Zhang L., Russell S.J. Pathogenic measles viruses cannot evolve to bypass vaccine-induced neutralizing antibodies. bioRxiv. 2020 doi: 10.1101/2020.10.22.351189. [DOI] [Google Scholar]
- Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.-Y., Rustagi A., Rogers A.J., et al. Human B Cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski J., McCandlish D.M., Plotkin J.B. Inferring the shape of global epistasis. Proc. Natl. Acad. Sci. USA. 2018;115:E7550–E7558. doi: 10.1073/pnas.1804015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Public Health England Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01. 2020. https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E., et al. Virological; 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. [Google Scholar]
- Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J., Thomas P., Graham C., Seow J., Lee N., et al. The impact of Spike mutations on SARS-CoV-2 neutralization. bioRxiv. 2021 doi: 10.1101/2021.01.15.426849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.-H., Feng J., et al. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021 doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen T.L., Taylor Stone E., Hassert M., Geerling E., Grimberg B.T., Espino A.M., Pantoja P., Climent C., Hoft D.F., George S.L., et al. The receptor binding domain of SARS-CoV-2 spike is the key target of neutralizing antibody in human polyclonal sera. bioRxiv. 2020 doi: 10.1101/2020.08.21.261727. [DOI] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. The circulating SARS-CoV-2 spike variant N439K maintains fitness while evading antibody-mediated immunity. bioRxiv. 2020 doi: 10.1101/2020.11.04.355842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toelzer C., Gupta K., Yadav S.K.N., Borucu U., Davidson A.D., Kavanagh Williamson M., Shoemark D.K., Garzoni F., Staufer O., Milligan R., et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370:725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloch C.M., Ronaldo da Silva F., Jr., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimaraes A.P.d.C., Mariani D., da Costa R.M., Ferreira O.C., Jr., et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. medRxiv. 2020 doi: 10.1101/2020.12.23.20248598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss W.N., Hou Y.J., Johnson N.V., Kim J.E., Delidakis G., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., Abbasi S.A., et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes in COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.20.423708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv. 2021 doi: 10.1101/2021.01.15.426911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentz A.E., Shusta E.V. A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl. Environ. Microbiol. 2007;73:1189–1198. doi: 10.1128/AEM.02427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Lambson B.E., Vermeulen M., van den Berg K., Rossouw T., Boswell M., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv. 2021 doi: 10.1101/2021.01.18.427166. [DOI] [PubMed] [Google Scholar]
- Wong A.H.M., Tomlinson A.C.A., Zhou D., Satkunarajah M., Chen K., Sharon C., Desforges M., Talbot P.J., Rini J.M. Receptor-binding loops in alphacoronavirus adaptation and evolution. Nat. Commun. 2017;8:1735. doi: 10.1038/s41467-017-01706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A Highly Conserved Cryptic Epitope in the Receptor Binding Domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table contains the sample metadata from the second supplementary file of Crawford et al., (2020b) as well as the neutralization titers measured in the present study. The columns titled "NT50_pre" and "NT50_post" give the pre- and post-depletion neutralization measurements, respectively. The table is available online at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/experimental_validations/results/rbd_absorptions/TableS1.csv
The file gives the “escape fraction” for each mutation, as well as the total escape fraction at each site and the maximum escape fraction for any mutation at the site. The table is available online at https://raw.githubusercontent.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/main/results/supp_data/human_sera_raw_data.csv.
Data Availability Statement
We provide data and code in the following ways:
The complete code for the full computational data analysis pipeline of the mapping experiments is available on GitHub at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera
A Markdown summary of the analysis workflow with renderings of all the code is on GitHub at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/results/summary/summary.md
The escape fraction measured for each mutation in Table S3 and also at https://raw.githubusercontent.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/main/results/supp_data/human_sera_raw_data.csv
The barcode counts for each RBD variant in each mapping condition are at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/results/counts/variant_counts.csv
All raw sequencing data are available on the NCBI Short Read Archive at BioProject: PRJNA639956 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA639956), BioSample: SAMN17185313 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN17185313).
The patient metadata and neutralization titers of convalescent plasma pre- and post-depletion of RBD-binding antibodies is available in Table S1 and online at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_HAARVI_sera/blob/main/experimental_validations/results/rbd_absorptions/TableS1.csv