Abstract
Aim
To investigate the sources of infection among healthcare workers (HCWs) and patients in a teaching hospital in the Netherlands during the early stages of the coronavirus disease 2019 (COVID-19) pandemic using epidemiological and whole-genome sequencing data.
Methods
From 3rd April to 11th May 2020, 88 HCWs and 215 patients were diagnosed with COVID-19. Whole-genome sequences were obtained for 30 HCWs and 20 patients.
Results
Seven and 11 sequence types were identified in HCWs and patients, respectively. Cluster A was the most common sequence type, detected in 23 (77%) HCWs; of these, 14 (61%) had direct patient contact and nine (39%) had indirect patient contact. In addition, seven patients who were not hospitalized in the COVID-19 cohort isolation ward who became positive during their admission were infected with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) cluster A. Following universal masking of all HCWs and emphasis on physical distancing during meals and breaks, no further evidence was found for patient-to-HCW or HCW-to-HCW transmission or vice versa.
Conclusion
The finding that patients and HCWs were infected with SARS-CoV-2 cluster A suggests both HCW-to-HCW and HCW-to-patient transmission.
Keywords: SARS-CoV-2, Outbreak, COVID-19, Molecular typing, WGS, Healthcare worker
Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), emerged in Wuhan in December 2019 and has since spread across the world [1,2]. The first patient with COVID-19 in the Netherlands was identified on 27th February 2020, and the virus has since spread rapidly throughout the country [3].
Prompt recognition, isolation and management of suspected cases are key factors to prevent the transmission of SARS-CoV-2 in hospitals. At the start of the pandemic, the focus was on the prevention of patient-to-healthcare worker (HCW) transmission, and HCWs used personal protective equipment (PPE), including N95 masks, eye protection, disposable gowns and gloves, while caring for patients with suspected or confirmed COVID-19.
Isolation wards were set up in the study hospital for suspected and confirmed cases of COVID-19 on 2nd March 2020. Hand shaking is no longer allowed in the hospital, social distancing is implemented, the staff restaurant is closed, and gatherings and meetings for HCWs are prohibited. Hand, coughing and sneezing hygiene are promoted. No visitors are allowed. Furthermore, a strategy for low-threshold testing for HCWs with mild respiratory symptoms without fever, management and follow-up has been implemented.
Despite the implementation of these stringent infection prevention control measures, a nosocomial outbreak of SARS-CoV-2 amongst HCWs and patients was suspected when a cluster of HCWs from a single non-COVID ward tested positive for SARS-CoV-2. To investigate this outbreak, epidemiological and whole-genome sequencing (WGS) data for infected HCWs and patients were compared.
Methods
Study site
This study was performed in a teaching hospital with nursing facilities in Rotterdam and Schiedam, the Netherlands with approximately 45,000 admissions per year. On 21st April 2020, an epidemiological investigation was initiated when eight HCWs were diagnosed with COVID-19 in a single non-COVID ward at the Schiedam site.
Infection control measures
In response, universal use of surgical masks for HCWs working in all clinical wards, as well as outpatient clinics, was implemented immediately in order to prevent further spread amongst HCWs and from HCWs to hospitalized patients. HCWs were no longer allowed to work in both locations during the outbreak. Social distancing during meals and coffee breaks was emphasized. All SARS-CoV-2-positive HCWs were asked to self-isolate until at least 24 h after resolution of symptoms, in line with the guidelines of the National Institute of Public Health. HCWs and admitted patients with respiratory symptoms were tested for SARS-CoV-2.
Sample and data collection
Nasopharyngeal swabs (ESwab medium, Copan Diagnostics Inc., Brescia, Italy) from 632 HCWs and 5448 patients with mild respiratory symptoms were tested between 3th April and 11th May 2020. All HCWs who tested positive for SARS-CoV-2 received a voluntary questionnaire regarding symptoms and possible source of the infection. HCWs from the non-COVID wards were divided into three categories: HCWs with direct patient contact, including medical doctors and nursing staff; HCWs with indirect contact, such as individuals who work on the wards without touching the patients (e.g. cleaners and food distributors); and HCWs without patient contact. Reverse transcription polymerase chain reaction assays were performed using MagnaPure96 and LightCycler 480 II (Roche, Basel, Switzerland), as described by Corman et al. [4].
Whole-genome sequencing and comparative genome analysis
Fifty-nine SARS-CoV-2-positive nasopharyngeal swabs (cycle threshold <32) from HCWs and patients involved in this outbreak were selected for WGS analysis. Sequencing was performed using an amplicon-based approach on the Nanopore platform [5]. The consensus sequence was determined using reference-based alignment. The sequences were aligned with all other sequences from the Netherlands generated on 4th June 2020 using muscle [6], and phylogenetic analysis was performed using IQ-TREE [7]. A general time-reversible model of nucleotide substitution with estimated base frequencies, proportion of invariant sites, and γ distribution of rates across sites was used in this maximum-likelihood analysis, as described previously [6]. Clusters were defined as sequences with a maximum of two nucleotide differences. The consensus sequences have been submitted to GISAID under accession numbers EPI_ISL_461312-EPI_ISL_461333, EPI_ISL_461340-EPI_ISL_461343 and EPI_ISL_461367. Visualization of the phylogenetic data was performed using Interactive Tree Of Life v4 [8].
Ethical approval
The institutional review board (IRB) waived the need for informed consent as tests were performed on samples that were required for routine clinical care (IRB Protocol No. 2020-086).
Results
Between 3rd April and 11th May 2020, 88 of 632 (14%) tested HCWs and 215 of 5448 (4%) patients tested positive for SARS-CoV-2. Samples from 59 cases were referred for sequencing, yielding 50 whole-genome sequences from 30 HCWs and 20 patients (Table I ).
Table I.
Distribution of sequence clusters among healthcare workers (HCWs) and patients
| Sequence cluster |
||||||
|---|---|---|---|---|---|---|
| Total | U | A | B | C | D | |
| Total | 50 | 12 (24%) | 31 (62%) | 3 (6%) | 2 (4%) | 2 (4%) |
| HCWs | 30 | 4 (13%) | 23 (77%) | 1 (3%) | 2 (7%) | |
| With direct patient contact | 19 | 3 (16%) | 14 (74%) | 2 (11%) | ||
| With indirect patient contact | 10 | 1 (10%) | 9 (90%) | |||
| No patient contact | 1 | 0 | 1 (100%) | |||
| Patients | 20 | 8 (40%) | 8 (40%) | 2 (10%) | 2 (10%) | 0 |
| Positive at admission | 13 | 8 (62%) | 1 (8%) | 2 (15%) | 2 (15%) | 0 |
| Positive during admission | 7 | 0 | 7 (100%) | 0 | 0 | 0 |
U, unique.
Four sequence clusters were observed, and 12 samples did not cluster with other sequences from the facility (Figure 1 ). Cluster A was the predominant cluster, with 31 (62%) sequences belonging to this cluster, while seven (14%) sequences were part of clusters B (N=3), C (N=2) and D (N=2). Cluster A was also the predominant cluster detected during the outbreak in HCWs on the non-COVID ward, found in 23 (77%) HCWs.
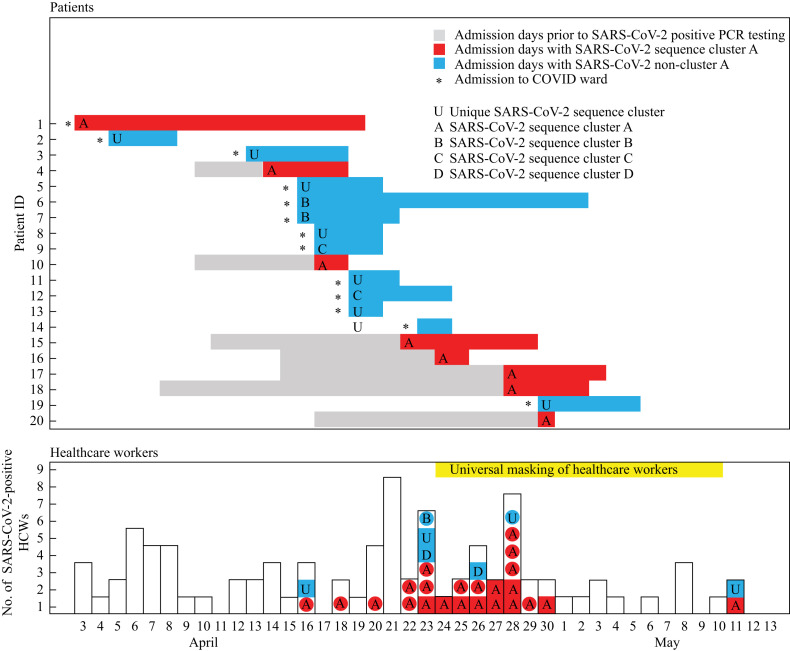
Figure 1.
Timeline of 88 healthcare workers (HCWs) and 20 admitted patients with laboratory-confirmed severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) with their corresponding sequence cluster type. PCR, polymerase chain reaction; COVID-19, coronavirus disease 2019.
The incidence of cluster A was higher among HCWs with indirect patient contact; among this group, nine of 10 (90%) sequenced isolates belonged to cluster A. Among the infected HCWs with direct patient contact, 14 of 19 (74%) sequenced isolates belonged to cluster A.
Figure 2 shows the epidemic curve combined with WGS sequencing of SARS-CoV-2-infected HCWs. Thirteen of 20 (65%) SARS-CoV-2-positive patients presented to the emergency ward with symptoms, and were transferred to the COVID-19 cohort isolation ward (Figure 2). Patient 1 was infected with SARS-CoV-2 cluster A. Patients 5 and 6 were admitted on the same day with SARS-CoV-2 cluster B. No direct link was found, but these two patients resided only 2 km apart. The remaining sequences of patients sampled on admission were unique. Patient 14 had been sampled 4 days prior to admission. None of these sequences were identified in infected HCWs, showing that the infection prevention policy was effective in this patient group.
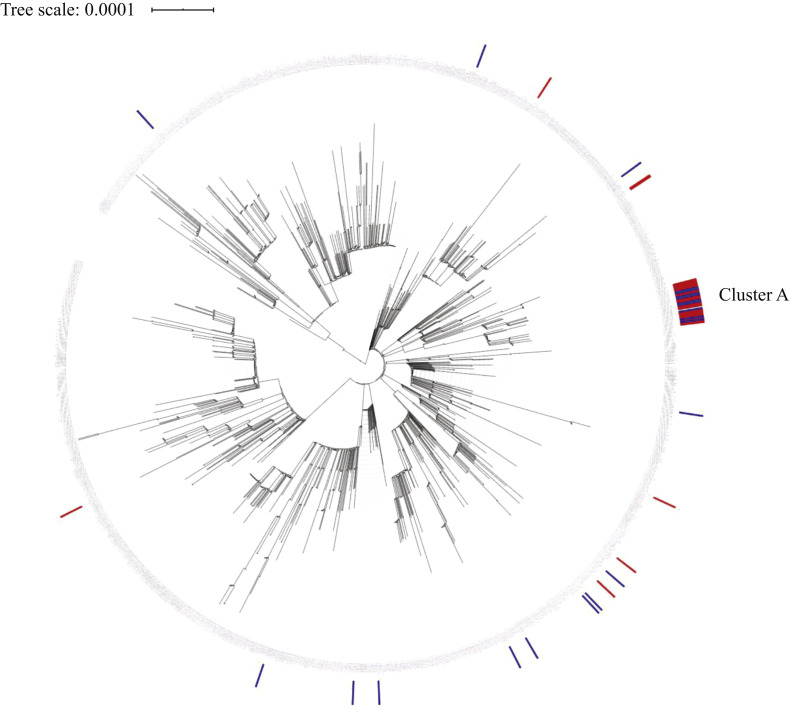
Figure 2.
Maximum-likelihood phylogeny of all complete genomes of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) strains in the Netherlands (5th June 2020) and focusing on the SARS-CoV-2 strains described in this study. The scale indicates the number of nucleotide substitutions per site. Patient sequences are shown in blue and healthcare worker sequences are shown in red.
In the subsequent period, seven patients on different non-COVID wards who became positive during hospital admission were infected with SARS-CoV-2 cluster A (Figure 2). Patient 20 tested positive on 30th April 2020, 1 day after transfer to a nursing home. The intervals between hospital admission, positive sampling (4–19 days after admission) and symptom onset, as recorded in their medical records (data not shown), suggest that COVID-19 was acquired during hospital admission.
Eight HCWs working on the same non-COVID ward tested positive on 21st April 2020. In response to this, all HCWs who worked on this ward were instructed to wear surgical masks at all times. Over the following days, multiple HCWs tested positive.
WGS included all SARS-CoV-2 sequences detected in the Netherlands to date (N=1477). Cluster A was phylogenetically related in HCWs and patients from the teaching hospital (Figure 1). Only one other person from the Rotterdam area was detected with SARS-CoV-2 cluster A in the same time period. Epidemiological data of this case were not available.
After the analysis, it became clear that on the ward with the cluster of HCWs that prompted the investigation of transmission, only one patient was sequence typed as cluster A. As such, it is less likely that patient-to-HCW transmission played a significant role on this ward.
The voluntary questionnaire was completed by 16 of the 30 SARS-CoV-2-positive HCWs. HCWs mentioned that they worked in both locations, and questionnaire responses indicated that food and cleaning staff worked in different departments in the hospital. Multiple patient transfers occurred between wards. Seven HCWs reported that they had been in close contact with colleagues during breaks who subsequently became symptomatic and tested positive.
Three HCWs reported unprotected care for a patient who later turned out to be positive (cluster A) during admission. Over 3 weeks of follow-up, no additional positive HCWs were detected. The combination of epidemiological and WGS analysis was highly suggestive of HCW-to-HCW and HCW-to-patient transmission.
Discussion
In this study, epidemiological data were combined with WGS data to analyse the extent of intrahospital spread of SARS-CoV-2 in a teaching hospital. Despite all efforts to implement strict infection control measures, HCWs and patients acquired COVID-19 in hospital. The identification of SARS-CoV-2 cluster A in HCWs and patients hospitalized for other conditions suggests transmission between HCWs, and also from HCWs to patients on the non-COVID ward. Identical or near-identical sequences were not found with any of the SARS-CoV-2 sequences detected in the Netherlands to date, indicating that this specific cluster was linked to this hospital outbreak.
A previous study using WGS found widespread community transmission of SARS-CoV-2 among HCWs in a very early phase of the epidemic in the Netherlands. No evidence of widespread nosocomial transmission was found [9,10]. However, during ongoing community transmission, there is potential risk of unnoticed introduction of SARS-CoV-2 to hospitals through HCWs and subsequent transmission to other HCWs and patients nursed outside of dedicated COVID-19 wards.
HCWs are at increased risk of exposure to unsuspected cases of COVID-19 within hospitals [11]. However, they can also become sources of onward transmission. Maltezou et al. reported that in almost 50% of HCWs infected with SARS-CoV-2, a colleague was the source of exposure [12]. During the outbreak, the study hospital followed the national policy at that time (i.e. HCWs with mild respiratory symptoms should be tested for SARS-CoV-2). Positive HCWs were not allowed to work and were placed on mandatory medical leave for ≥7 days until all symptoms resolved completely. However, recent work from the Netherlands showed that 63% of HCWs continued to work despite mild symptoms in the early stages of the epidemic [13].
In addition, screening of HCWs in a large UK hospital highlighted the role of asymptomatic carriage in SARS-CoV-2 transmission [14]. Analysis of this particular outbreak and the knowledge that asymptomatic and presymptomatic carriers can be contagious motivated the study hospital to change its SARS-CoV-2 testing policy. In the case of an unexpected SARS-CoV-2-positive result in a HCW or patient after admission, it is now recommended that all close contacts should be tested immediately and then subsequently on days 3 and 7, regardless of symptoms. Patients are isolated pending test results. HCWs are allowed to work with surgical masks while awaiting their test results.
The main route of transmission of SARS-CoV-2 is considered to be from person-to-person via droplet transmission, but other routes such as aerosol transmission via hospital ventilation systems have been suggested [15]. Some ventilation systems recirculate indoor air or mix air in common ventilation ducts to reduce energy costs. At the study hospital, it was established that the ventilation system worked properly, did not have recirculation routes and had well-separated air intakes and outlets.
This study had several limitations. Firstly, it was not possible to sequence all HCWs and patients involved in this outbreak. In the analysis, the timing of diagnosis of Patient 1 and the subsequent finding of (near-)identical genomes in patients and HCWs suggested transmission from Patient 1 to HCWs and patients after admission. However, the analysis was incomplete and did not cover the full development of transmission, so another scenario cannot be excluded. Furthermore, the authors were unable to track the ward location of all HCWs during their shifts because some worked simultaneously in other departments or locations. Nevertheless, the authors were able to identify unprotected contact moments between infected HCWs and infected patients, and did observe that cleaners and food distributors did not adhere to social distancing rules during coffee breaks and meals. Although these are potential routes of transmission, this study cannot confirm these routes with certainty. In addition, a sequence related to cluster A was also observed in the general population, detected due to random regional sampling efforts. This indicates that diversity in viral genomes is still limited, and clusters based on WGS have to be supported by epidemiological information.
HCWs can play an important role in nosocomial transmission. This study highlighted that effective strategies are necessary to contain and prevent the spread of COVID-19 within hospitals and amongst HCWs.
Acknowledgements
The authors wish to thank the laboratory staff of the Department of Medical Microbiology and Infection Control and the Occupational Health Service, Franciscus Gasthuis & Vlietland Hospital and ErasmusMC, Department of Viroscience, for processing the diagnostic samples.
Conflict of interest statement
None declared.
Funding sources
BO, RS and MK received funding from the European Union's Horizon 2020 Research and Innovation Programme [Grant Nos. 874735 (VEO), 848096 (SHARP JA) and 101003589 (RECoVER)].
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinross P., Suetens C., Gomes Dias J., Alexakis L., Wijermans A., Colzani E. Rapidly increasing cumulative incidence of coronavirus disease (COVID-19) in the European Union/European Economic Area and the United Kingdom, 1 January to 15 March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.11.2000285. pii=2000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alderweireld C.E.A., Buiting A.G.M., Murk J.-L.A.N., Verweij J.J., Berrevoets M.A.H., van Kasteren M.E.E. COVID-19: patient zero in the Netherlands. Ned Tijdschr Geneeskd. 2020;164:D4962. [PubMed] [Google Scholar]
- 4.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. pii=2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oude Munnink B.B., Nieuwenhuijse D.F., Stein M., O’Toole Á., Haverkate M., Mollers M. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med. 2020;26:1405–1410. doi: 10.1038/s41591-020-0997-y. [DOI] [PubMed] [Google Scholar]
- 6.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kluytmans-van den Bergh M.F.Q., Buiting A.G.M., Pas S.D., Bentvelsen R.G., van den Bijllaardt W., van Oudheusden A.J.G. Prevalence and clinical presentation of health care workers with symptoms of coronavirus disease 2019 in 2 Dutch hospitals during an early phase of the pandemic. JAMA Netw Open. 2020;3:e209673. doi: 10.1001/jamanetworkopen.2020.9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sikkema R.S., Pas S.D., Nieuwenhuijse D.F., O’Toole Á., Verweij J., van der Linden A. COVID-19 in health-care workers in three hospitals in the south of the Netherlands: a cross-sectional study. Lancet Infect Dis. 2020;20:1273–1280. doi: 10.1016/S1473-3099(20)30527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmann S., Rubin Z., Sato H., Yong K.O., Terashita D., Balter S. Coronavirus 2019 (COVID-19) infections among healthcare workers, Los Angeles County, February–May 2020. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maltezou H.C., Dedoukou X., Tseroni M., Tsonou P., Raftopoulos V., Papadima K. SARS-CoV-2 infection in healthcare personnel with high-risk occupational exposure: evaluation of 7-day exclusion from work policy. Clin Infect Dis. 2020;71:3182–3187. doi: 10.1093/cid/ciaa888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reusken C.B., Buiting A., Bleeker-Rovers C., Diederen B., Hooiveld M., Friesema I. Rapid assessment of regional SARS-CoV-2 community transmission through a convenience sample of healthcare workers, the Netherlands, March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.12.2000334. pii=2000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivett L., Sridhar S., Sparkes D., Routledge M., Jones N.K., Forrest S. Screening of healthcare workers for SARS-CoV-2 highlights the role of asymptomatic carriage in COVID-19 transmission. Elife. 2020;9 doi: 10.7554/eLife.58728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amoatey P., Omidvarborna H., Baawain M.S., Al-Mamun A. Impact of building ventilation systems and habitual indoor incense burning on SARS-CoV-2 virus transmissions in Middle Eastern countries. Sci Total Environ. 2020;733:139356. doi: 10.1016/j.scitotenv.2020.139356. [DOI] [PMC free article] [PubMed] [Google Scholar]