Abstract
Background
Validated biomarkers to evaluate HIV-1 cure strategies are currently lacking, therefore requiring analytical treatment interruption (ATI) in study participants. Little is known about the safety of ATI and its long-term impact on patient health.
Objectives
ATI safety was assessed and potential biomarkers predicting viral rebound were evaluated.
Methods
PBMCs, plasma and CSF were collected from 11 HIV-1-positive individuals at four different timepoints during ATI (NCT02641756). Total and integrated HIV-1 DNA, cell-associated (CA) HIV-1 RNA transcripts and restriction factor (RF) expression were measured by PCR-based assays. Markers of neuroinflammation and neuronal injury [neurofilament light chain (NFL) and YKL-40 protein] were measured in CSF. Additionally, neopterin, tryptophan and kynurenine were measured, both in plasma and CSF, as markers of immune activation.
Results
Total HIV-1 DNA, integrated HIV-1 DNA and CA viral RNA transcripts did not differ pre- and post-ATI. Similarly, no significant NFL or YKL-40 increases in CSF were observed between baseline and viral rebound. Furthermore, markers of immune activation did not increase during ATI. Interestingly, the RFs SLFN11 and APOBEC3G increased after ATI before viral rebound. Similarly, Tat-Rev transcripts were increased preceding viral rebound after interruption.
Conclusions
ATI did not increase viral reservoir size and it did not reveal signs of increased neuronal injury or inflammation, suggesting that these well-monitored ATIs are safe. Elevation of Tat-Rev transcription and induced expression of the RFs SLFN11 and APOBEC3G after ATI, prior to viral rebound, indicates that these factors could be used as potential biomarkers predicting viral rebound.
Introduction
Combination ART (cART) is able to suppress viral replication in HIV-1-infected individuals, but cannot eradicate HIV-1. Lifelong cART remains mandatory as the latent viral reservoir fuels viral replication after treatment interruption, rendering HIV-1 a chronic disease. This latent HIV-1 reservoir is the last hurdle towards an HIV-1 cure and consequently the target of cure strategies. Most of these cure strategies comprise a multitarget approach, both activating the latent reservoir and enhancing the immune response, e.g. shock and kill.1 The lack of biomarkers assessing reservoir reduction or virological control means that analytical treatment interruption (ATI) trials are becoming the gold standard to evaluate the potency of cure strategy interventions.2,3 Although these intensively monitored ATIs are well tolerated by most of the participants, little is known about ATI safety and its long-term impact on patient health.4,5 Recent reports demonstrated no changes in viral reservoir size [total HIV-1 DNA, intact HIV-1 DNA (near full-length) and cell-associated (CA) HIV-1 RNA] and little diversity in composition of virus populations pre- and post-ATI.6–8 These data are reassuring, but additional analyses evaluating virological and immunological characteristics in different body compartments pre- and post-ATI are crucial for further safety assessment. Neopterin, kynurenine and kynurenine/tryptophan ratio are commonly assessed immune activation markers in plasma and CSF and, further, YKL-40 and neurofilament light chain (NFL) are used as markers of neuroinflammation and neuronal injury, respectively.9,10 Increased levels of these markers have been detected in several diseases characterized by persistent inflammation (e.g. Alzheimer’s disease, cancer and viral infections, including HIV-1).10–12 Hence, assessing neopterin, kynurenine, kynurenine/tryptophan ratio, YKL-40 and NFL in CSF pre- and post-ATI could inform on the potential consequences of ATI on the brain.
Longer periods of treatment interruptions have been associated with increased CNS inflammation and NFL, as signs of neuronal injury.13
ATI, combined with extensive patient sampling, could give broader insights into the origin of viral rebound and can help identify potential biomarkers for predicting the time to viral rebound (TTVR) after treatment interruption.14
Because restriction factors (RFs), antiviral factors acting at specific stages of the viral life cycle, are induced early upon HIV-1 infection, after IFN production,15 these factors could be potential biomarkers for TTVR. Mechanisms that drive RF expression are not fully elucidated and measurement of RF levels in vivo before, during and after ATI could contribute to understanding the timing of RF induction. The best-characterized RFs include bone marrow stromal cell antigen 2 (BST2)/tetherin, apolipoprotein B mRNA-editing enzyme catalytic subunit 3G (APOBEC3G), SAM domain- and HD domain-containing protein 1 (SAMHD1) and tripartite motif-containing protein 5 (TRIM5), each performing its antiviral activity at different stages of the viral replication cycle (inhibition of virion release, viral hypermutation at reverse transcription, depletion of dNTP pool and targeting the viral capsid, respectively).16 Other RFs have been described recently, such as MX2, SLFN11 and PAF1, interfering with nuclear import, translation of viral RNA and other early events in the replication cycle.16–18 The HIV-1 genome encodes accessory proteins able to counteract RF activity, such as viral infectivity factor (Vif), viral protein U (Vpu), negative regulatory factor (Nef) and viral protein R (Vpr),19 allowing virus replication to continue.20In vivo RF data are limited, but their expression was found to be positively correlated with IFN-stimulated gene (ISG) levels and to viral load (VL) for some factors (APOBEC3G, TRIM5 and MX2).21,22 Additionally, APOBEC3G, TRIM5, BST2 and MX2 expression levels are increased in seroconverters (SRCVs),23 ART-naive acutely infected HIV-1 patients, confirming RF expression is induced as an early antiviral defence mechanism of the host immune system and suggesting that RF levels could be induced early post-ATI. In addition, the determination of different CA HIV-1 RNA transcripts during ATI can potentially give insights into RF activity. TAR, long LTR, polyA, Pol and Tat-Rev transcripts represent transcription initiation, elongation, termination, CA unspliced HIV-1 RNA (usRNA) and multiply spliced HIV-1 RNA (msRNA), respectively. Levels of HIV-1 msRNA indicate the ability to overcome blocks to transcription initiation, elongation and termination and were reported to increase at viral rebound.24,25
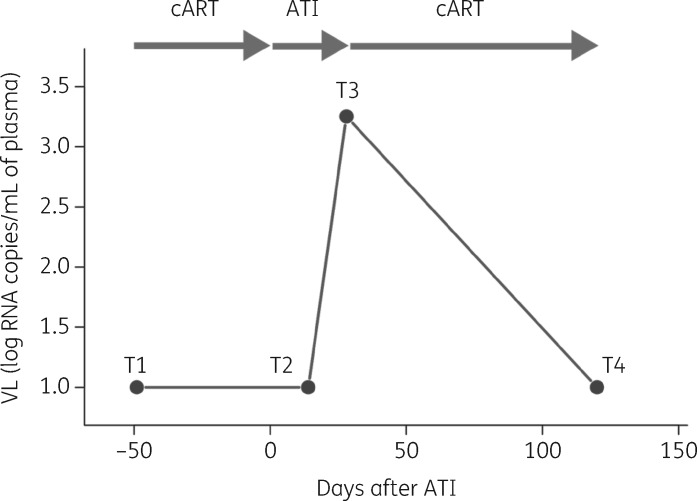
To assess ATI safety and to identify markers that could predict viral rebound, we explored markers of inflammation, viral reservoir size and RF expression levels at four different timepoints before, during and after ATI [baseline on cART (T1; undetectable VL), post-ATI (T2; undetectable VL), at viral rebound (T3; detectable VL) and 3 months after restart of cART (T4; undetectable VL)].
Methods
Patient cohort
The HIV-STAR ATI trial was designed to identify and characterize the relevant anatomical compartment(s) of the replication-competent HIV reservoir (trial registry number on clinicaltrials.gov: NCT02641756). The study was conducted between 1/1/2016 and 1/12/2017. The primary outcome of this study was published recently.14 The study protocol, experimental design and recruitment strategy were approved by the Ethics Committee of the University Hospital of Ghent (Belgian registration numbers: B670201525474 and B670201628230). All participants provided written consent. Participants were recruited from the AIDS reference centre at the Ghent University Hospital depending on strict inclusion criteria and after intensive counselling. Eleven HIV-1-infected, long-term-treated participants under chronic cART were included. In a second phase of the study, treatment was interrupted and participants were monitored twice a week with VL measurements and, upon viral rebound, treatment was reinitiated immediately at T3, which was defined by a VL >1000 copies/mL or two consecutive measurements >200 copies/mL. The study design allowed us to further investigate virological and immunological characteristics and their relationship with viral rebound. All participants are included in the current substudy.
RF and viral reservoir markers (total HIV-1 DNA, integrated HIV-1 DNA and different transcripts of CA HIV-1 RNA indicating specific transcriptional blocks: TAR, long LTR, Pol, polyA and Tat-Rev) were measured in PBMCs at T1 (on cART, undetectable VL), T2 (7–15 days after ATI, undetectable VL), T3 (viral rebound, detectable VL) and T4 (3 months after cART restart) (Figure 1). Additionally, inflammation markers were determined in plasma at these four different timepoints and for 10/11 participants at T1 and T3 in CSF.
Figure 1.
Schematic overview of the study set-up. PBMCs were collected at four different timepoints (T1, T2, T3 and T4). CSF was collected at two timepoints (T1 and T3). T1 represents the sampling under cART (baseline; undetectable VL). ATI occurs at Day 0. T2 represents 7–15 days after ATI (undetectable VL). At T3, viral rebound occurs (detectable VL). T4 represents a timepoint after cART restart (approximately 3 months; undetectable VL).
Quantitative real-time PCR
RNA was extracted from 107 PBMCs (RNA innuPREP Mini Kit, Analytik, Germany) and reverse-transcribed into cDNA using the qScript cDNA SuperMix (Quantabio, MA, USA) according to the manufacturer’s protocol. Real-time PCR was performed as previously described.23 Table S1 (available as Supplementary data at JAC Online) depicts used assays. Reference gene stability was assessed for ACTB, GAPDH, TBP, B2M, YWHAZ, PLOD1, HMBS and UBC on a subset of the samples. ACTB, GAPDH and YWHAZ were selected for normalization after analysis with the GeNorm algorithm.26 Expression analysis was performed with qbase+ software (Biogazelle).27
Neuroinflammation markers
NFL and YKL-40 protein were measured in CSF as markers of neuronal injury and neuroinflammation, respectively. In addition, neopterin, tryptophan and kynurenine were measured, both in plasma and CSF, as markers of immune activation. NFL concentration was measured using an in-house ELISA, as previously described in detail.28 YKL-40 concentration was measured using a commercially available ELISA according to the manufacturer’s instructions (R&D Systems, Minneapolis, MN, USA). NFL and YKL-40 measurements were performed by board-certified laboratory technicians in one round of experiments with baseline and follow-up samples side by side on the ELISA plates to minimize variation. Intra-assay coefficients of variation were below 8% for both NFL and YKL-40. Neopterin was measured by ELISA (B·R·A·H·M·S, Hennigsdorf, Germany) and kynurenine, for calculation of the kynurenine/tryptophan ratio, by HPLC.29
For details on the amplification of V1–V3 env from CSF and the phylogeny estimation, we refer to our recently published paper.14
Total and integrated HIV-1 DNA
Total genomic DNA was extracted from 107 PBMCs (DNeasy Blood & Tissue Kit, QIAGEN, The Netherlands). Restriction and digital droplet PCR (ddPCR) reactions were performed as described previously.30 Reference gene ribonuclease P/MRP subunit p30 (RPP30) was used for normalization and quantification of total HIV-1 DNA was performed with ddpcRquant, an in-house-developed software.31 Integrated HIV-1 DNA was quantified using the repetitive sampling Alu-HIV PCR method involving a nested-PCR approach.32 This method was performed as described previously.23 Briefly, in the first PCR, a forward primer and a reverse primer, targeting a human Alu fragment and the HIV-1 gag region, respectively, were used to quantify 40 replicates. Additionally, 20 replicates of background quantification were performed using only the HIV-1 gag primer.23
CA HIV-1 RNA transcripts
RNA was extracted from 107 million PBMCs (RNA innuPREP Mini Kit). TAR, long LTR, polyA, Pol and Tat-Rev CA HIV-1 RNA transcripts were quantified as described previously by Yukl et al.24 Input for reverse transcription (RT) reactions was maximized based on RNA mass recovery: a median of 0.385 and 3.286 μg for (i) TAR and (ii) long LTR, Pol, polyA and Tat-Rev RT, respectively, was used. Normalization was performed depending on input RNA mass as previously described.24
Statistical analysis
Non-parametric Friedman test with post hoc Dunn statistical analysis was performed. Holm P value adjustment method was used for multiple comparisons. Spearman correlation analysis was performed to define significant correlations between RFs, neuroinflammation markers and immunological and virological reservoir parameters at single timepoints. Additionally, repeated measurements correlation analysis was performed to detect similar or antagonistic profiles for these markers at the different timepoints. Statistical analyses and graphing was performed with R software using the following packages: PMCMR, Hmisc, graphics, ggplot2, corrplot and rmcorr.33
Results
Study participants
Eleven HIV-1-infected individuals were included in this study and sampled at four timepoints pre- and post-ATI (Figure 1). Participants were on stable cART for ≥3 years and did not have significant comorbidities at the time of inclusion. CD4 nadir values were >300 cells/mm3 for all but one participant (Table 1). Participants received a standard cART regimen involving an integrase inhibitor for at least 3 months before study inclusion. The median age of the participants was 40 years (range 32–56) (Table 1). Median TTVR was 21 days (range 15–36) with a median HIV-1 RNA level of 3.29 log copies/mL (range: 3.23–3.64) at viral rebound. Demographic variables are available in Table 1.
Table 1.
Clinical and virological characteristics of HIV-STAR ATI trial patients; n = 11. Values shown are median (IQR)
| Clinical characteristics | |
| age (years) | 40 (37.5–46.5) |
| time on cART (years) | 4 (3–7) |
| CD4 nadir (cells/mm3) | 378 (329–413) |
| CD4 count at T1 (cells/mm3) | 688 (616.5–872) |
| CD4/CD8 at T1 | 1.05 (0.935–1.33) |
| Virological characteristics | |
| VL zenith (log copies/mL) | 4.8 (4.48–5.05) |
| VL at rebound (log copies/mL) | 3.29 (3.23–3.64) |
| TTVR (>1000 copies/mL) (days) | 21 (20.5–26.5) |
Quantification of viral reservoir change during ATI: size and activity
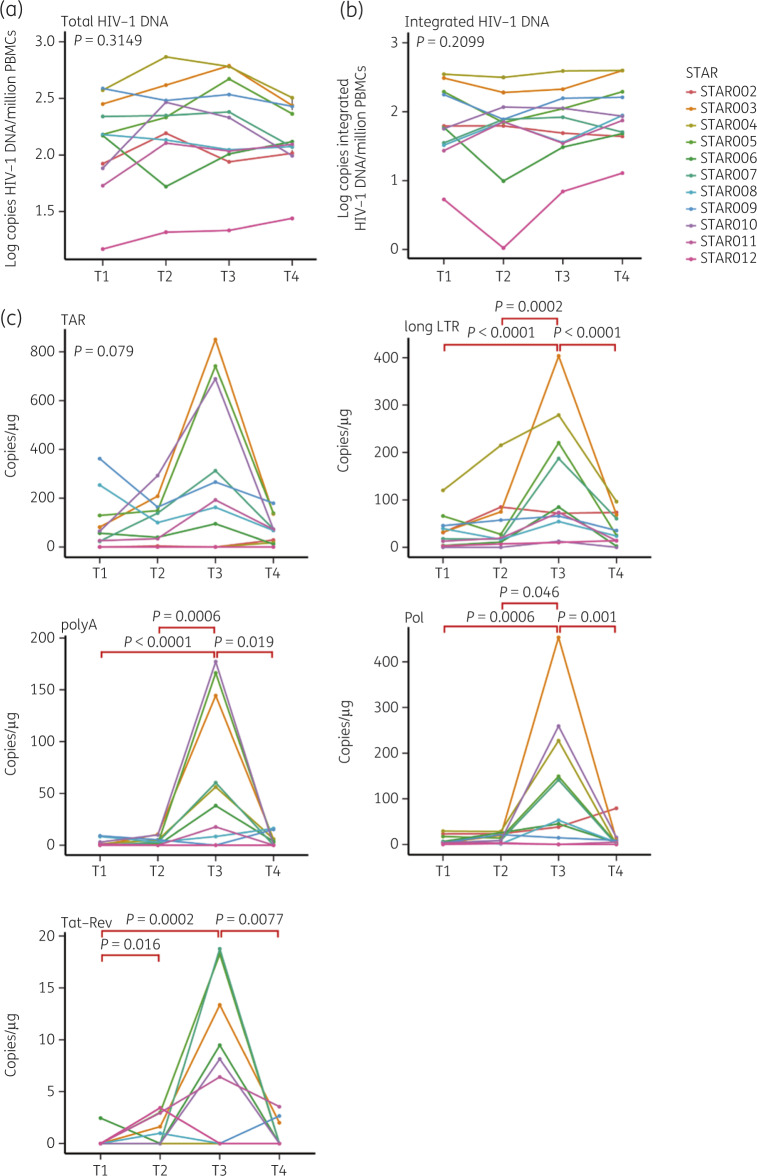
Levels of total and integrated HIV-1 DNA were determined at the four timepoints during ATI to further assess the stability of the viral reservoir and ATI safety. No remarkable changes at the different timepoints were detected for either marker (Figure 2a and b). Spearman correlation analysis at T1, T2, T3 and T4 revealed a highly significant positive correlation between total and integrated HIV-1 DNA levels (Spearman R = 0.75, R = 0.90, R = 0.90 and R = 0.90, respectively).
Figure 2.
Viral reservoir size quantification at the four different timepoints during ATI. Levels of total HIV-1 DNA (a) and integrated HIV-1 DNA (b) at T1, T2, T3 and T4. Levels for the different transcripts (TAR, long LTR, polyA, Pol and Tat-Rev) of CA HIV-1 RNA (c) at T1, T2, T3 and T4. Friedman statistical analysis with post hoc Dunn test was performed. Significant P values are indicated in red. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
TAR, long LTR, polyA, Pol and Tat-Rev HIV-1 RNA transcripts were determined at the different timepoints and exhibited peak levels at T3. Significant increases were detected for long LTR, polyA, Pol and Tat-Rev at T3 compared with T1 and T4 (T1: P < 0.0001, P < 0.0001, P = 0.0006 and P = 0.0002, respectively; T4: P < 0.0001, P = 0.019, P = 0.001 and P = 0.0077, respectively; Figure 2c). Repeated measurement correlations indeed indicated highly significant positive correlation between TAR, long LTR, polyA, Pol and Tat-Rev RNA transcription levels at the four timepoints. Interestingly, Tat-Rev levels were also significantly elevated at T2 compared with T1 (P = 0.016) (Figure 2c).
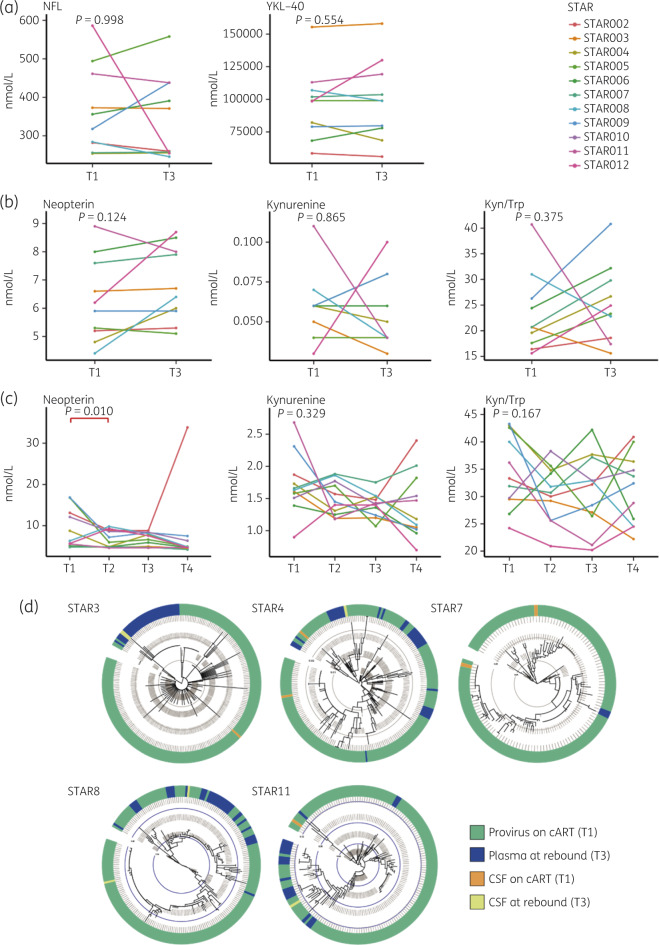
No increased inflammation in plasma and CSF after ATI
No significant increase in NFL or YKL-40 in CSF was observed between baseline (T1) and viral rebound (T3) (Figure 3a). Furthermore, markers of immune activation (neopterin, kynurenine and kynurenine/tryptophan ratio) were also found to be stable in CSF (Figure 3b). Similarly, plasma levels of neuroinflammatory markers did not increase during viral rebound (Figure 3c). Interestingly, neopterin levels in CSF pre-ATI were negatively correlated with time on treatment (P = 0.004), suggesting that CNS inflammation decreases with time on cART.
Figure 3.
Inflammation marker levels in CSF and plasma during ATI. (a) Levels of NFL and YKL-40 in CSF at T1 and T3. (b) Levels of immune activation markers neopterin, kynurenine and kynurenine/tryptophan in CSF at T1 and T3. (c) Levels of neopterin, kynurenine and kynurenine/tryptophan in plasma at T1, T2, T3 and T4. Statistical Wilcoxon signed rank (a and b) and Friedman test with post hoc Dunn analysis (c) were performed. Significant P values are indicated in red. Kyn/Trp, kynurenine/tryptophan. (d) Maximum-likelihood phylogenetic trees representing CSF and plasma sequences at T1 and T3 for five participants from whom we obtained CSF sequences. The coloured strip represents sampling origin for each sequence as indicated by the legend. The trees are drawn to scale and the grey circles represent the branch length from the root expressed as the number of substitutions per site. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
To further investigate viral replication in the CNS, the CSF-derived virus at T1 and T3 was sequenced for five patients. There was no clear sign for compartmentalization in the CNS (Figure 3d). Rebound lineages in CSF were more closely related to rebound lineages found in plasma compared with those found in CSF on cART, rendering it unlikely that these emerged from pre-existing CSF lineages. Rather, these most probably represent rebounding virus that crossed the blood–brain barrier, which is also indicated by the fact that at least one of the T3 CSF sequences was identical to a rebound virus in 2/5 patients (Figure 3d).
SLFN11 and APOBEC3G expression elevation prior to viral rebound
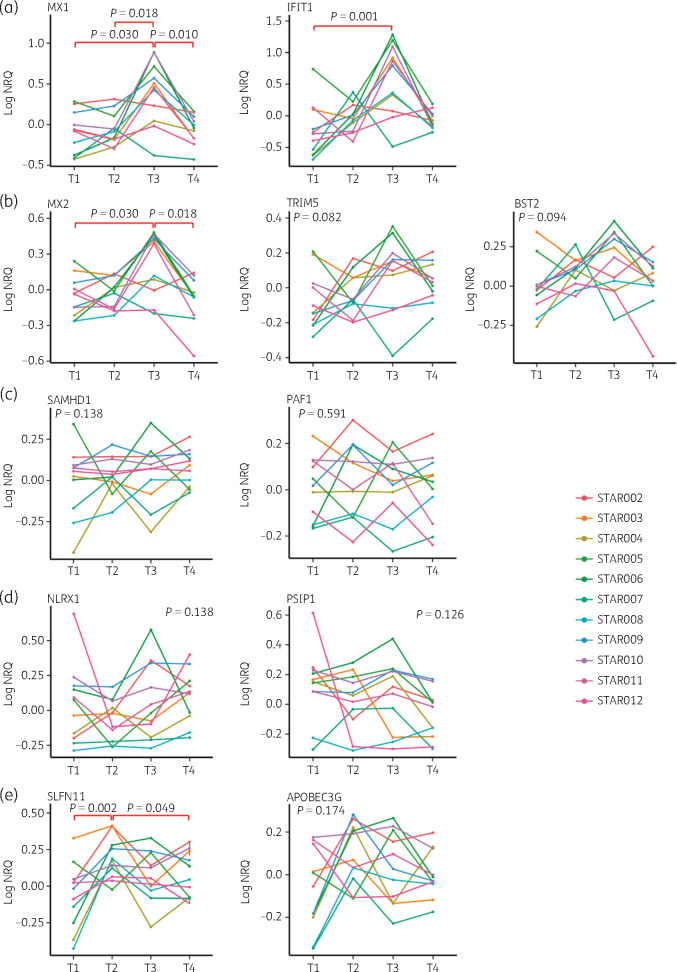
IFIT1 and MX1 are well-characterized ISGs and were used as markers for IFN exposure. They do not specifically interfere with HIV. Significant MX1 up-regulation was detected at viral rebound (T3) compared with timepoints with undetectable VL (T1, T2 and T4; P = 0.03, P = 0.02 and P = 0.01, respectively; Figure 4a). Similarly, IFIT1 levels demonstrated up-regulation at viral rebound (T3) compared with T1 (P = 0.001), suggesting that IFN induction is linked to active replication (Figure 4a). As most RFs are also induced by IFN, we wanted to determine whether APOBEC3G, SAMHD1, BST2, TRIM5, MX2, SLFN11 and PAF1 expression profiles would follow IFIT1 and MX1 levels and therefore exhibit peak expression at viral rebound (T3). Additionally to RFs, we hypothesized that expression levels of two HIV-1 dependency factors (NLRX1 and PSIP1) could also be linked to VL levels.
Figure 4.
ISG, RF and HIV-1 dependency factor expression levels at four timepoints during ATI. Normalized relative quantity levels at T1, T2, T3 and T4 for: (a) MX1 and IFIT1; (b) MX2, TRIM5 and BST2; (c) SAMHD1 and PAF1; and (d) NLRX1 and PSIP1. Friedman statistical analysis with post hoc Dunn test was performed. Significant P values are indicated in red. NRQ, normalized relative quantity. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Levels of the RFs MX2, TRIM5 and BST2 followed this assumption and on average reached their highest level at T3 although not reaching statistical significance for TRIM5 and BST2 (Figure 4b). SAMHD1 and PAF1 expression did not significantly change during ATI (Figure 4c). Similarly, HIV-1 dependency factors, NLRX1 and PSIP1, involved in inhibition of IFN response and encoding the integrase cofactor LEDGF/75, demonstrated no significant differences in expression levels between the different timepoints during ATI (Figure 4d). Surprisingly, SLFN11 demonstrated significant up-regulation at T2, which is post-ATI, but with still undetectable VL, compared with T1 and T4 (P = 0.002 and P = 0.049, respectively). APOBEC3G followed a similar pattern to SLFN11, although up-regulation at T2 was not significant (Figure 4e). Repeated measurements correlation analysis demonstrated a highly significant correlation between SLFN11 and APOBEC3G over the four timepoints (R = 0.80 and P < 0.0001), indicating their similar expression profiles (Figure S1).
RF expression levels are not correlated with TTVR
Because SLFN11 and APOBEC3G levels are up-regulated before plasma VL becomes detectable, we tested whether their expression was linked to TTVR and therefore could be used as a predictive marker. However, no association between expression levels of SLFN11, APOBEC3G or other antiviral factors and TTVR was detected. Interestingly, levels of total and integrated HIV-1 DNA at T1 and T2 were positively correlated with VL levels at rebound (T3) (T1: R = 0.52 and R = 0.70, respectively; T2: R = 0.9 and R = 0.85, respectively; Figure S2), suggesting that a larger viral reservoir gives rise to higher plasma VL levels.
Discussion
ATIs remain the gold standard to evaluate new possible HIV-1 cure strategies as we lack surrogate biomarkers that are able to assess the efficiency of cure strategies and their impact on TTVR after treatment interruption. In addition, the safety of these ATIs and the long-term effect on the HIV reservoir in the blood or other compartments is unclear. The primary research objective of the HIV-STAR ATI trial was to identify the origin of viral rebound after ATI by comparing proviral V1–V3 env sequences from different reservoirs [blood, lymph node, gut-associated lymphoid tissue (GALT), brain] with viral sequences found in plasma after ATI.14 We could not find any sign of compartmentalization by analysing more than 4000 sequences in these 11 participants and concluded that rebound is a heterogeneous process, involving different anatomical reservoirs and cellular subsets. In the present substudy, the effect of ATI on the viral reservoir size and inflammation status was evaluated at different timepoints before, during and after ATI to further address these safety concerns. Additionally, our study investigated the expression levels of HIV-1 RFs, dependency factors and the expression of different CA viral RNA transcripts defining distinct blocks to transcription at these timepoints to identify possible biomarkers for viral rebound.
Safety concerns in ATI trials: reservoir size, activity and inflammation
ATI did not increase levels of total and integrated HIV-1 DNA over the four different timepoints or of the different CA HIV-1 RNA transcripts pre- and post-ATI (T1 versus T4). These data are consistent with previous findings and indicate a stable viral reservoir after a short period of treatment interruption.6,7 Interestingly, levels of total and integrated HIV-1 DNA at T1 and T2 were positively correlated with VL levels at rebound (T3), suggesting that HIV-1-positive individuals with a larger viral reservoir before ATI have higher plasma VL levels at viral rebound.
We further assessed the effect of ATI on the HIV brain compartment. No detectable signs of neuronal injury or major CNS inflammation during viral rebound in plasma after (short) ATI were found, indicating that it is very unlikely that extensive replication in CNS was ongoing this early after treatment cessation. This is in line with the limited sequencing data that we obtained from CSF, which showed no signs of strong compartmentalization or of early rebound by CSF lineages.
These data supplement previous published results depicting that ATI combined with close monitoring is safe for the patient.6–8 However, two cases of HIV sexual transmission during treatment discontinuation within an ATI were reported recently,34,35 suggesting that considering the use of pre-exposure prophylaxis in seronegative partners should be advised in future ATI studies.
RF analysis
Expression of RFs and ISGs at the different timepoints was evaluated to find possible biomarkers predicting viral rebound. Levels of ISGs and the RFs MX2, TRIM5 and BST2 had a similar profile over time, peaking at the time VL became detectable (T3), suggesting that VL drives ISG and RF expression. These findings are in line with previous reports linking the levels of several RFs and VL in different cohorts of HIV-1-positive individuals.21,23 Interestingly, we found that SLFN11 and APOBEC3G were induced prior to other RFs and prior to the time of viral rebound (T3) and therefore could have the potential to serve as biomarkers predicting viral rebound after ATI. Elevated SLFN11 expression has been linked to long-term non-progressor (LTNP) status and reduced viral reservoir size,21,23 suggesting a role for SLFN11 in VL control. Therefore, early induction of SLFN11 could be a host defence mechanism attempting to control the virus. Yet, we could not find a correlation between the up-regulation of SLFN11 and APOBEC3G and TTVR. We also did not identify significant links between TTVR and expression of other RFs, participant characteristics (time since primary infection, time on cART, time before cART initiation), virological (total HIV-1 DNA, integrated HIV-1 DNA, VL zenith) and immunological (CD4 nadir) parameters (data not shown). Surprisingly, at T1 (cART; undetectable VL), a strong negative correlation was detected between expression levels of antiviral factors (MX1, IFIT1, APOBEC3G, PAF1, SAMHD1 and SLFN11) and NK cell count. Lower NK count and higher RF expression levels in ART-treated individuals have been linked to lower immune activation and better preservation of immune responses, respectively.
Reservoir activity prior to detectable VL
Another finding that merits further investigation is the expression profile of CA HIV-1 RNA transcripts defining distinct blocks to transcription during ATI. Transcription of TAR, long LTR, polyA, Pol and Tat-Rev fragments tended to be up-regulated at viral rebound (T3). Interestingly, increased Tat-Rev expression preceded viral rebound, whereas for other transcripts the increase coincided with the timepoint of viral rebound (T3). Tat-Rev transcription indicates the ability to overcome blocks to transcription initiation, elongation and termination and therefore might be a suitable marker for forthcoming viral rebound.24,25 To note, although Tat-Rev transcript levels were at the lower end of detection, we were able to pick up quantifiable results by ddPCR, underlining the strength of this platform for nucleic acid quantification.
Limitations
The current study has several limitations. This ATI trial included only 11 participants, making it difficult to draw population-based conclusions. However, the design of ATIs, especially with this extensive tissue sampling, makes it challenging to include participants, both from a motivational and a financial point of view. Second, RF and HIV-1 RNA expression analyses were performed in PBMCs and not in CD4 T cells or T cell subsets, in contrast to other studies.8,36 Nonetheless, significant signals could be picked up. If CD4 T cells could be purified from the PBMC fraction, it can be expected that the observed signal of HIV-1 transcripts from PBMCs would increase.
Conclusions
Overall, our data support that ATI is safe and, if combined with close monitoring and immediate treatment reinitiation at the time of viral rebound, this intervention can be considered as the final and most comprehensive read-out of HIV cure trials. ATI did not increase viral reservoir size and it did not reveal signs of increased neuronal injury or inflammation. Furthermore, ATIs give rise to unique opportunities to further investigate the viral reservoir in blood and other anatomical compartments and should be used to investigate potential biomarkers for viral rebound. Elevation of Tat-Rev transcription and induced expression of the RFs SLFN11 and APOBEC3G rapidly after ATI prior to viral rebound indicates that these factors could be used as potential biomarkers predicting viral rebound. In this context, evaluating additional timepoints could be beneficial to see whether these transcripts can be detected even earlier than T2.
Supplementary Material
Acknowledgements
We would like to acknowledge the HIV-1-infected individuals who participated in this study, Ghent University Hospital and all other collaborators for providing the necessary support and resources. We would also like to acknowledge the doctors involved at the AIDS Reference Centre in Ghent for recruiting participants, all of the doctors involved in the sampling procedures, Dr D. Hemelsoet and Professor M. Coppens, and all the supporting nurses (especially E. Van Caelenbergh, A. Masset, E. Caluwé and S. Vanherreweghe) and facilities.
Funding
This work was supported by a Merck Sharp & Dohme (MSD) investigator grant (grant number: ISS 52777). M.-A.D.S. was granted a Research Foundation Flanders (FWO) travel grant and a Sofina Gustave-Boel fellowship. L.V. is supported by the FWO (1.8.020.09.N.00), S.P. is supported by the Delaney AIDS Research Enterprise (DARE) to Find a Cure (1U19AI096109 and 1UM1AI126611-01) and the Australian National Health and Medical Research Council (AAP1061681), C.V.H. and S.R. are funded by the FWO (FWO-SB grant ID: 1S 308 16 N and 1S 329 16 N, respectively). P.L. is supported by the Special Research Fund, KU Leuven (‘Bijzonder Onderzoeksfonds’, KU Leuven, OT/14/115), the FWO (G066215N) and the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 725422-ReservoirDOCS). B.V. is supported by a postdoctoral fellowship from the FWO. H.Z. is a Wallenberg Academy Fellow supported by grants from the Swedish Research Council (#2018–02532), the European Research Council (#681712) and Swedish State Support for Clinical Research (#ALFGBG-720931 and ALFGBG-717531) and the UK Dementia Research Institute at UCL. The funders had no role in the design of the study, the collection of data, analysis and interpretation, or the decision to submit the work for publication.
Transparency declarations
H.Z. has served on scientific advisory boards for Roche Diagnostics, Wave, Samumed and CogRx, has given lectures in symposia sponsored by Biogen and Alzecure, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg (all outside the submitted work). All other authors: none to declare.
Author contributions
M.-A.D.S. performed patient inclusion, sample collection, data analysis/interpretation and manuscript drafting. C.V.H. performed experimental work, data analysis, interpretation, visualization, statistical analyses and manuscript drafting. N.D.L. performed experimental work, data analysis and statistics. S.R. performed data analysis. B.V. performed data analysis. W.T. contributed to data interpretation. Y.N. performed experimental work. H.Z. generated the NFL and YKL-40 data, interpreted data and revised the manuscript for important intellectual content. D.F. performed the analysis on plasma immune markers. S.P., P.L., M.G. and L.V. supervised the experiments. B.V.D.G, J.P. and F.V.W participated in participant recruitment. All co-authors revised and edited the manuscript.
References
- 1. Sadowski I, Hashemi FB.. Strategies to eradicate HIV from infected patients: elimination of latent provirus reservoirs. Cell Mol Life Sci 2019; 76: 3583–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kutzler MA, Jacobson JM.. Treatment interruption as a tool to measure changes in immunologic response to HIV-1. Curr Opin HIV AIDS 2008; 3: 131–5. [DOI] [PubMed] [Google Scholar]
- 3. Siliciano JD, Siliciano RF.. Assays to measure latency, reservoirs, and reactivation. Curr Top Microbiol Immunol 2018; 417: 23–41. [DOI] [PubMed] [Google Scholar]
- 4. Henrich TJ, Hanhauser E, Marty FM. et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med 2014; 161: 319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. El-Sadr WM, Lundgren J, Neaton JD. et al. CD4+ count-guided interruption of antiretroviral treatment. N Engl J Med 2006; 355: 2283–96. [DOI] [PubMed] [Google Scholar]
- 6. Salantes DB, Zheng Y, Mampe F. et al. HIV-1 latent reservoir size and diversity are stable following brief treatment interruption. J Clin Invest 2018; 128: 3102–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clarridge KE, Blazkova J, Einkauf K. et al. Effect of analytical treatment interruption and reinitiation of antiretroviral therapy on HIV reservoirs and immunologic parameters in infected individuals. PLoS Pathog 2018; 14: e1006792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams JP, Hurst J, Stohr W. et al. HIV-1 DNA predicts disease progression and post-treatment virological control. eLife 2014; 3: e03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yilmaz A, Yiannoutsos CT, Fuchs D. et al. Cerebrospinal fluid neopterin decay characteristics after initiation of antiretroviral therapy. J Neuroinflammation 2013; 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gelpi M, Hartling HJ, Ueland PM. et al. Tryptophan catabolism and immune activation in primary and chronic HIV infection. BMC Infect Dis 2017; 17: 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hermansson L, Yilmaz A, Axelsson M. et al. Cerebrospinal fluid levels of glial marker YKL-40 strongly associated with axonal injury in HIV infection. J Neuroinflammation 2019; 16: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yilmaz A, Blennow K, Hagberg L. et al. Neurofilament light chain protein as a marker of neuronal injury: review of its use in HIV-1 infection and reference values for HIV-negative controls. Expert Rev Mol Diagn 2017; 17: 761–70. [DOI] [PubMed] [Google Scholar]
- 13. Gisslen M, Rosengren L, Hagberg L. et al. Cerebrospinal fluid signs of neuronal damage after antiretroviral treatment interruption in HIV-1 infection. AIDS Res Ther 2005; 2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Scheerder MA, Vrancken B, Dellicour S. et al. HIV rebound is predominantly fueled by genetically identical viral expansions from diverse reservoirs. Cell Host Microbe 2019; 26: 347–58 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doyle T, Goujon C, Malim MH.. HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol 2015; 13: 403–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merindol N, Berthoux L.. Restriction factors in HIV-1 disease progression. Curr HIV Res 2015; 13: 448–61. [DOI] [PubMed] [Google Scholar]
- 17. Ghimire D, Rai M, Gaur R.. Novel host restriction factors implicated in HIV-1 replication. J Gen Virol 2018; 99: 435–46. [DOI] [PubMed] [Google Scholar]
- 18. Liu L, Oliveira NM, Cheney KM. et al. A whole genome screen for HIV restriction factors. Retrovirology 2011; 8: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simon V, Bloch N, Landau NR.. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 2015; 16: 546–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malim MH, Bieniasz PD.. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 2012; 2: a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abdel-Mohsen M, Raposo RA, Deng X. et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology 2013; 10: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raposo RA, Abdel-Mohsen M, Deng X. et al. Dynamic regulation of host restriction factor expression over the course of HIV-1 infection in vivo. J Virol 2014; 88: 11624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Hecke C, Trypsteen W, Malatinkova E. et al. Early treated HIV-1 positive individuals demonstrate similar restriction factor expression profile as long-term non-progressors. EBioMedicine 2019; 41: 443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yukl SA, Kaiser P, Kim P. et al. HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med 2018; 10: eaap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fischer M, Joos B, Hirschel B. et al. Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef. J Infect Dis 2004; 190: 1979–88. [DOI] [PubMed] [Google Scholar]
- 26. Hellemans J, Vandesompele J.. Selection of reliable reference genes for RT-qPCR analysis. Methods Mol Biol 2014; 1160: 19–26. [DOI] [PubMed] [Google Scholar]
- 27. Hellemans J, Mortier G, De Paepe A. et al. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 2007; 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaetani L, Hoglund K, Parnetti L. et al. A new enzyme-linked immunosorbent assay for neurofilament light in cerebrospinal fluid: analytical validation and clinical evaluation. Alzheimers Res Ther 2018; 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laich A, Neurauter G, Widner B. et al. More rapid method for simultaneous measurement of tryptophan and kynurenine by HPLC. Clin Chem 2002; 48: 579–81. [PubMed] [Google Scholar]
- 30. Rutsaert S, De Spiegelaere W, Van Hecke C. et al. In-depth validation of total HIV-1 DNA assays for quantification of various HIV-1 subtypes. Sci Rep 2018; 8: 17274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trypsteen W, Mohammadi P, Van Hecke C. et al. Differential expression of lncRNAs during the HIV replication cycle: an underestimated layer in the HIV-host interplay. Sci Rep 2016; 6: 36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Spiegelaere W, Malatinkova E, Lynch L. et al. Quantification of integrated HIV DNA by repetitive-sampling Alu-HIV PCR on the basis of Poisson statistics. Clin Chem 2014; 60: 886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Faschingbauer M, Nelitz M, Urlaub S. et al. Return to work and sporting activities after high tibial osteotomy. Int Orthop 2015; 39: 1527–34. [DOI] [PubMed] [Google Scholar]
- 34. Lelievre JD, Hocqueloux L.. Unintended HIV-1 transmission to a sex partner in a study of a therapeutic vaccine candidate. J Infect Dis 2019; 220 Suppl 1: S5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ugarte A, Romero Y, Tricas A. et al. Unintended HIV-1 infection during analytical therapy interruption. J Infect Dis 2019; doi:10.1093/infdis/jiz611. [DOI] [PubMed] [Google Scholar]
- 36. Hurst J, Hoffmann M, Pace M. et al. Immunological biomarkers predict HIV-1 viral rebound after treatment interruption. Nat Commun 2015; 6: 8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.