Abstract
Background
Methionyl-tRNA synthetase (MetRS) inhibitors are under investigation for the treatment of intestinal infections caused by Giardia lamblia.
Objectives
To properly analyse the therapeutic potential of the MetRS inhibitor 1717, experimental tools including a robust cell-based assay and a murine model of infection were developed based on novel strains of G. lamblia that employ luciferase reporter systems to quantify viable parasites.
Methods
Systematic screening of Giardia-specific promoters and luciferase variants led to the development of a strain expressing the click beetle green luciferase. Further modifying this strain to express NanoLuc created a dual reporter strain capable of quantifying parasites in both the trophozoite and cyst stages. These strains were used to develop a high-throughput cell assay and a mouse infection model. A library of MetRS inhibitors was screened in the cell assay and Compound-1717 was tested for efficacy in the mouse infection model.
Results
Cell viability in in vitro compound screens was quantified via bioluminescence readouts while infection loads in mice were monitored with non-invasive whole-animal imaging and faecal analysis. Compound-1717 was effective in clearing mice of Giardia infection in 3 days at varying doses, which was supported by data from enzymatic and phenotypic cell assays.
Conclusions
The new in vitro and in vivo assays based on luciferase expression by engineered G. lamblia strains are useful for the discovery and development of new therapeutics for giardiasis. MetRS inhibitors, as validated by Compound-1717, have promising anti-giardiasis properties that merit further study as alternative therapeutics.
Introduction
Giardia lamblia infections are among the most common causes of chronic diarrhoea in children in resource-limited environments. New therapeutic agents are needed to address issues with existing therapies including resistance, toxicity and reduced efficacy. No vaccines have been developed for clinical use, so case management depends solely on antimicrobial chemotherapy.1 Current therapies approved by the FDA for giardiasis include metronidazole and tinidazole.2–4 However, approximately 20% of clinical cases involve metronidazole- (and presumably tinidazole-) resistant Giardia.5 Second-line drugs such as albendazole, nitazoxanide, furazolidone and paromomycin generally have lower efficacy rates and/or potentially dangerous side effects.6 These factors necessitate the development of a new therapeutic agent, which requires various experimental tools for screening and verification of potential leads.
While experimental methods suitable for compound screening against G. lamblia have been described in the literature,7 infection models in adult mice can benefit from an improved infection monitoring method. The deficiency of the in vivo model can be explained in part by earlier studies showing instability in the expression of transgenes in Giardia compared with many other eukaryotic cells.8 The instability leads to variable expression as Giardia, by an unknown mechanism, down-regulates transgenic markers.8 We recently described a reporter system in G. lamblia WBC6 trophozoites using a red-shifted firefly luciferase (PpyRE9h) under the control of β-tubulin promoter (pβTub).9,10 PpyRE is a red-shifted Ser284Thr mutant of the yellow–green emitting WT Ppy derived from Photinus pyralis (North American firefly).9,11 PpyRE9h was developed by altering nine amino acids in PpyRE to make it useful in pH-independent, thermostable luciferase applications, including in vivo implementations.9,12 While the bioluminescent output of G. lamblia WBC6:pβTub::PpyRE9h was sufficiently stable for 24 h in vitro assays, it lacked long-term stability over multiple passages and proved unsuitable for a mouse infection model. In this study, we describe the development of a new strain of G. lamblia using a click beetle green luciferase (CBG99) under a short glutamate dehydrogenase promoter (pGDHS) that can be used for in vitro and in vivo assays. A second strain was engineered by endogenously tagging one copy of the cyst-wall protein 1 gene (CWP1) with NanoLuc (Nluc) in the G. lamblia WBC6:pGDHS::CBG99 strain, establishing a second developmentally induced reporter that measures cyst quantity (pCWP1::CWP1-Nluc). The strains can be visualized in an animal model of infection using non-invasive imaging to evaluate experimental drug effects in Giardia-infected animals. The model was validated with metronidazole treatment and was subsequently used to support G. lamblia methionyl-tRNA synthetase (GlMetRS) enzyme inhibitor 1717 as a potential therapeutic agent for treatment of giardiasis.
Materials and methods
Plasmid construction and G. lamblia transfection
To optimize luminescence of Giardia for in vitro and in vivo experiments, various combinations of luciferases and Giardia-specific promoters were tested. Twelve gene constructs were generated: PpyRE9h driven by three Giardia promoters, five luciferases driven by the long glutamate dehydrogenase promoter (pGDHL) and CBG99 driven by four Giardia promoters.
The PpyRE9h constructs were driven by pGDHL, the short glutamate dehydrogenase promoter (pGDHS)13 and the ornithine carbamoyltransferase promoter (pOCT).14 These were amplified by PCR using the primers in Table 1 and digested with EcoRI and XbaI. The promoter fragments were cloned into the EcoRI-XbaI site of integration vector pPACV-integ.10,15
Table 1.
List of primers
| Sequence (5′–3′) | |
|---|---|
| pOCT | |
| forward | TTGCGGCCGCGAATTCTCAATTGTCAGCTTCATTTTTATTAT |
| reverse | TTTCTAGATTTAATTTTCAGCCTCTACTGTAGAGCGTTTA |
| pGDHS | |
| forward | TTGAATTCGACCACAAATAACGCCTTTAA |
| reverse | TTTCTAGATTTAAAATCTGGGGCGCCTGTAATTAAA |
| pGDHL | |
| forward | TTGCGGCCGCGAATTCAATGTCCACAAACTAAAAGTATAC |
| reverse | TTTCTAGATTTAAAATCTGGGGCGCCTGTAATTAAA |
| Fluc | |
| forward | AATCTAGAATGGAAGACGTCAAAAACATAAA |
| reverse | GCTTAATTAATTATTCTCGAGACACGGCGATCTTT |
| Luc2 | |
| forward | AATCTAGAATGGAAGATGCCAAAAACATTAAGAA |
| reverse | GCTTAATTAATTACACGGCGATCTTGCCGCCCTTCTT |
| CBR | |
| forward | AATCTAGAATGGTAAAGCGTGAGAAAAATGTCATCTATGG |
| reverse | GCTTAATTAATTATTTGTACAAACCGCCGGCCTTCACCAA |
| Eluc | |
| forward | AATCTAGAATGGAGAGAGAGAAGAACGTGGTGTA |
| reverse | GCTTAATTAACTACACATTGATCCTAGCAGAAGCACA |
| CBG99 | |
| forward | TTTCTAGAATGGTGAAGCGTGAGAAAAATGTCATCTATGG |
| reverse | GGTTAATTAATTATTTGTACAAACCGCCGGCCTTCTCCA |
| CWP1 | |
| forward | GCTCTAGACAACGGCTTACTAAATCATTCT |
| reverse | CCGGATCCactagtAGGCGGGGTGAGGCAGTACTCTCC |
| Nluc | |
| forward | GTGGATCCGGAGGCGGTTCAGGCGGAGGTGGCTCTGTCTTCACACTCGAAGATTTCGTT |
| reverse | AAGAATTCTTACGCCAGAATGCGTTCGCACAGCCGC |
For constructs driven by pGDHL, the coding regions of the standard firefly luciferase (Fluc), the firefly luciferase 2 (Luc2), the red click beetle (CBR), the enhanced green-emitting (Eluc) and the CBG99 genes were amplified with primers from Table 1 and digested with XbaI and PacI. The XbaI-PacI fragments were cloned downstream of the promoter in the pPACV-integ vector’s XbaI-PacI cloning site.
To generate the CBG99 constructs, pGDHL, pGDHS, pOCT and pβTub were amplified with primers from Table 1, digested using EcoRI and XbaI, and cloned into the EcoRI and XbaI site of vector pPACV-integ. The CWP1-Nluc construct was made from the PCR amplicon of CWP1 (GL50803_5638) and Nluc genes using primers from Table 1. The amplicons were digested with XbaI-BamHI and BamHI-EcoRI, respectively, then cloned into the XbaI and EcoRI site of the pKS-NEO vector.
For integration, 30 μg of pPACV-integ constructs was linearized with SwaI while the CWP1-Nluc/pKS-NEO construct was linearized with StuI overnight, at 25°C and 37°C respectively. They were precipitated with ethanol and incubated with 300 μL of chilled G. lamblia cells (∼13 × 106 cells/mL) for 30 min before electroporation (Bio-Rad GenePulser X at 375 V, 1000 μF, 750 ohms). After electroporation, cells were incubated on ice for 10 min, then transferred to fresh medium at 37°C. Transfectants were selected with puromycin dihydrochloride (Gibco, Ireland) after overnight recovery.
Parasite cultures
G. lamblia WBC6:pβTub::PpyRE9h and WT (WBC6, ATCC 50803) trophozoites were the starting strains and all strains were grown in conditions previously described.10,16 Antibiotic selection pressure of 32 mg/L puromycin was applied to transfected strains. Cultures were incubated at 37°C in 16 mL Falcon round-bottom polystyrene test tubes (Corning Inc., USA).
G. lamblia trophozoite in vitro bioluminescence optimization
The optimum concentration of luminescent substrate, d-luciferin (GoldBio, USA), required for cell identification was determined as described earlier.10 AkaLumine (a d-luciferin analogue that is not recognized by the CBG99 enzyme) was used as a control.17,18 Plates were incubated for time increments between 5 and 180 min at room temperature. Parasite numbers were correlated with bioluminescence intensity by plating 2-fold serial dilutions of cells.10 Plates were read with an EnVision Multilabel Plate Reader (Perkin Elmer, USA) after incubation at room temperature. Assays were repeated at least twice on different days.
G. lamblia luciferase expression stability
To evaluate the stability of G. lamblia WBC6:pGDHL::PpyRE9h and G. lamblia WBC6:pGDHL::CBG99 in the absence of puromycin selection, the strains were diluted (1:130 dilutions) twice a week with or without puromycin for 4 weeks. The luciferase activity proportional to cell concentration was measured as described above.
G. lamblia cell assay development
Assays were performed in clear, flat-bottom 96- or 384-well plates (Corning Inc., USA) in modified TYI-S-33 medium.10,16 Compound screens used one concentration between 20 μM and 1 μM, and dose–response assays employed 3-fold serial dilutions starting at concentrations up to 60 μM, depending on the compound’s potency. Growth was evaluated under a microscope before using vinyl stickers to obscure the clear bottoms for luminescence reading. Ten microlitres of 2.5 mg/mL d-luciferin was added to each well and allowed to incubate at room temperature on a shaker protected from light. Response curves and EC50 values (the concentration at which cell growth is inhibited by 50%) were calculated using Prism 6 (GraphPad, USA). Assays were repeated on different days.
Using the G. lamblia WBC6:pGDHS::CBG99 strain, a library of MetRS inhibitors was screened at a final concentration of 20 μM. G. lamblia WBC6:pGDHS::CBG99 and WBC6:pGDHS::CBG/Nluc trophozoites were used to confirm hits (compounds causing >80% growth inhibition) in a 2 μM screen.
Giardia murine model development and efficacy of Compound-1717
Female BALB/c mice, 8–12 weeks old and weighing approximately 20 g, were used for this study. To promote parasite colonization, mice were administered antibiotics (0.25 mg/mL ampicillin and neomycin) in drinking water for the duration of the experiment, starting 6 days before infection.19 A cocktail containing neomycin (15 mg/kg), ampicillin (50 mg/kg) and metronidazole (50 mg/kg) was dosed by oral gavage 3 days before infection. A second antibiotic dose containing ampicillin (50 mg/kg) and neomycin (15 mg/kg) was given 1 day before infection.
Mice were administered 1 × 107G. lamblia WBC6:pGDHS::CBG99 trophozoites by oral gavage. Infection was confirmed using non-invasive IVIS imaging, after intraperitoneal injection of 150 mg/kg d-luciferin and sedation under continuous administration of isoflurane anaesthetic.19
A pilot in vivo drug treatment analysis with metronidazole at 50 mg/kg and dosing vehicle (3% ethanol and 7% Tween 80 in normal saline) as negative control once daily (q24h) was used to validate the model. This was followed by studies with Compound-1717 at 50 mg/kg q24h. Once the model was functionally optimized, various doses of Compound-1717 were tested: 50 mg/kg twice daily (q12h), 25 mg/kg q12h and 50 mg/kg q24h. Control groups included treatment with dosing vehicle q12h and metronidazole at 50 mg/kg q12h. Treatments were introduced by oral gavage for 3 days. The course of infection was tracked by IVIS imaging.
Faecal samples were collected throughout the treatment timeline. DNA was extracted using the QIAamp Fast DNA Stool Mini Kit (QIAGEN, Germany). Cyst shedding in treated and untreated mice was verified by nested PCR analysis of the highly conserved GDH gene.
G. lamblia cyst in vitro bioluminescence
An encystation protocol was adapted from previous studies to demonstrate the signal intensity of G. lamblia WBC6:pGDHS::CBG/Nluc cysts.20,21 Confluent trophozoites were incubated with encystation media for 48 h, then pelleted and resuspended in growth media for 24 h. Cells were pelleted and resuspended in cold distilled water and stored at 4°C overnight. The amount of light emission was quantified as described above, with the modification that Nano-Glo® luciferase reagent (Promega, USA) was the luminescent substrate.
Ethics
All animal procedures were conducted in adherence with federal regulations and University of Washington’s IACUC guidelines under protocol number 2154-01.
Results
Promoter and luciferase gene selection
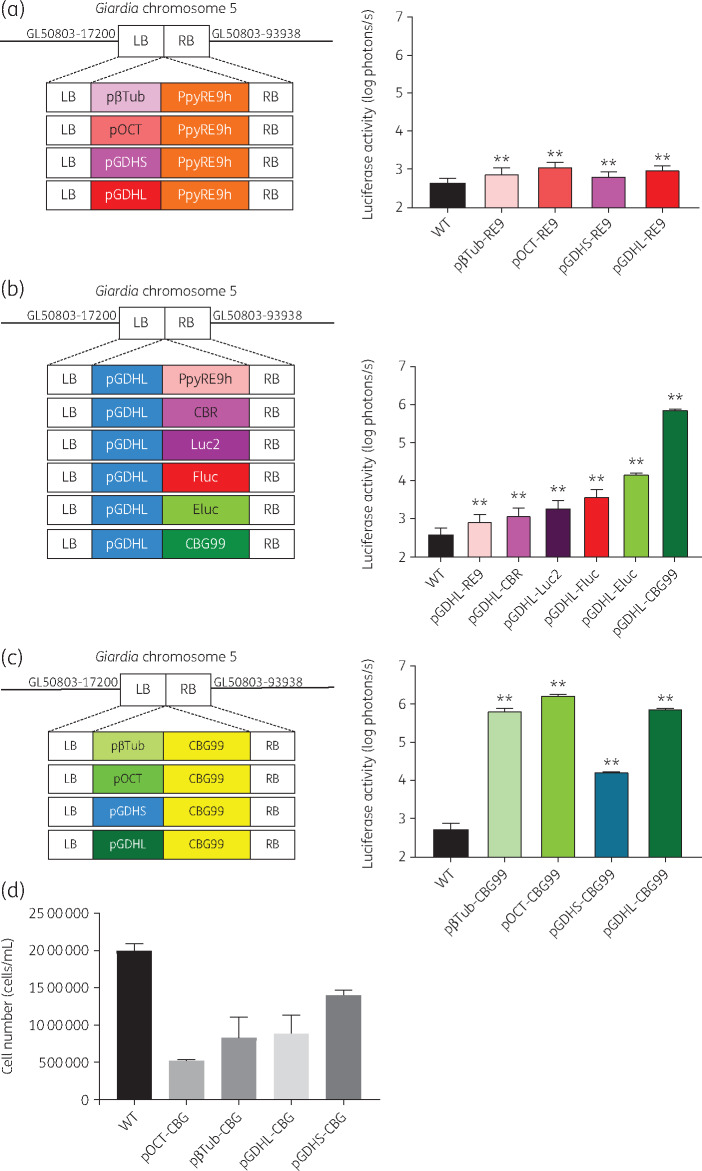
We previously used the β-tubulin promoter to drive red-shifted firefly luciferase PpyRE9h in G. lamblia WBC6 cells to assay growth inhibition by compounds in the MMV Pathogen Box.10 Further analysis showed that the luminescent signal diminished after 3 weeks of continuous incubation with or without antibiotic selection. To improve the expression level of PpyRE9h, the pβTub was replaced by three Giardia constitutive promoters: a 206 bp pOCT,14 a 44 bp pGDHS13 and a 165 bp pGDHL.13 The resultant G. lamblia trophozoites expressing pGDHL::PpyRE9h and pOCT::PpyRE9h had 2- and 3-fold increased luminescence levels relative to pβTub::PpyRE9h, respectively. There was no significant change for pGDHS::PpyRE9h relative to the pβTub::PpyRE9h strain. However, the highest luminescence value that any combination of promoter and PpyRE9h achieved was a modest 1200 photons/s for G. lamblia WBC6:pOCT::PpyRE9h (Figure 1a). We speculated that this would not be bright enough for a mouse infection model, where the luminescent signal must penetrate multiple layers of tissue to visualize infection non-invasively. A fitness cost also appeared for pOCT::PpyRE9h cells, exhibited by a decrease in growth rate. Thus, further work was needed to establish a brighter and more stable luciferase reporter system in G. lamblia. Earlier studies demonstrated that Fluc, Luc2 and CBR have higher sensitivity than PpyRE9h to report promoter activity in other systems, so these luciferases were assayed.22,23
Figure 1.
In vitro characterization of luciferase sensitivity. (a) Left: schematic of PpyRE9h driven by four Giardia specific promoters and homologous recombination. Right: in vitro comparison of PpyRE9h-dependent photon flux driven by four Giardia-specific promoters. (b) Left: schematic of six different luciferases driven by pGDHL and homologous recombination. Right: in vitro quantitative analysis of luciferase-dependent photon flux from six different luciferases driven by pGDHL. (c) Left: schematic of CBG99 driven by four Giardia-specific promoters and homologous recombination. Right: in vitro comparison of CBG99-dependent photon flux driven by four Giardia-specific promoters. LB, left border; RB, right border. (d) Effect of different promoters on growth rate. At Day 1, 150 μL (1 × 105 cells/mL) of culture was transferred to 13 mL TYI-S-33. Cell density measurement after 3 days incubation at 37°C showed a significant fitness loss for most of the luciferase strains relative to the parent WT parasite. G. lamblia WBC6:pGDHS::CBG99 showed the lowest fitness disadvantage. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The red luciferase genes for PpyRE9h, Fluc, Luc2 and CBR inserted downstream of pGDHL were transfected into G. lamblia WBC6 cells. Among the resultant strains, trophozoites expressing Fluc had the brightest signal intensity, followed by those expressing Luc2 (Figure 1b). Two green luciferase genes, CBG99 and Eluc, previously shown to exhibit higher levels of bioluminescence than Fluc,24,25 were also assayed under the control of pGDHL. G. lamblia WBC6:pGDHL::CBG99 was 100-fold brighter in signal intensity than G. lamblia WBC6:pGDHL::Eluc. We conclude that G. lamblia WBC6:pGDHL::CBG99 is the brightest reporter system among the red and green luciferases tested (Figure 1b).
Comparative analysis to determine the most suitable promoter for CBG99 in Giardia was subsequently performed. The CBG99 gene in the transfection construct was placed under the control of four different Giardia promoters: pβTub, pOCT, pGDHS and pGDHL. All the transgenic G. lamblia WBC6 strains expressing CBG99 gave robust bioluminescence signals, with pGDHS::CBG99 being the lowest even on the log scale (Figure 1c). However, G. lamblia strains expressing CBG99 driven by pβTub, pOCT and pGDHL had substantial fitness disadvantages in their growth rate relative to the WT strain. The effect was least pronounced with G. lamblia WBC6:pGDHS::CBG99 (Figure 1d). Though it had the lowest signal of the promoters tested with CBG99, the signal intensity was still several-fold brighter than the one given by pGDHS::PpyRE9h and therefore more suitable for drug-screening assays and murine model development.
G. lamblia WBC6:pGDHS::CBG99 in vitro bioluminescence signal
Optimum luminescence was achieved after 5 min of incubation with d-luciferin at a final concentration of ∼8 mg/L (96-well). At this concentration and incubation time, the signal was high enough to distinguish living cells at 103 cells/mL concentration. The signal remained stable for up to 30 min after the addition of substrate, but the luminescence decreased after longer incubation. The amount of bioluminescence activity was directly proportional to the number of viable transfected G. lamblia trophozoites per well, as determined by a comparative analysis with direct cell count (Figure 2a). The blank medium control wells showed no luminescence when compared with the empty background wells and the sample wells. AkaLumine showed no signal at any timepoint, as expected. The Z′ factor, a screening window coefficient that provides insight into the dynamic range and data variation associated with signal measurements, was used to monitor the quality of each experiment.26,27 The value ranges from 0 to 1 and is inversely proportional to the probability of false negatives or positives. The average Z′ of this assay was 0.6.
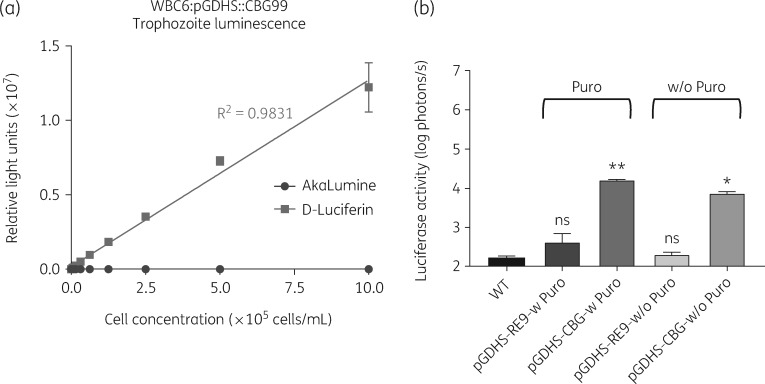
Figure 2.
Parasite cell count versus relative light units. (a) The plot shows the linearity of bioluminescence readouts from the G. lamblia WBC6:pGDHS::CBG99 strain. Readout follows 5 min of incubation at room temperature. AkaLumine showed no signal at any timepoint. (b) In vitro comparison of PpyRE9h-dependent and CBG99-dependent photon flux in the presence (Puro) or absence (w/o Puro) of selection antibiotic (puromycin) for 4 weeks. Level of significance is indicated by ns = not significant, *P ≤ 0.001 and **P ≤ 0.0001.
G. lamblia luciferase expression stability
The bioluminescent signal of G. lamblia WBC6:pβTub::PpyRE9h was the same as the WT G. lamblia WBC6 after 3 weeks of incubation, suggesting a loss of the luciferase activity. Therefore, we tested for signal stability in the G. lamblia WBC6:pGDHS::CBG99 cells in the absence of the selection antibiotic. After 4 weeks without antibiotic, the bioluminescent signal of G. lamblia WBC6:pGDHS::CBG99 was detectable at sufficiently high levels. The signal was 35 times brighter than G. lamblia WBC6:pβTub::PpyRE9h cell bioluminescence after the same amount of time (Figure 2b). This is especially important to guarantee robust detection in a chronic mouse infection model lasting over 3 weeks.
Screening of MetRS inhibitors including Compound-1717
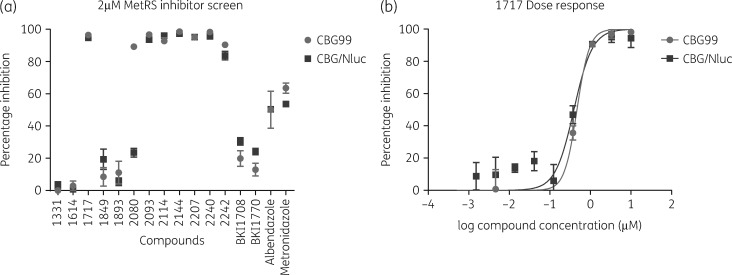
The initial MetRS inhibitor screen at 20 μM proved to be highly potent against the parasites, with inhibition rates >98% across most compounds. The concentration was subsequently dropped to 2 μM, where most compounds still had potent inhibition (Figure 3a, Table 2). Compounds BKI1708 and BKI1770, which are inhibitors of apicomplexan kinases not found in G. lamblia, were included as negative controls.28 The low inhibition rates of BKI1708 and BKI1770 at both concentrations demonstrate the specificity of the assay. Percentage inhibition data from the compound screens are presented in Table 2.
Figure 3.
(a) MetRS inhibitor screening of G. lamblia WBC6:pGDHS::CBG99 versus WBC6:pGDHS::CBG/Nluc. (b) Dose–response plot of Compound-1717 inhibition of G. lamblia WBC6:pGDHS::CBG99 versus WBC6:pGDHS::CBG/Nluc.
Table 2.
Percentage inhibition of luciferase G. lamblia cells by MetRS inhibitors (at inhibitor final concentration of 2 µM)
| Compound | Chemical structure | Percentage inhibition |
|||
|---|---|---|---|---|---|
| CBG99 |
CBG/Nluc |
||||
| mean | SEM | mean | SEM | ||
| 1331 |
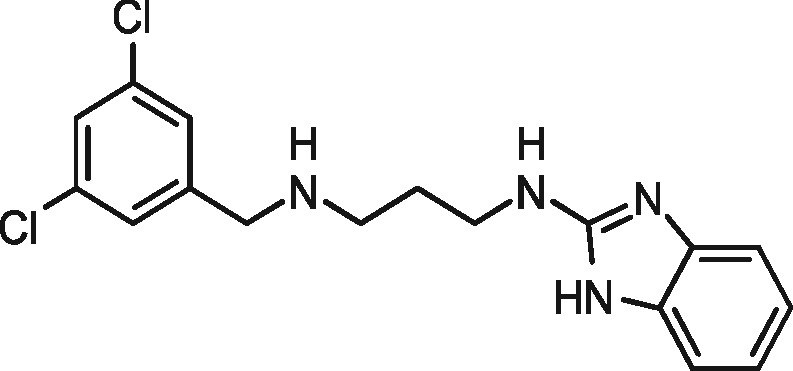
|
no inhibition | — | no inhibition | — |
| 1614 |
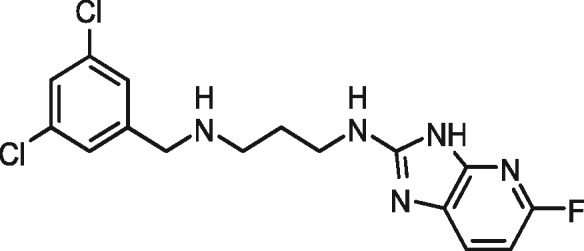
|
no inhibition | — | no inhibition | — |
| 1717 |
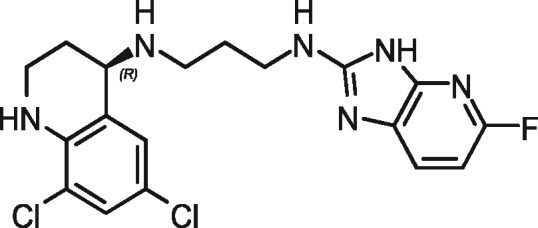
|
96 | 0 | 96 | 0 |
| 1849 |
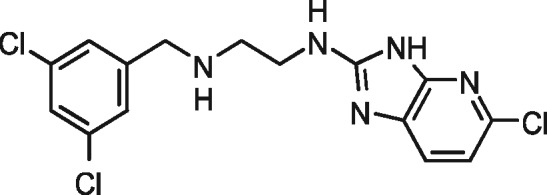
|
4 | 8 | 8 | 7 |
| 1893 |
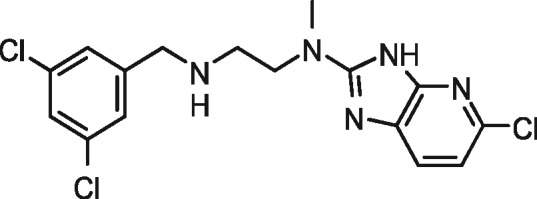
|
8 | 9 | no inhibition | — |
| 2080 |
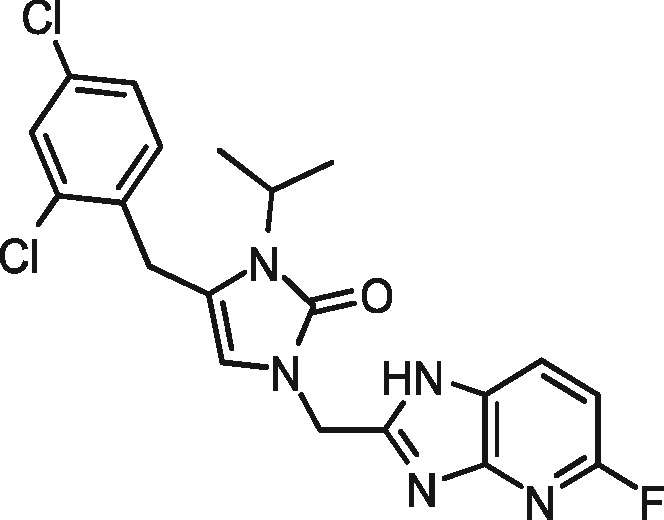
|
89 | 1 | 83 | 0 |
| 2093 |
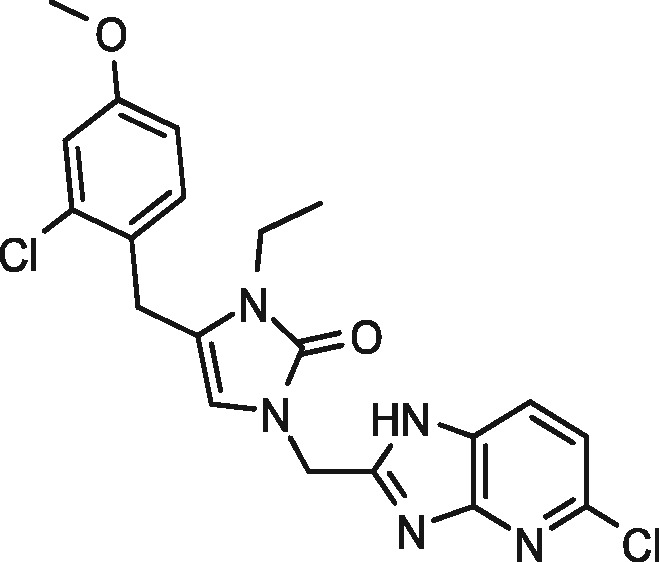
|
97 | 0 | 96 | 0 |
| 2114 |
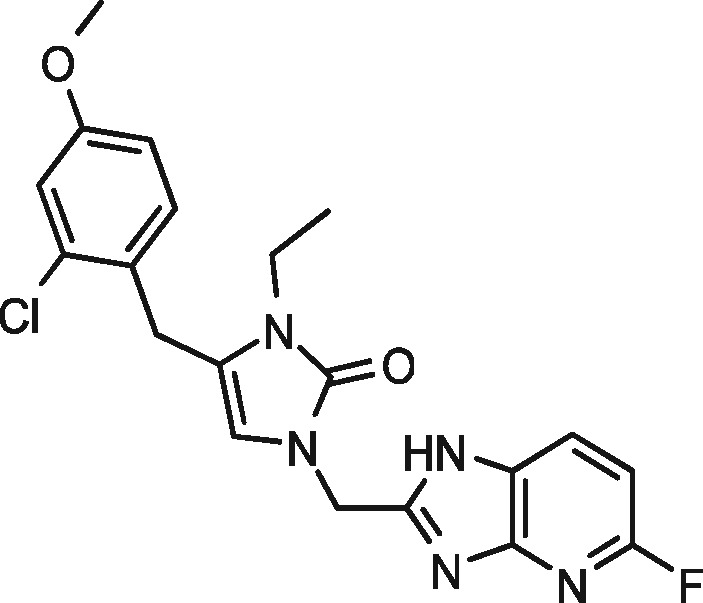
|
93 | 2 | 97 | 0 |
| 2144 |
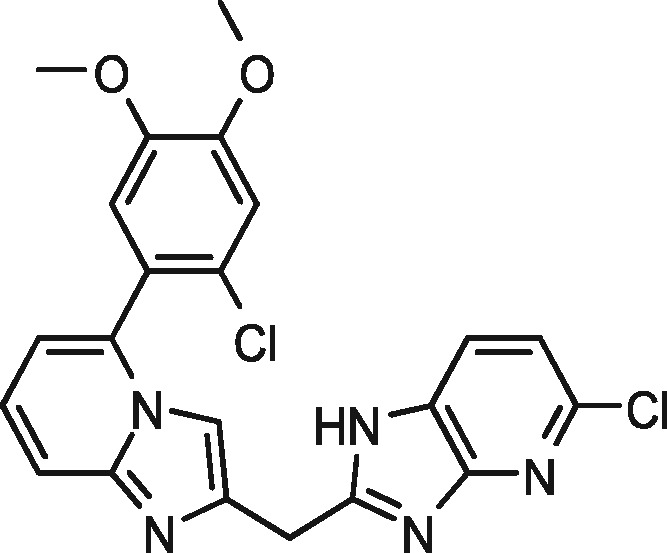
|
99 | 0 | 99 | 0 |
| 2207 |
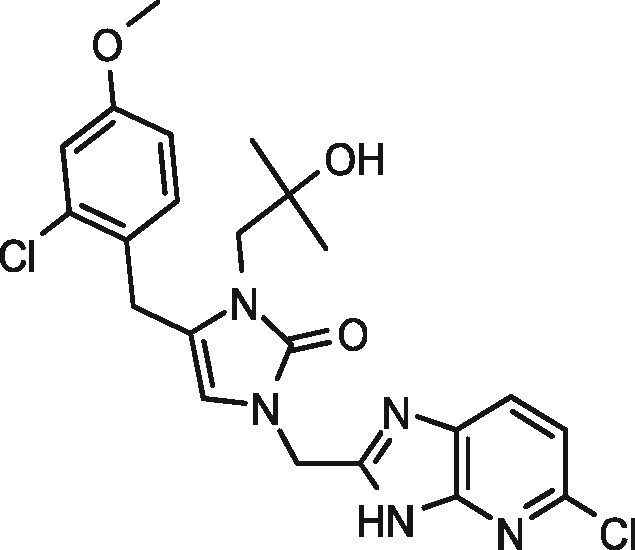
|
95 | 1 | 97 | 1 |
| 2240 |
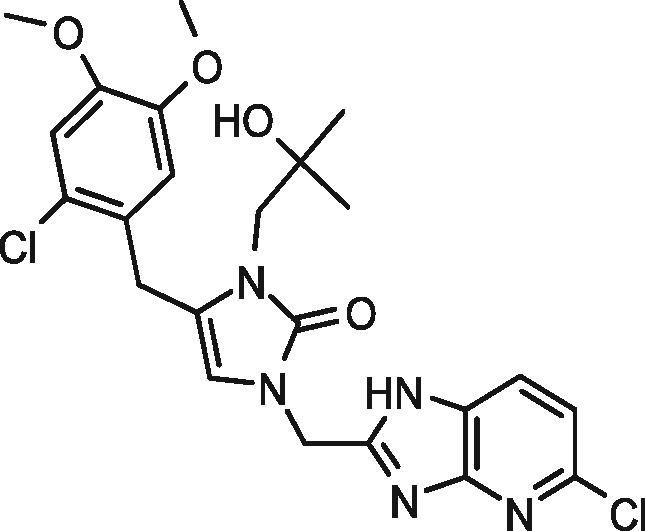
|
98 | 1 | 96 | 1 |
| 2242 |
|
90 | 1 | 93 | 1 |
The 2 μM screen in 384-well plates with G. lamblia WBC6:pGDHS::CBG99 and G. lamblia WBC6:pGDHS::CBG/Nluc had average Z′ values of 0.7 and 0.6, respectively. The screening data show that these compounds exhibit a similar inhibition profile against both strains (Figure 3a). Compound-1717 has previously shown potent inhibition of GlMetRS activity and its EC50 against WT WBC6 cells was determined to be 453 nM.29 The EC50 of Compound-1717 for G. lamblia WBC6:pGDHS::CBG99 and WBC6:pGDHS::CBG/Nluc was found to be 465 nM and 392 nM, respectively (Figure 3b). The metronidazole EC50 values of 2.867 μM and 3.114 μM obtained for G. lamblia WBC6:pGDHS::CBG99 and WBC6:pGDHS::CBG/Nluc strains, respectively, are within the range of previously reported literature values.10,29–31
Efficacy of metronidazole and Compound-1717 in the Giardia murine model
Infection in mice was established within 5 days and held for more than 2 weeks while antibiotics were maintained in drinking water. The rate of G. lamblia WBC6:pGDHS::CBG99 infection of BALB/c female mice was around 90%, which is consistent with earlier reports.32 IVIS image analysis revealed infection signals of up to 100-fold higher radiance values than background luminescence from uninfected mice.
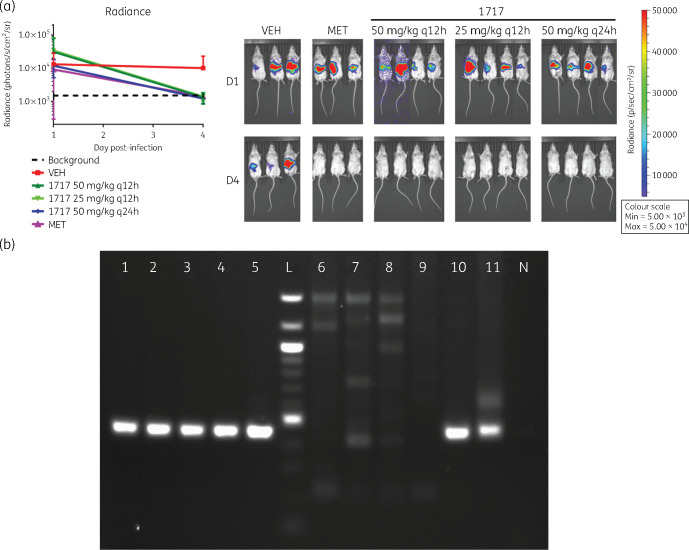
The infection model using the G. lamblia WBC6:pGDHS::CBG99 strain was validated by metronidazole, the currently available therapeutic agent. At 50 mg/kg q24h, treated mice showed no luminescent infection signal above background levels after 4 days. In the subsequent experiments, the efficacy of Compound-1717 was determined. All mice were cleared of infection after 3 days of dosing with 50 mg/kg q12h, 25 mg/kg q12h and 50 mg/kg q24h. Imaging 24 h after the final dose showed that the drug cleared the infection compared with the vehicle controls (Figure 4a). Molecular detection analysis with PCR confirmed the absence or presence of Giardia cysts expelled in the faeces of treated and untreated mice (Figure 4b). The lack of visible luminescence above background levels as confirmed by the radiance plot (Figure 4) a week after the last dose further corroborates this analysis.
Figure 4.
(a) Left: radiance plot and mouse images before and after treatment. Right: non-invasive imaging of trophozoite growth in mice, 5 days post-infection (D1) with the G. lamblia WBC6:pGDHS::CBG99 strain before the start of treatment. Mice were treated with 50 mg/kg metronidazole q12h (MET), Compound-1717 dosed at 50 mg/kg q12h, 25 mg/kg q12h and 50 mg/kg q24h. Treatment duration was 3 days. Mice in panel VEH were dosed with the dosing vehicle (3% ethanol, 7% Tween 80 in normal saline) as a control. Imaging on Day 8 post-infection (D4) showed that Compound-1717 and metronidazole cleared the infection relative to the untreated controls (VEH). The plot of the intensity/radiance data showed a significant drop after the conclusion of treatment relative to untreated controls. Note that images for some groups of mice on D1 have been cut together. This is due to differential intensities of the luminescence signal obtained from some mice causing differences in the length of time needed to acquire images. All images were taken during the same session and were scaled together. (b) Gel electrophoresis of cyst detection PCR. The cyst detection PCR was run on pooled samples at the start and end of treatment. Samples are: 1717 50 mg/kg q12h before treatment (BT) (1), 1717 25 mg/kg q12h BT (2), 1717 50 mg/kg q24h BT (3), metronidazole 50 mg/kg q12h BT (4), vehicle BT (5), 1717 50 mg/kg q12h after treatment (AT) (6), 1717 25 mg/kg q12h AT (7), 1717 50 mg/kg q24h AT (8), metronidazole 50 mg/kg q12h AT (9), vehicle AT (10), extracted DNA from G. lamblia cysts (11) and negative control (N). These were run with a 100 bp ladder (L). KOD Hot Start (Sigma–Aldrich, St. Louis, MO, USA) was used for all PCR steps. The initial PCR amplified the complete coding region of the GDH gene (GL50803_21942). The amplicon was used as the template for the nested PCR, which used a combination of previously described primers: GDHeF and gdhR1.38,39 The expected product size was 449 bp.
G. lamblia WBC6:pGDHS::CBG99 versus WBC6:pGDHS::CBG/Nluc
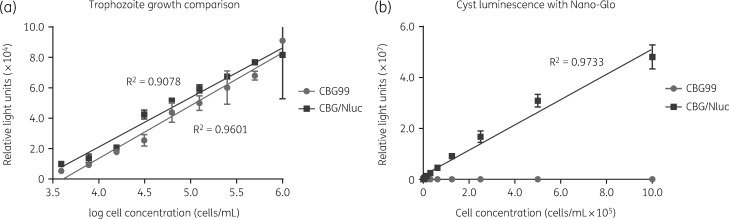
To further confirm infection and clearance during in vivo assays, we attempted to measure luminescence in G. lamblia WBC6:pGDHS::CBG99 cysts from faecal samples. There was no detectable luminescence even in untreated infected controls. This is likely because the cyst wall, which protects cysts from rupturing in water, prevents the uptake of d-luciferin. To improve upon this reporter strain, we endogenously tagged the CWP1 gene of G. lamblia WBC6:pGDHS::CBG99 strain with Nluc. Figure 5(a) is a plot of bioluminescence outputs from cysts of G. lamblia WBC6:pGDHS::CBG99 versus WBC6:pGDHS::CBG/Nluc using Nano-Glo. The growth and proliferation of the two strains was compared and shown to be similar (Figure 5b).
Figure 5.
G. lamblia WBC6:pGDHS::CBG99 versus G. lamblia WBC6:pGDHS::CBG/Nluc. (a) Plot comparing the growth of G. lamblia WBC6:pGDHS::CBG99 versus G. lamblia WBC6:pGDHS::CBG/Nluc. The same serial concentrations of trophozoites from both strains were incubated at 37°C in growth medium. Cell count versus luminescence was plotted after 44 h. (b) Plots of bioluminescence readouts from G. lamblia WBC6:pGDHS::CBG/Nluc cysts showed a linear relationship to cell count relative to no signal from the WBC6:pGDHS::CBG99 strain.
Discussion
Diarrhoeal syndromes are among the leading causes of morbidity and mortality by infectious diseases. G. lamblia, a significant contributor to the overall global diarrhoeal burden,33 poses a public health challenge due to diminishing treatment options.5 The luminescent G. lamblia strains described here could be important tools in the search for new giardiasis treatments. G. lamblia WBC6:pGDHS::CBG99 and WBC6:pGDHS::CBG/Nluc were developed by a process of iterative refinement that led to the most suitable luciferase and promoter combination. The bioluminescent signal remained stable for weeks in vitro and in vivo without concurrent administration of selective pressure. We earlier determined that Nluc is approximately 17 times brighter than CBG99 when driven by the same promoter (not shown). Since mice shed few cysts during infection,32 a more sensitive luciferase is required to validate infection or clearance in experimental drug studies. CBG99 is a better reporter of cell viability as it requires the presence of ATP for luminescence and therefore will not produce light in dead cells. That Nluc does not require ATP makes it a viable reporter for the cyst stage when cells are relatively dormant. Hence, Nluc was not considered as an alternative to CBG99 but rather as a supplement. Additionally, fusing Nluc to CWP1, which is secreted to form the cyst wall, eliminates the need for luminescent substrate to travel through the cyst-wall barrier to reach the luciferase. This modification will provide the ability to follow total parasite load in animals as well as easily quantify cyst production. The G. lamblia WBC6:pGDHS::CBG/Nluc strain was tested in the infection model and proven to have similar characteristics to the parent G. lamblia WBC6:pGDHS::CBG99 strain. Ongoing experiments are geared toward optimizing the Nluc detection system for stable and dependable readouts due to the usual low cyst output in mice.32
We demonstrated the usefulness of the luciferase reporter system to evaluate treatment with Compound-1717 in a mouse model of giardiasis. Compound-1717, a fluoro-imidazopyridine, is one of a new class of inhibitors that stop protein synthesis by targeting parasitic MetRS, as previously described.29 These inhibitors are lethal to G. lamblia parasites but non-toxic to mammalian cells in cell-based assays.29 Chemical synthesis, pharmacokinetics, cytotoxicity and inhibitory activity of Compound-1717 on the WT G. lamblia WBC6 strain and the GlMetRS enzyme were previously described.29,34,35 Analysis of Compound-1717’s pharmacokinetic profile showed that a single 50 mg/kg oral dose in mice would have sufficient gut and systemic levels to be effective for treatment of mouse giardiasis.35 The compound is about eight times more potent than metronidazole, which has an EC50 of 5 μM.29 It has solubilities of 52 μM (pH 7.4), 96 μM (pH 6.5) and 100 μM (pH 2). Compound-1717 satisfies Lipinski’s rule of five for druggability with a mol. wt of 409.29 g/mol, a log P of 3.11, 4 H-bond donors and 5 H-bond acceptors. Earlier experiments suggest that MetRS inhibitor 1717 has ‘cidal’ anti-Giardia activity.29 All of these factors led to the use of Compound-1717 in the present study as a potential therapeutic agent. Our results further support Compound-1717, as its efficacy in clearing Giardia infections in mice has been demonstrated. Compound-1717, developed as a trypanosomal agent, was used as a proof-of-principle molecule to demonstrate that inhibitors of GlMetRS could be a starting point for the development of new anti-giardiasis therapeutic agents. Further development of this series of inhibitors could facilitate innovative therapeutic options for giardiasis whose aetiological agent, G. lamblia, is rapidly evolving past currently available therapies. Since there is no overlap in the mechanism of action with any currently available drugs, cross-resistance is unlikely.
It is a widely accepted school of thought that there are significant practical limitations to the use of green-light-emitting reporters for in vivo studies because of increased attenuation of shorter wavelength emissions.36 Hence, red-shifted luminescence is believed to be better for whole-animal imaging because it penetrates deeper into living tissues.36,37 We report here a new approach for in vivo infection and efficacy studies of potential therapeutic agents using G. lamblia expressing the green-luminescence-emitting CBG99 gene. The technique enables successful infection of G. lamblia in mice after antibiotic alteration of the microbiome, plus parasite localization using marker gene expression for gauging the impact of experimental drugs in vivo. It is noteworthy that while infection is visible by IVIS imaging, the plot of infection intensity/radiance starts declining after 2 weeks post-infection. A chronic-infection model using IFN-γ-knockout B6 mice is currently being optimized to overcome this.
Acknowledgements
We would like to thank Dr Kelly M. Hennessey, Matthew A. Hulverson, Nora Molasky, Dr Wesley C. Van Voorhis and Ryan Choi for helpful discussions.
Funding
This study was supported by grants from National Institute of Allergy and Infectious Diseases and National Institute of Health under award numbers R01AI110708, R01AI097177, R21AI119715, R21AI127493, R21AI123690 and R21AI140881.
Transparency declarations
None to declare.
References
- 1. Serradell MC, Saura A, Rupil LL. et al. Vaccination of domestic animals with a novel oral vaccine prevents Giardia infections, alleviates signs of giardiasis and reduces transmission to humans. NPJ Vaccines 2016; 1: 16018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nabarro LE, Lever RA, Armstrong M. et al. Increased incidence of nitroimidazole-refractory giardiasis at the Hospital for Tropical Diseases, London: 2008-2013. Clin Microbiol Infect 2015; 21: 791–6. [DOI] [PubMed] [Google Scholar]
- 3. Farthing MJ. Giardiasis. Gastroenterol Clin North Am 1996; 25: 493–515. [DOI] [PubMed] [Google Scholar]
- 4. Tejman-Yarden N, Eckmann L.. New approaches to the treatment of giardiasis. Curr Opin Infect Dis 2011; 24: 451–6. [DOI] [PubMed] [Google Scholar]
- 5. Lalle M. Giardiasis in the post genomic era: treatment, drug resistance and novel therapeutic perspectives. Infect Disord Drug Targets 2010; 10: 283–94. [DOI] [PubMed] [Google Scholar]
- 6. Hill DR, Nash TE.. Giardia lamblia In: Mandell GL, Bennett JE, Dolin R, eds. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Churchill Livingstone/Elsevier, 2010; 3527–34. [Google Scholar]
- 7. Muller J, Nillius D, Hehl A. et al. Stable expression of Escherichia coli β-glucuronidase A (GusA) in Giardia lamblia: application to high-throughput drug susceptibility testing. J Antimicrob Chemother 2009; 64: 1187–91. [DOI] [PubMed] [Google Scholar]
- 8. Muller J, Ley S, Felger I. et al. Identification of differentially expressed genes in a Giardia lamblia WB C6 clone resistant to nitazoxanide and metronidazole. J Antimicrob Chemother 2008; 62: 72–82. [DOI] [PubMed] [Google Scholar]
- 9. Branchini BR, Ablamsky DM, Davis AL. et al. Red-emitting luciferases for bioluminescence reporter and imaging applications. Anal Biochem 2010; 396: 290–7. [DOI] [PubMed] [Google Scholar]
- 10. Hennessey KM, Rogiers IC, Shih HW. et al. Screening of the Pathogen Box for inhibitors with dual efficacy against Giardia lamblia and Cryptosporidium parvum. PLoS Negl Trop Dis 2018; 12: e0006673.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Branchini BR, Southworth TL, Khattak NF. et al. Red- and green-emitting firefly luciferase mutants for bioluminescent reporter applications. Anal Biochem 2005; 345: 140–8. [DOI] [PubMed] [Google Scholar]
- 12. Liang Y, Walczak P, Bulte JW.. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. J Biomed Opt 2012; 17: 016004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yee J, Mowatt MR, Dennis PP. et al. Transcriptional analysis of the glutamate dehydrogenase gene in the primitive eukaryote, Giardia lamblia. Identification of a primordial gene promoter. J Biol Chem 2000; 275: 11432–9. [DOI] [PubMed] [Google Scholar]
- 14. Jerlstrom-Hultqvist J, Stadelmann B, Birkestedt S. et al. Plasmid vectors for proteomic analyses in Giardia: purification of virulence factors and analysis of the proteasome. Eukaryot Cell 2012; 11: 864–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stefanic S, Morf L, Kulangara C. et al. Neogenesis and maturation of transient Golgi-like cisternae in a simple eukaryote. J Cell Sci 2009; 122: 2846–56. [DOI] [PubMed] [Google Scholar]
- 16. Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg 1983; 77: 487–8. [DOI] [PubMed] [Google Scholar]
- 17. Kuchimaru T, Iwano S, Kiyama M. et al. A luciferin analogue generating near-infrared bioluminescence achieves highly sensitive deep-tissue imaging. Nat Commun 2016; 7: 11856.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hall MP, Woodroofe CC, Wood MG. et al. Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging. Nat Commun 2018; 9: 132.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pham JK, Nosala C, Scott EY. et al. Transcriptomic profiling of high-density Giardia foci encysting in the murine proximal intestine. Front Cell Infect Microbiol 2017; 7: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boucher SE, Gillin FD.. Excystation of in vitro-derived Giardia lamblia cysts. Infect Immun 1990; 58: 3516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kane AV, Ward HD, Keusch GT. et al. In vitro encystation of Giardia lamblia: large-scale production of in vitro cysts and strain and clone differences in encystation efficiency. J Parasitol 1991; 77: 974–81. [PubMed] [Google Scholar]
- 22. Gil JS, Machado HB, Herschman HR.. A method to rapidly and accurately compare the relative efficacies of non-invasive imaging reporter genes in a mouse model and its application to luciferase reporters. Mol Imaging Biol 2012; 14: 462–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ur Rahman S, Stanton M, Casey PG. et al. Development of a click beetle luciferase reporter system for enhanced bioluminescence imaging of Listeria monocytogenes: analysis in cell culture and murine infection models. Front Microbiol 2017; 8: 1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakajima Y, Yamazaki T, Nishii S. et al. Enhanced beetle luciferase for high-resolution bioluminescence imaging. PLoS One 2010; 5: e10011.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu T, Close D, Handagama W. et al. The expanding toolbox of in vivo bioluminescent imaging. Front Oncol 2016; 6: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang JH, Chung TDY, Oldenburg KR.. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999; 4: 67–73. [DOI] [PubMed] [Google Scholar]
- 27. Kummel A, Gubler H, Gehin P. et al. Integration of multiple readouts into the Z′ factor for assay quality assessment. J Biomol Screen 2010; 15: 95–101. [DOI] [PubMed] [Google Scholar]
- 28. Huang W, Hulverson MA, Choi R. et al. Development of 5-aminopyrazole-4-carboxamide-based bumped-kinase inhibitors for cryptosporidiosis therapy. J Med Chem 2019; 62: 3135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ranade RM, Zhang Z, Gillespie JR. et al. Inhibitors of methionyl-tRNA synthetase have potent activity against Giardia intestinalis trophozoites. Antimicrob Agents Chemother 2015; 59: 7128–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen CZ, Kulakova L, Southall N. et al. High-throughput Giardia lamblia viability assay using bioluminescent ATP content measurements. Antimicrob Agents Chemother 2011; 55: 667–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hennessey KM, Smith TR, Xu JW. et al. Identification and validation of small-gatekeeper kinases as drug targets in Giardia lamblia. PLoS Negl Trop Dis 2016; 10: e0005107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Byrd LG, Conrad JT, Nash TE.. Giardia lamblia infections in adult mice. Infect Immun 1994; 62: 3583–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crannell ZA, Cabada MM, Castellanos-Gonzalez A. et al. Recombinase polymerase amplification-based assay to diagnose Giardia in stool samples. Am J Trop Med Hyg 2015; 92: 583–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Faghih O, Zhang Z, Ranade RM. et al. Development of methionyl-tRNA synthetase inhibitors as antibiotics for Gram-positive bacterial infections. Antimicrob Agents Chemother 2017; 61: e00999-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Z, Koh CY, Ranade RM. et al. 5-Fluoroimidazo[4,5-b]pyridine is a privileged fragment that conveys bioavailability to potent trypanosomal methionyl-tRNA synthetase inhibitors. ACS Infect Dis 2016; 2: 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Colin M, Moritz S, Schneider H. et al. Haemoglobin interferes with the ex vivo luciferase luminescence assay: consequence for detection of luciferase reporter gene expression in vivo. Gene Ther 2000; 7: 1333–6. [DOI] [PubMed] [Google Scholar]
- 37. Deliolanis NC, Kasmieh R, Wurdinger T. et al. Performance of the red-shifted fluorescent proteins in deep-tissue molecular imaging applications. J Biomed Opt 2008; 13: 044008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Read CM, Monis PT, Thompson RC.. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol 2004; 4: 125–30. [DOI] [PubMed] [Google Scholar]
- 39. Yang R, Jacobson C, Gardner G. et al. Development of a quantitative PCR (qPCR) for Giardia and analysis of the prevalence, cyst shedding and genotypes of Giardia present in sheep across four states in Australia. Exp Parasitol 2014; 137: 46–52. [DOI] [PubMed] [Google Scholar]