Abstract
Identification of matrix-bound nanovesicles (MBV) as ubiquitous components of the extracellular matrix (ECM) raises questions regarding their biologic functions and their potential theranostic application. Unlike liquid-phase extracellular vesicles (e.g., exosomes), MBV are tightly bound to the ECM, which makes their isolation and harvesting more challenging. The indiscriminate use of different methods to harvest MBV can alter or disrupt their structural and/or functional integrity. The objective of the present study was to compare the effect of various MBV harvesting methods upon yield, purity, and biologic activity. Combinations of four methods to solubilize the ECM (collagenase [COL], liberase [LIB], or proteinase K [PK] and nonenzymatic elution with potassium chloride) and four isolation methods (ultracentrifugation, ultrafiltration [UF], density barrier, and size exclusion chromatography [SEC]) were used to isolate MBV from urinary bladder-derived ECM. All combinations of solubilization and isolation methods allowed for the harvesting of MBV, however, distinct differences were noted. The highest yield, purity, cellular uptake, and biologic activity were seen with MBV isolated by a combination of liberase or collagenase followed by SEC. The combination of proteinase K and UF was shown to have detrimental effects on bioactivity. The results show the importance of selecting appropriate MBV harvesting methods for the characterization and evaluation of MBV and for analysis of their potential theranostic application.
Impact statement
Identification of matrix-bound nanovesicles (MBV) as ubiquitous components of the extracellular matrix (ECM) has raised questions regarding their biologic functions and their potential theranostic application. This study demonstrates that the harvesting methods used can result in samples with physical and biochemical properties that are unique to the isolation and solubilization methods used. Consequently, developing harvesting methods that minimize sample contamination with ECM remnants and/or solubilization agents will be essential in determining the theranostic potential of MBV in future studies.
Keywords: extracellular matrix, extracellular vesicles, matrix-bound nanovesicles
Introduction
The International Society for Extracellular Vesicles (ISEV) has proposed the term Extracellular Vesicle (EV) to describe “particles naturally released from the cell that are delimited by a lipid bilayer and cannot replicate.”1 Although a consensus has not been reached on specific EV markers to describe different subpopulations, efforts have been made to categorize them by characteristics such as size, lipid, and protein composition and biogenesis.2–7 EV are produced by many cell types and protect a luminal cargo of signaling molecules, including proteins, signaling lipids, cytokines, miRNA, mRNA, making them important mediators of intercellular communication.1,3,8–10
EV are almost exclusively described in body fluids and cell culture supernatants.11–16 These vesicles are commonly referred to as exosomes, which are of great interest for their potential use as disease biomarkers and therapeutic agents.8,9,17–19 EV have also been described in bone and cartilage with speculation upon their role in bone formation and calcification.20–28 More recently, EV have been identified within the extracellular matrix (ECM) of soft tissues and termed matrix-bound nanovesicles (MBV).29,30 MBV are a distinct subset of EV and differ from exosomes both in lipid membrane composition and luminal cargo.31 Considering the ubiquitous distribution of MBV,30 and their potential role in development, homeostasis, wound healing, tissue regeneration, and neoplasia,29–34 it is important that effective methods are used to harvest the EV without disrupting their structural and functional integrity.
Whereas the isolation of exosomes from body fluids has been well described and faithfully repeated,35–37 MBV must first be dissociated from the parent ECM before isolation for subsequent investigation or use. The harvesting of ECM from source tissues typically involves the use of detergents, enzymes, and/or mechanical forces to disrupt and remove cells and cell remnants38–41 with subsequent dissolution of the remaining ECM to release and isolate the MBV. The MBV must survive these biophysical manipulations with preservation of functional surface moieties, lipid membrane integrity, and intravesicular cargo. The objective of the present study was to compare the effects of ECM solubilization by collagenase (COL), liberase (LIB), proteinase K (PK), or elution with potassium chloride (KCL) followed by MBV isolation by ultracentrifugation (UC), ultrafiltration (UF), density barrier (DB), or size exclusion chromatography (SEC) upon purity, yield, and biologic activity of MBV from urinary bladder-derived ECM.
Materials and Methods
ECM preparation
ECM was prepared from porcine urinary bladder (Tissue Source LLC, Lafayette, IN) as previously described.42 Briefly, bladders were mechanically scraped to remove the tunica serosa, tunica muscularis externa, tunica submucosa, and tunica muscularis mucosa. The urothelial cells on the surface of the tunica mucosa were removed by rinsing the tissue in deionized water. The remaining tissue, consisting of the basement membrane and lamina propria of the tunica mucosa, was decellularized by exposure to a solution of peracetic acid (0.1%) and ethanol (4%) for 2 h with agitation in a shaker at 300 rpm. The resulting urinary bladder ECM (UBM-ECM) was then thoroughly rinsed with phosphate-buffered saline (PBS) and sterile water. The tissue was lyophilized and milled into powder form using a Wiley Mill with a #60 mesh screen.
ECM solubilization
Powdered UBM-ECM (100 mg) was solubilized by treatment with one of three enzymatic methods: collagenase (0.1 mg/mL, type XI; Sigma-Aldrich), liberase (0.01 mg/mL, Liberase TH; Sigma-Aldrich), or proteinase K (0.1 mg/mL; Invitrogen) in buffer (50 mM tris [pH 8], 5 mM CaCl2, and 200 mM NaCl). For every 100 mg of UBM-ECM powder, 10 mL of the enzymatic buffer was used. The mixture was vortexed at maximum speed for 10 s and then incubated overnight at room temperature under constant agitation. For the nonenzymatic elution method, 100 mg of UBM-ECM powder was diluted in 10 mL of 0.1 M KCL (Sigma-Aldrich) in PBS and incubated at 37°C for 30 min followed by incubation at 4°C for 2 h, always with constant agitation.
MBV isolation
Immediately following solubilization, each of the UBM-ECM samples was centrifuged sequentially at 500 g for 10 min, 2500 g for 20 min, and 10,000 g for 30 min (three times) to separate and remove insoluble collagen fibrils and other nonsoluble remnants. The supernatant was recovered between each centrifugation step, and the pellet was discarded. Following centrifugation, the supernatant was filter-sterilized with a 0.22 μm PES filter (Millipore) and frozen at −80°C until ready for further processing. Three samples of each solubilization method were thawed and subjected to one of four MBV isolation methods: UC, UF, DB, or SEC.
Ultracentrifugation
Samples isolated using UC were subjected to 100,000 g (Beckman Coulter Optima L-90K ultracentrifuge, SW32Ti) for 2 h at 4°C and the pellets were resuspended in 500 μL of 1 × PBS.
Ultrafiltration
Samples isolated using the UF method were placed in 100 kDa Amicon filter (Millipore) and then centrifuged at 4000 g for 20 min or until <500 μL of sample remained in the filter. The concentrated samples were recovered and taken to a final volume of 500 μL with 1 × PBS.
Density barrier
For the DB method, 2 mL of 50% OptiPrep™ (Sigma) was placed at the bottom of an ultraclear tube (Beckman Coulter) followed by 10 mL of 2% OptiPrep (not allowing them to mix) and lastly by the sample. The samples were centrifuged at 100,000 g (Beckman Coulter Optima L-90K ultracentrifuge, SW32Ti) for 2 h at 4°C. The fraction between the 2% and 50% OptiPrep was recovered (∼3 mL), diluted in 1 × PBS, concentrated using the UF protocol described above, and recovered to a final volume of 500 μL of 1 × PBS.
Size exclusion chromatography
Samples subjected to SEC were processed as described previously.43 Briefly, frozen samples were lyophilized and resuspended in 1 mL of particle-free water. A 1.5 × 12 cm minicolumn (Econo-Packcolumns; Bio-Rad, Hercules, CA) was packed with Sepharose 2B (Sigma-Aldrich, St. Louis, MO) having a column bed of 10 mL. The column was washed with 20 mL of 1 × PBS and a porous frit was placed on top of the bead column to avoid disturbing the beads during sample elution with PBS. The resuspended sample (1 mL) was loaded onto the column, and 10 fractions of 1 mL were collected. All fractions were tested for particle and protein concentration to determine the fractions in which MBV were present. For all SEC samples, fractions 3, 4, and 5 were collected and combined due to high particle concentration and low protein content (Supplementary Fig. S1). Combined fractions were concentrated using the UF method as described above and recovered to a final volume of 500 μL of 1 × PBS.
Experiment
Overview of experimental design
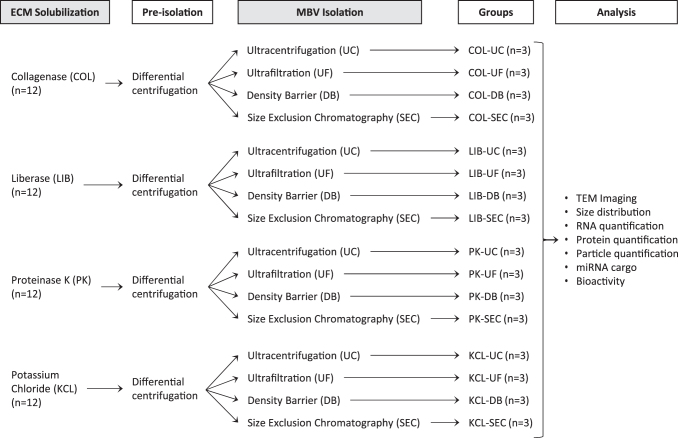
MBV were harvested by using a combination of common methods of ECM solubilization and subsequent vesicle isolation techniques generally used for exosome isolation. Four different methods of ECM solubilization were investigated, three of which were enzymatic (COL, LIB, and PK); and one nonenzymatic (KCL elution). After each solubilization method, released MBV were isolated by UC, UF, DB, or SEC (Fig. 1). The efficiency of each combination of ECM solubilization and MBV isolation methods was evaluated by MBV imaging, analysis of size distribution and particle concentration, protein quantification, and miRNA quantification. The potential bioactivity of the collected MBV upon cells was evaluated by quantification of MBV uptake and a cell proliferation assay.
FIG. 1.
Overview of experimental design. Powder UBM-ECM (0.1 g) was solubilized with one of three enzymatic buffers (COL, LIB, or PK) or with KCL overnight at room temperature. The solubilized sample went through a preisolation step in which differential centrifugation was performed by spinning at 500 g for 10 min (1 × ), discarding the pellet and spinning again at 2500 g for 20 min (1 × ), and finally at 10,000 g for 30 min (3 × ) followed by sterile filtration (0.2 μm). The resultant supernatant of each sample was used to isolate the MBV with one of four methods (UC, UF, DB, or SEC), aliquoted, and stored at −80°C until analysis. COL, collagenase; DB, density barrier; ECM, extracellular matrix; KCL, potassium chloride; LIB, liberase; MBV, matrix-bound nanovesicles; PK, proteinase K; SEC, size exclusion chromatography; UC, ultracentrifugation; UBM, urinary bladder; UF, ultrafiltration.
MBV imaging
Images of isolated MBV were obtained using transmission electron microscopy (TEM). Briefly, 7 μL of sample was placed on carbon-coated grids for 2 min and then decanted with filter paper to remove excess liquid. The grid was left to dry for a minute and then a drop of 1% uranyl acetate was added. The stain was immediately decanted using filter paper and the grid was left to dry for at least 1 min. The stained MBV were imaged at 80 kV with a JEOL JEM-1011TEM at a magnification of 80,000 × .
MBV size distribution and concentration
MBV size and concentration were determined using nanoparticle tracking analysis (NTA)44 as previously described.45 Samples of harvested MBV were diluted (1:100 to 1:1000) with particle-free water and injected into the sample cubicle of a NanoSight LM10 (Malvern Panalytical) at an infusion rate of “50.” The rate of Brownian motion of the particles was measured to determine their size distribution with three replicates of 60-s videos. Particle size was described as the mode ± SD (standard deviation) of the distribution, while the particle concentration was described as mean particles per milliliter ± SD.
Protein quantification
Protein concentration of the MBV samples was determined using the bicinchoninic acid (BCA) assay quantification kit (Pierce Chemical) following the manufacturer's instructions.
Silver stain of sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels
Equal volumes (4 μL) of MBV samples were resuspended in Laemmli buffer (R&D Systems) containing 5% β-mercaptoethanol (Sigma-Aldrich) and loaded on a 4% to 20% gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; Bio-Rad). The gels were run using Mini-PROTEAN electrophoresis module assembly (Bio-Rad) at 150 mV in running buffer (25 mM tris base, 192 mM glycine, and 0.1% SDS). Silver staining of gels was performed using the Silver Stain Plus Kit (Bio-Rad) according to the manufacturer's instructions. Briefly, after electrophoresis, the gel was placed in a fixative solution for 20 min with gentle agitation, followed by a rinse with water for 20 min and then staining and developing until bands were visualized. After the desired staining was reached, the membranes were placed in 5% acetic acid to stop the reaction. Images were taken in a ChemiDoc Touch instrument (Bio-Rad).
Isolation and quantification of miRNA in MBV samples
MBV miRNA was isolated using the Exiqon miRCURY™ RNA Isolation kit (Qiagen) following the manufacturer's instructions. Before RNA isolation, all MBV samples were treated with RNase A (1 μg/mL; Thermo Scientific) and DNase (500 U) (RQ1 Promega) at 37°C for 30 min to degrade free nucleic acid remaining from the tissue decellularization process. Following lysis of the MBV membrane with a lysis buffer provided in the kit, the sample was added to spin columns and washed several times according to the manufacturer's protocol to purify the RNA. Isolated RNA was collected in 30 μL of water and quantified using a NanoDrop spectrophotometer (NanoDrop).
miRNA quantification by ddPCR
MBV miRNA was isolated as described above, using the miRNeasy Mini Kit (Qiagen) following the manufacturer's instructions. The cDNA templates were prepared from 10 ng of RNA using the TaqMan Advanced miRNA cDNA Synthesis Kit (Applied Biosystems) following the manufacturer's instructions. Three different TaqMan assays were evaluated using ddPCR:has-miR-145-5p (VIC), has-miR-125-5p (FAM), and mmu-miR-451 (FAM) (all from Thermo Fisher Scientific).
Droplet digital polymerase chain reaction (ddPCR™) was used because of its high precision and absolute quantification of nucleic acid targets without the need for external calibrators or endogenous controls. Quantification was performed using a Bio-Rad's QX200 ddPCR system following the manufacturer's instructions. Briefly, samples were processed using an Automated Droplet generator (Bio-Rad) to create uniform nanoliter-sized droplets. The droplets were transferred to a 96-well plate for PCR in a thermocycler (C1000 Touch™ Thermal Cycler; Bio-Rad) and later transferred to the QX200 Droplet Reader to evaluate and measure the fluorescence level of individual droplets. The data analysis was performed using the QuantaSoft™ software to determine the starting concentration of the target molecule in units of copies per microliter.
Perivascular stem cell culture
Human perivascular stem cells (PVSC) derived from skeletal muscle46 were a gift from Dr. Bruno Péault (University of Pittsburgh, currently University of California, Los Angeles). PVSC were expanded in growth media, which consisted of Dulbecco's modification of Eagle's medium (DMEM) supplemented with 20% fetal bovine serum (FBS) and 1% penicillin–streptomycin. Cells were cultured at 37°C and 5% CO2.
Cell uptake of MBV
The uptake of MBV by PVSC was determined as follows. The membrane of the MBV was labeled with PKH67 (LOT# MINI67; Sigma) according to the manufacturer's instructions. For ease of labeling and to eliminate unbound PKH67 that would potentially label the PVSC, all four solubilization methods were evaluated, but only SEC was used to isolate the MBV. Briefly, solubilized and preisolated ECM samples (Fig. 1) were lyophilized and later resuspended in a solution consisting of 1 mL of Diluent C and 6 μL of PKH67 cell linker provided by the kit. The resuspended samples were incubated at room temperature for 5 min protected from the light and then loaded into a column with Sepharose 2B for isolation using SEC (as described in the MBV Isolation section). Fractions 2 to 5 were collected, combined, and concentrated to 250 μL using 100 kDa cutoff columns. NTA was used to determine the concentration of each sample.
PVSC were cultured overnight in cover glass chambers (155382; LAB-TEK) with 1 mL of growth media at a density of 7000 cells/cm2. The next morning, PVSC were treated with of 3E9 particles per milliliter for 2 h at 37°C. The particle concentration used for this treatment was determined by conducting a dose/response assay in which several concentrations were evaluated to determine if a significant effect on proliferation was observed and potential toxicity (data not shown). After the incubation period, cells were fixed with 2% paraformaldehyde for 20 min at room temperature followed by three washes with 1 × PBS. Fixed cells were counterstained with DAPI for 5 min to identify the nuclei and stained with phalloidin (A22283; Invitrogen) for 20 min in the dark at room temperature to visualize the cell cytoskeleton. Images were taken at 20 × and 40 × using a Zeiss Axio Observer microscope. The percentage of cells with visible intracellular green PKH67-labeled particles was counted using seven independent 20 × fields of view per treatment group (n = 3). CellProfiler™ Cell Image Analysis Software47 was used to quantify the images.
MBV bioactivity
PVSC were expanded as described above and later plated in 96-well plates at 5000 cells per well (n = 4). Cells were allowed to attach for ∼8 h and then serum-starved overnight with exosome-free starvation media (DMEM supplemented with 0.5% FBS and 1% penicillin–streptomycin, previously ultracentrifuged overnight at 100,000 g to eliminate exosomes present in the serum).
The effect of MBV on proliferation of PVSC was determined by the Cell Proliferation ELISA, BrdU colorimetric assay (Roche) as directed by the manufacturer. Briefly, the overnight serum-starved cells were treated with 4E9 particles per milliliter of each MBV group for 24 h, followed by incubation with the BrdU labeling reagent for additional 24 h at 37°C. Cells were fixed and incubated with the anti-BrdU antibody for 90 min at room temperature, and washed and incubated with substrate solution for 20 min. Absorbance of all wells was analyzed using a plate reader at 450 nm (reference wavelength: 690 nm).
Remnant enzymatic activity in MBV samples
As remnant enzymes derived from the solubilization process could be coisolated with the MBV, this influences their bioactivity. Therefore, the presence of active enzyme in the harvested samples was evaluated. Briefly, a 1.5-mm-thick gel consisting of 10% acrylamide and 4 mg/mL of gelatin was prepared following the guidelines of Cold Spring Harbor Protocols for SDS-PAGE.48 After casting, the gels were removed from the casting glass cassettes and placed on a flat surface. Sixteen evenly distributed 2 mm holes were created in each gel, followed by placement of 3 μL of neat or diluted MBV samples (30% MBV/PBS +70% media, same dilutions used for the bioactivity assay) into the holes. These gels were incubated overnight at 37°C. The next day, the gels were stained with Imperial™ Protein Stain (24615; Thermo Scientific) and incubated at room temperature for 1 h in an orbital shaker (50 rpm). After staining, the gels were washed twice with deionized water at room temperature for 1 h each under constant orbital shaking, and immediately thereafter images of the gel were taken in a ChemiDoc imager (Bio-Rad).
Statistical analysis
Two-way ANOVA was performed to compare the concentration of RNA, protein, particles, and mode in all the MBV samples isolated by different ECM solubilization methods (COL, LIB, PK, KCL) and different isolation methods (UF, UC, DB, SEC). For the bioactivity assay, the absorbance was scaled to the control group (PBS) and differences between each normalized group and the control were evaluated using the BootstRatio, a web-based statistical analysis to compare fold-change.49 The results of the MBV uptake experiment were analyzed using one-way ANOVA. Significance is based on p-values <0.05. All experiments were performed in triplicate unless stated otherwise.
Experimental Results
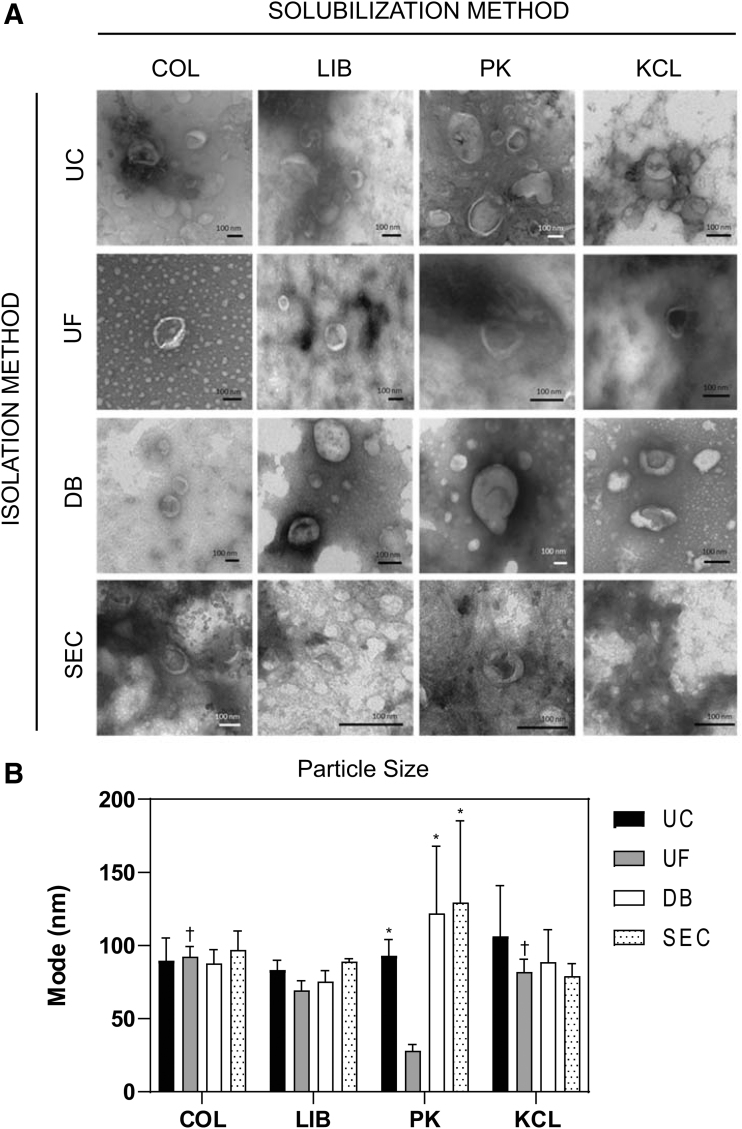
MBV morphology is unaffected but particle size may be influenced by different harvesting methods
Representative TEM images of MBV isolated by the 16 different methods showed no notable morphologic differences between groups (Fig. 2A). Quantitative evaluation by NTA showed MBV size in the range between ∼30 and 150 nm (Supplementary Fig. S1). The mode was used compare the size of the MBV between groups (Fig. 2B). There was no significant difference between the mode of any of the samples evaluated except for the PK-UF group (28.0 ± 4.3 nm), which was significantly smaller than the four groups: COL-SEC (97.0 ± 13.0 nm, p < 0.05), KCL-UC (106.3 ± 34.6 nm, p < 0.05), PK-DB (122.0 ± 45.9 nm, p < 0.01), and PK-SEC (129.3 ± 55.9 nm, p < 0.001). Complete statistical analysis is available in the Supplementary Data.
FIG. 2.
Morphology of MBV. (A) Transmission electron micrographs of MBV harvested with different solubilization and isolation methods. (B) Modal particle size for MBV across all harvesting methods (n = 3). *Significantly different to UF grouped by the PK solubilization method. †Significantly different to PK grouped by the UF isolation method.
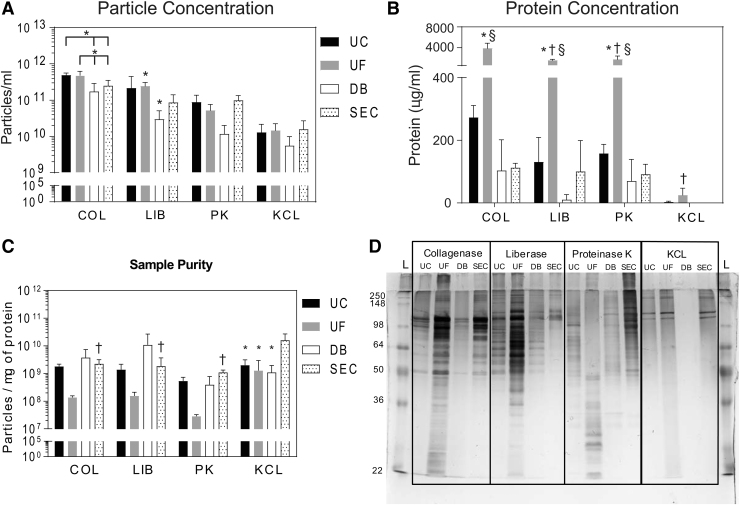
Quantity of MBV is influenced by the MBV harvesting method
The number of MBV isolated was influenced by both the solubilization (p < 0.0001) and the isolation method (p < 0.01) (Fig. 3A). Particle yield in samples solubilized with COL and isolated with UC or UF was higher than samples isolated with DB (p < 0.01), while SEC showed no significant differences with any of these groups. Within the LIB solubilization method, UF had a higher particle yield than the DB method. For the other solubilization methods (PK and KCL), there was no significant difference in the particle yield when isolation methods were compared. Comparing across solubilization methods, when COL was used in combination with UC or UF, higher particle yield was obtained compared with samples solubilized with LIB (p < 0.01), PK (p < 0.0001), or KCL (p < 0.0001). Samples solubilized with KCL were almost always significantly lower than the samples solubilized with the three enzymatic methods. Complete statistical analysis is available in the Supplementary Data. Thus, the highest particle concentration was obtained with COL and LIB solubilization methods when combined with UC, UF, or SEC isolation methods. The lowest particle yield was obtained in samples solubilized with KCL and/or isolated using the DB method.
FIG. 3.
Quantification of MBV samples. (A) Particle yield after harvesting MBV using different solubilization and isolation methods, calculated by NTA. *A significant difference between marked groups when grouped by the solubilization method. (B) Protein concentration in MBV samples harvested using different solubilization and isolation methods, calculated by BCA assay. *Difference to all other isolation methods when grouped by the solubilization method; †a difference compared with COL-UF when grouped by the isolation method; and §significantly different to KCL-UF when grouped by the isolation method. No other differences were seen. (C) Sample purity expressed as particles per mg protein. *Differences within groups sorted by the solubilization method; †differences within groups sorted by the isolation method. Differences were only seen in the KCL-SEC group and between KCL-SEC and COL-, LIB-, and DB-SEC samples. (D) Silver stain showing unique protein profiles generated by each MBV harvesting method (samples were equally loaded by volume). BCA, bicinchoninic acid; NTA, nanoparticle tracking analysis.
Protein concentration is influenced by the MBV harvesting method
Protein concentration (Fig. 3B) was found to be significantly influenced by both the solubilization (p < 0.0001) and the isolation method (p < 0.0001), and the interaction of both (p < 0.0001). Within the COL, LIB, and PK solubilization methods, the protein concentration obtained in the samples that were isolated using UF was significantly higher than in samples isolated using UC, DB, and SEC (p < 0.0001 for all). Comparing across solubilization methods, there was no significant difference in the protein content for MBV isolated by UC, DB, or SEC; however, within the UF groups, COL-UF had significantly more protein content than LIB-UF, PK-UF, and KCL-UF (p < 0.0001 for all). In addition, protein content was significantly higher in LIB-UF and PK-UF samples compared with KCL-UF samples (p < 0.0001 for both). Thus, the use of collagenase and UF method yields the highest protein concentration of all methods evaluated to harvest MBV.
MBV harvesting methods create distinct protein peptide profiles
The ratio between particle and protein concentration is a commonly reported method of MBV quantification.45 Calculating this ratio showed that all methods had similar particle:protein ratios. Within the COL, LIB, and PK groups, no differences were seen between isolation methods (Fig. 3C). Only KCL-SEC presented a significantly higher ratio compared with KCL-UC, KCL-UF, and KCL-DB (p < 0.01). Similarly, comparing isolation methods showed no differences with respect to solubilization method except for KCL-SEC, which had a higher ratio than COL-SEC (p < 0.05), LIB-SEC (p < 0.01), and PK-SEC (p < 0.01). Thus, while there were differences in MBV yield and protein concentration, using the ratio between particle and protein concentration as a metric may be misleading as it masks the differences in protein content generated by the different harvesting methods.
Silver staining of MBV samples subjected to SDS-PAGE showed that each harvesting method generated a unique protein profile (Fig. 3D). Samples isolated by UF had the highest protein content, while SEC had the lowest. UF samples showed a wide range of protein sizes ranging from 250 to 22 kDa, while samples isolated by SEC contained only a few distinct bands. Importantly, each method had a unique pattern of bands that was generated by the combination of solubilization method and isolation method that was not apparent using the BCA assay.
Quantification of miRNA in MBV is influenced by the harvesting method
Preliminary studies31 identified three miRNAs (miR125b-5p, miR145–5p, and miR451a) that were highly expressed in MBV. Absolute quantification of miRNA copies performed by ddPCR (Fig. 4) showed that the concentration of miRNA copies in each sample was influenced by the harvesting methods used to obtain the MBV.
FIG. 4.
MicroRNA content in MBV samples. Differential expression of the three miRNAs analyzed was seen in MBV samples harvested using different solubilization and isolation methods.
Samples solubilized with PK had more copies of miRNA than solubilized with COL, LIB, or KCL. Comparison of the number of copies identified between MBV isolation methods, groups isolated using UF generally had the highest number of miRNA copies across all solubilization methods, except in COL. Samples that were isolated using SEC consistently had the lowest miRNA concentration.
Cellular uptake of MBV is influenced by the solubilization method used
Fluorescent images taken of PVSC (Fig. 5) incubated with MBV from the COL group showed the highest percentage of cells positive for intracellular MBV (55.2% ± 17.9%), followed by the LIB group (38.2% ± 12.1%), the PK group (23.7% ± 7.8%), and finally by the KCL group (19.1% ± 7.4%). The percentage of positive cells was significantly higher in the COL group compared with the percentage of positive cells in the PK and KCL groups (p < 0.05).
FIG. 5.
Uptake of MBV by perivascular progenitor cells. (A) Images of PVSC (40× magnification) after 2 h at 37°C incubation, with labeled MBV (3 × 109 particles/mL) showing the presence of MBV (green) within the cells. (B) Quantification of the percentage of cells (N = 7 20 × fields of view) that contained MBV following incubation with MBV. *Significant difference compared with collagenase samples. PVSC, perivascular stem cells. Color images are available online.
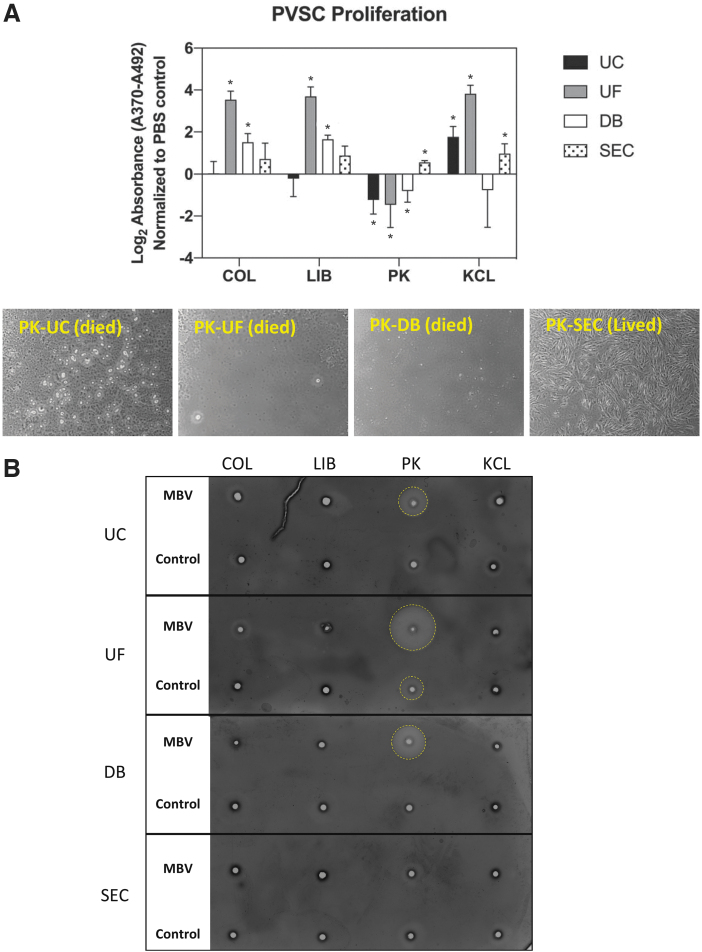
MBV effects on PVSC proliferation are influenced by the harvesting methods
The effect of MBV upon PVSC proliferation was quantified after treating the cells for 24 h with each of the 16 MBV groups (n = 3) (Fig. 6A). The proliferative response of PVSC was distinctly affected by the methods used to harvest the MBV. Samples isolated using COL and LIB as solubilization methods showed similar patterns of proliferative activity across all the isolation methods. Samples solubilized with COL and LIB and isolated using UC or SEC did not affect proliferation. Samples isolated using UF showed the highest proliferative activity (p < 0.0001 for both COL and LIB), followed by DB (p < 0.01 for COL and p < 0.0001 for LIB) compared with the control. MBV samples prepared using the PK solubilization method all showed cell death rate during the assay (Fig. 6B) (addressed in the following section), except in the samples isolated using SEC, which induced a significant increase in PVSC proliferative activity (p < 0.0001). Finally, samples solubilized with KCL all induced a significant increase in PVSC proliferation (p < 0.01 for UC, p < 0.0001 for UF, and p < 0.05 for SEC) except for the samples isolated with DB, which had no effect. In general, MBV samples isolated using UF resulted in the most proliferation, followed by SEC, DB, and finally by UC. Only KCL-solubilized samples promoted an increase in cell proliferation when isolated by UC (p < 0.01).
FIG. 6.
Cell proliferation induced by MBV. (A) Quantification of BrdU uptake (normalized to control) by PVSC following treatment with MBV samples for 24 h. BrdU uptake was measured by colorimetric absorbance assay. *Significant difference compared with control (B) Images (20 × ) of PVSC that were treated with samples solubilized with PK showed decreased viability in all groups except where MBV were isolated using SEC. (C) Gelatin zymogram shows residual proteolytic activity (yellow circles) present in all MBV samples solubilized with PK, except PK-SEC MBV samples that were prepared at the same dilution as the proliferation. Color images are available online.
Harvesting methods affect removal of residual enzymes
The possibility that active enzymes from the solubilization process were present in the MBV samples and potentially influenced the bioactivity assays was tested using a modified zymogram (Fig. 6C). In samples prepared at the same dilution, only samples harvested with PK as the solubilization method and UC, UF, or DB as the isolation method showed evidence of proteolytic activity. The only isolation method that consistently removed active enzyme from the MBV samples was SEC.
Discussion
Unlike exosomes, which are found in physiological fluids and have well-documented isolation methods, MBV must be liberated from tightly bound structural components of the ECM and then purified from the solubilized ECM components. Results of the present study show that the methods used to harvest MBV from ECM have no detectable effect on morphology but may alter the size distribution, particle concentration, coisolated protein concentration, cellular uptake, and bioactivity of the MBV. Of the methods investigated, the combination of liberase or collagenase as an ECM solubilization method followed by SEC as the isolation method provided the highest MBV yield, purity (more MBV with less remnant ECM protein and enzymes), cellular uptake, and bioactivity. In contrast, the combination of PK and UF methods resulted in MBV samples with low particle yield, high protein content, and most notably high remnant enzyme activity that affected subsequent cellular activity assays. Due to the high contaminant protein, PK-UF samples were visibly more viscous than the rest of the MBV samples. It is likely that the high viscosity affected the particle quantification and size analysis in the NanoSight, resulting in unreliable data such as low particle yield and low modal size (Supplementary Fig. S1).
Three common enzymatic methods of ECM digestion (collagenase, liberase, and proteinase K) were investigated along with one nonenzymatic elution method KCL. Collagenase is a relatively crude combination of collagenase isoforms (approximately between 68 and 130 kDa) digesting collagen fibers and disaggregating connective tissue.50 Liberase is a highly purified preparation of collagenases I and II, blended in a precise ratio with a nonclostridial protease, thermolysin.51 Proteinase K is a smaller (28.9 kDa) and very stable protease that degrades a broad spectrum of proteins.52 The nonenzymatic method, KCL, supported protein solubility53 and the release embedded MBV. Downstream analysis of the MBV harvested using these ECM solubilization methods showed that enzymatic methods were more efficient at liberating MBV since they increased MBV yield, compared with KCL, regardless of the subsequent isolation method used. However, because the ECM was degraded by the enzymatic methods, protein fragments were created that were then coisolated in the final MBV preparation. While the MBV samples obtained with the nonenzymatic method had a lower yield of MBV, these samples had the highest purity, particularly using the combination of KCL and SEC. Further investigation is needed to determine if modifications to the nonenzymatic method, such as increasing incubation time, salt molarity, or even the use of different salt solutions, could increase the yield of MBV without adversely affecting sample purity.
Purification of MBV from the solubilized ECM was achieved using common exosome purification methods such as UC, DB, UF, and SEC. UC is considered the gold standard to isolate EV and consists of subjecting a solution to centrifugate a high g-force (100,000 g) to pellet the vesicles.54 Although UC is widely used, it is sensitive to different parameters that can affect purification such as the g force used, the rotor type (fixed angle or swinging bucket),55 and the viscosity of the sample.56 UC also allows remnant ECM proteins to pellet with the isolated EV. UF can result in low yield due to vesicles adhering to the membrane and the coisolation of proteins that have similar sizes as the EV of interest.57 DB is a method that can result in high-purity EV samples,58,59 and shares the same issues as UC. Moreover, there is no standard regarding the number of layers of cushion solution to use. There are reports using only one layer59 up to four layers.58 SEC is a method that separates particles by size as the sample flows through a column of beads. SEC provides very good separation of different sized particles, with good yield and purity.60 Yield and purity are further improved when combined with other techniques such as UC.61 However, coisolation of proteins in the fractions containing EV has been reported.62 Thus, SEC may effectively enrich for a specific subset of protein fragments that elute in the same fractions as the EV.
The present study has clearly shown that the use of UC, UF, DB, and SEC results in notable differences in MBV yield, purity, and bioactivity. Studies with exosomes have also shown similar differences in isolation efficiency and concentration of coisolated proteins61–63 and also different miRNA profiling.64 It is clear that methods used to isolate EV, including MBV, have quantifiable effects that can lead to misinterpretation of results.
Understanding the potential downstream effects of ECM solubilization methods and subsequent MBV isolation methods is necessary for the investigation of MBV biology and their potential theranostic use. Previous work has established that MBV influence cellular behavior.29,30,32,65,66 The molecular mechanisms by which MBV contribute to cell phenotype and potentially constructive therapeutic outcomes are yet to be determined, and effective methods for harvesting MBV will be required for such studies. Previous studies have shown the ability of MBV to alter the inflammatory response and promote healing of damaged tissues,29,34 but equally, harvesting MBV from chronic wounds and examining their miRNA or protein cargo could help identify mediators of the injury process. Exosomes have been shown to have diagnostic potential for diseases, including cancer, neurodegenerative disease, and pathologic infections.67–69 It is conceivable that MBV would have the same potential. Differences in ECM between source tissues and between tissues harvested from different age pigs have been described.70–72 It is likely that similar differences exist in MBV isolated from different tissues or animals of different ages. These differences in MBV cargo or bioactivity will also influence the theranostic potential of MBV.
All the combinations of solubilization and isolation methods evaluated were able to isolate MBV embedded within UBM-ECM. Importantly, each method used produced samples with distinct characteristics. Consequently, assessment of sample purity is important to demonstrate that functional properties of a sample are associated with vesicles and not with the coisolated contaminants.45 A simple quantification of purity such as the ratio of particles to protein that has been used by others45 does not provide any details about the type or distribution of the coisolated protein contaminants.
The different solubilization methods tested produced a unique protein profile, including methods that resulted in only a few prominent bands (e.g., KCL-SEC) to others that produced multiple low- and high-molecular-weight proteins (e.g., Col-UF). It is possible that some methods such as SEC may enrich for proteins of a specific molecular weight that are filtered by the column and coisolated in the same fractions as the MBV.57,61 Although the present study did not characterize these protein fragments, it is possible that these cryptic peptides may influence bioactivity, affecting MBV uptake and cell proliferation results. Previous studies have shown that enzymatic digests of ECM bioscaffolds can generate cryptic peptides that show bioactivity in vitro and in vivo73–77 and it is possible that cryptic peptides were generated by the enzymes used in the present study. Moreover, uptake efficiency could have been affected by these remnant proteins associated with the MBV. In several harvesting methods, particularly those using proteinase K, residual enzyme from the ECM digestion was found to be retained in the MBV samples following purification. The results of the PVSC proliferation assay showed that the presence of active, residual PK enzyme resulted in cell death. The presence of residual enzymes obviously has critical implications for downstream assays measuring bioactivity since the residual enzyme can not only denature soluble proteins and growth factors from culture media, but potentially damage cell:matrix interactions that keep the cells anchored to the tissue culture plastic as well.
The three miRNAs used in the present study were selected based on previously obtained RNA-seq data that showed them to be among the most abundant miRNAs in MBV.31 While their biologic relevance with respect to macrophage polarization,78,79 tissue remodeling,80–82 and regeneration83 has been shown, miRNA results are included herein to emphasize the potential impact of the different isolation methods upon the properties of harvested MBV.
Limitations of the present study include the possible overestimation of particle concentration measurement since NTA may not adequately discriminate between MBV and nonvesicular particulate material. There are alternative isolation methods such as tunable microfluidic systems,84 tangential flow filtration,85 and precipitation-based methods,86 among others, that may be effective for MBV isolation but are beyond the scope of the present study.
Conclusions
Although all the methods tested were effective at isolating MBV, each method resulted in samples with physical and biochemical properties that were unique to the solubilization and isolation methods used. Suitable MBV harvesting methods that maintain vesicles integrity and functionality while minimizing sample contamination with ECM remnants and/or solubilization agents will be essential in determining MBV physiologic function and therapeutic utility in future studies.
Supplementary Material
Disclosure Statement
S.F.B. is the chief scientific officer and equity holder in ECM Therapeutics. Inc., which has license rights to MBV technology from the University of Pittsburgh. The authors declare that they have no other competing interests.
Funding Information
Lina M. Quijano, Juan D. Naranjo, Salma O. El-Mossier, Neill J. Turner, and Stephen F. Badylak were supported by NIH grant R01AR073527 and grant 2019-447-002 from the Medical Technology Enterprise Consortium.
Supplementary Material
Particle concentration analysis results
Particle size analysis results
References
- 1. Thery, C., Witwer, K.W., Aikawa, E., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7, 1535750, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yanez-Mo, M., Siljander, P.R., Andreu, Z., et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4, 27066, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Niel, G., D'Angelo, G., and Raposo, G.. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19, 213, 2018 [DOI] [PubMed] [Google Scholar]
- 4. Latifkar, A., Cerione, R.A., and Antonyak, M.A.. Probing the mechanisms of extracellular vesicle biogenesis and function in cancer. Biochem Soc Trans 46, 1137, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abels, E.R., and Breakefield, X.O.. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol 36, 301, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colombo, M., Raposo, G., and Thery, C.. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30, 255, 2014 [DOI] [PubMed] [Google Scholar]
- 7. Dreyer FaB, A Biogenesis and functions of exosomes and extracellular vesicles. In: Federico, M., ed. Lentiviral Vectors and Exosomes as Gene and Protein Delivery Tools. New York: Humana Press, 2016, pp. 201–216 [DOI] [PubMed] [Google Scholar]
- 8. Gurunathan, S., Kang, M.H., Jeyaraj, M., Qasim, M., and Kim, J.H.. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 8, 307, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andaloussi, S.E., Mager, I., Breakefield, X.O., and Wood, M.J.. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 12, 347, 2013 [DOI] [PubMed] [Google Scholar]
- 10. Malda, J., Boere, J., van de Lest, C.H., van Weeren, P., and Wauben, M.H.. Extracellular vesicles—new tool for joint repair and regeneration. Nat Rev Rheumatol 12, 243, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Asea, A., Jean-Pierre, C., Kaur, P., et al. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J Reprod Immunol 79, 12, 2008 [DOI] [PubMed] [Google Scholar]
- 12. Caby, M.P., Lankar, D., Vincendeau-Scherrer, C., Raposo, G., and Bonnerot, C.. Exosomal-like vesicles are present in human blood plasma. Int Immunol 17, 879, 2005 [DOI] [PubMed] [Google Scholar]
- 13. Ogawa, Y., Miura, Y., Harazono, A., et al. Proteomic analysis of two types of exosomes in human whole saliva. Biol Pharm Bull 34, 13, 2011 [DOI] [PubMed] [Google Scholar]
- 14. Pisitkun, T., Shen, R.F., and Knepper, M.A.. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A 101, 13368, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haraszti, R.A., Miller, R., Stoppato, M., et al. Exosomes produced from 3D cultures of MSCs by tangential flow filtration show higher yield and improved activity. Mol Ther 26, 2838, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phinney, D.G., and Pittenger, M.F.. Concise review: MSC-derived exosomes for cell-free therapy. Stem Cells 35, 851, 2017 [DOI] [PubMed] [Google Scholar]
- 17. Gyorgy, B., Hung, M.E., Breakefield, X.O., and Leonard, J.N.. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu Rev Pharmacol Toxicol 55, 439, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quesenberry, P.J., Aliotta, J., Camussi, G., et al. Potential functional applications of extracellular vesicles: a report by the NIH Common Fund Extracellular RNA Communication Consortium. J Extracell Vesicles 4, 27575, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barile, L., and Vassalli, G.. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther 174, 63, 2017 [DOI] [PubMed] [Google Scholar]
- 20. Ali, S.Y., Sajdera, S.W., and Anderson, H.C.. Isolation and characterization of calcifying matrix vesicles from epiphyseal cartilage. Proc Natl Acad Sci U S A 67, 1513, 1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anderson, H.C. Vesicles associated with calcification in the matrix of epiphyseal cartilage. J Cell Biol 41, 59, 1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bab, I.A., Muhlrad, A., and Sela, J.. Ultrastructural and biochemical study of extracellular matrix vesicles in normal alveolar bone of rats. Cell Tissue Res 202, 1, 1979 [DOI] [PubMed] [Google Scholar]
- 23. Dereszewski, G., and Howell, D.S.. The role of matrix vesicles in calcification. Trends Biochem Sci 3, 151, 1978 [Google Scholar]
- 24. Hsu, H.H., and Anderson, H.C.. Calcification of isolated matrix vesicles and reconstituted vesicles from fetal bovine cartilage. Proc Natl Acad Sci U S A 75, 3805, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Landis, W.J., Paine, M.C., Hodgens, K.J., and Glimcher, M.J.. Matrix vesicles in embryonic chick bone: considerations of their identification, number, distribution, and possible effects on calcification of extracellular matrices. J Ultrastruct Mol Struct Res 95, 142, 1986 [DOI] [PubMed] [Google Scholar]
- 26. Muhlrad, A., Bab, I.A., Deutsch, D., and Sela, J.. Occurrence of actin-like protein in extracellular matrix vesicles. Calcif Tissue Int 34, 376, 1982 [DOI] [PubMed] [Google Scholar]
- 27. Sela, J., Bab, I., and Deol, M.S.. Patterns of matrix vesicle calcification in osteomalacia of Gyro mice. Metab Bone Dis Relat Res 4, 129, 1982 [DOI] [PubMed] [Google Scholar]
- 28. Sela, J., and Bab, I.A.. The relationship between extracellular matrix vesicles and calcospherities in primary mineralization of neoplastic bone tissue. TEM and SEM studies on osteosarcoma. Virchows Arch A Pathol Anat Histol 382, 1, 1979 [DOI] [PubMed] [Google Scholar]
- 29. Huleihel, L., Bartolacci, J.G., Dziki, J.L., et al. Matrix-bound nanovesicles recapitulate extracellular matrix effects on macrophage phenotype. Tissue Eng Part A 23, 1283, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huleihel, L., Hussey, G.S., Naranjo, J.D., et al. Matrix-bound nanovesicles within ECM bioscaffolds. Sci Adv 2, e1600502, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hussey, G.S., Pineda Molina, C., Cramer, M.C., et al. Lipidomics and RNA sequencing reveal a novel subpopulation of nanovesicle within extracellular matrix biomaterials. Sci Adv 6, eaay4361, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Faust, A., Kandakatla, A., van der Merwe, Y., et al. Urinary bladder extracellular matrix hydrogels and matrix-bound vesicles differentially regulate central nervous system neuron viability and axon growth and branching. J Biomater Appl 31, 1277, 2017 [DOI] [PubMed] [Google Scholar]
- 33. Hussey, G.S., Cramer, M.C., and Badylak, S.F.. Extracellular matrix bioscaffolds for building gastrointestinal tissue. Cell Mol Gastroenterol Hepatol 5, 1, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Merwe, Y., Faust, A.E., Sakalli, E.T., et al. Matrix-bound nanovesicles prevent ischemia-induced retinal ganglion cell axon degeneration and death and preserve visual function. Sci Rep 9, 3482, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma, S., Scholz-Romero, K., Rice, G.E., and Salomon, C.. Methods to enrich exosomes from conditioned media and biological fluids. Methods Mol Biol 1710, 103, 2018 [DOI] [PubMed] [Google Scholar]
- 36. Vaswani, K., Koh, Y.Q., Almughlliq, F.B., Peiris, H.N., and Mitchell, M.D.. A method for the isolation and enrichment of purified bovine milk exosomes. Reprod Biol 17, 341, 2017 [DOI] [PubMed] [Google Scholar]
- 37. Thery, C., Amigorena, S., Raposo, G., and Clayton, A.. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3,22, 2006 [DOI] [PubMed] [Google Scholar]
- 38. White, L.J., Taylor, A.J., Faulk, D.M., et al. The impact of detergents on the tissue decellularization process: a ToF-SIMS study. Acta Biomater 50, 207, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang, J., Hu, Z.Q., Turner, N.J., et al. Perfusion-decellularized skeletal muscle as a three-dimensional scaffold with a vascular network template. Biomaterials 89, 114, 2016 [DOI] [PubMed] [Google Scholar]
- 40. Zambaiti, E., Scottoni, F., Rizzi, E., et al. Whole rat stomach decellularisation using a detergent-enzymatic protocol. Pediatr Surg Int 35, 21, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosario, D.J., Reilly, G.C., Ali Salah, E., Glover, M., Bullock, A.J., and Macneil, S.. Decellularization and sterilization of porcine urinary bladder matrix for tissue engineering in the lower urinary tract. Regen Med 3, 145, 2008 [DOI] [PubMed] [Google Scholar]
- 42. Mase, V.J.Jr., Hsu, J.R., Wolf, S.E., et al. Clinical application of an acellular biologic scaffold for surgical repair of a large, traumatic quadriceps femoris muscle defect. Orthopedics 33, 511, 2010 [DOI] [PubMed] [Google Scholar]
- 43. Hong, C.S. FS, and Whiteside, T.L. Isolation of biologically active exosomes from plasma of patients with cancer. Methods Mol Biol 1633, 257, 2017 [DOI] [PubMed] [Google Scholar]
- 44. Filipe, V., Hawe, A., and Jiskoot, W.. Critical evaluation of nanoparticle tracking analysis (NTA) by nanosight for the measurement of nanoparticles and protein aggregates. Pharm Res 27, 796, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Webber, J., and Clayton, A.. How pure are your vesicles? J Extracell Vesicles 2, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Crisan, M., Yap, S., Casteilla, L., et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3, 301, 2008 [DOI] [PubMed] [Google Scholar]
- 47. McQuin, C., Goodman, A., Chernyshev, V., et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol 16, e2005970, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. SDS-PAGE Gel. Cold Harbor Spring Protocols. 2015. Available at: http://cshprotocols.cshlp.org/content/2015/7/pdb.rec087908.short (accessed October12, 2020)
- 49. Cleries, R., Galvez, J., Espino, M., Ribes, J., Nunes, V., and de Heredia, M.L.. BootstRatio: a web-based statistical analysis of fold-change in qPCR and RT-qPCR data using resampling methods. Comput Biol Med 42, 438, 2012 [DOI] [PubMed] [Google Scholar]
- 50. Salamone, M., Cuttitta, A., Bertuzzi, F., Ricordi, C., Ghersi, G., and Seidita. G.. Biochemical comparison between clostridium hystoliticum collagenases G and H obtained by DNA recombinant and extractive procedures. Chem Eng Trans 27, 259, 2012 [Google Scholar]
- 51. Linetsky, E., Bottino, R., Lehmann, R., Alejandro, R., Inverardi, L., and Ricordi, C.. Improved human islet isolation using a new enzyme blend, liberase. Diabetes 46, 1120, 1997 [DOI] [PubMed] [Google Scholar]
- 52. Ebeling, W., Hennrich, N., Klockow, M., Metz, H., Orth, H.D., and Lang, H.. Proteinase K from Tritirachium album Limber. Eur J Biochem 47, 91, 1974 [DOI] [PubMed] [Google Scholar]
- 53. Zhang, J. Protein-protein interactions. In: Cai, W., ed. Protein-Protein Interactions—Computational and Experimental Tools. London, United Kingdom: IntechOpen, 2012, pp. 359–376 [Google Scholar]
- 54. Momen-Heravi, F. Isolation of extracellular vesicles by ultracentrifugation. Methods Mol Biol 1660, 25, 2017 [DOI] [PubMed] [Google Scholar]
- 55. Cvjetkovic, A., Lotvall, J., and Lasser, C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J Extracell Vesicles 3, 2014. DOI: 10.3402/jev.v3.23111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Momen-Heravi, F., Balaj, L., Alian, S., et al. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front Physiol 3, 162, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Taylor, D.D., and Shah, S.. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 87, 3, 2015 [DOI] [PubMed] [Google Scholar]
- 58. Kalra, H., Adda, C.G., Liem, M., et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 13, 3354, 2013. 24115447 [Google Scholar]
- 59. Gupta, S., Rawat, S., Arora, V., et al. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res Ther 9, 180, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lozano-Ramos, I., Bancu, I., Oliveira-Tercero, A., et al. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J Extracell Vesicles 4, 27369, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. An, M., Wu, J., Zhu, J., and Lubman, D.M.. Comparison of an optimized ultracentrifugation method versus size-exclusion chromatography for isolation of exosomes from human serum. J Proteome Res 17, 3599, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baranyai, T., Herczeg, K., Onodi, Z., et al. Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS One 10, e0145686, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gheinani, A.H., Vogeli, M., Baumgartner, U., et al. Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery. Sci Rep 8, 3945, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rekker, K., Saare, M., Roost, A.M., et al. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem 47, 135, 2014 [DOI] [PubMed] [Google Scholar]
- 65. Hynes, R.O. Integrins: bidirectional, allosteric signaling machines. Cell 110, 673, 2002 [DOI] [PubMed] [Google Scholar]
- 66. Hynes, R.O. The extracellular matrix: not just pretty fibrils. Science 326, 1216, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. He, C., Zheng, S., Luo, Y., and Wang, B.. Exosome theranostics: biology and translational medicine. Theranostics 8, 237, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Panagiotara, A., Markou, A., Lianidou, E.S., Patrinos, G.P., and Katsila, T.. Exosomes: a cancer theranostics road map. Public Health Genomics 20, 116, 2017 [DOI] [PubMed] [Google Scholar]
- 69. Terrasini, N., and Lionetti, V.. Exosomes in critical illness. Crit Care Med 45, 1054, 2017 [DOI] [PubMed] [Google Scholar]
- 70. Sicari, B.M., Johnson, S.A., Siu, B.F., et al. The effect of source animal age upon the in vivo remodeling characteristics of an extracellular matrix scaffold. Biomaterials 33, 5524, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brown, B.N., Barnes, C.A., Kasick, R.T., et al. Surface characterization of extracellular matrix scaffolds. Biomaterials 31, 428, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brown, B.N., Londono, R., Tottey, S., et al. Macrophage phenotype as a predictor of constructive remodeling following the implantation of biologically derived surgical mesh materials. Acta Biomater 8, 978, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Banerjee, P., and Shanthi, C.. Cryptic peptides from collagen: a critical review. Protein Pept Lett 23, 664, 2016 [DOI] [PubMed] [Google Scholar]
- 74. Brennan, E.P., Tang, X.H., Stewart-Akers, A.M., Gudas, L.J., and Badylak, S.F.. Chemoattractant activity of degradation products of fetal and adult skin extracellular matrix for keratinocyte progenitor cells. J Tissue Eng Regen Med 2, 491, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Agrawal, V., Tottey, S., Johnson, S.A., Freund, J.M., Siu, B.F., and Badylak, S.F.. Recruitment of progenitor cells by an extracellular matrix cryptic peptide in a mouse model of digit amputation. Tissue Eng Part A 17, 2435, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Agrawal, V., Kelly, J., Tottey, S., et al. An isolated cryptic peptide influences osteogenesis and bone remodeling in an adult mammalian model of digit amputation. Tissue Eng Part A 17, 3033, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Swinehart, I.T., and Badylak, S.F.. Extracellular matrix bioscaffolds in tissue remodeling and morphogenesis. Dev Dyn 245, 351, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Essandoh, K., Li, Y., Huo, J., and Fan, G.C.. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock 46, 122, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang, Y., Zhang, M., Zhong, M., Suo, Q., and Lv, K.. Expression profiles of miRNAs in polarized macrophages. Int J Mol Med 31, 797, 2013 [DOI] [PubMed] [Google Scholar]
- 80. Waki, T., Lee, S.Y., Niikura, T., et al. Profiling microRNA expression during fracture healing. BMC Musculoskelet Disord 17, 83, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cheng, Z., Dai, L.L., Wang, X., et al. MicroRNA-145 down-regulates mucin 5AC to alleviate airway remodeling and targets EGFR to inhibit cytokine expression. Oncotarget 8, 46312, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82. Higashi, K., Yamada, Y., Minatoguchi, S., et al. MicroRNA-145 repairs infarcted myocardium by accelerating cardiomyocyte autophagy. Am J Physiol Heart Circ Physiol 309, H1813, 2015 [DOI] [PubMed] [Google Scholar]
- 83. Diaz Quiroz, J.F., Tsai, E., Coyle, M., Sehm, T., and Echeverri, K.. Precise control of miR-125b levels is required to create a regeneration-permissive environment after spinal cord injury: a cross-species comparison between salamander and rat. Dis Model Mech 7, 601, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shin, S., Han, D., Park, M.C., et al. Separation of extracellular nanovesicles and apoptotic bodies from cancer cell culture broth using tunable microfluidic systems. Sci Rep 7, 9907, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Busatto, S., Vilanilam, G., Ticer, T., et al. Tangential flow filtration for highly efficient concentration of extracellular vesicles from large volumes of fluid. Cells 7, 273, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Soares Martins, T., Catita, J., Martins Rosa, I., and O ABdCES, Henriques, A.G.. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS One 13, e0198820, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.