Abstract
Starting with the launch of the Human Genome Project three decades ago, genomics has become progressively entrenched within the bedrock of the biomedical research enterprise. Capitalizing on the momentum of the project’s successful completion in 2003, genomics now regularly plays a central and catalytic role in basic and translational research, and studies increasingly demonstrate the vital role that genomic information can play in clinical care. Looking ahead, the anticipated advances in technology development, biological insights, and clinical applications (among others) will lead to more widespread integration of genomics into virtually all areas of biomedical research, the adoption of genomics into mainstream medical and public-health practices, and an increasing relevance of genomics in everyday life. On behalf of the research community, the National Human Genome Research Institute recently completed a multi-year process of strategic engagement to capture input about the future research priorities and opportunities in human genomics, with an emphasis on health applications. Here we articulate the highest-priority elements envisioned for the cutting-edge of human genomics going forward – that is, at “The Forefront of Genomics.”
Introduction
Three decades ago this month, a pioneering group of international researchers began an audacious journey to generate the first map and sequence of the human genome, marking the start of a 13-year odyssey called the Human Genome Project1–3. The project’s successful and early completion in 2003, which included parallel studies of a set of model organism genomes, catalyzed enormous progress in genomics research. Leading the signature advances has been a greater than one million-fold reduction in the cost of DNA sequencing4. This decrease has allowed the generation of innumerable genome sequences, including hundreds of thousands of human genome sequences (both in research and clinical settings), and the continuous development of new assays for identifying and characterizing functional genomic elements5,6. With these new tools, coupled with increasingly sophisticated statistical and computational methods, researchers have been enabled to create rich catalogs of human genomic variants7,8, to gain an ever-deepening understanding of the functional complexities of the human genome5, and to elucidate the genomic bases of thousands of human diseases9,10. In turn, the last decade has brought the initial realization of genomic medicine11, as research successes have been converted into powerful tools for use in healthcare, including somatic genome analysis for cancer (enabling development of targeted therapeutics)12, noninvasive prenatal genetic screening13, and genomics-based tests for a growing set of pediatric conditions and rare disorders14, among others.
In essence, with growing insights about the structure and function of the human genome and ever-improving laboratory and computational technologies, genomics has become increasingly woven into the fabric of biomedical research, medical practice, and society. The scope, scale, and pace of genomic advances to date were nearly unimaginable when the Human Genome Project began; even today, such advances are yielding scientific and clinical opportunities beyond our initial expectations, with many more anticipated in the decade ahead.
Embracing its leadership role in genomics, the National Human Genome Research Institute (NHGRI) has developed strategic visions for the field at key inflection points, in particular at the end of the Human Genome Project in 200315 and then again at the beginning of the last decade in 201116. These visions outlined the most compelling opportunities for human genomics research, in each case informed by a multi-year engagement process. NHGRI endeavored to start the new decade with an updated strategic vision for human genomics research. Through a planning process that involved over 50 events (e.g., dedicated workshops, conference sessions, and webinars) over the last two years (see http://genome.gov/genomics2020), the institute collected input from a large number of stakeholders, with the resulting input catalogued and synthesized using the framework depicted in Fig. 1.
Fig. 1 |. Four-area strategic framework at The Forefront of Genomics.

Together, the indicated progressive and interrelated areas serve to organize the major elements in the strategic vision described here.
Unlike the past, this round of strategic planning was significantly influenced by the now widely disseminated nature of genomics across biomedicine. A representative glimpse into this historic phenomenon is illustrated in Fig. 2. During the Human Genome Project, NHGRI was the primary funder of human genomics research at the U.S. National Institutes of Health (NIH), but the past two decades have brought a greater than ten-fold increase in the relative fraction of funding coming from other parts of NIH.
Fig. 2 |. Funding trends of NIH and NHGRI over the past 30 years.
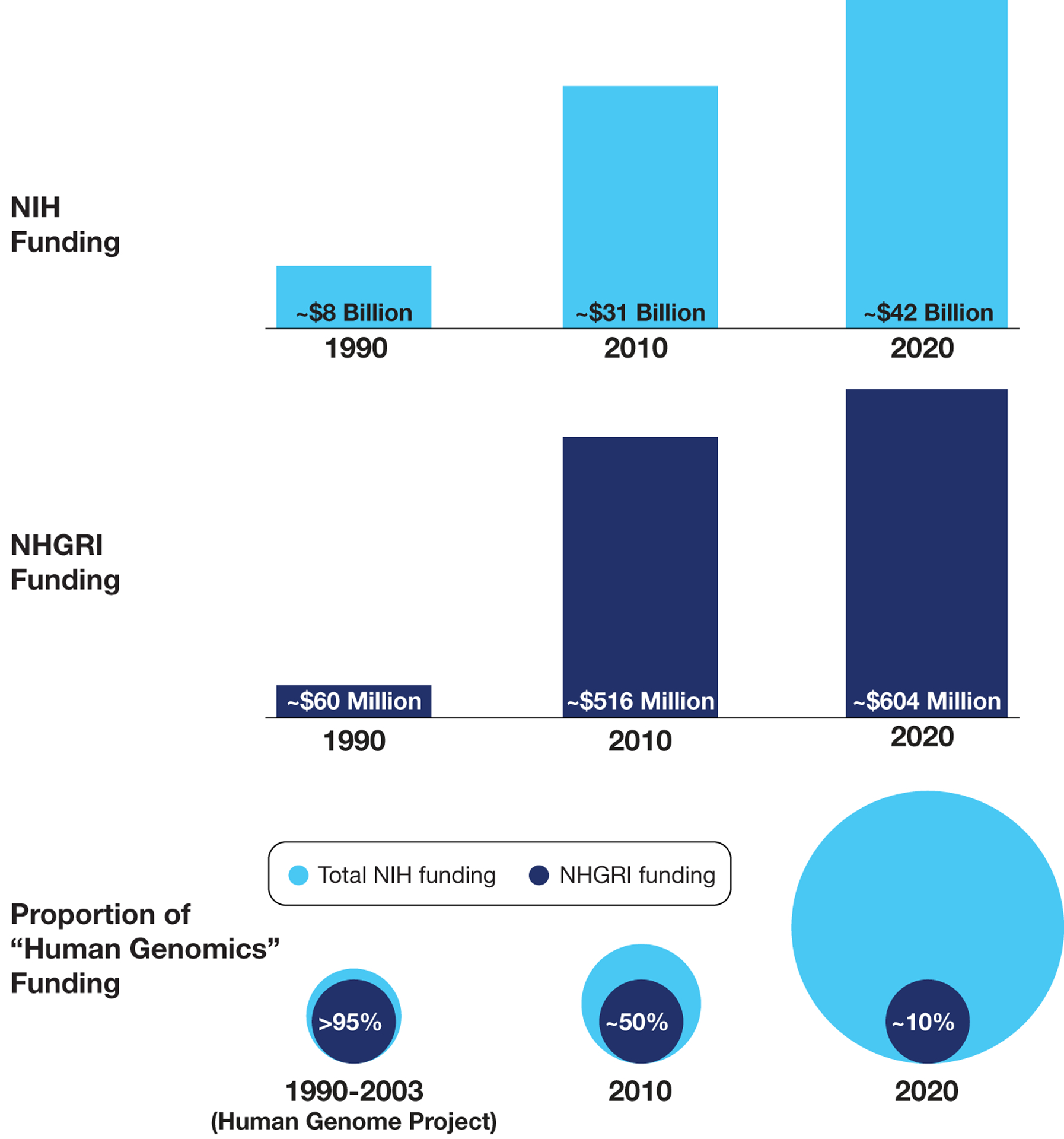
The total funding levels for NIH (top panel) and NHGRI (middle panel) are indicated for 1990, 2010, and 2020, respectively. Also shown (bottom panel) is the relative proportion of funds supporting human genomics research provided by NHGRI versus all of NIH for the three corresponding time intervals (as derived from queries of the internal NIH Research, Condition and Disease Categorization database for funds assigned to the “human genome” category). During the 30-year period when the NHGRI budget increased roughly ten-fold (middle panel), the proportion of total NIH funding for human genomics research actually increased more dramatically, from <5% during the Human Genome Project to ~90% at the beginning of the current decade (bottom panel). In essence, these trends reflect a leveraging of NHGRI’s funds that increased NIH’s overall human genomics research funding by greater than ten-fold.
The planning process continually encountered the realities associated with the broad and extensive use of genomics and the impracticality of being comprehensive, which together served to focus attention on the most cutting-edge opportunities in human genomics. This experience affirmed NHGRI’s recently rearticulated role in providing genomics leadership at NIH, embodied by our newly conceived organizational mantra: “The Forefront of Genomics.” We ultimately linked this mantra to the strategic planning process to help guide the formulation of input. From the ensuing discussions, it became apparent that responsible stewardship is a central aspect of being at (and pushing forward) The Forefront of Genomics, specifically in the four major areas detailed in Fig. 1, Boxes 1, 2, 3, and 4, and below.
Box 1. Guiding principles and values for human genomics.
Maintain an overarching focus on using genomics to understand biology, to enhance knowledge about disease, and to improve human health – genomics is now foundational across the entire continuum of biomedical research, from deciphering fundamental principles of biology to translating that knowledge into disease prevention and medical advances.
Strive for global diversity in all aspects of genomics research, committing to the systematic inclusion of ancestrally diverse and underrepresented individuals in major genomic studies – attention to diversity in genomics research is both socially just and scientifically essential, including meaningful, sustained partnerships with diverse communities in the design and implementation of research studies, the propagation of research findings, and the development and use of new technologies.
Maximize the utility of genomics for all members of the public, including the ability to access genomics in healthcare – engagement, inclusion, and understanding the needs of diverse and medically underserved groups are required to ensure that all members of society benefit equitably from genomic advances, with particular attention given to the equitable use of genomics in healthcare that avoids exacerbating and strives towards reducing health disparities.
Champion a diverse genomics workforce – the promise of genomics cannot be fully achieved without attracting, developing, and retaining a diverse workforce, including individuals from groups currently underrepresented in the genomics enterprise.
Provide a conceptual research framing that consistently examines the role of both genomic and non-genomic contributors to health and disease – routinely considering the importance of social and environmental factors influencing human health (and the interactions among those components and genomics) will be important for the comprehensive understanding of most human diseases.
Promote robust and consistently applied standards in genomics research – the use of carefully defined standards (e.g., those for generating, analyzing, storing, and sharing data) has benefited genomics in significant ways, and this must include appropriate privacy and data-security protections for those participating in genomics research.
Embrace the interdisciplinary and team-oriented nature of genomics research – starting with the Human Genome Project, some of the most challenging genomics endeavors have benefited from the creation and management of large, interdisciplinary research collaborations.
Adhere to the highest expectations and requirements related to open science, responsible data sharing, and rigor and reproducibility in genomics research – the genomics enterprise has a well-respected history of leading in these areas, and that commitment must be built upon and continually reaffirmed.
Pursue advances in genomics as part of a vibrant global community of genomics researchers and funders – the challenges in genomics require the collective energies and creativity of a collaborative international ecosystem that includes partnerships among researchers, funders, and other stakeholders from academia, government, and the commercial sector.
Box 2. Sustaining and improving a robust foundation for genomics.
Genome structure and function
Enable the routine generation and analysis of increasingly complex genomic data
Use evolutionary and comparative genomic data to maximize understanding of genome function
Genomic data science
Develop novel methods and build sustainable data resources for genomics research
Ensure facile storing, sharing, and computing on large genomic datasets
Develop integrated knowledgebases and informatics methods for genomic medicine
Genomics and society
Understand the interrelationships between genomics and the social and environmental factors influencing human health
Empower people to make well-informed decisions about genomic data and develop data-stewardship systems that reinforce their choices
Increase the genomic literacy of all sectors of society
Training and genomics workforce development
Ensure that the next generation of genomic scientists are sufficiently trained in data science
Train healthcare providers to integrate genomics into the clinical workflow
Foster a diverse genomics workforce
Box 3. Breaking down barriers that impede progress in genomics.
Laboratory and computational technologies
Transform the study of the functional consequences of genomic variation by enhancing the scale of DNA synthesis and editing
Maximally leverage the usability and utility of emerging datasets for genomic studies of human health and disease
Biological insights
Establish the means to determine the functional consequences of genomic variants affecting human health and disease
Characterize intraindividual genomic variation and understand its role in human disease
Implementation science
Develop and assess strategies for implementing the use of genomic information in clinical care
Test public health approaches for implementing population-wide genomic screening
Box 4. Compelling genomics research projects in biomedicine.

Acquire an increasingly comprehensive view of the roles and relationships of genes and regulatory elements in pathways and networks
Elucidate the genetic architecture of the majority of human diseases and traits
Design studies that include diverse ancestral populations to enable scientific discoveries and genomic medicine for all
Understand how the use of genomics can influence concepts of health, disease, responsibility, identity, family, and community
Extend multi-omic studies of human disease and health into clinical settings
Design and utilize genomic learning healthcare systems for knowledge generation and clinical care improvement
Principles and values for human genomics
As genomics has matured as a discipline, the field has embraced a growing set of fundamental principles and values that together serve as a guiding compass for the research efforts – some of these emerged organically within the field, whereas others have been adopted from the broader scientific community. The growing complexities of human genomics and its many applications (especially in medicine) at The Forefront of Genomics make it imperative to reaffirm, sharpen, and even extend these tenets, such as those highlighted in Box 1.
Many of these principles and values have been informed by the recognized area of genomics that focuses on ethical, legal, and social implications (ELSI) research17, which was established at the beginning of the Human Genome Project to help guarantee that the eugenics movement and other misuses of genetics are not repeated. ELSI research has since grown to encompass a broad portfolio of studies examining issues at the interface of genomics and society, the results of which have informed policies and laws related to genetic discrimination, intellectual property, data sharing, and informed consent18. Similar efforts seek to ensure that the benefits of genomics are available to all members of society19. Genomics, like all fields of science, must reckon with systematic injustices and biases, fully cognizant of their criticality for health equity. Looking ahead, ELSI research needs to focus on aspects of genomic medicine implementation that present challenging questions about legal boundaries, study governance, data control, privacy, and consent. Complex societal issues must also be studied, including the expanded application of genomics in non-medical realms (e.g., ancestry testing, law enforcement, and genetics-based marketing of consumer goods)20. Finally, ELSI research could also examine the implications of studying genetic associations with bio-behavioral traits (e.g., intelligence, sexual behavior, social status, and educational attainment)21 and of a future where machine learning and artificial intelligence are used to tailor risk communication and clinical decisions based on analyzing an individual’s genome sequence22.
Robust foundation for genomics
Genomics is now routinely and broadly utilized throughout biomedical research, with widespread reliance on a robust foundation for facilitating genomic advances. The foundation’s integrity depends on a number of key elements, including infrastructure, resources, and dynamic areas of technology development and research. Sustaining and improving that foundation are key responsibilities at The Forefront of Genomics, the major elements of which are highlighted in Box 2 and detailed in corresponding paragraphs below.
Genome structure and function
The last two decades have brought a greater than million-fold reduction in the cost of DNA sequencing23 along with an explosion in technologies for functional genomics6,24,25 (i.e., the study of how elements in the genome contribute to biological processes). Additional opportunities are poised to be unlocked as the generation and analysis of genomic data become even faster, cheaper, and more accurate. Near-term expectations include enhanced capabilities for generating high-quality and complete (e.g., telomere-to-telomere and phased) genome sequences26,27 and continued refinement and enhanced utility of a human genome reference sequence(s) that increasingly reflects human variation and diversity on a global scale28 and that serves as a substrate for genome annotation29. Technologies for generating DNA sequence and other data types (e.g., transcriptomic data, epigenomic data, and functional readouts of DNA sequences) need to be enabled at orders-of-magnitude lower costs, at single-cell resolution, at distinct spatial locations within tissues, and longitudinally over time30–32. These genomic data should be integrated with other multi-omic data (e.g., proteomes, metabolomes, lipidomes, and/or microbiomes) in sophisticated ways, including novel methods that collect multiple data types from a single sample32. Transformative approaches will become increasingly vital for assimilating, sharing, and analyzing these complex and heterogeneous data types33 and must expand to include the integration of environmental, lifestyle, clinical, and other phenotypic data. These capabilities should be incorporated into browsers, portals, and visualization tools for use by a broadening community of researchers and clinicians.
Genome sequences have now been generated for over 1,000 vertebrate species and are increasingly accompanied by multi-species annotations34. Understanding natural genomic variation, conservation of genomic elements, and the rapid evolutionary changes in genomic regions associated with specific traits is critical for attaining a comprehensive view of genome structure and function. The study of a wide range of organisms continues to be instrumental for elucidating the impact of genomic variation on biological processes and phenotypes, providing insights about the interplay of genomic variants and environmental pressures35 and the relevance of putative pathogenic variants identified in clinical studies36. It is essential that the generation of high-quality multi-species genomic data be accompanied by community-accepted standards for data, metadata, and data interoperability. New groundbreaking methods would allow for integrating functional data from diverse species with human data and visualizing increasingly complex comparative genomic datasets. Continued progress in this area would move the field closer to the long-term aspirational goal of understanding the evolutionary history of every base in the human genome.
Genomic data science
All major genomics breakthroughs to date have been accompanied by the development of groundbreaking statistical and computational methods. Accordingly, continued innovations in both traditional and advanced methods (including machine learning and artificial intelligence) should be prioritized37. These approaches must be considered from the early stages of study planning and data collection in ways that complement and enhance, rather than inhibit, technical progress. Further, the biomedical research community requires accurate, curated, accessible, secure, and interoperable genomic data repositories and informatics platforms that benefit all populations. Approaches for improving the efficiency of such resources include the use of shared storage and computing infrastructure, the adoption of common data-management processes, and the development of increasingly automated data-curation methods38. Carefully considered funding strategies must be designed to support these methods and resources, including a global, multi-funder model that ensures their development, enhancements, and long-term sustainability39.
Recent progress has brought substantial transformations in how the petabytes of genomic data being generated each year are assimilated and analyzed, including the emergence of cloud-based and federated approaches. Effective and efficient management of increasingly complex genomic datasets requires addressing challenges with these emerging approaches as well as innovations in the use of hardware, algorithms, software, standards, and platforms40. Current barriers include the lack of interoperable genomic data resources (which limits downstream access, integration, and analyses) and the absence of controlled and consistently adopted data and metadata vocabularies and ontologies41,42. User-friendly systems that capture metadata in a scalable, intelligent, and cost-effective fashion and that allow for intuitive data visualizations are essential. Ever-improving routines and guidelines should be formulated to continue and even enhance responsible data sharing, requiring the collective efforts of researchers, funders, and publishers alike; similar attention should focus on ensuring the use of FAIR (findable, accessible, interoperable, and reusable) data standards and the reproducibility of data analyses38. Innovations in technology and policy must be integrated to develop data-stewardship models that ensure open science and reduce data-access burdens to advance research, including the use of optimally balanced and ethically sound approaches for respecting participant preferences and consent as well as engaging communities. Such developments should be done in an open-source culture to build consensus and enable the development, maintenance, and utilization of best-in-class tools, pipelines, and platforms that can be applied to all datasets.
Fully integrating genomics into medical practice will require informatics and data-science advances that effectively connect the growing body of genomic knowledge to clinical decision-making. To make genomic information readily accessible and broadly useful to clinicians, user-friendly electronic health record-based clinical decision support tools must be created to interact with a variety of clinical data from electronic health record and other data systems (e.g., laboratory, pharmacy, and radiology) as well as non-computable reports, such as those provided as portable document format (PDF) files43,44. These efforts require well-curated, highly integrated, and up-to-date knowledgebases that connect genomic information to clinical characteristics, other phenotypic data, and information on family health history45. Also needed are reliable risk-stratification and prevention algorithms, including polygenic risk scores (PRSs)46, that incorporate both common and rare genomic variants from a broad range of population subgroups, phenotypic data, and environmental information into the risk modeling47. Such algorithms should be evaluated both for their validity across multiple populations and for their impact on patient outcomes and subsequent healthcare utilization. Finally, it will be important to evaluate new genomics-oriented clinical decision support tools to ensure that they are acceptable to practitioners across the spectrum of clinical disciplines.
Genomics and society
Understanding the role of genomics in human health requires knowledge and insights about how social, environmental, and genomic risk factors interact to produce health outcomes48,49 (Box 1). Given that such interactions are, in general, poorly understood, it is critical that studies of genomic risk (particularly of common, complex diseases) account for the social and environmental factors that influence health and disease50. These factors must be properly described, measured, and incorporated in genomics studies51. Optimal implementation of genomic medicine will require understanding how the intersectional aspects of people’s social and political identities influence the ways in which populations are described in research. Such knowledge will, in turn, provide clarity about the interrelationships among these multiple influences on health and disease.
People want to be able to make well-informed decisions about their genomic data, leading to the engagement efforts in initiatives such as the UK Biobank52 and the All of Us Research Program53. Partnering with communities and individuals is fundamental to engaging participants in such large-scale research. Genomic researchers must incorporate models and methods of community engagement in their experimental design. Such studies must be appropriately tailored for different cultures and designed to reduce inequities and healthcare disparities; they must also be accompanied by effective information dissemination54. An unrelenting focus on the optimal ways to conduct research in partnership with data stakeholders and communities would ensure the identification of the key issues and values influencing peoples’ choices about the provision of personal data for research55,56. Data-stewardship infrastructures that integrate appropriate policies, technologies57, and governance and legal frameworks must be developed and assessed to ensure alignment between communities’ and individuals’ decisions about their data and the practices of researchers and clinicians.
To fully realize the fruits of genomic advances, a working understanding of the basic concepts of genomics will be important for science educators58, healthcare professionals59, policymakers, and the public60. Multiple educational strategies will inevitably be required for enhancing the genomic literacy of these heterogeneous groups, which points to the need for innovative approaches that are shared, assessed, and improved over time58. A growing evidence base shows that increasing the understanding of key genomics concepts and applications attracts students to careers in genomics61, assists with the use of genomics for addressing health disparities62, and facilitates the uptake of genomic medicine63. Curricula for enhancing genomic literacy must be designed to be accessible, effective, and scalable for use in the full range of settings where genomics education is provided – including primary and secondary schools, science museums, and informal science-education venues. Researchers and educators must also disseminate information about both the science of genomics as well as the key ethical and societal implications of genomics64.
Training and genomics workforce development
Appropriate skills in data science and data stewardship are now prerequisites for becoming a genomics researcher65. Furthermore, given the ever-expanding use of genomics in basic, translational, social, behavioral, and clinical research, a greater number of scientists will require fundamental data-science skills appropriate for the genomic applications being utilized66. Establishing and maintaining data-science competencies for conducting genomics research requires a series of interrelated educational and training efforts67, including the recruitment of a cadre of data scientists into genomics and the reciprocal exchange of expertise between genomics researchers and data scientists.
Moving into healthcare, providers must be poised to manage questions from patients who receive genomic information, including that from direct-to-consumer testing, and this applies to the full spectrum of medical professionals (including nurses, pharmacists, physicians, and other clinicians)68. Education modules tailored to specific user groups should be designed to adapt rapidly to advances in genomics and data-science technologies; these should be available on demand and, where appropriate, integrated into existing clinical systems69. Research on the methodologies for train-the-trainer approaches, implementation of standards and competency-based education, and strategies for enhancing genomic literacy among all healthcare providers at all career stages70 should also be pursued. The involvement of patients, caregivers, educators, professional organizations71, and accreditation boards will be critical to ensure success. Importantly, cross-training in relevant aspects of genomics must also be available for specialists working in or around healthcare systems, including (but not limited to) those involved in health services research, health economics, law, bioethics, and social and behavioral sciences.
Both in research and clinical settings, the global genomics workforce – as with the general biomedical research workforce – falls considerably short of reflecting the diversity of the world’s population (a vivid example of this is seen in the U.S.72), which limits the opportunity of those systematically excluded to bring their unique ideas to scientific and clinical research73. To attain a diverse genomics workforce, new strategies and programs to reduce impediments to career opportunities in genomics are required, as are creative approaches to promote workforce diversity, leadership in the field, and inclusion practices. Efforts must intentionally include women, underrepresented racial and ethnic groups, disadvantaged populations, and individuals with disabilities. Initiatives should not focus exclusively on early-stage recruitment; rather, they must also include incentives to recruit and retain a diverse workforce at all career stages74 as well as novel approaches for cultivating the next generation of genomics practitioners.
Breaking down barriers in genomics
Genomics has benefited enormously from the proactive identification of major obstacles impeding progress and the subsequent focused efforts to break down those barriers. Prototypic successes include the call for a “$1,000 human genome sequence” following completion of the Human Genome Project15 and proposed actions to facilitate genomic medicine implementation in 201116; in these cases, both the risks of failure and the benefits of success were high. Once again, breaking down barriers, as highlighted in Box 3 and detailed below, would accelerate progress and create new research and clinical opportunities at The Forefront of Genomics.
Laboratory and computational technologies
Advances in DNA synthesis and genome editing allow the field of genomics to progress from largely observational (“reading DNA”) to more experimental (“writing” and “editing” DNA) approaches. Enabling true “synthetic genomics” (i.e., the synthesis, modification, and perturbation of nucleic acid sequences at any scale) will allow for more powerful experimental testing of hypotheses about genome variation and function and improve opportunities for linking genotypes to phenotypes75. Genome editing is increasingly being used for practical applications in medicine (e.g., in gene therapy76), biotechnology, agriculture, and other areas. Despite recent triumphs, however, the current approaches are limited in their ability to interrogate genome function at the pathway or network level and to study gene regulation, chromosome organization and mechanics, and other important phenomena that involve factors acting across large chromosomal (or genomic) distances. Furthermore, radically new capabilities for understanding how the full complement of genomic variation within any individual genome contributes to phenotype should be pursued. Innovative approaches for generating nucleic acid molecules with defined sequences and of any size, coupled with technologies that allow for the concurrent and large-scale perturbation of multiple genes or simultaneous examination of multiple genomic variants, would be transformative. These advances would benefit from the development of methods for introducing large synthetic constructs into mammalian cells.
In recent years, large human genomics projects have often relied on data generated as part of existing research studies, and emerging approaches involve developing biobanks and organized cohorts77–79. Meanwhile, direct-to-consumer (DTC) companies are generating substantial amounts of genomic data, and those efforts are rapidly being eclipsed by that being generated in the clinical care setting80. Properly leveraged, these DTC and clinical data offer opportunities for genomics-based studies at unprecedented scales; however, these data are often heavily fragmented, siloed, and mostly outside the purview of genomics researchers and their typical funders81. Eliminating the barriers to accessing these sources of data for conducting research is essential, but this will require resolving issues related to governance, policy infrastructure, and informatics and workflow solutions. Approaches are needed to mitigate the resulting gaps, limitations, and biases within this highly distributed data environment (e.g., with regards to population diversity, data-collection strategies, data standards, and data privacy), all while addressing concerns of the patients, participants, and groups. These challenges must be addressed globally81 (Box 1), so as to accommodate differences in healthcare systems and views about data privacy. In addition, the healthcare stakeholders should take advantage of opportunities offered by genomics, thereby enabling virtuous-cycle routes between genomic learning healthcare systems and basic genomics research82 (Fig. 3).
Fig. 3 |. Virtuous cycles in human genomics research and clinical care.
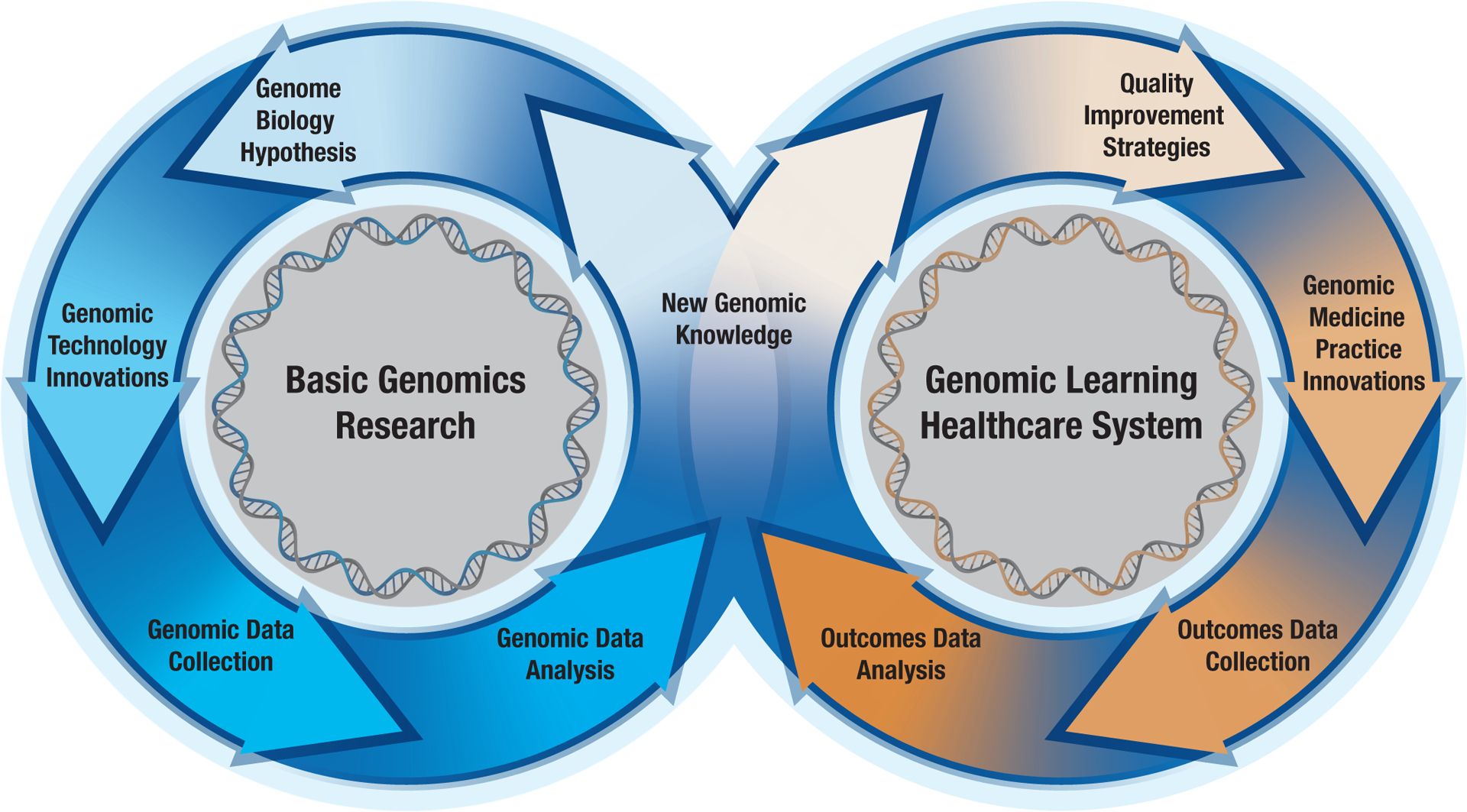
As human genomics has matured as a discipline, productive and connected virtuous cycles of activity have emerged, each self-improving with successive rounds of new advances. The cycle on the left reflects basic genomics research, in which technology innovations spur the collection and analysis of genomics research data, often yielding new knowledge and additional hypotheses for testing. The cycle on the right reflects a genomic learning healthcare system, in which the implementation of new genomic medicine practice innovations allows for the collection and analysis of outcomes data, often yielding new genomic knowledge and additional genomics-based strategies for improving the quality of clinical care. Note that the new knowledge emerging from either the left or right cycle has the potential to feed into the other, creating opportunities for “bench to bedside” and “bedside back to bench” progressions82 – both of which are expected to grow in the coming decade.
Biological insights
Despite progress in identifying genomic variants that cause monogenic traits or are statistically associated with complex phenotypes, connecting specific variants to phenotypes remains challenging83. Systematic approaches, including new tactics that connect high-throughput molecular readouts of functional genomic assays to organismal phenotypes, are required for establishing the phenotypic consequences of all genomic variants – individually and in combinations – in a cell-type context across the life span84. Progress in this area requires global collaboration85, advances in integrating multiple data types and performing perturbation assays, protein localization/interaction experiments, and animal models, as well as resources cataloging information about the fitness consequences of de novo mutations and the clinical relevance of genomic variants83. Because it is not possible to directly test every variant in all cell types and states, developmental stages, and disease processes, new data-collection strategies and analytical approaches are needed that can generalize and adapt predictions to new contexts, handle sparse data, and prioritize variants for experimental follow-up.
Recent advances have led to a greater appreciation of the extent of mosaicism – i.e., genomic variation among cells (both somatic and germline) within an individual. While there have been remarkable advances in understanding the somatic genomic changes encountered in cancer86, there is a paucity of detailed knowledge about other impacts of mosaicism beyond a few well-studied examples87. Important areas of future research include investigating the prevalence and extent of different forms of mosaic variation in both nuclear and mitochondrial DNA, the mechanisms that generate mosaicism, and the roles of mosaicism in physiology and human disease. Such efforts might reveal if this form of genomic variation contributes to variable penetrance and expressivity, comprises a form of genetic epistasis, explains any currently undiagnosed diseases or sporadic cases (or apparent phenocopies) of known inherited diseases9, and can inform the design of therapies for genetic diseases. Single-cell genomic technologies have extended knowledge about the functional impact of mosaicism in multiple experimental systems88,89, with the next challenge being to translate such single-cell understanding to in vivo settings. The development of new laboratory and clinical approaches for readily detecting genomic mosaicism at high spatial and temporal resolution, especially in ways that are relatively non-invasive (e.g., requiring minimal amounts of tissue), would be catalytic.
Implementation science
A critical barrier to using genomics for improving health and preventing disease is the lack of clinical uptake of proven genomic interventions. Implementation science approaches are needed to identify the most effective methods and strategies for facilitating the use of evidence-based genomic applications, most notably pharmacogenomics-based selection of medications90, in routine clinical care. Novel experimental designs, such as genotype-specific participant recruitment91 or integration of patient-provided genomic data92 (captured during previous healthcare encounters or from direct-to-consumer sources), should be explored for their potential to speed adoption and limit costs. The effectiveness of centralized resources for genomic referrals [e.g., genomic medicine specialists, consult services93,94, centers of excellence in undiagnosed diseases (akin to transplantation centers or cancer centers)] should be explored as potential steppingstones to the more generalized uptake of genomics in clinical care. Strategies for deploying the limited workforce of highly trained genetics/genomics specialists (e.g., systematic referral networks or telemedicine/telecounseling) should also be evaluated for their effectiveness at increasing the availability of services broadly – as opposed to being limited to select, highly specialized centers.
Universal newborn genetic screening may represent the most visible and successful approach to population-based identification of serious and treatable inherited conditions, but population screening across the lifespan for other genetic conditions is less widely accepted. Standard public health screening approaches for the U.S. Centers for Disease Control and Prevention Tier 1 conditions95,96 (e.g., Lynch syndrome, hereditary breast and ovarian cancer, and familial hypercholesterolemia) identify people at risk through blood relatives of affected individuals (referred to as “cascade testing” by geneticists97). Implementation research methods, coupled with effective science communication, are primed for optimizing approaches for engaging individuals in genetic testing for these disorders and also other emerging indications, such as genetic predisposition to adverse drug effects (pharmacogenomics), carrier testing of prospective parents, use of PRSs in disease detection and prevention46, and genomic indicators (e.g., gene-expression and epigenetic patterns) of exposure to infectious pathogens98 and other environmental agents.
Compelling genomics research projects
The field of genomics has routinely benefited from a willingness to articulate ambitious – often audacious – research efforts that aim to address questions and acquire knowledge that (at the time) may seem out of reach. Such boldness has served to stimulate interest in emerging opportunities, recruit new expertise, galvanize international collaborations involving multiple funders, and propel the field forward. While by no means comprehensive, the areas highlighted in Box 4 and detailed below illustrate the broadening range of compelling research projects that are ripe for pursuit at The Forefront of Genomics.
Advances in understanding gene regulation5,24, the myriad functional roles of RNA99, and the multi-dimensional nature of the nucleome100 – coupled with the utility of single-cell genomic approaches30,31 and anticipated new technological and computational capabilities for analyzing genomic datasets and variants – provide an unprecedented opportunity for deciphering the individual and combined roles of each gene and regulatory element. This must start with establishing the function of each human gene, including the phenotypic impact of human gene knockouts. Because genes and regulatory elements do not function in isolation, it is imperative to build robust experimental and computational models that deduce causal relationships and accurately predict cellular and organismal phenotypes using pathway and network models101,102. Novel analysis methods must address functional redundancy as well as the nearly boundless experimental space and complexity, including cell states and fates, temporal relationships, environmental conditions, and individual genetic background.
Building on the recent successes in unraveling the genetic underpinnings of rare and undiagnosed diseases9, the field is poised to gain a more comprehensive understanding of the genetic architecture of all human diseases and traits10,85. However, myriad complexities can be anticipated. For example, any given genomic variant(s) may affect more than one disease or trait (i.e., pleiotropy); can confer disease risk or reduce it; and can act additively, synergistically, and/or through intermediates. New methods for analyzing data that account for human diversity103, coupled with a growing clarity about genotype-phenotype relationships, must be developed to deduce associations and interactions among genomic variants and environmental factors, improve estimates of penetrance and expressivity, and enhance the clinical utility of genomic information for predicting risk, prognosis, treatment response, and, ultimately, clinical outcomes.
Prioritizing the generation of genomic and corresponding phenotypic data from ancestrally diverse participants is a scientific imperative104 and essential for achieving equitable benefits from genomic advances105 (Box 1). However, this is an area in which genomics has repeatedly fallen short19, leading to missed opportunities for understanding genome structure and function, identifying variants conferring risk for common diseases106, and implementing genomic medicine for the benefit of all107–109. Ideally, studies should be designed for different groups, tailored for local sensibilities and situations, and consistent in capturing key information beyond participants’ ancestry (e.g., the physical and social environments in which they live and receive healthcare110). Leveraging new insights from studies of diverse populations will require the development of new and robust methods for identifying novel signatures of natural selection, performing genotype imputation, mapping disease loci, characterizing genomic variant pathogenicity, and calculating PRSs103,109. Success in these efforts will yield a more-complete understanding of how the human genome functions in different environments and offer benefit to those participating in genomics research. Attaining the level of population diversity that will truly benefit all people requires bold scientific and community-based leadership, dedicated resources from funders, highly committed researchers, and effective partnerships that earn the trust of diverse groups of participants and their communities.
As genomics has grown in medicine and society, its potential to influence people’s actions has also expanded. Increasingly, genomics has affected concepts of health, disease, responsibility, family, identity, and community, raising a number of important and changing questions. When and how is genomic information shared and communicated within families111? Will the identification of a strong genetic risk for a disease change a person’s perception of their health or others’ perception of that person? With some genetic risks being more common in certain identifiable populations, what role does group affiliation play in how risk is communicated and perceived, including potential group stigmatization? Research that catalogs, analyzes, and measures the impact of genomics on individuals, families, and communities is important for providing a more informed context to avoid future misrepresentations, misunderstandings, and misuses of genomics54. Finally, researchers must appreciate how their own backgrounds and experiences shape their interpretations of genomic data112.
Extending genomics research in clinical settings beyond DNA sequence to include other multi-omic data, together with clinical variables and outcomes, would advance understanding of disease onset and progression and may also prove important for drug-discovery efforts113,114. This would require tissue- and cell-specific analyses that integrate these data, providing real-time snapshots of biological and disease processes. For clinical applicability and adoption, these high-dimensional, multi-omic data should be integrated with clinical decision support tools and electronic health records. Ultimately, such efforts could reveal important relationships among genomic, environmental, and behavioral variation and facilitate a transition of the use of genomics in medicine from diagnosing and treating disease to maintaining health.
Sharp barriers between research and clinical care obstruct the virtuous cycle of moving scientific discoveries rapidly into clinical care and bringing clinical observations back to the research setting82 (Fig. 3). Learning healthcare systems – in which real-time data on outcomes of healthcare delivery are accessed and used to enhance clinical practice – can lead to continuous care improvement, but only if the barriers between research and clinical care are reduced115. For example, offering genome sequencing to all members of a healthcare system, performed in conjunction with research and participant engagement and provided in real time81, could help to assess the clinical utility of genomic information and may allow providers to improve disease diagnosis and management. System-wide implementation of such an experiment requires not only extensive patient and provider education, sophisticated informatics capabilities, and genomics-based clinical decision support, but also the development and evaluation of data security and privacy protections to ensure patient confidentiality116. Patients should be engaged in the design of such systems and informed at entry to them (and periodically thereafter), so as to be fully aware of the nature of the ongoing research with their clinical data and the goals and potential risks of their participation117. Extending such studies across multiple healthcare systems should reveal common challenges and solutions118,119, thereby enhancing the learning healthcare model for genomic medicine more broadly (Fig. 3).
Concluding thoughts
The dawn of genomics featured the launch of the Human Genome Project in October 19901. Three decades later, the field’s journey has included stunning technological advances and high-profile programmatic successes, which in turn have led to the widespread infusion of genomic methods and approaches across the life sciences and, increasingly, into medicine and society.
NHGRI has for the third time15,16 since the Human Genome Project undergone an extensive horizon-scanning process to capture, synthesize, and articulate the most compelling strategic opportunities for the next phase of genomics – with particular attention to those elements most relevant to human health. The now near-ubiquitous nature of genomics (including in the complex healthcare ecosystem) presented practical challenges for attaining a holistic assessment of the field. Another reality was that NHGRI’s investment in genomics has now been multiplied many-fold by the seeding of human genomics throughout the broader research community. These changes reflect a continued maturation of both the field (in general) and NHGRI (more specifically), nicely aligning with the institute’s evolving leadership role at The Forefront of Genomics.
Embracing that role, NHGRI formulated the strategic vision described here, which provides an optimistic outlook that the successes in human genomics over the past three decades will be amplified in the coming decade. Many of the details about what is needed to fulfill the promise of genomics have now come into focus. Major unsolved problems remain – among them determining the role for the vast majority of functional elements in the human genome (especially those outside of protein-coding regions), understanding the full spectrum of genomic variation (especially that implicated in human disease), developing data-science capabilities (especially those that keep pace with data generation), and improving healthcare through the deployment of genomic medicine (especially in the areas of prevention, diagnosis, and therapeutic development). The new decade also brings new research questions related to the societal implications of genomics, including those related to social inequities, pointing to the continued importance of investigating the ethical, legal, and social issues related to genomics. But now more than ever, solutions to these problems seem to be within striking distance. Towards that end (and with the characteristic spirit of genomics audacity), we offer ten bold predictions of what might be realized in human genomics by 2030 (Box 5).
Box 5: Bold Predictions for Human Genomics by 2030.
Some of the most impressive genomics achievements, when viewed in retrospect, could hardly have been imagined ten years earlier. Here are ten bold predictions for human genomics that might come true by 2030. While most are unlikely to be fully attained, achieving one or more of these would require individuals to strive for something that currently seems out of reach. These predictions were crafted to be both inspirational and aspirational in nature, provoking discussions about what might be possible at The Forefront of Genomics in the coming decade.
Generating and analyzing a complete human genome sequence will be routine for any research laboratory, becoming as straightforward as carrying out a DNA purification.
The biological function(s) of every human gene will be known; for non-coding elements in the human genome, such knowledge will be the rule rather than the exception.
The general features of the epigenetic landscape and transcriptional output will be routinely incorporated into predictive models of the impact of genotype on phenotype.
Research in human genomics will have moved beyond population descriptors based on historic social constructs such as race.
Studies involving analyses of genome sequences and associated phenotypic information for millions of human participants will be regularly featured at school science fairs.
The regular use of genomic information will have transitioned from boutique to mainstream in all clinical settings, making genomic testing as routine as complete blood counts (CBCs).
The clinical relevance of all encountered genomic variants will be readily predictable, rendering the diagnostic designation “variant of uncertain significance (VUS)” obsolete.
A person’s complete genome sequence along with informative annotations can be securely and readily accessible on their smartphone.
Individuals from ancestrally diverse backgrounds will benefit equitably from advances in human genomics.
Genomic discoveries will lead to curative therapies involving genomic modifications for dozens of genetic diseases.
The strategic vision articulated here was crafted on behalf of the field of human genomics and emphasized broad strategic goals opposed to implementation tactics. The realization of these goals will require additional planning in conjunction with the collective creativity, energies, and resources of the global community of scientists, funders, and research participants. NHGRI has taken some initial steps for implementing this vision, although these will inevitably need to be adapted as advances occur and circumstances change. Indeed, the final words of this strategic vision were formulated as the world moved urgently to deal with the COVID-19 pandemic (see Epilogue), providing a vivid reminder of the need to be nimble and the importance of nurturing all parts of the research continuum – from basic to translational to clinical – for protecting public health and advancing medical science.
Despite the seismic changes seen in genomics since the inception of the field, the fundamental sense of curiosity, marvel, and purpose associated with genome science seems to be timeless. In concluding NHGRI’s previous strategic vision16 – published just under a decade ago – the then-envisioned opportunities and challenges were provided with “… a continuing sense of wonder, a continuing need for urgency, a continuing desire to balance ambition with reality, and a continuing responsibility to protect individuals while maximizing the societal benefits of genomics….” With the 2020 strategic vision described here providing a thoughtful guide and with enduring feelings of wonder, urgency, ambition, and social consciousness providing unfettered momentum, we are ready to embark on the next exciting phase of the human genomics journey.
Epilogue: COVID-19 and genomics
SARS-CoV-2 emerged as global threat to public health at the end of the multi-year process that generated the above strategic vision. Nonetheless, the pandemic provides a potent lesson about how a tiny string of nucleic acids can wreak global havoc on humankind. Understanding the mechanisms involved in the virus’s transmission, invasion, and clearance, as well as the highly variable and at times disastrous physiologic responses to it, are fertile grounds for genomics research. Indeed, genomics rapidly assumed critical roles in COVID-19 research and clinical care in areas such as the (1) deployment of DNA- and RNA-sequencing technologies for diagnostics, viral isolate tracking, and environmental monitoring; (2) use of synthetic nucleic acid technologies for studying SARS-CoV-2 virulence and facilitating vaccine development; (3) examination of how human genomic variation influences infectivity, disease severity, vaccine efficacy, and treatment response; (4) adherence to principles and values related to open science, data sharing, and consortia-based collaborations; and (5) provision of genomic data science tools for studying COVID-19 pathophysiology. The growing adoption of genomic approaches and technologies into myriad aspects of the global response to the COVID-19 pandemic serves as another important and highly visible example of the integral and vital nature of genomics in modern research and medicine.
Acknowledgements
The strategic vision described here was formulated on behalf of NHGRI. We are grateful to the many members of the institute staff for their contributions to the associated planning process (see http://genome.gov/genomics2020 for details) as well as to the numerous external colleagues who provided input to the process and draft versions of this strategic vision. The National Advisory Council for Human Genome Research (current members are Jeffrey Botkin, Trey Ideker, Sharon Plon, Jonathan Haines, Stephen Fodor, Rafael Irizarry, Patricia Deverka, Wendy Chung, Mark Craven, Hal Dietz, Stephen Rich, Howard Chang, Lisa Parker, Len Pennacchio, and Olga Troyanskaya) ratified the strategic planning process, themes, and priorities associated with this strategic vision.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.The Human Genome Project. Available at: https://www.genome.gov/human-genome-project. (Accessed: 28th June 2020)
- 2.Lander ES et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 3.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004). [DOI] [PubMed] [Google Scholar]
- 4.The Cost of Sequencing a Human Genome | NHGRI. Available at: https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost. (Accessed: 12th June 2020)
- 5.Moore JE et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shema E, Bernstein BE & Buenrostro JD Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet 51, 19–25 (2019). [DOI] [PubMed] [Google Scholar]
- 7.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; Analysis of a large dataset of exome sequences, yielding important descriptions of the extent and nature of human genomic variation and new insights into protein evolution.
- 9.Posey JE et al. Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med 21, 798–812 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claussnitzer M et al. A brief history of human disease genetics. Nature 577, 179–189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolio TA et al. Opportunities, resources, and techniques for implementing genomics in clinical care. Lancet 394, 511–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mardis ER The Impact of Next-Generation Sequencing on Cancer Genomics: From Discovery to Clinic. Cold Spring Harb. Perspect. Med 9, a036269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianchi DW & Chiu RWK Sequencing of Circulating Cell-free DNA during Pregnancy. N. Engl. J. Med 379, 464–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright CF, FitzPatrick DR & Firth HV Paediatric genomics: diagnosing rare disease in children. Nat. Rev. Genet 19, 253–268 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Collins FS, Green ED, Guttmacher AE & Guyer MS A vision for the future of genomics research. Nature 422, 835–847 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Green ED & Guyer MS Charting a course for genomic medicine from base pairs to bedside. Nature 470, 204–213 (2011). [DOI] [PubMed] [Google Scholar]
- 17.McEwen JE et al. The Ethical, Legal, and Social Implications Program of the National Human Genome Research Institute: Reflections on an Ongoing Experiment. Annu. Rev. Genomics Hum. Genet 15, 481–505 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Burke W et al. The translational potential of research on the ethical, legal, and social implications of genomics. Genet. Med 1–9 (2014). doi: 10.1038/gim.2014.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popejoy AB & Fullerton SM Genomics is failing on diversity. Nature 538, 161–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive analysis of genome-wide association studies, demonstrating continued severe under-representation of individuals of African and Latin American ancestry and indigenous peoples.
- 20.Wolf SM et al. Integrating Rules for Genomic Research, Clinical Care, Public Health Screening and DTC Testing: Creating Translational Law for Translational Genomics. J. Law, Med. Ethics 48, 69–86 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adam D The promise and peril of the new science of social genomics. Nature 574, 618–620 (2019). [DOI] [PubMed] [Google Scholar]; Summary of recent studies examining the genetics of biobehavioral traits, highlighting dangers to groups and society of overinterpreting results in this new field.
- 22.Dias R & Torkamani A Artificial intelligence in clinical and genomic diagnostics. Genome Medicine 11, 70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss JA, Gibbs RA, Makhijani VB & Marziali A Cultivating DNA Sequencing Technology After the Human Genome Project. Annu. Rev. Genomics Hum. Genet 21, annurev-genom-111919–082433 (2020). [DOI] [PubMed] [Google Scholar]; Retrospective overview of the NHGRI program for advancing DNA sequencing technologies, the goal of which was to reduce the cost of sequencing a human genome to $1,000.
- 24.ENCODE: Encyclopedia of DNA Elements – ENCODE. Available at: https://www.encodeproject.org/. (Accessed: 24th June 2020)
- 25.Risca VI & Greenleaf WJ Unraveling the 3D genome: Genomics tools for multiscale exploration. Trends in Genetics 31, 357–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logsdon GA, Vollger MR & Eichler EE Long-read human genome sequencing and its applications. Nature Reviews Genetics 1–18 (2020). doi: 10.1038/s41576-020-0236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miga KH et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 1–21 (2020). doi: 10.1038/s41586-020-2547-7 [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of the use of emerging DNA sequencing technologies, analysis methods, and validation routines to produce the first gapless de novo assembly of a human chromosome sequence.
- 28.Human Pangenome Reference Consortium – Diverse Human References Drive Genomic Discoveries for Everyone. Available at: https://humanpangenome.org/. (Accessed: 29th June 2020)
- 29.Zerbino DR, Frankish A & Flicek P Progress, Challenges, and Surprises in Annotating the Human Genome. Annu. Rev. Genomics Hum. Genet 21, annurev-genom-121119–083418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rood JE et al. Toward a Common Coordinate Framework for the Human Body. Cell 179, 1455–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart T & Satija R Integrative single-cell analysis. Nature Reviews Genetics 20, 257–272 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Mimitou EP et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schreiber J, Durham T, Bilmes J & Noble WS Avocado: A multi-scale deep tensor factorization method learns a latent representation of the human epigenome. Genome Biol. 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham F et al. Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewin HA et al. Earth BioGenome Project: Sequencing life for the future of life. Proceedings of the National Academy of Sciences of the United States of America 115, 4325–4333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindblad-Toh K What animals can teach us about evolution, the human genome, and human disease. Ups. J. Med. Sci 125, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schatz MC Biological data sciences in genome research. Genome Research 25, 1417–1422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson MD et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; Description of foundational principles to improve data sharing and stewardship by ensuring that biomedical research data (including genomic data) is findable, accessable, interoperable, and reusable.
- 39.Anderson W et al. Towards Coordinated International Support of Core Data Resources for the Life Sciences. bioRxiv 110825 (2017). doi: 10.1101/110825 [DOI] [Google Scholar]
- 40.Grossman RL Data Lakes, Clouds, and Commons: A Review of Platforms for Analyzing and Sharing Genomic Data. Trends Genet. 35, 223–234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haendel MA, Chute CG & Robinson PN Classification, ontology, and precision medicine. N. Engl. J. Med 379, 1452–1462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martínez-Romero M et al. Using association rule mining and ontologies to generate metadata recommendations from multiple biomedical databases. Database 2019, 59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy KD et al. Opportunities to implement a sustainable genomic medicine program: lessons learned from the IGNITE Network. Genet. Med 21, 743–747 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams MS et al. Genomic information for clinicians in the electronic health record: Lessons learned from the clinical genome resource project and the electronic medical records and genomics network. Front. Genet 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemke AA et al. Primary care physician experiences utilizing a family health history tool with electronic health record–integrated clinical decision support: an implementation process assessment. J. Community Genet 11, 339–350 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khera AV et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 1 (2018). doi: 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]; Development and validation of genome-wide polygenic scores that identify population subsets with risk levels equivalent to monogenic genomic variants that are commonly reported and acted upon.
- 47.Zeggini E, Gloyn AL, Barton AC & Wain LV Translational genomics and precision medicine: Moving from the lab to the clinic. Science (80-. ). 365, 1409–1413 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Koehly LM et al. Social and behavioral science at the forefront of genomics: Discovery, translation, and health equity. Soc. Sci. Med 112450 (2019). doi: 10.1016/j.socscimed.2019.112450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan SS, Cooper R & Greenland P Do Polygenic Risk Scores Improve Patient Selection for Prevention of Coronary Artery Disease? JAMA 323, 614–615 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Mostafavi H et al. Variable prediction accuracy of polygenic scores within an ancestry group. Elife 9, 1–52 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris TT, Davies NM, Hemani G & Smith GD Population phenomena inflate genetic associations of complex social traits. Sci. Adv 6, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Investigators T “All of U. R. P. The “All of Us” Research Program. N. Engl. J. Med 381, 668–676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrison NA et al. Genomic Research Through an Indigenous Lens: Understanding the Expectations. Annu. Rev. Genomics Hum. Genet 20, 495–517 (2019). [DOI] [PubMed] [Google Scholar]; Discussion of issues related to conducting genomics research with indigenous peoples, coupled with suggestions for respecting tribal governance and protecting native people from group harms.
- 55.Sanderson SC et al. Public Attitudes toward Consent and Data Sharing in Biobank Research: A Large Multi-site Experimental Survey in the US. Am. J. Hum. Genet 100, 414–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; Survey results from 13,000 individuals regarding participation in research in which their data are shared with others, yielding insight into factors that predict a willingness of people to participate in research and concerns about data privacy.
- 56.Milne R et al. Trust in genomic data sharing among members of the general public in the UK, USA, Canada and Australia. Hum. Genet 138, 1237–1246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grishin D, Obbad K & Church GM Data privacy in the age of personal genomics. Nat. Biotechnol 37, 1115–1117 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Literacy Genomic, Education and Engagement Initiative. Available at: https://www.genome.gov/leadership-initiatives/Genomic-Literacy-Education-Engagement-Initiative. (Accessed: 29th June 2020)
- 59.Manolio TA & Murray MF The growing role of professional societies in educating clinicians in genomics. Genetics in Medicine 16, 571–572 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Krakow M, Ratcliff CL, Hesse BW & Greenberg-Worisek AJ Assessing Genetic Literacy Awareness and Knowledge Gaps in the US Population: Results from the Health Information National Trends Survey. Public Health Genomics 20, 343–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.LaRue KM, McKernan Mm. P., Bass KM & Wray CG Teaching the Genome Generation: Bringing Modern Human Genetics into the Classroom Through Teacher Professional Development. J. STEM Outreach 1, 48–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mboowa G & Sserwadda I Role of genomics literacy in reducing the burden of common genetic diseases in Africa. Mol. Genet. Genomic Med 7, e776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veilleux S, Bouffard M & Bourque Bouliane M Patient and Health Care Provider Needs and Preferences in Understanding Pharmacogenomic and Genomic Testing: A Meta-Data Analysis. Qual. Health Res 30, 43–59 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Kung J & Wu C-T Leveling the Playing Field: Closing the Gap in Public Awareness of Genetics between the Well Served and Underserved. Hastings Cent. Rep 46, 17–20 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Stephens ZD et al. Big data: Astronomical or genomical? PLoS Biol. 13, e1002195 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Attwood TK, Blackford S, Brazas MD, Davies A & Schneider MV A global perspective on evolving bioinformatics and data science training needs. Brief. Bioinform 20, 398–404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Genomics Education Partnership | Genomics Research. Available at: http://gep.wustl.edu/. (Accessed: 16th June 2020)
- 68.Campion MA, Goldgar C, Hopkin RJ, Prows CA & Dasgupta S Genomic education for the next generation of health-care providers. Genetics in Medicine 21, 2422–2430 (2019). [DOI] [PubMed] [Google Scholar]
- 69.McClaren BJ et al. Development of an Evidence-Based, Theory-Informed National Survey of Physician Preparedness for Genomic Medicine and Preferences for Genomics Continuing Education. Front. Genet 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dougherty MJ, Wicklund C & Johansen Taber KA Challenges and opportunities for genomics education: Insights from an Institute of Medicine Roundtable Activity. J. Contin. Educ. Health Prof 36, 82–85 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Inter-Society Coordinating Committee for Practitioner Education in Genomics | NHGRI. Available at: https://www.genome.gov/For-Health-Professionals/Inter-Society-Coordinating-Committee-for-Practitioner-Education-in-Genomics. (Accessed: 16th June 2020)
- 72.Valantine HA, Collins FS & Verma IM National Institutes of Health addresses the science of diversity. Proceedings of the National Academy of Sciences of the United States of America 112, 12240–12242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hofstra B et al. The diversity–innovation paradox in science. Proc. Natl. Acad. Sci. U. S. A 117, 9284–9291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; Study of the U.S. doctorate recipients from 1977 to 2015, identifiying novel contributions by gender and racial/ethnic minority scholars, evidence for lower rates of recognition by majority scholars, and the resulting diversity–innovation paradox in science.
- 74.Martinez LR, Boucaud DW, Casadevall A & August A Factors Contributing to the Success of NIH-Designated Underrepresented Minorities in Academic and Nonacademic Research Positions. CBE—Life Sci. Educ 17, ar32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schindler D, Dai J & Cai Y Synthetic genomics: a new venture to dissect genome fundamentals and engineer new functions. Curr. Opin. Chem. Biol 46, 56–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doudna JA The promise and challenge of therapeutic genome editing. Nature 578, 229–236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; Review of the scientific, technical, and ethical aspects of using CRISPR technology for therapeutic applications in humans.
- 77.UK Biobank. Available at: https://www.ukbiobank.ac.uk/. (Accessed: 14th June 2020)
- 78.National Institutes of Health (NIH) — All of Us. Available at: https://allofus.nih.gov/. (Accessed: 14th June 2020)
- 79.International HundredK+ Cohorts Consortium (IHCC) – Linking cohorts, understanding biology, improving health. Available at: https://ihccglobal.org/. (Accessed: 14th June 2020)
- 80.Birney E, Vamathevan J & Goodhand P Genomics in healthcare: GA4GH looks to 2022. bioRxiv (2017). doi: 10.1101/203554 [DOI] [Google Scholar]
- 81.Stark Z et al. Integrating Genomics into Healthcare: A Global Responsibility. American Journal of Human Genetics (2019). doi: 10.1016/j.ajhg.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manolio TA et al. Bedside Back to Bench: Building Bridges between Basic and Clinical Genomic Research. Cell 169, 6–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rehm HL et al. ClinGen - The clinical genome resource. N. Engl. J. Med 372, 2235–2242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Starita LM et al. Variant Interpretation: Functional Assays to the Rescue. American Journal of Human Genetics 101, 315–325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Homepage | International Common Disease Alliance. Available at: https://www.icda.bio/. (Accessed: 24th June 2020)
- 86.Welcome to the Pan-Cancer Atlas. Available at: https://www.cell.com/pb-assets/consortium/PanCancerAtlas/PanCani3/index.html. (Accessed: 19th June 2020)
- 87.Steensma DP et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baslan T & Hicks J Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat. Rev. Cancer 17, 557–569 (2017). [DOI] [PubMed] [Google Scholar]
- 89.D’Gama AM & Walsh CA Somatic mosaicism and neurodevelopmental disease. Nature Neuroscience 21, 1504–1514 (2018). [DOI] [PubMed] [Google Scholar]
- 90.Roden DM et al. Pharmacogenomics. The Lancet 394, 521–532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Corbin LJ et al. Formalising recall by genotype as an efficient approach to detailed phenotyping and causal inference. Nat. Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Savatt JM et al. ClinGen’s GenomeConnect registry enables patient-centered data sharing. Hum. Mutat 39, 1668–1676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eadon MT et al. Implementation of a pharmacogenomics consult service to support the INGENIOUS trial. Clinical Pharmacology and Therapeutics 100, 63–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Darnell AJ et al. A Clinical Service to Support the Return of Secondary Genomic Findings in Human Research. Am. J. Hum. Genet 98, 435–441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tier Table Database|Home|PHGKB. Available at: https://phgkb.cdc.gov/PHGKB/tierStartPage.action. (Accessed: 17th June 2020)
- 96.Dotson WD et al. Prioritizing Genomic Applications for Action by Level of Evidence: A Horizon-Scanning Method. Clin. Pharmacol. Ther 95, 394–402 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hopkins PN Genotype-guided diagnosis in familial hypercholesterolemia: population burden and cascade screening. Curr. Opin. Lipidol 28, 136–143 (2017). [DOI] [PubMed] [Google Scholar]
- 98.Bierne H, Hamon M & Cossart P Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med 2, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bhat AA et al. Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. Mol. Cancer 19, 57 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sparks TM, Harabula I & Pombo A Evolving methodologies and concepts in 4D nucleome research. Curr. Opin. Cell Biol 64, 105–111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Young AI, Benonisdottir S, Przeworski M & Kong A Deconstructing the sources of genotype-phenotype associations in humans. Science 365, 1396–1400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mitra K, Carvunis A-R, Ramesh SK & Ideker T Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet 14, 719–732 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bien SA et al. The Future of Genomic Studies Must Be Globally Representative: Perspectives from PAGE. Annu. Rev. Genomics Hum. Genet 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bentley AR, Callier SL & Rotimi CN Evaluating the promise of inclusion of African ancestry populations in genomics. npj Genomic Med. 5, 5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hindorff LA et al. Prioritizing diversity in human genomics research. Nat. Rev. Genet 19, 175–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wojcik GL et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570, 514–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Landry LG, Ali N, Williams DR, Rehm HL & Bonham VL Lack of diversity in genomic databases is a barrier to translating precision medicine research into practice. Health Aff. 37, 780–785 (2018). [DOI] [PubMed] [Google Scholar]
- 108.Manrai AK et al. Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med 375, 655–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of frequent erroneous classification of genomic variants as pathogenic among patients of African or unspecified ancestry that were subsequently recategorized as benign, with significant health implications of those misclassifications.
- 109.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Horowitz CR et al. Successful recruitment and retention of diverse participants in a genomics clinical trial: a good invitation to a great party. Genet. Med 0, 1–7 (2019). [DOI] [PubMed] [Google Scholar]
- 111.Botkin JR, Mancher M, Busta ER & Downey AS Returning Individual Research Results to Participants. (National Academies Press, 2018). doi: 10.17226/25094 [DOI] [PubMed] [Google Scholar]
- 112.Lázaro-Muñoz G et al. International Society of Psychiatric Genetics Ethics Committee: Issues facing us. Am. J. Med. Genet. Part B Neuropsychiatr. Genet 180, 543–554 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lloyd-Price J et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hasin Y, Seldin M & Lusis A Multi-omics approaches to disease. Genome Biol. 18, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chambers DA, Feero WG & Khoury MJ Convergence of implementation science, precision medicine, and the learning health care system: A new model for biomedical research. JAMA - J. Am. Med. Assoc 315, 1941–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sugano S International code of conduct for genomic and health-related data sharing. Hugo J. 8, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Clayton EW, Halverson CM, Sathe NA & Malin BA A systematic literature review of individuals’ perspectives on privacy and genetic information in the United States. PLoS ONE 13, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cavallari LH et al. Multi-site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genet. Med 21, 2255–2263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ginsburg GS A Global Collaborative to Advance Genomic Medicine. Am. J. Hum. Genet 104, 407–409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


