Abstract
Objectives
Repeat-positive tests for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in individuals with coronavirus disease 2019 (COVID-19) were common. We aimed to investigate the rate and risk factors of recurrent positive detection of SARS-CoV-2 in hospitalized individuals with COVID-19.
Methods
Oropharyngeal and nasopharyngeal swabs (n = 3513) were collected to detect SARS-CoV-2 during the hospitalization. We analysed the recurrent positive rate after consecutive negative results and its relationship to demographic characteristics.
Results
Among 599 enrolled individuals with COVID-19, the median time for viral RNA shedding was 24 days (interquartile range 19–33 days). The positive rates of RT-PCR were 35.9% (215/599), 17.0% (65/383) and 12.4% (23/185) after one, two and three consecutive negative RT-PCR test results, respectively. Medians of Ct values of initial positive test, rebound positive test after two consecutive negative results, and rebound positive after three consecutive negative results were 28.8, 32.8 and 36.1, respectively. Compared with male patients, females had a significantly higher rate of recurrent positive RT-PCR after three consecutive negative results (18.2%, 18/99, versus 5.8%, 5/86; p 0.013). Older individuals (≥55 years) had a significantly higher rate of recurrent positive RT-PCR after one negative result (42.3%, 165/390, versus 23.9%, 50/209; p < 0.001). Nasopharyngeal swab tests produced a higher positive rate than oropharyngeal swab tests (37.3%, 152/408, versus 35.8%, 1111/3105).
Conclusion
Our study revealed the prevalence and dynamic characteristics of recurrent positive RT-PCR to SARS-CoV-2. We showed that around 17.0% (65/383) of patients tested positive for SARS-CoV-2 after two consecutive negative results. Patients with a rebound positive RT-PCR test had a low viral load. Older age and being female were risk factors for recurrent positive results.
Keywords: Coronavirus disease 2019, Recurrent positive, RT-PCR, Severe acute respiratory syndrome coronavirus 2, Viral RNA shedding
Introduction
To date, coronavirus disease 2019 (COVID-19) associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic [1,2]. At the time of drafting the manuscript (Jan 5th, 2021), over 85 500 000 cases were confirmed worldwide with a case fatality rate of approximately 2.2% [3].
The Chinese National Health Committee (seventh version) updated their guidelines regarding discharge and discontinuing isolation to include resolution of fever, improvement in respiratory symptoms and radiography findings, and with two consecutive negative real-time RT-PCR test results for SARS-COV-2 RNA (from two consecutive respiratory specimens collected ≥24 hours apart) [4]. The US CDC updated its criteria for discontinuation of transmission-based precautions for individuals with confirmed COVID-19; a negative RT-PCR test is no longer recommended [5]. Nonetheless, increasing reports on recurrent positive RT-PCR tests for SARS-CoV-2 have aroused wide concern [[6], [7], [8]]. Prolonged viral RNA shedding in certain infected individuals and relatively high false negatives for the viral test by RT-PCR may be responsible for the recurrence. A false-negative RT-PCR result is defined as the negative test result followed by a recurrent positive test. False-negative results were mainly caused by errors in sampling and detection methods.
The prevalence of recurrent positive results of RT-PCR tests to SARS-CoV-2 were reported in several studies. Here, we conducted a retrospective study to investigate the dynamic viral RNA shedding and impact factors for recurrent positive test results of RT-PCR to SARS-CoV-2.
Materials and methods
Study design and participants
We performed a retrospective study with 599 hospitalized individuals with COVID-19 in Tongji Hospital of Huazhong University of Science and Technology in Wuhan, China. All the patients enrolled were confirmed as being diagnosed with COVID-19 according to the diagnosis and treatment guideline for SARS-CoV-2 from the Chinese National Health Committee (seventh version) [4]. The inclusion criteria of this study were as follows: (a) laboratory-confirmed diagnosis of COVID-19; (b) moderate or severe illness; and (c) receive at least one RT-PCR test after two consecutive negative RT-PCR test results during hospitalization. Patients included for analysis in our previous publication were excluded to avoid duplication of data [9] (see Supplementary material, Fig. S1). This study was approved by the ethics committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. Written informed consent was waived by the ethics commission of the designated hospital for emerging infectious disease. All data were collected up to the final follow-up date (21 April 2020).
Data collection and definitions
We collected demographic and clinical data of patients from the electronic medical record system. We included patients with moderate or severe illness according to the guidelines—Moderate: individuals who have evidence of lower respiratory disease by clinical assessment or imaging and a saturation of oxygen (Spo 2) ≥94% on room air at sea level and Severe: individuals who have respiratory frequency >30 breaths per minute, Spo 2 <94% on room air at sea level (or, for patients with chronic hypoxemia, a decrease from baseline of >3%), ratio of arterial partial pressure of oxygen to fraction of inspired oxygen (Pao 2/Fio 2) <300 mmHg, or lung infiltrates >50%) [4,5]. Oropharyngeal and nasopharyngeal swabs were collected both upon admission and during hospitalization by qualified medical professionals through standard procedures under level 3 biosafety protection. Real-time RT-PCR assay were performed on swabs to detect SARS-CoV-2 using COVID-19 test kits (Shanghai Huirui Biotechnology Co., Ltd, Shanghai, China). Once two consecutive negative results were collected, the period between symptom onset and the date of first negative RT-PCR test result was considered as the length of viral RNA shedding.
RT-PCR assay for SARS-CoV-2
Oropharyngeal swab samples or nasopharyngeal swab samples were collected to extract RNA to confirm the diagnosis of COVID-19. Total RNA was extracted using magnetic beads (Tianlong, Xi'an, China). Two target genes, including open reading frame 1ab (ORF1ab) and nucleocapsid protein (N), were simultaneously amplified and tested during the real-time RT-PCR assay. The real-time RT-PCR assay was performed using a COVID-19 nucleic acid detection kit according to the manufacturer's protocol (Shanghai Huirui Biotechnology Co., Ltd). The results of the RT-PCR assay were expressed as the cycle threshold (Ct) value. As recommend by the instruction, a Ct value < 37 was defined as a positive test result, and a Ct value ≥ 39.2 was considered as a negative test. Ct values between 37 and 39.2 suggested that a confirmatory RT-PCR should be obtained using the same sample.
Ethical approval
This study was approved by the ethics committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. All procedures followed in this study were in accordance with the 1964 Helsinki Declaration and later versions. Written informed consent was waived by the ethics commission of the designated hospital for emerging infectious disease.
Statistical analysis
We performed statistical analyses using SPSS version 24.0 (IBM, Armonk, NY, USA). Continuous variables were present as mean ± standard error of the mean or medians (interquartile range, IQR) and were analysed with Mann–Whitney U test. We reported categorical variables as whole numbers and percentages. The cut-off value for age was determined from the receiver operating characteristic curve according to prolonged viral shedding (>24 days). A p value < 0.05 was considered statistically significant.
Results
Demographic and clinical characteristics of patients with recurrent positive RT-PCR results
We included a total of 599 individuals with a confirmed diagnosis of COVID-19 in our study (Table 1 ). In detail, the median age of the included patients was 61 years (IQR 49–68 years) with 291 men (48.6%, 291/599) and 308 women (51.4%, 308/599). A total of 569 (95.0%, 569/599) were classified as having moderate disease. No patient was transferred to an intensive care unit. The median time from onset of symptoms to admission was 10 days (IQR 7–14 days) and the median length of hospital stay was 30 days (23–40 days). The median numbers of RT-PCR tests per patient was 5 (IQR 4–8). The median length for viral shedding was 24 days (IQR 19–33 days). The percentages of patients who shed virus for less than 3 weeks, between 3 and 6 weeks and more than 6 weeks were 40.4%, 48.1% and 11.5%, respectively (see Supplementary material, Fig. S2).
Table 1.
The demographic and clinical characteristics of patients with COVID-19
| Variables | All patients (n = 599) |
|---|---|
| Age (years), median (IQR) | 61 (49–68) |
| Gender, male, n (%) | 291 (48.6%) |
| Smoking, Yes, n (%) | 24 (4.0%) |
| Co-morbidities, n (%) | |
| Hypertension | 122 (20.3%) |
| Diabetes | 68 (11.3%) |
| Pulmonary diseases | 13 (2.2%) |
| Malignancy | 5 (0.8%) |
| Renal failure | 6 (1.0%) |
| Cerebrovascular diseases | 9 (1.5%) |
| Severity on admission, n (%) | |
| Moderate | 569 (95.0%) |
| Severe | 30 (5.0%) |
| RT-PCR test, median (IQR) | |
| Onset of symptom to first RT-PCR test (days) | 7 (4–11) |
| Length of viral RNA shedding (days) | 24 (19–33) |
| No. of RT-PCR tests of each patients | 5 (4–8) |
| Clinical characteristics, median (IQR) | |
| Onset of symptom to admission (days) | 10 (7–14) |
| Length of hospital stay (days) | 30 (23–40) |
| Positive rate of SARS-CoV-2 RT-PCR (weeks after onset) | |
| Week 2 | 90.2% |
| Week 3 | 59.6% |
| Week 4 | 35.4% |
| Week 5 | 20.7% |
| Week 6 | 11.5% |
| Week 7 | 5.3% |
| Week 8 | 1.0% |
| Week 9 | 0.5% |
| ≥Week 10 | 0% |
Abbreviations: COVID-19, coronavirus disease 2019; IQR, interquartile range; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
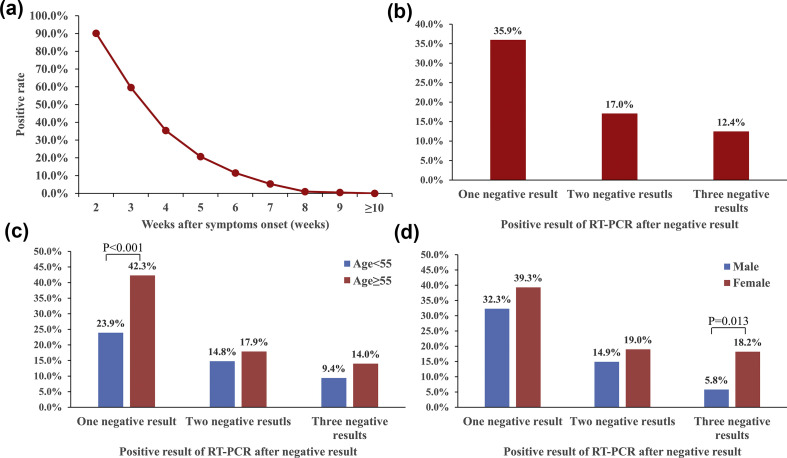
Positive RT-PCR rates were 90.2% (540/599), 59.6% (357/599), 35.4% (212/599), 20.7% (124/599), 11.5% (68/599), 5.3% (32/599), 1.0% (6/599), 0.5% (3/599) and 0% (0/599) on weeks 2, 3, 4, 5, 6, 7, 8, 9 and 10 or later after symptoms onset, respectively (Fig. 1 a).
Fig. 1.
Positive rate of RT-PCR test and recurrent positive result. (a) Positive rate of RT-PCR to SARS-CoV-2 from symptom onset. (b) Recurrent positive rate of RT-PCR after consecutive negative results. (c) Impact of age on recurrent positive rate of RT-PCR. (d) Impact of gender on recurrent positive rate of RT-PCR.
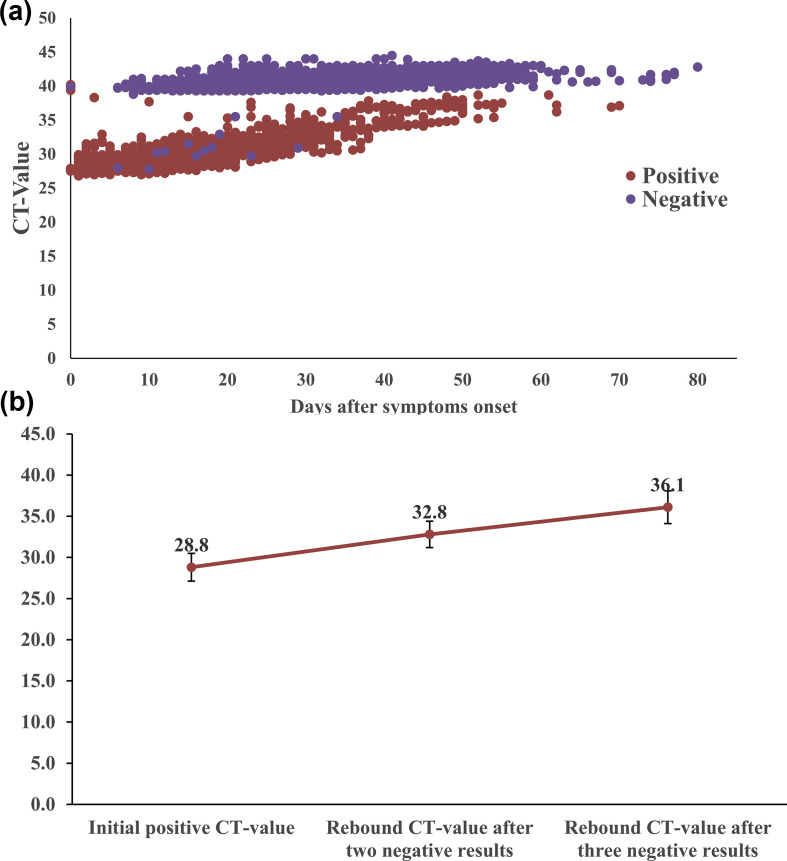
Table 2 shows the demographic characteristics of individuals with recurrent positive RT-PCR results; 35.9% (215/599) of patients had positive RT-PCR results after one negative result. Among 383 individuals with two consecutive negative RT-PCR results, 17.0% (65/383) had recurrent positive test results. Among 185 individuals with three consecutive negative RT-PCR results, 12.4% (23/185) had a recurrent positive test result (Fig. 1b). As demonstrated in Fig. 1c, individuals older than 55 years had a significantly higher recurrence rate after one negative result (42.3%, 165/390, versus 23.9%, 50/209; p < 0.001); however, no significant difference in recurrent positive rate was found after two or three consecutive negative RT-PCR results regardless of age. Women had a significantly higher rate of recurrent positive RT-PCR after three consecutive negative results than men (18.2%, 18/99, versus 5.8%, 5/86; p 0.013) (Fig. 1d). The rate of recurrent positive tests between genders showed no significant difference after one or two consecutive negative RT-PCR results. Fig. 2 shows the dynamic of RT-PCR Ct values. Medians of Ct values of initial positive test, rebound positive test after two consecutive negative results and rebound positive test after three consecutive negative results were 28.8 (95% CI 28.0–29.7), 32.8 (95% CI 32.0–34.4) and 36.1 (95% CI 33.6–37.1), respectively.
Table 2.
The demographic characteristics of patients with recurrent positive RT-PCR result for SARS-CoV-2
| Variables | RT-PCR after one negative result (n = 599) |
p value | RT-PCR after two consecutive negative results (n = 383) |
p value | RT-PCR after 3 consecutive negative results (n = 185) |
p value | |||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | ||||
| Age (years) | <0.001 | 0.553 | 0.484 | ||||||
| <55 | 50/209 (23.9%) | 159/209 (76.1%) | 17/115 (14.8%) | 98/115 (85.2%) | 6/64 (9.4%) | 58/64 (90.6%) | |||
| ≥55 | 165/390 (42.3%) | 225/390 (57.7%) | 48/268 (17.9%) | 220/268 (82.1%) | 17/121 (14.0%) | 104/121 (86.0%) | |||
| Gender | 0.088 | 0.341 | 0.013 | ||||||
| Male | 94/291 (32.3%) | 197/291 (67.7%) | 28/188 (14.9%) | 160/188 (85.1%) | 5/86 (5.8%) | 81/86 (94.2%) | |||
| Female | 121/308 (39.3%) | 187/308 (60.7%) | 37/195 (19.0%) | 158/195 (81.0%) | 18/99 (18.2%) | 81/99 (81.8%) | |||
| Total | 215/599 (35.9%) | 384/599 (64.1%) | 65/383 (17.0%) | 318/383 (83.0%) | 23/185 (12.4%) | 162/185 (87.6%) | |||
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Fig. 2.
Dynamic characteristics of Ct value by RT-PCR (a), and initial and rebound Ct value (medians, interquartile range) after negative RT-PCR tests (b).
Comparison of viral RNA shedding time in individuals with COVID-19 by age and gender
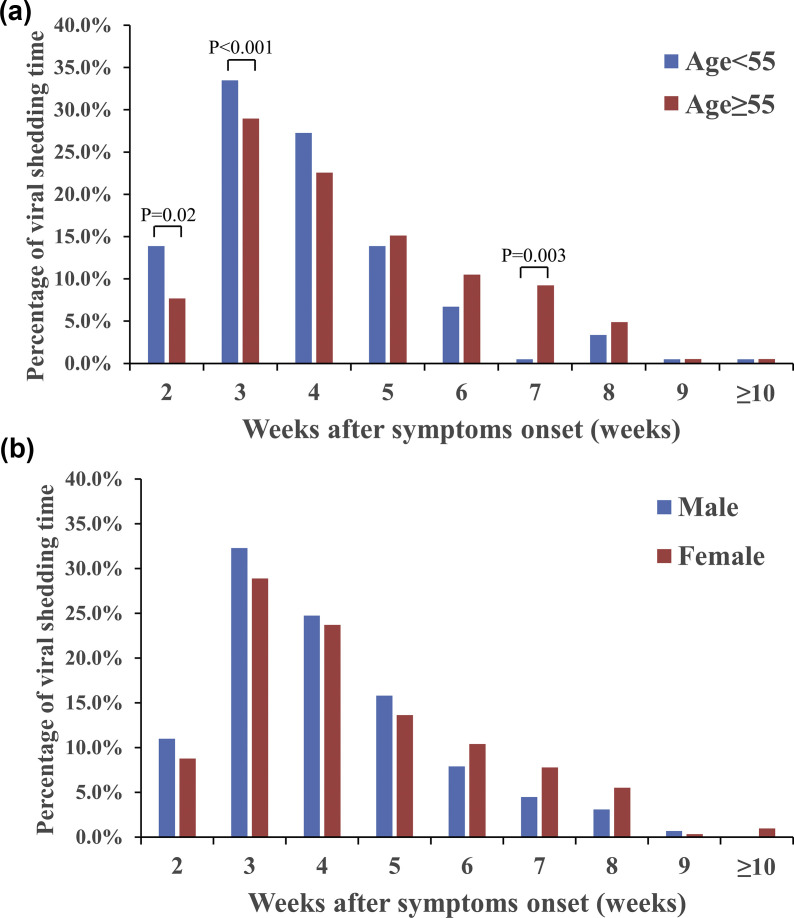
We analysed the viral RNA shedding time of 599 individuals grouped by age and gender (Table 3 ). As demonstrated in Fig. 3 a, individuals <55 years had a shorter period of viral RNA shedding than those aged ≥55 years, with a significantly higher percentage of end of viral shedding on week 2 (13.9%, 29/209, versus 7.7%, 30/390; p 0.02) and week 3 (33.5%, 70/209, versus 29.0%, 113/390; p < 0.01) after symptom onset. Individuals aged ≥55 years had a significantly higher percentage for length of viral shedding on week 7 after symptom onset (9.2%, 36/390, versus 0.5%, 1/209; p 0.03). Men had a shorter period of viral RNA shedding than women, with a higher percentage of viral shedding during the early stages (from week 2 to 5 after symptom onset) (Fig. 3b).
Table 3.
Viral RNA shedding time of patients with COVID-19
| Viral RNA shedding time | Age |
p value | Gender |
p value | ||
|---|---|---|---|---|---|---|
| <55 years | ≥55 years | Male | Female | |||
| Week 2 | 29/209 (13.9%) | 30/390 (7.7%) | 0.020 | 32/291 (11.0%) | 27/308 (8.8%) | 0.36 |
| Week 3 | 70/209 (33.5%) | 113/390 (29.0%) | <0.001 | 94/291 (32.3%) | 89/308 (28.9%) | 0.37 |
| Week 4 | 57/209 (27.3%) | 88/390 (22.6%) | 0.200 | 72/291 (24.7%) | 73/308 (23.7%) | 0.77 |
| Week 5 | 29/209 (13.9%) | 59/390 (15.1%) | 0.680 | 46/291 (15.8%) | 42/308 (13.6%) | 0.45 |
| Week 6 | 14/209 (6.7%) | 41/390 (10.5%) | 0.130 | 23/291 (7.9%) | 32/308 (10.4%) | 0.29 |
| Week 7 | 1/209 (0.5%) | 36/390 (9.2%) | 0.003 | 13/291 (4.5%) | 24/308 (7.8%) | 0.1 |
| Week 8 | 7/209 (3.3%) | 19/390 (4.9%) | 0.390 | 9/291 (3.1%) | 17/308 (5.5%) | 0.15 |
| Week 9 | 1/209 (0.5%) | 2/390 (0.5%) | 0.950 | 2/291 (0.7%) | 1/308 (0.3%) | 0.54 |
| ≥Week 10 | 1/209 (0.5%) | 2/390 (0.5%) | 0.950 | 0/291 (0.0%) | 3/308 (1.0%) | 0.21 |
Abbreviations: COVID-19, coronavirus disease 2019.
Fig. 3.
Dynamic characteristics of viral RNA shedding by RT-PCR grouped by (a) age and (b) gender.
Comparison of positive rate for SARS-CoV-2 detection by oropharyngeal and nasopharyngeal swabs
From week 1 to week 6 after symptom onset, nasopharyngeal swabs produced a higher positive rate for SARS-CoV-2 than oropharyngeal swabs (see Supplementary material, Fig. S3a). In weeks 3, 4 and 5 after symptom onset, positive rates from nasopharyngeal swabs were significantly higher than those from oropharyngeal swabs (65.0%, 13/20, versus 30.1%, 195/647, 44.6%, 37/83, versus 22.9%, 155/677, and 36.3%, 33/91, versus 17.9%, 85/474; p < 0.05). Medians of the Ct values of initial positive test, rebound positive test after two consecutive negative results and rebound positive test after three consecutive negative results from nasopharyngeal swabs were higher than those from oropharyngeal swabs (see Supplementary material, Fig, S3b). From week 6 to week 10 and beyond after onset of symptoms, positive rates for viral detection using nasopharyngeal and oropharyngeal tests were not significantly different (see Supplementary material, Table S1).
Discussion
With more than 85 500 000 infections, the world is now facing a pandemic of COVID-19. In this study, we collected data from 599 individuals with COVID-19 and summarized the duration of viral RNA shedding. Meanwhile, we analysed the rate of recurrent positive RT-PCR results and their impact factors.
According to various reports, the median duration of viral RNA shedding ranged from 12 to 31 days from illness onset [[10], [11], [12]]. These reports had a small sample size or a relatively short observation period. In our study, we demonstrated the dynamics of viral RNA shedding after symptom onset in 599 individuals with COVID-19. We found that the median time from symptom onset to the end of viral RNA shedding was 24 days (IQR 19–33 days); 40.4% (242/599) of patients had viral shedding within 3 weeks after symptom onset, and 11.5% (69/599) of patients for over 6 weeks (see Supplementary material, Fig. S2).
Prolonged viral RNA shedding of COVID-19 was reported in various studies [7,13,14]. Although the potential mechanism for prolonged viral shedding was not yet elucidated, studies showed that impaired immune function and a high level of pro-inflammatory cytokines in the serum of individuals with COVID-19 were risk factors for prolonged shedding [13,14]. Our previous study also revealed distinct characteristics between patients with or without prolonged viral RNA shedding. We found that hypertension, older age, lymphopenia and high levels of interleukin-2 receptor were independent risk factors for prolonged viral RNA shedding [15]. Patients with prolonged RNA shedding were generally asymptomatic. However, a recent study reported a case of recurrent symptomatic pneumonia with rebound positive RT-PCR test after discharge from hospital. The authors speculate that low titres of anti-SARS-CoV-2 antibodies may be the reason for COVID-19 relapse [16].
Recurrent detection of SARS-CoV-2 viral RNA by RT-PCR was reported in recent studies [[6], [7], [8],17,18]. The large-scale study including 257 individuals with COVID-19 by Zou et al. revealed that 53 patients (20.6%) suffered with recurrence of positive SARS-CoV-2 RNA detection after two consecutive negative results [6]. Zou et al. also demonstrated that the positive rate of RT-PCR test for SARS-CoV-2 was 5.4% after three negative detections. Another study, by Hao et al. [14] including 104 individuals with COVID-19 found that the positive rates of RT-PCR viral detection were 30.5%, 16.4% and 4.8% after one, two and three consecutive negative RT-PCR test results, respectively. In our study we found that the positive rates of RT-PCR tests were 35.9%, 17.0% and 12.4% after one, two and three consecutive negative RT-PCR tests. The positive rate (17.0%) after two negative tests was close to those of previous reports.
The causes of recurrent positive tests for SARS-CoV-2 were unclear. Currently, false-negative RT-PCR tests were considered as the main reason for recurrent positivity, and they could result from errors in sampling or non-infectious viral RNA remnants. However, the false-negative results did not include the negative results achieved when the viral load was very low or reached the limit of the detection assay. Recurrent positives of COVID-19 have raised increasing public concern regarding the infectivity of individuals with recurrent positive tests. In theory, viral transmission is determined by viral replication. Recurrent-positive patients may be a potential source of transmission. However, accumulating evidence has suggested that patients with COVID-19 who have re-tested positive are barely infectious. In late-phase, non-infectious remnant viral RNA may still be detectable by RT-PCR. According to different reports, the Ct values retrieved from re-positive patients were generally higher than those from initial positive tests. A viral dynamics study by He et al. revealed that the highest viral load was detected at the time of symptom onset. The authors inferred that transmissibility of COVID-19 peaked on or even before symptom onset [19]. The viral load in nasopharyngeal swabs decreased to extremely low limits on day 21 after infection [19]. A recent report by Bullard et al. indicated no viral growth in samples with a Ct value > 24 or symptom onset to test of >8 days [20]. Another study also proved that samples retrieved from positive RT-PCR tests following negative results with Ct values > 29.5 were not associated with virus culture in vitro [21]. A study from England showed that the probability of culturing virus declined to 8% in samples with Ct value > 35 and to 6% 10 days after symptom onset [22]. In a large-scale multicentre investigation, researchers found that none of the 209 close contacts of 69 re-positive patients developed COVID-19 [23]. Ct values are inversely related to viral RNA copy number. According to the literature, Ct values of 35 may correspond to a viral load between 1.0 × 102 and 1.0 × 103 copies/mL [24]. In our study, we also noted a trend of increased Ct values in individuals with re-positive tests after two (32.8) and three (36.1) negative tests. At this point, without standardization of different RT-PCR assays, we do not have a precise threshold value of infectivity. Nonetheless, according to the above reports, we can deduce that patients with Ct values > 35 (viral load <103 copies/ml) were hardly infectious. In our study, the median Ct values of recurrent positive (32.8 and 36.1) suggested that the transmission risk of these patients was low, even if viral shedding could still be detected by RT-PCR in late phase. Until now, no evidence indicating aggressive treatment or public isolation were benefit for these patients. Our results suggested that patients with Ct value > 36 may be considered for discharge.
Furthermore, we investigate the impact of age and gender on false-negative rates of RT-PCR to SARS-CoV-2. We found that older individuals and women tended to have a higher false-negative rate of RT-PCR tests. Notably, female patients had a significantly higher rate of recurrent positive RT-PCR after three consecutive negative results than male patients (18.2%, 18/99, versus 5.8%, 5/86; p 0.013). In clinical practice, the above findings may provide information for repeat viral testing in selected patients. Studies also showed that older age, co-morbidity, low level of antibody response and host immunity status may be potential risk factors for recurrent positivity of RT-PCR tests for SARS-CoV-2, while comprehensive intervention may be a protective factor [25,26].
Various studies investigate the dynamics of viral shedding in individuals with COVID-19. Patients with severe disease had a longer duration of viral shedding [9,10]. Our study showed that 36.7% of patients aged ≥55 years, compared with 47.4% of younger patients (age <55 years), had a median viral shedding time of <3 weeks. Male patients had a shorter viral RNA shedding period than females (43.3%, 126/291 versus 37.7%, 116/308 in 3 weeks) (Table 3). In our study, samples from nasopharyngeal swabs produced a higher positive rate than samples from oropharyngeal swabs. The mean RT-PCR Ct value was higher from oropharyngeal specimens than from nasopharyngeal specimens (see Supplementary material, Fig. S3). Hence, a false-negative test result of RT-PCR may be related to the sampling site. We suggest that nasopharyngeal swabs should be the standard procedure to obtain respiratory specimens for viral tests.
This large-scale study revealed the false-negative rate and dynamic profile of RT-PCR tests for SARS-CoV-2. We also investigated the different viral shedding dynamics of gender and age. The present study has several limitations. First, some data, such as laboratory characteristics, serological data and treatments, were incomplete and not included for analysis. Second, we only included symptomatic hospitalized patients with moderate to severe illness. The results may not be applicable for asymptomatic infected individuals or critical cases. Third, patients included in our study had not received identical treatments, which might have an impact on the duration of viral shedding.
Conclusions
In summary, in this study we for the first time performed a large-scale investigation of the dynamic characteristics of viral RNA shedding in 599 hospitalized individuals with COVID-19. We showed that the median length of viral RNA shedding was 24 days from symptom onset. The positive rate of RT-PCR tests to SARS-CoV-2 was 17% after two consecutive negative results. Recurrent positive patients with a relatively high Ct value were unlikely to be infectious.
Transparency declaration
All authors declare that there are no conflicts of interest.
Acknowledgement
We thank Cheng Chen for correcting the English in this manuscript.
Editor: Laurent Kaiser
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.01.028.
Author contributions
All authors participated in the study design. CG and SZ contributed to conceptualization and writing the original draft. LZ and ATX contributed to formal analysis and project administration. CCJ contributed to data curation, methodology and software. YXT contributed to methodology and software. All authors have agreed on the final version and meet the major criteria recommended by the ICMJE (http://www.icmje.org/).
Availability of data and materials
The database used and/or analysed during the current study is not publicly available (to maintain privacy) but can be available from the corresponding author on reasonable request.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Comparison of viral RNA detection rate between nasopharyngeal or oropharyngeal swabs samples in patients with COVID-19.
Flow chart of the retrospective observational study.
Dynamic profile of viral RNA shedding in all included patients.
Comparison of positive rate (a) and rebound Ct value (b) for SARS-CoV-2 detection by nasopharyngeal or oropharyngeal swabs.
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zunyou W., Jennifer M.G. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 3.WHO . 2020. Coronavirus disease (COVID-2019) situation reports.https://covid19.who.int/ Available from: [Google Scholar]
- 4.China National Health Commission . 2020. Diagnosis and treatment of 2019-nCoV pneumonia in China.https://www.chinadaily.com.cn/pdf/2020/1.Clinical.Protocols.for.the.Diagnosis.and.Treatment.of.COVID-19.V7.pdf (Version 7) Available from: [Google Scholar]
- 5.Centers for Disease Control and Prevention . 2020. Coronavirus disease 2019 (COVID-19). Discontinuation of transmission-based precautions and disposition of patients with COVID-19 in healthcare settings (interim guidance)https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html Updated. Available from: [Google Scholar]
- 6.Zou Y., Wang B.R., Sun L., Xu S., Kong Y.G., Shen L.J. The issue of recurrently positive patients who recovered from COVID-19 according to the current discharge criteria: investigation of patients from multiple medical institutions in Wuhan, China. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa301. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao A.T., Tong Y.X., Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020 doi: 10.1002/jmv.25855. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan J., Kou S., Liang Y., Zeng J., Pan Y., Liu L. PCR assays turned positive in 25 discharged COVID-19 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa398. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao A.T., Tong Y.X., Gao C., Zhu L., Zhang Y.J., Zhang S. Dynamic profile of RT-PCR findings from 301 COVID-19 patients in Wuhan, China: a descriptive study. J Clin Virol. 2020;127:104346. doi: 10.1016/j.jcv.2020.104346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou F., Yu T., Du R.H., Fan G.H., Liu Y., Liu Z.B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou B., She J., Wang Y., Ma X. The duration of viral shedding of discharged patients with severe COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa451. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian G.Q., Chen X.Q., Lv D.F., Ma H.Y., Wang L.P., Yang N.B. Duration of SARS-CoV-2 viral shedding during COVID-19 infection. Infect Dis (Lond) 2020;52:511–512. doi: 10.1080/23744235.2020.1748705. [DOI] [PubMed] [Google Scholar]
- 13.Xu K.J., Chen Y.F., Yuan J., Yi P., Ding C., Wu W.R. Factors associated with prolonged viral RNA shedding in patients with COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa351. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao S.R., Lian J.S., Lu Y.F., Jia H.Y., Hu J.H., Yu G.D. Decreased B cells on admission was associated with prolonged viral RNA shedding from respiratory tract in coronavirus disease 2019: a case control study. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa311. jiaa311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao C., Zhu L., Jin C.C., Tong Y.X., Xiao A.T., Zhang S. Proinflammatory cytokines are associated with prolonged viral RNA shedding in COVID-19 patients. Clin Immunol. 2020;221:108611. doi: 10.1016/j.clim.2020.108611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X., Zhou J., Zhao J. Recurrent pneumonia in a patient with new coronavirus infection after discharge from hospital for insufficient antibody production: a case report. BMC Infect Dis. 2020;20:500. doi: 10.1186/s12879-020-05231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan L., Xu D., Ye G.M., Xia C., Wang S.K., Li Y.R. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.2783. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J.F., Yan K., Ye H.H., Lin J., Zheng J.J., Cai Ting. SARS-CoV-2 turned positive in a discharged patient with COVID-19 arouses concern regarding the present standard for discharge. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.007. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X., Lau E., Wu P., Deng X.L., Wang J., Hao X.X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 20.Bullard J., Dust K., Funk D., Strong James, Alexander David, Garnett Lauren. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020:ciaa638. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gniazdowski V., Morris C.P., Wohl S., Mehoke T., Ramakrishnan S., Thielen P. Repeat COVID-19 molecular testing correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1616. ciaa1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singanayagam A., Patel M., Charlett A., Bernal J., Saliba V., Ellis J. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S.L., Xu H., Feng H.Y., Sun J.F., Li X., Zhou L. Epidemiological and clinical findings of short-term recurrence of severe acute respiratory syndrome coronavirus 2 ribonucleic acid polymerase chain reaction positivity in 1282 discharged coronavirus disease 2019 cases: a multicenter, retrospective, observational study. Open Forum Infect Dis. 2020;7:ofaa432. doi: 10.1093/ofid/ofaa432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lirong Z., Feng R., Mingxing H., Lijun L., Huitao H., Zhongsi H. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye G., Pan Z., Pan Y., Deng Q., Chen L., Li J. Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. J Infect. 2020;80:e14–e17. doi: 10.1016/j.jinf.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He S., Tian J., Li X., Zhou Y., Xiao M., Zhang Y. Positive RT-PCR test results in 420 patients recovered from COVID-19 in Wuhan: an observational study. Front Pharmacol. 2020;11:549117. doi: 10.3389/fphar.2020.549117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of viral RNA detection rate between nasopharyngeal or oropharyngeal swabs samples in patients with COVID-19.
Flow chart of the retrospective observational study.
Dynamic profile of viral RNA shedding in all included patients.
Comparison of positive rate (a) and rebound Ct value (b) for SARS-CoV-2 detection by nasopharyngeal or oropharyngeal swabs.
Data Availability Statement
The database used and/or analysed during the current study is not publicly available (to maintain privacy) but can be available from the corresponding author on reasonable request.