Abstract
Diabetic kidney disease (DKD) is the most common diabetic complication and is a leading cause of end-stage kidney disease. Increasing evidence shows that DKD is regulated not only by many classical signaling pathways but also by epigenetic mechanisms involving chromatin histone modifications, DNA methylation, and non-coding RNA (ncRNAs). In this review, we focus on our current understanding of the role and mechanisms of ncRNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) in the pathogenesis of DKD. Of them, the regulatory role of TGF-β/Smad3-dependent miRNAs and lncRNAs in DKD is highlighted. Importantly, miRNAs and lncRNAs as biomarkers and therapeutic targets for DKD are also described, and the perspective of ncRNAs as a novel therapeutic approach for combating diabetic nephropathy is also discussed.
Keywords: diabetic kidney disease, micro RNAs, long non-coding RNAs, TGF-β, fibrosis, inflammation, biomarker, therapeutic target
Introduction
Diabetic kidney disease (DKD) is one of the most predominant diabetic complications and is a leading cause of chronic kidney disease (CKD). It is reported that up to 20–50% of living diabetes, including type 1 (T1DM) and type 2 (T2DM) diabetes, would eventually develop into DKD (Selby and Taal, 2020), which contributes to the high mortality of patients with DKD (Braunwald, 2019). The established DKD is characterized by the onset of persistent albuminuria and progressive decline of estimated glomerular filtration rate (eGFR) (Magee et al., 2017). Pathologically, the histological features of DKD include the thickening of the glomerular basement membrane (GBM), glomerular capillary hypertension, mesangial expansion, nodular sclerosis, glomerulosclerosis, interstitial fibrosis, inflammation, and tubular atrophy (Raval et al., 2020).
In patients with diabetes, hyperglycemia may trigger oxidative stress, renal inflammation, and fibrosis in kidneys (Matoba et al., 2019; Patel et al., 2020). Among those pathogenic factors, renal fibrogenesis is the major driving force in the development of DKD (Hills and Squires, 2011; Lan, 2012a). It is well-established that transforming growth factor β (TGF-β) as the master regulator for the fibrotic and inflammatory process in CKD (Meng et al., 2016). Hyperglycemic factors such as advanced glycation end products (AGEs) and angiotensin II (AngII) may trigger the activation of TGF-β signaling via Smad dependent or independent pathway, therefore promoting fibrosis in kidneys (Lan, 2011; Meng et al., 2016; Gu et al., 2020) (Figure 1).
FIGURE 1.
The crosstalk of canonical and noncanonical TGF-β signaling pathways associated with renal fibrosis and inflammation in diabetic nephropathy. TGF-β/Smad and NF-κB signaling pathway are highly activated under hyperglycemic conditions. AGEs, Ang II, IL-1β and TNF-α etc, may trigger these two pathways to promote fibrosis and inflammation in diabetic kidneys. Abbreviations: AGEs, advanced glycation end products; RAGE, receptor for AGE; Ang II, angiotensin II; AT1/2, Ang II receptor 1 and 2; BMP, bone morphogenic protein; TNF-α, tumor necrosis factor α; TNFR, TNF receptor; IL-1β, interleukin 1β; IL-1R, IL-1 receptor; ERK, extracellular-signal regulated kinase; IκBα, nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor α. (Figure created with BioRender.com).
The emerging field of epigenetic regulation by ncRNAs has focused on the pathogenic pathways to halt the progression of DKD. With no function in protein-coding, ncRNAs were implicated as therapeutic targets or biomarkers for DKD (Loganathan et al., 2020). Interestingly, these ncRNAs could also be regulated by TGF-β (Meng et al., 2015). In this review, we will focus on the regulatory role of miRNAs and lncRNAs in the progression of DKD, and their potentials as therapeutic targets and biomarkers for DKD are highlighted. Moreover, the mechanisms of ncRNAs on renal fibrosis and inflammation in DKD based on the TGF-β/Smad-mediated signaling pathway will also be discussed.
The Emerging Role of Non-coding RNAs in DKD
miRNAs are single-stranded endogenous RNAs (20–22 nucleotides in length) that regulate gene expression on the post-transcriptional or transcriptional level (Wahid et al., 2010). LncRNAs are RNA transcripts over 200 nucleotides in length, which are able to modulate gene expression by binding to either DNAs, RNAs, or proteins (Yao et al., 2019). The roles of miRNAs and lncRNAs in kidney development and disease have been reviewed (Kaucsár et al., 2010; Moghaddas Sani et al., 2018; Zhou et al., 2019). Thus, we mainly focus on the roles and underlying mechanisms of miRNAs and lncRNAs relevant to DKD pathogenesis (as shown in Figure 2).
FIGURE 2.
Potential role of miRNAs and lncRNAs in the pathogenesis of diabetic kidney disease. Under hyperglycemic conditions, the expression of TGF-β, growth factors such as CTGF, FGF, and cytokines may induce ECM accumulation, EMT, ER stress, oxidative stress, insulin resistance, glucose toxicity, fibrosis, and inflammatory response. These pathogenic processes are positively or negatively regulated by ncRNAs (miRNAs and lncRNAs) to promote cell apoptosis, autophagy, hypertrophy, fibrosis, inflammation in the diabetic kidney. Abbreviations: GFs, growth factors; ECM, extracellular matrix; EMT, epithelial-mesenchymal transition; ER, endoplasmic reticulum. (Figure created with BioRender.com).
Non-Smad-dependent miRNAs in DKD
The functional relevance of miRNA in renal diseases has caught our attention since the rapid development of RNA sequencing strategy. In most cases, miRNAs hybridize to the 3’UTRs (untranslated regions) of the target mRNAs and hence silencing the expression of target genes. Up to date, the function and underlying mechanisms of many miRNAs in renal diseases have been well-demonstrated and reviewed (Hou and Zhao, 2013). These miRNAs are of great importance to the epigenetic regulation on DKD.
Renal tubulointerstitial fibrosis (TIF) is one of the predominant features of DKD. A group of miRNAs have been shown to be profibrotic in DKD ( Table 1). The expression of miR-22 was increased in streptozotocin (STZ)-induced DKD model and in high glucose (HG)-treated tubular epithelial cells (TECs). miR-22 targets phosphatase and tensin homolog (PTEN), therefore suppressing autophagy and inducing the expression of collagen IV and α-smooth muscle actin (α-SMA) (Zhang et al., 2018). A high level of miR-23a was also observed in diabetic patients and HG-cultured TECs. It directly targets the nuclear transcription co-repressor Ski-related novel protein N (SnoN) (Tan et al., 2006), a crucial negative regulator to TGF-β/Smad3-mediated signaling pathway, to induce fibrosis in DKD (Xu et al., 2018a). Sirtuin 1 (SIRT1) expression in the nucleus and the cytoplasm has also been shown as a renoprotective regulator by inhibiting TGF-β/Smad-induced fibrosis and downstream hypoxia-inducible factor-1α (HIF-1α). miR-34a-5p, miR-217, miR-133b, and miR-199b may dcirectly or indirectly target and suppress the expression of SIRT1 under hyperglycemic conditions (Shao et al., 2016; Sun et al., 2018b; Xue et al., 2018). The transient receptor potential cation channel subfamily C member 1 (TRPC1) is downregulated in diabetic patients and animal models, which may contribute to the development of DKD (Zhang et al., 2009a). miR-135a targets TRPC1 to promote the fibrotic process in diabetic renal injury (He et al., 2014). Interestingly, diabetic-induced albumin triggers the expression of miR-184 in the tubular cells to promote TIF, which is associated with decreased expression of lipid phosphate phosphatase 3 (LPP3) (Zanchi et al., 2017). The canopy 1 (CNPY1) is a target of miR-370 to modulate fibroblast growth element signaling (Matsui et al., 2011). Overexpression of miR-370 significantly increases the accumulation of extracellular matrix (ECM) and promotes the proliferation of mesangial cells (MCs) (Yu et al., 2019). On the other hand, the anti-fibrotic miR-342 binds to the 3’UTR of SRY-box 6 (SOX6), therefore inhibiting SOX6 expression and the level of fibrotic biomarkers (Jiang et al., 2020b). miR-379 is also involved in the pathogenesis of DKD. It is reported that miR-379 triggers miR-let-7, which prevents ECM accumulation and proliferation of MCs (Li et al., 2019b). Nevertheless, some miRNAs exert protective effects by inhibiting the epithelial-to-mesenchymal transition (EMT). Notably, miR-30c, miR-98-5p and miR-302a-3p target the fibrosis-related JAK1, Snail1, HMGA2, and ZEB1, respectively, thus blocking the fibrotic process in DKD by inhibiting EMT (Zhao et al., 2017; Tang et al., 2018b; Zhu et al., 2019c; Gao et al., 2020). Furthermore, miR-455-3p also inhibits renal fibrosis by targeting ROCK2, together with the reduction of anti-inflammatory cytokines such as tumor necrosis factor-α (TNF-α) and monocyte chemotactic protein 1 (MCP-1) (Wu et al., 2018a). Interestingly, miR-455-3p also serves as a sponge for pathogenic lncRNA Hottip. Hottip is upregulated under HG conditions, while miR-455-3p may reverse Hottip-mediated fibrosis and inflammation (Zhu et al., 2019b). Fatty acid accumulation (FAC) was also induced by DKD, fatty acid synthase (FASN) is not only the vital lipogenic enzyme to FAC, but also an upregulated molecule that contributes to glomerulosclerosis and renal inflammation. miR-544 binds to the 3’UTR of FASN thus attenuating the infiltration of inflammatory cells, the activation of NF-κB signaling and renal fibrosis (Sun et al., 2020). All these findings have suggested a crucial role of miRNAs in DKD-induced renal fibrosis based on the epigenetic regulation level.
TABLE 1.
Non-Smad-dependent miRNAs in DKD.
| miRNA | Target | Pathological output(s) | References |
|---|---|---|---|
| miR-22 | PTEN | Pro-fibrosis | (Zhang et al., 2018) |
| miR-23a | SnoN | (Xu et al., 2018a) | |
| miR-34a-5p | SIRT1 | (Xue et al., 2018) | |
| miR-133b | SIRT1 | (Sun et al., 2018b) | |
| miR-199b | |||
| miR-135a | TRPC1 | (He et al., 2014) | |
| miR-184 | LPP3 | (Zanchi et al., 2017) | |
| miR-370 | CNPY1 | (Yu et al., 2019) | |
| miR-30c | JAK1; Snail1 | Anti-fibrosis | (Zhao et al., 2017; Gao et al., 2020) |
| miR-98-5p | HMGA2 | (Zhu et al., 2019c) | |
| miR-302a-3p | ZEB1 | (Tang et al., 2018b) | |
| miR-342 | SOX6 | (Jiang et al., 2020b) | |
| miR-379-5p | LIN28B | (Li et al., 2019b) | |
| miR-455-3p | ROCK2 | Anti-fibrosis | (Wu et al., 2018a; Zhu et al., 2019b) |
| miR-544 | FASN | Anti-inflammation | (Sun et al., 2020) |
| miR-217 | SIRT1/HIF-1α | Pro-fibrosis | (Shao et al., 2016) |
| Pro-inflammation | |||
| miR-770-5p | TIMP3 | Pro-inflammation | (Zhang et al., 2019c; Wang and Li, 2020) |
| miR-15b-5p | Sema3A | Anti-inflammation | (Fu et al., 2019) |
| miR-34b | IL-6R | (Lv et al., 2019) | |
| miR-140-5p | TLR4 | (Su et al., 2020) | |
| miR-146a | NOX4 | (Wan and Li, 2018) | |
| miR-218 | IKK-β | (Li et al., 2020a) | |
| miR-374a | MCP-1 | (Yang et al., 2018) | |
| miR-423-5p | NOX4 | (Xu et al., 2018c) | |
| miR-451 | LMP7 | (Sun et al., 2016b) | |
| miR-485 | NOX5 | (Wu et al., 2020) | |
| miR-874 | TLR4 | (Yao et al., 2018) |
Hyperglycemia triggers the inflammatory response by recruiting immune infiltration and inducing the production of pro-inflammatory cytokines. Of note, podocyte is the barrier to maintain glomerular filtration, and it also functions as the receptor and producer of various cytokines. The dysfunction of podocyte is an essential event in lesion development and glomerulonephritis. This process promotes the progression of DKD (Lal and Patrakka, 2018). Stimulated by HG, miR-770-5p is upregulated and promotes podocyte injury by targeting metalloproteinase 3 (TIMP3), and Tp53 regulated inhibitor of apoptosis 1 (TRIAP1), knocking down of miR-770-5p reverse the apoptosis and inflammation induced by HG in kidney biopsy and mouse podocytes (Zhang et al., 2019c; Wang and Li, 2020). On the other hand, more anti-inflammation related miRNAs have been identified. Overexpression of miR-15b-5p significantly restrained HG-induced apoptosis, oxidative stress and inflammation in podocytes, it also directly targets Sema3A, suggesting that miR-15b-5p could be a therapeutic target for DKD (Fu et al., 2019). miR-34b targets to the interleukin-6 (IL-6) receptor and downstream JAK2/STAT3 signaling, thus reducing the expression of TNF-α, IL-6, interleukin-1β (IL-1β), and caspase-3 in TECs (Lv et al., 2019). The nicotinamide adenine dinucleotide phosphate (NAPDH) oxidase (NOX)-derived reactive oxygen species (ROS) may induce inflammation, implying that NOX enzymes as novel targets for DKD (Lambeth et al., 2008). Of note, miR-146a (Wan and Li, 2018), miR-423-5p (Xu et al., 2018c), and miR-485 (Wu et al., 2020) target NOX4 and NOX5, respectively, to reduce the production of pro-inflammatory cytokines. NF-κB signaling pathway is the classical player in inflammation, which is activated in a wide range of kidney diseases, including DKD. miR-218 targets the IKK-β to regulate NF-κB signaling, as well as reducing the expression of TNF-α, IL-6, IL-1β, and MCP-1 (Li et al., 2020a). miR-451 also targets large multifunctional protease (LMP7) to modulate NF-κB-mediated renal inflammation, which is confirmed by the downregulating level of pro-inflammatory molecules (Sun et al., 2016a). In addition, miR-140-5p and miR-874 also function as anti-inflammatory modulators in suppressing the expression of TNF-α, IL-6, IL-1β in TECs by directly binding to toll like receptor 4 (TLR4), the upstream molecule of NF-κB signaling (Yao et al., 2018; Su et al., 2020). These reports suggest that miRNA-mediated renal fibrosis and inflammation have critical functions in DKD.
Non-Smad-dependent lncRNAs in DKD
As promising candidates, some miRNA drugs have been approved to proceed toward phase III or IV trials in the coming future. However, the toxicity and off-target effects of miRNA are somehow inevitable (Seok et al., 2018; Hanna et al., 2019). The emerging studies on lncRNAs have shed light on their characteristics of tissue-and-cell-type-specificity and regulation on both transcriptional and translational levels, making lncRNA as the promising therapeutic targets and attractive drugs for DKD treatment ( Table 2) (Kato, 2018; Guo et al., 2019).
TABLE 2.
Non-Smad-dependent lncRNAs and their mechanisms in DKD.
| lncRNA | Target | Pathological output(s) | References |
|---|---|---|---|
| ZEB1-AS1 | miR-216a-5p; MLL1; p53 | Anti-fibrosis | (Wang et al., 2018a; Meng et al., 2020) |
| NR_038323 | miR-324-3p; DUSP1 | (Ge et al., 2019b) | |
| 1700020I14Rik | miR-34a-5p | (Li et al., 2018a) | |
| CYP4B1-PS1-001 | Nucleolin | (Wang et al., 2016b; Wang et al., 2018c) | |
| ENSMUST00000147869 | Cyp4a12a | (Wang et al., 2016c) | |
| XIST | miR-93-5p; CDKN1A | Pro-fibrosis | (Yang et al., 2019a) |
| PVT1 | miR-23b-3p; WT1 | (Zhong et al., 2020) | |
| SNHG16 | miR-141-3p; CCND1 | (Jiang et al., 2020a) | |
| OIP5-AS1 | miR-30c-5p | (Fu et al., 2020) | |
| LINC00968 | p21/EZH2 | (Li et al., 2018b) | |
| ASncmtRNA-2 | ROS | (Gao et al., 2017a) | |
| MEG3 | miR-181a; Egr-1; TLR4; miR-145 | Pro-fibrosis | (Li et al., 2019a; Zha et al., 2019) |
| BLNC1 | NRF2/HO-1; NF-κB | (Feng et al., 2019) | |
| NEAT1 | Klotho/ERK1/2; miR-23c; Akt/mTOR; miR-27b-3p/ZEB1 | (Ma et al., 2019a; Huang et al., 2019b; Wang et al., 2019b; Li et al., 2020b; Yang et al., 2020) | |
| MALAT1 | Wnt/β-catenin; miR-145/ZEB2; SRSF1; IL-6; TNF-α | (Puthanveetil et al., 2015; Hu et al., 2017; Liu et al., 2019a; Zhang et al., 2019a) | |
| Hottip | miR-455-3p; Wnt2B | (Zhu et al., 2019b) | |
| Gm4419 | NF-κB/NLRP3; p50 | (Yi et al., 2017) | |
| GAS5 | MMP9; miR-221; SIRT1 | Anti-fibrosis | (Ge et al., 2019a; Zhang et al., 2020) |
| Anti-inflammation | |||
| Rpph1 | Gal-3/Mek/Erk | Pro-inflammation | (Zhang et al., 2019b) |
| HOXA-AS2 | miR-302b-3p; TIMP3 | Anti-inflammation | (Li and Yu, 2020) |
LncRNA zinc finger E-box binding homeobox 1 antisense 1 (ZEB1-AS1) plays a protective role in DKD by targeting profibrotic miR-216a-5p to inhibit HK-induced EMT and renal fibrosis. Besides, the anti-fibrotic function of ZEB1-AS1 is also verified that it may bind to H3K4 methyltransferase myeloid and lymphoid or mixed-lineage leukemia 1 (MLL1) and p53 in patients with DKD (Wang et al., 2018a; Meng et al., 2020). lncRNA NR_038323 exerts an anti-fibrotic effect by interacting with miR-324-3p. miR-324-3p is verified to induce dual-specificity protein phosphatase-1 (DUSP1) and the activation of p38/MAPK and ERK1/2 signaling (Ge et al., 2019b). Moreover, the expression of lncRNA 1700020I14Rik is decreased in db/db mice. Bioinformatic method and RNA binding protein immunoprecipitation assay have confirmed the interaction of 1700020I14Rik and miR-34a-5p, which may then modulate the SIRT1/HIF-1α signaling to prohibit renal fibrosis (Li et al., 2018a). Nucleolin is a nuclear protein that expresses on the surface of endothelial cells. CYP4B1-PS1-001 is the lncRNA that upregulated in early DKD. By direct interaction with Nucleolin, CYP4B1-PS1-001 inhibits fibrosis in MCs (Wang et al., 2016b; Wang et al., 2018c). Nevertheless, some lncRNAs interact with miRNAs to trigger and promote the fibrotic process. (Yang et al., 2019a; Jiang et al., 2020a; Fu et al., 2020; Zhong et al., 2020). Interestingly, LINC00968 inhibits p21 by recruiting EZH2 to enhance proliferation and fibrosis of MCs (Li et al., 2018b). ASncmtRNA-2 is upregulated by ROS, and it promotes the expression of TGF-β1 and other fibrotic factors (Gao et al., 2017b).
As shown in Table 2, by direct interaction with miRNAs or inflammatory molecules, lncRNAs play as sponges, inhibitors, or activators to influence either fibrogenesis or inflammatory response. All these findings have demonstrated a critical role of lncRNAs therapeutic targets in the pathogenesis of DKD.
TGF-β/Smad-dependent Non-coding RNAs in DKD
TGF-β signaling is highly activated under diabetic conditions and has been shown to be a major pathway leading to DKD. It has been well established that DKD-associated fibrosis and inflammation are mediated by TGF-β via Smad-dependent or -independent signaling pathways (Chung and Lan, 2015; Tang et al., 2018a). Active TGF-β1 binds and activates TGF-β receptor II (TβRII) and receptor I (TβRI) which induces phosphorylation of Smad2/3 to form a complex with Smad4 that translocate into the nucleus to regulate transcription of target genes. In general, Smad3 is pathogenic, while Smad2 and Smad7 are protective. Smad4 plays diverse roles in renal fibrosis and inflammation, suggesting Smad4 may not serve as the ideal therapeutic target for DKD (Chung et al., 2013; Li et al., 2014). Many ncRNAs are induced by TGF-β to regulate renal fibrosis and inflammation via Smad-dependent mechanisms in DKD as highlighted in Table 3.
TABLE 3.
TGF-β/Smad3-dependent miRNAs and lncRNAs in renal fibrotic and inflammatory response of DKD.
TGF-β/Smad-dependent miRNAs in Renal Fibrosis and Inflammation in DKD
miR-192 is the first landmark found in DKD (Kato et al., 2007). TGF-β upregulated miR-192 in MCs and glomeruli from db/db mice, STZ-induced mice model as well as in DKD patients (Kato et al., 2007; Krupa et al., 2010; Putta et al., 2012; Ma et al., 2016; Liu et al., 2018). Indeed, these studies have shown the high correlation between miR-192 and diabetic kidneys. Mechanistically, miR-192 may promote the expression of collagens by targeting the E-box repressor Smad-1 interacting protein (SIP1 or Zeb2) (Kato et al., 2007; Putta et al., 2012). Also, activation of Akt may lead to MCs proliferation and hypertrophy in DKD. miR-192 upregulates miR-216a and miR-217, inhibiting PTEN to induce Akt activation under diabetic conditions. Nevertheless, miR-192 also plays a complex and diverse role in DKD depending on different models or time points. One study has observed a correlation between miR-192 level, tubulointerstitial fibrosis, and eGFR. TGF-β treatment decreases the expression of miR-192 in TECs, resulting in the promotion of fibrosis and the decline of eGFR (Krupa et al., 2010). Similarly, by targeting Egr1, miR-192 decreases the expression of TGF-β1 and fibronectin in glucose-treated TECs and Otsuka-Long-Evans-Tokushima-Fatty rats, a diabetic murine model (Liu et al., 2018). These studies have reported the complexity of miRNA in mediating the fibrotic process in DKD.
miR-200 family (miR-200a, miR-200b, miR-200c) is well-studied miRNA clusters that maintain the epithelial differentiation in cells. Induced by TGF-β or hyperglycemia, the expression of miR-200a are downregulated in TECs. miR-200a functions as a suppressor to EMT, thus protecting kidney from diabetic insults by inhibiting the TGF-β-mediated fibrotic process. Mechanistic study has further revealed that miR-200a downregulates TGF-β2 expression by directly targeting the 3’UTR of TGF-β2 (Wang et al., 2011). However, the expression of miR-200b/c are elevated in glomeruli from type 1 diabetes (T1DM) and type 2 diabetes (T2DM) mice model and in MCs treated with TGF-β1 (Kato et al., 2011b), implying that difference on the miR-200 expression may due to cell type specificity and individual variability. miR-200 family may serve as the therapeutic targets specific to certain cell types response to DKD process.
miR-21 is another well-studied miRNA in renal disease. Although the expression of miR-21 is downregulated in early DKD (Zhang et al., 2009b), it is upregulated in TECs and MCs stimulated by TGF-β1 or HG and in the renal biopsies of DKD patients (Zhong et al., 2011; Wang et al., 2013; Zhong et al., 2013; Wang et al., 2014; Lai et al., 2015; Mcclelland et al., 2015; Kölling et al., 2017; Chen et al., 2018). The mechanism of miR-21 participates in DKD may be related to its activation on both canonical and noncanonical TGF-β signaling. miR-21 not only suppresses the inhibitory Smad7 of TGF-β signaling to promote fibrosis (Zhong et al., 2013; Wang et al., 2014) but also targeting the Sprouty (SPRY) to activate the Ras/MEK/ERK signaling to activate fibrogenesis of TGF-β signaling (Xu et al., 2014). In addition, miR-21 also exerts profibrotic and pro-inflammatory effects by targeting PTEN, tissue inhibitor of matrix metalloproteinases (TIMPs), and other molecules, as shown in Table 3.
miR-29 family is demonstrated to be protective miRNAs that are highly expressed in kidneys but significantly reduced under diabetic conditions. The expression of miR-29 family in various renal cells is decreased when they are stimulated with TGF-β1 or treated with HG (Qin et al., 2011; Chen et al., 2014). The protective role of miR-29 family has been supported by the evidence that overexpression of miR-29 may inhibit the transcription of collagen genes while suppression of miR-29 promotes ECM accumulation. Many studies have identified fibrosis-related targets of miR-29 under hyperglycemic conditions, demonstrating the anti-fibrotic role of miR-29 in DKD. Insterestingly, studies also revealed that miR-29c, serves as a signature miRNA that promotes the progression of DN and fibrosis (Long et al., 2011; Shao et al., 2019; Huang et al., 2020). More and more studies are revealing the functions and mechanisms of miRNAs in fibrosis and inflammation during diabetic conditions, these miRNAs may play as potential therapeutic targets to combat DKD.
TGF-β/Smad-Dependent lncRNAs in Renal Fibrosis and Inflammation in DKD
Under hyperglycemic condition, the expression of profibrotic and pro-inflammatory lncRNAs are usually upregulated, suggesting their regulatory role in DKD. TGF-β-mediated lncRNA Erbb4-IR is highly expressed in diabetic db/db mice and AGEs-treated MCs. It is regulated by Smad3 as Smad3 deficiency inhibits the transcription of Erbb4 (Feng et al., 2018; Xu et al., 2020). The upregulation of Erbb4-IR is consistent with the elevation of albuminuria, serum creatinine, and fibrotic biomarkers. The mechanistic role of Erbb4-IR may be the binding of Erbb4-IR with the 3’UTR of miR-29b, therefore suppressing anti-fibrotic miR-29b expression. Moreover, Erbb4-IR may also bind with Smad7 to promote renal fibrosis (Sun et al., 2018a; Feng et al., 2018).
lncRNA NR_033515 is found to be significantly increased in the serum of DKD patients, which has shown a positive correlation with KIM-1 and NGAL, diagnostic markers of DKD. The mechanistic study has further confirmed the fibrotic role of NR_033515 by revealing the binding of NR_033515 and miR-743b-5p, resulting in the proliferation, EMT, and fibrosis increasing level of proliferation-related proliferating cell nuclear antigen (PCNA), Cyclin D1, and the fibrotic proteins during DKD (Gao et al., 2018).
Arid2-IR is regulated by Smad3, knockdown of Arid2-IR in TECs has no effect on TGF-β/Smad-mediated fibrosis but promotes IL-1β-induced NF-κB-driven renal inflammation in obstructive kidney disease (Zhou et al., 2015). However, a recent study has reported the profibrotic effect of Arid2-IR by interacting with early growth response protein-1 (Egr1) in high-fat-diet and STZ-induced mice. Arid2-IR induces the expression of collagens and α-SMA in mouse MCs, contributing to the ECM accumulation in DKD (Yang et al., 2019c).
Interestingly, lncRNA NONHSAG053901 also targets Egr1 in mouse MCs, but their interaction has promoted inflammation by upregulating pro-inflammatory cytokines (Peng et al., 2019). The pathogenic role of Smad3-regulated LRNA9884 is observed in db/db mice with more severe albuminuria, histological injuries, and a decline of eGFR. LRNA9884 is induced by AGEs, and it targets MCP-1 to promote MCP-1-driven renal inflammation (Zhang et al., 2019d).
lncRNAs taurine upregulated gene 1 (TUG1) is an anti-fibrotic lncRNA mediated by TGF-β with multiple functions in DKD. In response to metabolic alterations of DKD, the expression of TUG1 is downregulated in podocytes. Overexpression of TUG1 can reverse the mitochondrial dysfunction in podocytes by targeting the transcription factor peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α) (Li and Susztak, 2016; Shen et al., 2019). In consistence with previous results, TUG1 can also modulate mitochondrial bioenergetics in podocytes by binding with PGC-1α (Long et al., 2016). These findings have highlighted the connection between lncRNAs and DKD. By interacting with TNF receptor-associated factor 5 (TRAF5), TUG1 can suppress TRAF5-mediated podocyte apoptosis (Lei et al., 2018) and negatively downregulate the PI3K/Akt signaling to inhibit proliferation and ECM deposit in MCs (Zang et al., 2019). TUG1 is also able to interact with miR-21, thus promoting the expression of TIMP3 to alleviate renal fibrosis in HG-stimulated TECs and in db/db mice (Wang et al., 2019a). Furthermore, TUG1 sponges for miR-377 to regulate PPARγ and ECM in MCs (Duan et al., 2017). All these protective effects of lncRNA TUG1 in various cell types has supported its therapeutic potential in treating DKD. Besides, some lncRNAs may play diverse roles in the pathogenesis of DKD. lncRNA psoriasis-susceptibility related RNA gene induced by stress (PRINS) may exert both anti-fibrotic, anti-inflammatory but pro-apoptotic effects by regulating Smad7 in DKD. It has been demonstrated that there is a positive correlation between PRINS and Smad7 in DKD patients. As overexpression of Smad7 inhibits renal fibrosis and inflammation but also induces apoptosis in podocytes (Schiffer et al., 2001; Ka et al., 2012), thus, overexpression of PRINS upregulates Smad7 expression and promotes apoptosis in mouse podocytes (Jiao et al., 2019). lncRNA PRINS may be a therapeutic target of DKD-induced renal fibrosis and inflammation. But the underlying mechanisms of interaction between PRINS and Smad7 remain unexplored. In conclusion, the connection of TGF-β-mediated lncRNA and DKD is well-defined. Further studies on revealing the therapeutic targets and underlying mechanisms of these lncRNAs remain to be further explored.
Non-coding RNAs as Novel Biomarkers for DKD
The diagnosis and monitoring of renal injuries in DKD are now dependent on the detection of urinary albumin or serum creatinine. However, some patients may not present microalbuminuria or creatinine alterations during the progression of DKD, suggesting that none of these measures can accurately indicate the severity and type of injury induced by hyperglycemia (Magee et al., 2017; Lin et al., 2018). In addition, urinary albumin is not specific to DKD, which may also occur in other diseases. Besides, the diagnostic and prognostic test of renal biopsy is invasive and may not be a reliable way to establish the full patterns of DKD. Thus, the availability of sensitive and specific biomarkers will provide therapeutic benefits in the control of DKD.
Non-coding RNAs in body fluids could facilitate communication between cells. Non-coding RNAs may exist in a stable form in serum and urine. As biomarkers, they may form a complex with proteins or be stored in transporters, including exosomes, microparticles, and apoptotic bodies. Based on the tissue- and cell type-specific characteristics of lncRNAs, significant differences in expression of novel lncRNAs in DKD (as shown in Tables 2 and 3) have mapped the signaling pathways in the pathogenesis of diabetic nephropathy (Guo et al., 2019). Indeed, a recent study has reported a novel lncRNA, PANDAR, related to T2DM DKD patients. The expression of PANDAR is upregulated in diabetic patients and higher in DKD patients with massive proteinuria, demonstrating its potential as biomarker and predictor for prognosis and progression of DKD (Zhao et al., 2020). The expression of lncRNA CASC2 is downregulated in T2DM patients with chronic renal failure but not T2DM patients with other complications, suggesting that lncRNA cancer susceptibility candidate 2 (CASC2) could also serve as a renal specific biomarker for DKD. Moreover, the study has further followed up for 5 years and found out that serum level of lncRNA CASC2 is negatively correlated with the incidence of chronic renal failure, supporting that serum level of lncRNA CASC2 may be a specific and reliable biomarker for diagnosis in DKD progression (Wang et al., 2018b). These studies have shown that lncRNAs are of high relevance in the development and progression of DKD, however, further mechanistic investigations on lncRNAs as therapeutic targets are warranted.
Some circulating miRNAs may also serve as sensitive and useful biomarkers for early detection and diagnosis for DKD (Zhang et al., 2016; Nascimento and Domingueti, 2019). For instance, in the early stage of T2DM DKD, the expression of miR-377 is positive, while miR-192 is negatively correlated with renal function (Tayel et al., 2020). In addition, circulating miRNA of miR-1246, miR-642a-3p, let-7c-5p, miR-1255b-5p, let-7i-3p, miR-5010-5p, and miR-150-3p are significantly upregulated in DKD patients compared with healthy volunteers (Kim et al., 2019). Moreover, the expression of miR-126 is decreased in DKD patients, which is negatively associated with albuminuria, level of fasting glucose, and glycated hemoglobin but positively correlated with eGFR (Al-Kafaji et al., 2016). The level of serum miR-21 is also consistent with tissue miR-21 that closely reflects renal function in DKD (Wang et al., 2016a). Up to date, many studies have reported the expression profiles of circulating miRNAs in diabetic nephropathy, making miRNAs as one of the promising candidates for DKD diagnosis and therapeutic targets.
The urinary exosomal miRNAs are called as “liquid biopsy” (La Marca and Fierabracci, 2017), which are typically secreted by cells from renal segments. They carry proteins, RNAs, and biomarkers that may reflect renal injury and dysfunction (Xu et al., 2018b). For example, miR-200b is a novel urinary biomarker that negatively correlates with the degree of renal fibrosis in CKD and DKD (Yu et al., 2018). One study has suggested that the expression of miR-27b-3p and miR-1228-3p in urine may be useful indicators for the degrees of renal fibrosis of DKD patients (Conserva et al., 2019). Notably, the expression of miR-126 in urine is increased in DKD patients compared to diabetic patients without renal disease. Interestingly, the urinary level of miR-126 is significantly decreased in DKD patients with a better diabetic control, implying that miR-126 may be a biomarker in DKD and monitor for DKD treatment response (Liu et al., 2014).
Although the clinical relevance in urinary miRNAs have been well studied (Lv et al., 2013; Cheng et al., 2014), there is still no consensus on the normalization of miRNAs isolated from urine, as the levels of urinary miRNAs may be high veriable and affected by urinary contents and concentrations. Neverthless, the better normalizer strategies should be encouraged (Blondal et al., 2013; Lekchnov et al., 2016; Corral-Vazquez et al., 2017), as the normalization of the validated data may help to provide statistically significant results without causing unwanted bias.
Non-coding RNAs as Promising Therapeutic Targets for DKD
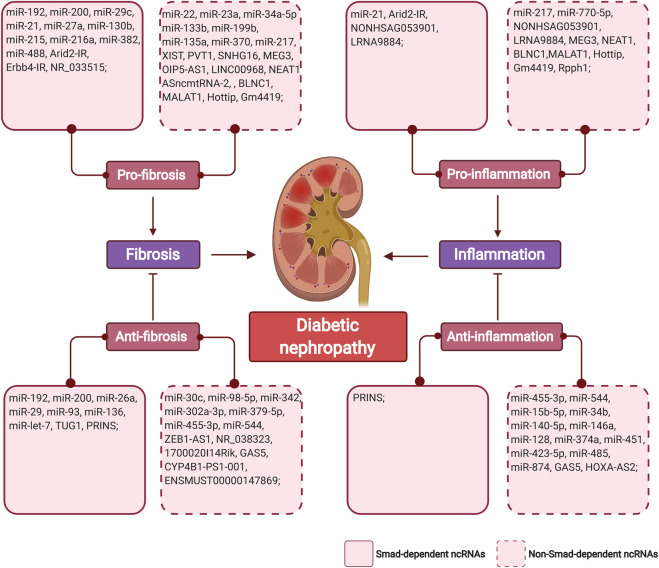
The regulatory role of non-coding RNAs in the pathogenesis of DKD has highlighted their potential as therapeutic targets for DKD. Restoring expression or inhibition of non-coding RNAs in renal or inflammatory cells under diabetic conditions may halt renal fibrosis and inflammation (Figure 3). Besides, rebalancing the overactivated TGF-β signaling induced by hyperglycemia could be another strategy that controls renal complication.
FIGURE 3.
The summary of TGF-β/Smad-dependent and non-TGF-β/Smad-dependent miRNAs and lncRNAs in diabetic renal fibrosis and inflammation. Non-coding RNAs are classified as pro/anti-fibrosis, pro/anti-inflammation in regard with their mechanistic funcions in diabetic nephropathy. (Figure created with BioRender.com).
The delivery of synthetic non-coding RNA oligonucleotides, plasmids, or inhibitors may alter pathogenic signaling pathways related to DKD. Antagonism of miR-21 not only reduces the loss of podocytes and albuminuria but also inhibits renal fibrotic response by inhibition of collagen and fibronectin in vivo and in vitro (Wang et al., 2013; Kölling et al., 2017; Roy et al., 2020). Silencing miR-215 with specific antagomir increases the expression of CTNNBIP1, reduces of β-catenin activity, and accumulation of fibrotic proteins in db/db mice (Mu et al., 2013b). We have established the non-invasive ultrasound microbubble-mediated gene transfer to knock down renal expression of miR-21, thus suppressing the activation of the TGF-β and NF-κB signaling pathways by targeting Smad7 in the diabetic mouse model (Zhong et al., 2013). In addition, restoring the expression of miR-29b by delivery of doxycycline-inducible pre-miR-29b into the kidney, could significantly reverse the pathological changes of progressive DKD (Chen et al., 2014). Moreover, kidney-specific silencing of lncRNA Erbb4-IR and LRNA9884 with ultrasound technique can convert plasmids into the damaged kidney to ameliorate injuries, albuminuria, fibrosis, and inflammation (Sun et al., 2018a; Zhang et al., 2019d). Notably, exosomes secreted by cells contain non-coding RNAs that may have a regulatory role in DKD. Injection of exosomes from HG-treated macrophages induces MCs proliferation, fibrotic, and inflammatory factors activation in vivo as well as in vitro. Intriguingly, exosomes from TGF-β1 knockdown macrophages may reverse pathogenic changes in MCs (Zhu et al., 2019a), underscoring the importance of TGF-β signaling in the pathogenesis of DKD.
The rapid development of the field of non-coding RNAs has helped these RNA-based biopharmaceuticals to enter clinical trials before market approval. However, non-coding RNA treatments remain to be explored. The low expression, low conservation between species, time specificity, toxicity, and off-target effect of non-coding RNA are obstacles waiting to be solved in the development of RNA therapy (Yang et al., 2014; Ard et al., 2017). Up to date, the number of non-coding RNAs related to clinical trials on DKD is limited (Sankrityayan et al., 2019). Nevertheless, some ongoing miRNA-based therapies may be the potential next-generation medicine for DKD (Chakraborty et al., 2017). For example, Remlarsen, a miR-29 mimic that is undergoing in the clinical test (https://clinicaltrials.gov/ct2/show/NCT03601052) and could be the promising drug to combat renal fibrosis in DKD. Hopefully, new technologies such as clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) gene editing may represent novel strategies to modulate the expression and function of non-coding RNAs in DKD (Miano et al., 2019). Further studies are needed to reveal the therapeutic potential of ncRNAs in the clinical treatment of DKD.
Conclusion and Future Perspectives
Non-coding RNAs have garnered the major attention of researchers in the past few decades. We are now shifting toward their regulatory role and mutual relationship in the pathogenesis of DKD. Reports in this review and available literature have drawn the patterns of ncRNAs profiles in the process of diabetic nephropathy, but further investigation into the crucial mechanisms of ncRNAs in epigenetic regulation is warranted. Moreover, as biomarkers, the expression of renal ncRNAs may reflect the cellular response to hyperglycemic injuries, thus contributing to the early diagnosis and prognosis of DKD. The discovery of miRNAs and lncRNAs also represents a new field of molecular therapy into DKD treatment. Together these findings are expected to yield novel insights into the complex pathogenesis of DKD and could be incorporated in the clinical settings.
Author Contributions
Y-YG wrote and revised the manuscript. F-HL, X-RH, WM, and LZ revised the manuscript. X-SL, X-QY, and H-YL revised and edited the manuscript. All authors contributed to the manuscript conception development, data collection and analysis, and discussion on the manuscript writing and revising.
Funding
This work was supported by the Guangdong-Hong Kong-Macao-Joint Labs Program from Guangdong Science and Technology (2019B121205005); the Research Grants Council of Hong Kong (Grants GRF 14163317, 14117418, 14104019, R4012-18, and C7018-16G); Lui Che Woo Institute of Innovative Medicine (CARE); the Health and Medical Research Fund of Hong Kong (Grants HMRF 05161326, 06173986, and 14152321); the National Natural Science Foundation of China (No.81873261 and No. 81903956), the Project of Guangdong Province Administration of Traditional Chinese Medicine (No. 20201133). Project from the State Key Laboratory of Dampness Syndrome of Chinese Medicine (SZ2020ZZ22).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Al-Kafaji G., Al-Mahroos G., Al-Muhtaresh H. A., Skrypnyk C., Sabry M. A., Ramadan A. R. (2016). Decreased expression of circulating microRNA-126 in patients with type 2 diabetic nephropathy: a potential blood-based biomarker. Exp. Ther. Med. 12, 815–822. 10.3892/etm.2016.3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ard R., Allshire R. C., Marquardt S. (2017). Emerging properties and functional consequences of noncoding transcription. Genetics 207, 357–367. 10.1534/genetics.117.300095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondal T., Jensby Nielsen S., Baker A., Andreasen D., Mouritzen P., Wrang Teilum M., et al. (2013). Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59, S1–S6. 10.1016/j.ymeth.2012.09.015 [DOI] [PubMed] [Google Scholar]
- Braunwald E. (2019). Diabetes, heart failure, and renal dysfunction: the vicious circles. Prog. Cardiovasc. Dis 62, 298–302. 10.1016/j.pcad.2019.07.003 [DOI] [PubMed] [Google Scholar]
- Cai Z. G., Wu H. B., Xu X. P., Li W. (2018). Down-regulation of miR-26 plays essential roles in TGFβ-induced EMT. Cell Biol. Int. 15, 33–39. 10.1002/cbin.11029 [DOI] [Google Scholar]
- Castro N. E., Kato M., Park J. T., Natarajan R. (2014). Transforming growth factor β1 (TGF-β1) enhances expression of profibrotic genes through a novel signaling cascade and microRNAs in renal mesangial cells. J. Biol. Chem. 289, 29001–29013. 10.1074/jbc.M114.600783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A. R., Sharma G., Doss C. G. P., Lee S. S. (2017). Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids 8, 132–143. 10.1016/j.omtn.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Y., Zhong X., Huang X. R., Meng X. M., You Y., Chung A. C., et al. (2014). MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol. Ther. 22, 842–853. 10.1038/mt.2013.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhao L., Xing Y., Lin B. (2018). Down-regulation of microRNA-21 reduces inflammation and podocyte apoptosis in diabetic nephropathy by relieving the repression of TIMP3 expression. Biomed. Pharmacother. 108, 7–14. 10.1016/j.biopha.2018.09.007 [DOI] [PubMed] [Google Scholar]
- Cheng L., Sun X., Scicluna B. J., Coleman B. M., Hill A. F. (2014). Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 86, 433–444. 10.1038/ki.2013.502 [DOI] [PubMed] [Google Scholar]
- Chung A. C., Huang X. R., Meng X., Lan H. Y. (2010). miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J. Am. Soc. Nephrol. 21, 1317–1325. 10.1681/ASN.2010020134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung A. C., Lan H. Y. (2015). MicroRNAs in renal fibrosis. Front. Physiol. 6, 50 10.3389/fphys.2015.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung A. C., Yu X., Lan H. Y. (2013). MicroRNA and nephropathy: emerging concepts. Int. J. Nephrol. Renovascular Dis. 6, 169–179. 10.2147/IJNRD.S37885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conserva F., Barozzino M., Pesce F., Divella C., Oranger A., Papale M., et al. (2019). Urinary miRNA-27b-3p and miRNA-1228-3p correlate with the progression of kidney fibrosis in diabetic nephropathy. Sci. Rep. 9, 11357 10.1038/s41598-019-47778-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Vazquez C., Blanco J., Salas-Huetos A., Vidal F., Anton E. (2017). Normalization matters: tracking the best strategy for sperm miRNA quantification. Mol. Hum. Reprod. 23, 45–53. 10.1093/molehr/gaw072 [DOI] [PubMed] [Google Scholar]
- Deshpande S. D., Putta S., Wang M., Lai J. Y., Bitzer M., Nelson R. G., et al. (2013). Transforming growth factor-β-induced cross talk between p53 and a microRNA in the pathogenesis of diabetic nephropathy. Diabetes 62, 3151–3162. 10.2337/db13-0305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N. (2019). Long noncoding RNA MALAT1 acts as a competing endogenous RNA to regulate TGF-. BioMed Res. Int. 2019, 1569638 10.1155/2019/1569638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L. J., Ding M., Hou L. J., Cui Y. T., Li C. J., Yu D. M. (2017). Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARγ in diabetic nephropathy. Biochem. Biophys. Res. Commun. 484, 598–604. 10.1016/j.bbrc.2017.01.145 [DOI] [PubMed] [Google Scholar]
- Fang Y., Xie T., Xue N., Kuang Q., Wei Z., Liang M., et al. (2017). miR-382 contributes to renal tubulointerstitial fibrosis by downregulating HSPD1. Oxid Med Cell Longev. 23, 4708516 10.1155/2017/4708516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M., Tang P. M., Huang X. R., Sun S. F., You Y. K., Xiao J., et al. (2018). TGF-beta mediates renal fibrosis via the Smad3-Erbb4-IR long noncoding RNA Axis. Mol. Ther. 26, 148–161. 10.1016/j.ymthe.2017.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Zhao J., Ding J., Shen X., Zhou J., Xu Z. (2019). LncRNA Blnc1 expression and its effect on renal fibrosis in diabetic nephropathy. Am. J. Tourism Res. 11, 5664–5672. 10.5772/36398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J. X., Sun G. Q., Wang H. L., Jiang H. X. (2020). LncRNA OIP5-AS1 induces epithelial-to-mesenchymal transition and renal fibrosis in diabetic nephropathy via binding to miR-30c-5p. J. Biol. Regul. Homeost. Agents 34 10.23812/20-199-A-68 [DOI] [PubMed] [Google Scholar]
- Fu Y., Wang C., Zhang D., Chu X., Zhang Y., Li J. (2019). miR-15b-5p ameliorated high glucose-induced podocyte injury through repressing apoptosis, oxidative stress, and inflammatory responses by targeting Sema3A. J. Cell. Physiol. 234, 20869–20878. 10.1002/jcp.28691 [DOI] [PubMed] [Google Scholar]
- Gao B. H., Wu H., Wang X., Ji L. L., Chen C. (2020). MiR-30c-5p inhibits high glucose-induced EMT and renal fibrogenesis by down-regulation of JAK1 in diabetic nephropathy. Eur. Rev. Med. Pharmacol. Sci. 24, 1338–1349. 10.26355/eurrev_202002_20191 [DOI] [PubMed] [Google Scholar]
- Gao J., Wang W., Wang F., Guo C. (2018). LncRNA-NR_033515 promotes proliferation, fibrogenesis and epithelial-to-mesenchymal transition by targeting miR-743b-5p in diabetic nephropathy. Biomed. Pharmacother. 106, 543–552. 10.1016/j.biopha.2018.06.104 [DOI] [PubMed] [Google Scholar]
- Gao Y., Chen Z.-Y., Wang Y., Liu Y., Ma J.-X., Li Y.-K. (2017a). Long non-coding RNA ASncmtRNA-2 is upregulated in diabetic kidneys and high glucose-treated mesangial cells. Experimental and therapeutic medicine 13, 581–587. 10.3892/etm.2017.4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Chen Z. Y., Wang Y., Liu Y., Ma J. X., Li Y. K. (2017b). Long non-coding RNA ASncmtRNA-2 is upregulated in diabetic kidneys and high glucose-treated mesangial cells. Exp. Ther. Med. 13, 581–587. 10.3892/etm.2017.4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Zhang R., Dai S., Zhang X., Li X., Bai C. (2019). Role of TGF-β/smad pathway in the transcription of pancreas-specific genes during beta cell differentiation. Front Cell Dev Biol. 7, 351 10.3389/fcell.2019.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Xu B., Xu W., Xia L., Xu Z., Shen L., et al. (2019a). Long noncoding RNA GAS5 inhibits cell proliferation and fibrosis in diabetic nephropathy by sponging miR-221 and modulating SIRT1 expression. Aging (Albany NY). 11, 8745–8759. 10.18632/aging.102249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Wang J., Wu D., Zhou Y., Qiu S., Chen J., et al. (2019b). lncRNA NR_038323 suppresses renal fibrosis in diabetic nephropathy by targeting the miR-324-3p/DUSP1 Axis. Mol. Ther. Nucleic Acids. 17, 741–753. 10.1016/j.omtn.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y.-Y., Liu X.-S., Huang X.-R., Yu X.-Q., Lan H.-Y. (2020). Diverse role of TGF-β in kidney disease. Frontiers in cell and developmental biology 8, 123 10.3389/fcell.2020.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Liu Z., Gong R. (2019). Long noncoding RNA: an emerging player in diabetes and diabetic kidney disease. Clin Sci (Lond). 133, 1321–1339. 10.1042/CS20190372 [DOI] [PubMed] [Google Scholar]
- Hanna J., Hossain G. S., Kocerha J. (2019). The potential for microRNA therapeutics and clinical research. Front. Genet. 10 10.3389/fgene.2019.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Peng F., Xia X., Zhao C., Luo Q., Guan W., et al. (2014). MiR-135a promotes renal fibrosis in diabetic nephropathy by regulating TRPC1. Diabetologia. 57, 1726–1736. 10.1007/s00125-014-3282-0 [DOI] [PubMed] [Google Scholar]
- Hills C. E., Squires P. E. (2011). The role of TGF-β and epithelial-to mesenchymal transition in diabetic nephropathy. Cytokine Growth Factor Rev. 22, 131–139. 10.1016/j.cytogfr.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Hou J., Zhao D. (2013). MicroRNA regulation in renal pathophysiology. Int. J. Mol. Sci. 14, 13078–13092. 10.3390/ijms140713078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Tian J., Geng J., Li X., Tang X., Zhang J., et al. (2016). MicroRNA-27a promotes renal tubulointerstitial fibrosis via suppressing PPARγ pathway in diabetic nephropathy. Oncotarget 7, 47760–47776. 10.18632/oncotarget.10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Wang R., Li X., Fan M., Lin J., Zhen J., et al. (2017). LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J. Cell Mol. Med. 21, 2732–2747. 10.1111/jcmm.13189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Zheng Y., Chen Y., Cheng Y., Jiang Y., Cai M., et al. (2019a). miR-216a-5p promotes mesangial cell proliferation by targeting FoxO1 in diabetic nephropathy. Int. J. Clin. Exp. Pathol. 12, 344–355. [PMC free article] [PubMed] [Google Scholar]
- Huang H., Huang X., Luo S., Zhang H., Hu F., Chen R., et al. (2020). The MicroRNA MiR-29c alleviates renal fibrosis via TPM1-mediated suppression of the Wnt/β-catenin pathway. Front. Physiol. 11, 331 10.3389/fphys.2020.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Xu Y., Ge X., Xu B., Peng W., Jiang X., et al. (2019b). Long noncoding RNA NEAT1 accelerates the proliferation and fibrosis in diabetic nephropathy through activating Akt/mTOR signaling pathway. J. Cell. Physiol. 234, 11200–11207. 10.1002/jcp.27770 [DOI] [PubMed] [Google Scholar]
- Jiang X., Ru Q., Li P., Ge X., Shao K., Xi L., et al. (2020a). LncRNA SNHG16 induces proliferation and fibrogenesis via modulating miR-141-3p and CCND1 in diabetic nephropathy. Gene Therapy 27, 557–566. 10.1038/s41434-020-0160-x [DOI] [PubMed] [Google Scholar]
- Jiang Z. H., Tang Y. Z., Song H. N., Yang M., Li B., Ni C. L. (2020b). miRNA-342 suppresses renal interstitial fibrosis in diabetic nephropathy by targeting SOX6. Int. J. Mol. Med. 45, 45–52. 10.3892/ijmm.2019.4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H., Xie D., Qiao Y. (2019). LncRNA PRINS is involved in the development of nephropathy in patients with diabetes via interaction with Smad7. Exp. Ther. Med. 17, 3203–3208. 10.3892/etm.2019.7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka S. M., Yeh Y. C., Huang X. R., Chao T. K., Hung Y. J., Yu C. P., et al. (2012). Kidney-targeting Smad7 gene transfer inhibits renal TGF-β/MAD homologue (SMAD) and nuclear factor κB (NF-κB) signalling pathways, and improves diabetic nephropathy in mice. Diabetologia 55, 509–519. 10.1007/s00125-011-2364-5 [DOI] [PubMed] [Google Scholar]
- Kato M., Arce L., Wang M., Putta S., Lanting L., Natarajan R. (2011a). A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney International. 80, 358–368. 10.1038/ki.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Arce L., Wang M., Putta S., Lanting L., Natarajan R. (2011b). A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int. 80, 358–368. 10.1038/ki.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M. (2018). Noncoding RNAs as therapeutic targets in early stage diabetic kidney disease. Kidney Res Clin Pract. 37, 197–209. 10.23876/j.krcp.2018.37.3.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Zhang J., Wang M., Lanting L., Yuan H., Rossi J. J., et al. (2007). MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. U. S. A. 104, 3432–3437. 10.1073/pnas.0611192104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaucsár T., Rácz Z., Hamar P. (2010). Post-transcriptional gene-expression regulation by micro RNA (miRNA) network in renal disease. Adv. Drug Deliv. Rev. 62, 1390–1401. 10.1016/j.addr.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Kim H., Bae Y. U., Jeon J. S., Noh H., Park H. K., Byun D. W., et al. (2019). The circulating exosomal microRNAs related to albuminuria in patients with diabetic nephropathy. J. Transl. Med. 17, 236 10.1186/s12967-019-1983-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga K., Yokoi H., Mori K., Kasahara M., Kuwabara T., Imamaki H., et al. (2015). MicroRNA-26a inhibits TGF-β-induced extracellular matrix protein expression in podocytes by targeting CTGF and is downregulated in diabetic nephropathy. Diabetologia. 58, 2169–2180. 10.1007/s00125-015-3642-4 [DOI] [PubMed] [Google Scholar]
- Kölling M., Kaucsar T., Schauerte C., Hübner A., Dettling A., Park J. K., et al. (2017). Therapeutic miR-21 silencing ameliorates diabetic kidney disease in mice. Mol. Ther. 25, 165–180. 10.1016/j.ymthe.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupa A., Jenkins R., Luo D. D., Lewis A., Phillips A., Fraser D. (2010). Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J. Am. Soc. Nephrol. 21, 438–447. 10.1681/ASN.2009050530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Marca V., Fierabracci A. (2017). Insights into the diagnostic potential of extracellular vesicles and their miRNA signature from liquid biopsy as early biomarkers of diabetic micro/macrovascular complications. Int. J. Mol. Sci. 18 10.3390/ijms18091974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J. Y., Luo J., O'connor C., Jing X., Nair V., Ju W., et al. (2015). MicroRNA-21 in glomerular injury. J. Am. Soc. Nephrol. 26, 805–816. 10.1681/ASN.2013121274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal M. A., Patrakka J. (2018). Understanding podocyte biology to develop novel kidney therapeutics. Front. Endocrinol. 9, 409 10.3389/fendo.2018.00409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth J. D., Krause K.-H., Clark R. A. (2008). NOX enzymes as novel targets for drug development. Semin. Immunopathol. 30, 339–363. 10.1007/s00281-008-0123-6 [DOI] [PubMed] [Google Scholar]
- Lan H. Y. (2011). Diverse roles of TGF-beta/Smads in renal fibrosis and inflammation. Int. J. Biol. Sci. 7, 1056–1067. 10.7150/ijbs.7.1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H. Y. (2012a). Transforming growth factor-beta/Smad signalling in diabetic nephropathy. Clin. Exp. Pharmacol. Physiol. 39, 731–738. 10.1111/j.1440-1681.2011.05663.x [DOI] [PubMed] [Google Scholar]
- Lan H. Y. (2012b). Transforming growth factor-β/Smad signalling in diabetic nephropathy. Clin. Exp. Pharmacol. Physiol. 39, 731–738. 10.1111/j.1440-1681.2011.05663.x [DOI] [PubMed] [Google Scholar]
- Lei X., Zhang L., Li Z., Ren J. (2018). Astragaloside IV/lncRNA-TUG1/TRAF5 signaling pathway participates in podocyte apoptosis of diabetic nephropathy rats. Drug Des. Dev. Ther. 12, 2785–2793. 10.2147/DDDT.S166525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekchnov E. A., Zaporozhchenko I. A., Morozkin E. S., Bryzgunova O. E., Vlassov V. V., Laktionov P. P. (2016). Protocol for miRNA isolation from biofluids. Anal. Biochem. 499, 78–84. 10.1016/j.ab.2016.01.025 [DOI] [PubMed] [Google Scholar]
- Li A., Peng R., Sun Y., Liu H., Peng H., Zhang Z. (2018a). LincRNA 1700020I14Rik alleviates cell proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1α signaling. Cell Death & Disease. 9, 461 10.1038/s41419-018-0527-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Jiang X., Duan L., Wang W. (2019a). Long non-coding RNA MEG3 impacts diabetic nephropathy progression through sponging miR-145. Am J Transl Res. 11, 6691–6698. [PMC free article] [PubMed] [Google Scholar]
- Li M., Guo Q., Cai H., Wang H., Ma Z., Zhang X. (2020a). miR-218 regulates diabetic nephropathy via targeting IKK-β and modulating NK-κB-mediated inflammation. J. Cell. Physiol. 235, 3362–3371. 10.1002/jcp.29224 [DOI] [PubMed] [Google Scholar]
- Li N., Jia T., Li Y. R. (2020b). LncRNA NEAT1 accelerates the occurrence and development of diabetic nephropathy by sponging miR-23c. Eur. Rev. Med. Pharmacol. Sci. 24, 1325–1337. 10.26355/eurrev_202002_20190 [DOI] [PubMed] [Google Scholar]
- Li N., Wang L. J., Xu W. L., Liu S., Yu J. Y. (2019b). MicroRNA-379-5p suppresses renal fibrosis by regulating the LIN28/let-7 axis in diabetic nephropathy. Int. J. Mol. Med. 44, 1619–1628. 10.3892/ijmm.2019.4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Chung A. C., Yu X., Lan H. Y. (2014). MicroRNAs in diabetic kidney disease. Internet J. Endocrinol. 2014, 593956 10.1155/2014/593956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. Y., Susztak K. (2016). The long noncoding RNA Tug1 connects metabolic changes with kidney disease in podocytes. J. Clin. Invest. 126, 4072–4075. 10.1172/JCI90828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yu H. M. (2020). Overexpression of HOXA-AS2 inhibits inflammation and apoptosis in podocytes via sponging miRNA-302b-3p to upregulate TIMP3. Eur. Rev. Med. Pharmacol. Sci. 24, 4963–4970. 10.26355/eurrev_202005_21187 [DOI] [PubMed] [Google Scholar]
- Li Z., Yu Z., Meng X., Yu P. (2018b). LncRNA LINC00968 accelerates the proliferation and fibrosis of diabetic nephropathy by epigenetically repressing p21 via recruiting EZH2. Biochem. Biophys. Res. Commun. 504, 499–504. 10.1016/j.bbrc.2018.08.048 [DOI] [PubMed] [Google Scholar]
- Lin Y.-C., Chang Y.-H., Yang S.-Y., Wu K.-D., Chu T.-S. (2018). Update of pathophysiology and management of diabetic kidney disease. Journal of the Formosan Medical Association = Taiwan yi zhi. 117, 662–675. 10.1016/j.jfma.2018.02.007 [DOI] [PubMed] [Google Scholar]
- Liu B., Qiang L., Wang G. D., Duan Q., Liu J. (2019a). LncRNA MALAT1 facilities high glucose induced endothelial to mesenchymal transition and fibrosis via targeting miR-145/ZEB2 axis. Eur. Rev. Med. Pharmacol. Sci. 23, 3478–3486. 10.26355/eurrev_201904_17713 [DOI] [PubMed] [Google Scholar]
- Liu F., Zhang Z. P., Xin G. D., Guo L. H., Jiang Q., Wang Z. X. (2018). miR-192 prevents renal tubulointerstitial fibrosis in diabetic nephropathy by targeting Egr1. Eur. Rev. Med. Pharmacol. Sci. 22, 4252–4260. 10.26355/eurrev_201807_15420 [DOI] [PubMed] [Google Scholar]
- Liu L., Pang X., Shang W., Feng G., Wang Z., Wang J. (2020). miR-136 improves renal fibrosis in diabetic rats by targeting down-regulation of tyrosine kinase SYK and inhibition of TGF-β1/Smad3 signaling pathway. Ren. Fail. 42, 513–522. 10.1080/0886022X.2020.1764854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gao G., Yang C., Zhou K., Shen B., Liang H., et al. (2014). Stability of miR-126 in urine and its potential as a biomarker for renal endothelial injury with diabetic nephropathy. Internet J. Endocrinol. 17, 393109 10.1155/2014/393109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yang Y., Wang Q., Kahaer A., Zhang J., Liao J., et al. (2019b). Regulatory effect of 1,25(OH)2D3 on TGF-β1 and miR-130b expression in streptozotocin-induced diabetic nephropathy in rats. Internet J. Endocrinol. 123, 1346 10.1155/2019/1231346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loganathan T. S., Sulaiman S. A., Abdul Murad N. A., Shah S. A., Abdul Gafor A. H., Jamal R., et al. (2020). Interactions among non-coding RNAs in diabetic nephropathy. Front. Pharmacol. 11, 191 10.3389/fphar.2020.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Badal S. S., Ye Z., Wang Y., Ayanga B. A., Galvan D. L., et al. (2016). Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Invest. 126, 4205–4218. 10.1172/JCI87927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Wang Y., Wang W., Chang B. H., Danesh F. R. (2011). MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J. Biol. Chem. 286, 11837–11848. 10.1074/jbc.M110.194969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv C., Zhou Y. H., Wu C., Shao Y., Lu C. L., Wang Q. Y. (2015). The changes in miR-130b levels in human serum and the correlation with the severity of diabetic nephropathy. Diabetes Metab Res Rev. 31, 717–724. 10.1002/dmrr.2659 [DOI] [PubMed] [Google Scholar]
- Lv L. L., Cao Y., Liu D., Xu M., Liu H., Tang R. N., et al. (2013). Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int. J. Biol. Sci. 9, 1021–1031. 10.7150/ijbs.6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv N., Li C., Liu X., Qi C., Wang Z. (2019). miR-34b alleviates high glucose-induced inflammation and apoptosis in human HK-2 cells via IL-6R/JAK2/STAT3 signaling pathway. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 25, 8142–8151. 10.12659/MSM.917128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Zhang L., Hao J., Li N., Tang J., Hao L. (2018). Up-regulation of microRNA-93 inhibits TGF-β1-induced EMT and renal fibrogenesis by down-regulation of Orai1. J. Pharmacol. Sci. 136, 218–227. 10.1016/j.jphs.2017.12.010 [DOI] [PubMed] [Google Scholar]
- Ma J., Zhao N., Du L., Wang Y. (2019a). Downregulation of lncRNA NEAT1 inhibits mouse mesangial cell proliferation, fibrosis, and inflammation but promotes apoptosis in diabetic nephropathy. Int. J. Clin. Exp. Pathol. 12, 1174–1183. [PMC free article] [PubMed] [Google Scholar]
- Ma X., Lu C., Lv C., Wu C., Wang Q. (2016). The expression of miR-192 and its significance in diabetic nephropathy patients with different urine albumin creatinine ratio. J Diabetes Res. 2016, 6789402 10.1155/2016/6789402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Shi J., Wang F., Li S., Wang J., Zhu C., et al. (2019b). MiR-130b increases fibrosis of HMC cells by regulating the TGF-β1 pathway in diabetic nephropathy. J. Cell. Biochem. 120, 4044–4056. 10.1002/jcb.27688 [DOI] [PubMed] [Google Scholar]
- Magee C., Grieve D. J., Watson C. J., Brazil D. P. (2017). Diabetic nephropathy: a tangled Web to unweave. Cardiovasc. Drugs Ther. 31, 579–592. 10.1007/s10557-017-6755-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba K., Takeda Y., Nagai Y., Kawanami D., Utsunomiya K., Nishimura R. (2019). Unraveling the role of inflammation in the pathogenesis of diabetic kidney disease. Int. J. Mol. Sci. 20 10.3390/ijms20143393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Thitamadee S., Murata T., Kakinuma H., Nabetani T., Hirabayashi Y., et al. (2011). Canopy1, a positive feedback regulator of FGF signaling, controls progenitor cell clustering during Kupffer's vesicle organogenesis. Proc. Natl. Acad. Sci. U. S. A. 108, 9881–9886. 10.1073/pnas.1017248108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcclelland A. D., Herman-Edelstein M., Komers R., Jha J. C., Winbanks C. E., Hagiwara S., et al. (2015). miR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin. Sci. (Lond.). 129, 1237–1249. 10.1042/CS20150427 [DOI] [PubMed] [Google Scholar]
- Meng Q., Zhai X., Yuan Y., Ji Q., Zhang P. (2020). lncRNA ZEB1-AS1 inhibits high glucose-induced EMT and fibrogenesis by regulating the miR-216a-5p/BMP7 axis in diabetic nephropathy. Braz. J. Med. Biol. Res. 53, e9288 10.1590/1414-431X20209288 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Meng X. M., Nikolic-Paterson D. J., Lan H. Y. (2016). TGF-beta: the master regulator of fibrosis. Nat. Rev. Nephrol. 12, 325–338. 10.1038/nrneph.2016.48 [DOI] [PubMed] [Google Scholar]
- Meng X. M., Tang P. M., Li J., Lan H. Y. (2015). TGF-beta/Smad signaling in renal fibrosis. Front. Physiol. 6, 82 10.3389/fphys.2015.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano J. M., Long X., Lyu Q. (2019). CRISPR links to long noncoding RNA function in mice: a practical approach. Vasc. Pharmacol. 114, 1–12. 10.1016/j.vph.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddas Sani H., Hejazian M., Hosseinian Khatibi S. M., Ardalan M., Zununi Vahed S. (2018). Long non-coding RNAs: an essential emerging field in kidney pathogenesis. Biomed. Pharmacother. 99, 755–765. 10.1016/j.biopha.2018.01.122 [DOI] [PubMed] [Google Scholar]
- Mu J., Pang Q., Guo Y.-H., Chen J.-G., Zeng W., Huang Y.-J., et al. (2013a). Functional implications of microRNA-215 in TGF-β1-induced phenotypic transition of mesangial cells by targeting CTNNBIP1. PloS One. 8, e58622 10.1371/journal.pone.0058622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Pang Q., Guo Y. H., Chen J. G., Zeng W., Huang Y. J., et al. (2013b). Functional implications of microRNA-215 in TGF-β1-induced phenotypic transition of mesangial cells by targeting CTNNBIP1. PLoS One. 8, e58622 10.1371/journal.pone.0058622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento L. R. D., Domingueti C. P. (2019). MicroRNAs: new biomarkers and promising therapeutic targets for diabetic kidney disease. Jornal brasileiro de nefrologia : 'orgao oficial de Sociedades Brasileira e Latino-Americana de Nefrologia. 41, 412–422. 10.1590/2175-8239-jbn-2018-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. M., Bose M., Cooper M. E. (2020). Glucose and blood pressure-dependent pathways-the progression of diabetic kidney disease. Int. J. Mol. Sci. 21 10.3390/ijms21062218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W., Huang S., Shen L., Tang Y., Li H., Shi Y. (2019). Long noncoding RNA NONHSAG053901 promotes diabetic nephropathy via stimulating Egr-1/TGF-β-mediated renal inflammation. J. Cell. Physiol. 234, 18492–18503. 10.1002/jcp.28485 [DOI] [PubMed] [Google Scholar]
- Puthanveetil P., Chen S., Feng B., Gautam A., Chakrabarti S. (2015). Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell Mol. Med. 19, 1418–1425. 10.1111/jcmm.12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putta S., Lanting L., Sun G., Lawson G., Kato M., Natarajan R. (2012). Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J. Am. Soc. Nephrol. 23, 458–469. 10.1681/ASN.2011050485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W., Chung A. C., Huang X. R., Meng X. M., Hui D. S., Yu C. M., et al. (2011). TGF-beta/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J. Am. Soc. Nephrol. 22, 1462–1474. 10.1681/ASN.2010121308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval N., Kumawat A., Kalyane D., Kalia K., Tekade R. K. (2020). Understanding molecular upsets in diabetic nephropathy to identify novel targets and treatment opportunities. Drug Discovery Today. 25, 862–878. 10.1016/j.drudis.2020.01.008 [DOI] [PubMed] [Google Scholar]
- Roy D., Modi A., Khokar M., Sankanagoudar S., Yadav D., Sharma S., et al. (2020). Microrna 21 emerging role in diabetic complications: a critical update. Curr. Diabetes Rev. 16, 21–29. 10.2174/1573399816666200503035035 [DOI] [PubMed] [Google Scholar]
- Sankrityayan H., Kulkarni Y. A., Gaikwad A. B. (2019). Diabetic nephropathy: the regulatory interplay between epigenetics and microRNAs. Pharmacol. Res. 141, 574–585. 10.1016/j.phrs.2019.01.043 [DOI] [PubMed] [Google Scholar]
- Schiffer M., Bitzer M., Roberts I. S., Kopp J. B., Ten Dijke P., Mundel P., et al. (2001). Apoptosis in podocytes induced by TGF-beta and Smad7. J. Clin. Invest. 108, 807–816. 10.1172/JCI12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby N. M., Taal M. W. (2020). An updated overview of diabetic nephropathy: diagnosis, prognosis, treatment goals and latest guidelines. Diabetes Obes. Metabol. 22 (Suppl. 1), 3–15. 10.1111/dom.14007 [DOI] [PubMed] [Google Scholar]
- Seok H., Lee H., Jang E.-S., Chi S. W. (2018). Evaluation and control of miRNA-like off-target repression for RNA interference. Cell. Mol. Life Sci. : CM. 75, 797–814. 10.1007/s00018-017-2656-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Huang Y., Hu H. L., Fan W. X., Yin X. N. (2019). Effect of miR-29c on renal fibrosis in diabetic rats via the AMPK/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 6250–6256. 10.26355/eurrev_201907_18445 [DOI] [PubMed] [Google Scholar]
- Shao Y., Lv C., Wu C., Zhou Y., Wang Q. (2016). Mir-217 promotes inflammation and fibrosis in high glucose cultured rat glomerular mesangial cells via Sirt1/HIF-1α signaling pathway. Diabetes Metab Res Rev. 32, 534–543. 10.1002/dmrr.2788 [DOI] [PubMed] [Google Scholar]
- Shen H., Ming Y., Xu C., Xu Y., Zhao S., Zhang Q. (2019). Deregulation of long noncoding RNA (TUG1) contributes to excessive podocytes apoptosis by activating endoplasmic reticulum stress in the development of diabetic nephropathy. J. Cell. Physiol. 234, 15123–15133. 10.1002/jcp.28153 [DOI] [PubMed] [Google Scholar]
- Shi M., Tian P., Liu Z., Zhang F., Zhang Y., Qu L., et al. (2020). MicroRNA-27a targets Sfrp1 to induce renal fibrosis in diabetic nephropathy by activating Wnt/β-Catenin signalling. Biosci. Rep. 40 10.1042/BSR20192794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. P., Goodwin J. E., Kanasaki K., Koya D. (2020). Inhibition of angiotensin-converting enzyme ameliorates renal fibrosis by mitigating DPP-4 level and restoring antifibrotic MicroRNAs. Genes. 11 10.3390/genes11020211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. P., Hedayat A. F., Kanasaki K., Goodwin J. E. (2019). microRNA crosstalk influences epithelial-to-mesenchymal, endothelial-to-mesenchymal, and macrophage-to-mesenchymal transitions in the kidney. Front. Pharmacol. 10, 904 10.3389/fphar.2019.00904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Ren J., Chen H., Liu B. (2020). MicroRNA-140-5p ameliorates the high glucose-induced apoptosis and inflammation through suppressing TLR4/NF-κB signaling pathway in human renal tubular epithelial cells. Biosci. Rep. 40 10.1042/BSR20192384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Yu P. F., Wang D., Teng J. (2019). MicroRNA-488 regulates diabetic nephropathy via TGF-β1 pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 4333–4340. 10.26355/eurrev_201905_17939 [DOI] [PubMed] [Google Scholar]
- Sun S. F., Tang P. M. K., Feng M., Xiao J., Huang X. R., Li P., et al. (2018a). Novel lncRNA Erbb4-IR promotes diabetic kidney injury in db/db mice by targeting miR-29b. Diabetes. 67, 731–744. 10.2337/db17-0816 [DOI] [PubMed] [Google Scholar]
- Sun T., Liu Y., Liu L., Ma F. (2020). MicroRNA-544 attenuates diabetic renal injury via suppressing glomerulosclerosis and inflammation by targeting FASN. Gene. 723, 143986 10.1016/j.gene.2019.143986 [DOI] [PubMed] [Google Scholar]
- Sun Y., Peng R., Peng H., Liu H., Wen L., Wu T., et al. (2016a). miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol. Cell. Endocrinol. 433, 75–86. 10.1016/j.mce.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Sun Y., Peng R., Peng H., Liu H., Wen L., Wu T., et al. (2016b). miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol. Cell. Endocrinol. 433, 75–86. 10.1016/j.mce.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Sun Z., Ma Y., Chen F., Wang S., Chen B., Shi J. (2018b). miR-133b and miR-199b knockdown attenuate TGF-β1-induced epithelial to mesenchymal transition and renal fibrosis by targeting SIRT1 in diabetic nephropathy. Eur. J. Pharmacol. 837, 96–104. 10.1016/j.ejphar.2018.08.022 [DOI] [PubMed] [Google Scholar]
- Tan R., Zhang J., Tan X., Zhang X., Yang J., Liu Y. (2006). Downregulation of SnoN expression in obstructive nephropathy is mediated by an enhanced ubiquitin-dependent degradation. J. Am. Soc. Nephrol. 17, 2781–2791. 10.1681/ASN.2005101055 [DOI] [PubMed] [Google Scholar]
- Tang P. M., Zhang Y. Y., Mak T. S., Tang P. C., Huang X. R., Lan H. Y. (2018a). Transforming growth factor-beta signalling in renal fibrosis: from Smads to non-coding RNAs. J. Physiol. 596, 3493–3503. 10.1113/JP274492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. B., Zheng L., Yan R., Yang J., Ning J., Peng L., et al. (2018b). miR302a-3p may modulate renal epithelial-mesenchymal transition in diabetic kidney disease by targeting ZEB1. Nephron. 138, 231–242. 10.1159/000481465 [DOI] [PubMed] [Google Scholar]
- Tayel S. I., Saleh A. A., El-Hefnawy S. M., Elzorkany K. M., Elgarawany G. E., Noreldin R. I. (2020). Simultaneous assessment of MicroRNAs 126 and 192 in diabetic nephropathy patients and the relation of these MicroRNAs with urinary albumin. Curr. Mol. Med. 20, 361–371. 10.2174/1566524019666191019103918 [DOI] [PubMed] [Google Scholar]
- Tung C. W., Ho C., Hsu Y. C., Huang S. C., Shih Y. H., Lin C. L. (2019). MicroRNA-29a attenuates diabetic glomerular injury through modulating cannabinoid receptor 1 signaling. Molecules. 24 10.3390/molecules24020264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid F., Shehzad A., Khan T., Kim Y. Y. (2010). MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta. 1803, 1231–1243. 10.1016/j.bbamcr.2010.06.013 [DOI] [PubMed] [Google Scholar]
- Wan R. J., Li Y. H. (2018). MicroRNA-146a/NAPDH oxidase4 decreases reactive oxygen species generation and inflammation in a diabetic nephropathy model. Mol. Med. Rep. 17, 4759–4766. 10.3892/mmr.2018.8407 [DOI] [PubMed] [Google Scholar]
- Wang B., Koh P., Winbanks C., Coughlan M. T., Mcclelland A., Watson A., et al. (2011). miR-200a Prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes. 60, 280–287. 10.2337/db10-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Komers R., Carew R., Winbanks C. E., Xu B., Herman-Edelstein M., et al. (2012). Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J. Am. Soc. Nephrol. 23, 252–265. 10.1681/ASN.2011010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Gao X., Zhang R., Zhao P., Sun Y., Li C. (2019a). LncRNA TUG1 ameliorates diabetic nephropathy by inhibiting miR-21 to promote TIMP3-expression. Int. J. Clin. Exp. Pathol. 12, 717–729. [PMC free article] [PubMed] [Google Scholar]
- Wang J., Duan L., Tian L., Liu J., Wang S., Gao Y., et al. (2016a). Serum miR-21 may be a potential diagnostic biomarker for diabetic nephropathy. Exp. Clin. Endocrinol. Diabetes. 124, 417–423. 10.1055/s-0035-1565095 [DOI] [PubMed] [Google Scholar]
- Wang J., Gao Y., Ma M., Li M., Zou D., Yang J., et al. (2013). Effect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochem. Biophys. 67, 537–546. 10.1007/s12013-013-9539-2 [DOI] [PubMed] [Google Scholar]
- Wang J., Pan J., Li H., Long J., Fang F., Chen J., et al. (2018a). lncRNA ZEB1-AS1 was suppressed by p53 for renal fibrosis in diabetic nephropathy. Mol. Ther. Nucleic Acids. 12, 741–750. 10.1016/j.omtn.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Y., Gao Y. B., Zhang N., Zou D. W., Wang P., Zhu Z. Y., et al. (2014). miR-21 overexpression enhances TGF-β1-induced epithelial-to-mesenchymal transition by target smad7 and aggravates renal damage in diabetic nephropathy. Mol. Cell. Endocrinol. 392, 163–172. 10.1016/j.mce.2014.05.018 [DOI] [PubMed] [Google Scholar]
- Wang L., Li H. (2020). MiR-770-5p facilitates podocyte apoptosis and inflammation in diabetic nephropathy by targeting TIMP3. Biosci. Rep. 40 10.1042/bsr20193653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Su N., Zhang Y., Wang G. (2018b). Clinical significance of serum lncRNA cancer susceptibility candidate 2 (CASC2) for chronic renal failure in patients with type 2 diabetes. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 24, 6079–6084. 10.12659/MSM.909510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wang S., Yao D., Yan Q., Lu W. (2016b). A novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation and fibrosis in diabetic nephropathy. Mol. Cell. Endocrinol. 426, 136–145. 10.1016/j.mce.2016.02.020 [DOI] [PubMed] [Google Scholar]
- Wang M., Yao D., Wang S., Yan Q., Lu W. (2016c). Long non-coding RNA ENSMUST00000147869 protects mesangial cells from proliferation and fibrosis induced by diabetic nephropathy. Endocrine. 54, 81–92. 10.1007/s12020-016-0950-5 [DOI] [PubMed] [Google Scholar]
- Wang S., Chen X., Wang M., Yao D., Chen T., Yan Q., et al. (2018c). Long non-coding RNA CYP4B1-PS1-001 inhibits proliferation and fibrosis in diabetic nephropathy by interacting with Nucleolin. Cell. Physiol. Biochem. 49, 2174–2187. 10.1159/000493821 [DOI] [PubMed] [Google Scholar]
- Wang S., Wen X., Han X. R., Wang Y. J., Shen M., Fan S. H., et al. (2018d). Repression of microRNA-382 inhibits glomerular mesangial cell proliferation and extracellular matrix accumulation via FoxO1 in mice with diabetic nephropathy. Cell Prolif. 51, e12462 10.1111/cpr.12462 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang X., Xu Y., Zhu Y. C., Wang Y. K., Li J., Li X. Y., et al. (2019b). LncRNA NEAT1 promotes extracellular matrix accumulation and epithelial-to-mesenchymal transition by targeting miR-27b-3p and ZEB1 in diabetic nephropathy. J. Cell. Physiol. 234, 12926–12933. 10.1002/jcp.27959 [DOI] [PubMed] [Google Scholar]
- Winbanks C. E., Wang B., Beyer C., Koh P., White L., Kantharidis P., et al. (2011). TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J. Biol. Chem. 286, 13805–13814. 10.1074/jbc.M110.192625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Liu J., Ding Y., Zhu M., Lu K., Zhou J., et al. (2018a). MiR-455-3p suppresses renal fibrosis through repression of ROCK2 expression in diabetic nephropathy. Biochem. Biophys. Res. Commun. 503, 977–983. 10.1016/j.bbrc.2018.06.105 [DOI] [PubMed] [Google Scholar]
- Wu J., Liu S., Yuan Z. W., Liu J. H., Li J. M., Chen T., et al. (2018b). MicroRNA-27a suppresses detrusor fibrosis in streptozotocin-induced diabetic rats by targeting PRKAA2 through the TGF-β1/Smad3 signaling pathway. Cell. Physiol. Biochem. 45, 1333–1349. 10.1159/000487560 [DOI] [PubMed] [Google Scholar]
- Wu J., Lu K., Zhu M., Xie X., Ding Y., Shao X., et al. (2020). miR-485 suppresses inflammation and proliferation of mesangial cells in an in vitro model of diabetic nephropathy by targeting NOX5. Biochem. Biophys. Res. Commun. 521, 984–990. 10.1016/j.bbrc.2019.11.020 [DOI] [PubMed] [Google Scholar]
- Xu B. H., Sheng J., You Y. K., Huang X. R., Ma R. C. W., Wang Q., et al. (2020). Deletion of Smad3 prevents renal fibrosis and inflammation in type 2 diabetic nephropathy. Metabolism. 103, 154013 10.1016/j.metabol.2019.154013 [DOI] [PubMed] [Google Scholar]
- Xu H.-F., Ding Y.-J., Zhang Z.-X., Wang Z.-F., Luo C.-L., Li B.-X., et al. (2014). MicroRNA-21 regulation of the progression of viral myocarditis to dilated cardiomyopathy. Mol. Med. Rep. 10, 161–168. 10.3892/mmr.2014.2205 [DOI] [PubMed] [Google Scholar]
- Xu H., Sun F., Li X., Sun L. (2018a). Down-regulation of miR-23a inhibits high glucose-induced EMT and renal fibrogenesis by up-regulation of SnoN. Hum. Cell. 31, 22–32. 10.1007/s13577-017-0180-z [DOI] [PubMed] [Google Scholar]
- Xu W. C., Qian G., Liu A. Q., Li Y. Q., Zou H. Q. (2018b). Urinary extracellular vesicle: a potential Source of early diagnostic and therapeutic biomarker in diabetic kidney disease. Chin. Med. J. (Engl.). 131, 1357–1364. 10.4103/0366-6999.232801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhang J., Fan L., He X. (2018c). miR-423-5p suppresses high-glucose-induced podocyte injury by targeting Nox4. Biochem. Biophys. Res. Commun. 505, 339–345. 10.1016/j.bbrc.2018.09.067 [DOI] [PubMed] [Google Scholar]
- Xue M., Li Y., Hu F., Jia Y. J., Zheng Z. J., Wang L., et al. (2018). High glucose up-regulates microRNA-34a-5p to aggravate fibrosis by targeting SIRT1 in HK-2 cells. Biochem. Biophys. Res. Commun. 498, 38–44. 10.1016/j.bbrc.2017.12.048 [DOI] [PubMed] [Google Scholar]
- Yang J., Shen Y., Yang X., Long Y., Chen S., Lin X., et al. (2019a). Silencing of long noncoding RNA XIST protects against renal interstitial fibrosis in diabetic nephropathy via microRNA-93-5p-mediated inhibition of CDKN1A. Am. J. Physiol. Ren. Physiol. 317, F1350–f1358. 10.1152/ajprenal.00254.2019 [DOI] [PubMed] [Google Scholar]
- Yang L., Froberg J. E., Lee J. T. (2014). Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem. Sci. 39, 35–43. 10.1016/j.tibs.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Chen X., Yang M., Zhao X., Chen Y., Zhao H., et al. (2019b). The variant at TGFBRAP1 is significantly associated with type 2 diabetes mellitus and affects diabetes-related miRNA expression. J. Cell Mol. Med. 23, 83–92. 10.1111/jcmm.13885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. L., Hu F., Xue M., Jia Y. J., Zheng Z. J., Li Y., et al. (2019c). Early growth response protein-1 upregulates long noncoding RNA Arid2-IR to promote extracellular matrix production in diabetic kidney disease. Am. J. Physiol. Cell Physiol. 316, C340–c352. 10.1152/ajpcell.00167.2018 [DOI] [PubMed] [Google Scholar]
- Yang Y. L., Xue M., Jia Y. J., Hu F., Zheng Z. J., Wang L., et al. (2020). Long noncoding RNA NEAT1 is involved in the protective effect of Klotho on renal tubular epithelial cells in diabetic kidney disease through the ERK1/2 signaling pathway. Exp. Mol. Med. 52, 266–280. 10.1038/s12276-020-0381-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Guo Z., Dong J., Sheng S., Wang Y., Yu L., et al. (2018). miR-374a regulates inflammatory response in diabetic nephropathy by targeting MCP-1 expression. Front. Pharmacol. 9, 900 10.3389/fphar.2018.00900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R.-W., Wang Y., Chen L.-L. (2019). Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551. 10.1038/s41556-019-0311-8 [DOI] [PubMed] [Google Scholar]
- Yao T., Zha D., Gao P., Shui H., Wu X. (2018). MiR-874 alleviates renal injury and inflammatory response in diabetic nephropathy through targeting toll-like receptor-4. J. Cell. Physiol. 234, 871–879. 10.1002/jcp.26908 [DOI] [PubMed] [Google Scholar]
- Yi H., Peng R., Zhang L.-Y., Sun Y., Peng H.-M., Liu H.-D., et al. (2017). LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 8, e2583 10.1038/cddis.2016.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. N., Hu M. L., Wang X. F., Li X. P., Zhang B. H., Lu X. Q., et al. (2019). Effects of microRNA-370 on mesangial cell proliferation and extracellular matrix accumulation by binding to canopy 1 in a rat model of diabetic nephropathy. J. Cell. Physiol. 234, 6898–6907. 10.1002/jcp.27448 [DOI] [PubMed] [Google Scholar]
- Yu Y., Bai F., Qin N., Liu W., Sun Q., Zhou Y., et al. (2018). Non-proximal renal tubule-derived urinary exosomal miR-200b as a biomarker of renal fibrosis. Nephron. 139, 269–282. 10.1159/000487104 [DOI] [PubMed] [Google Scholar]
- Zanchi C., Macconi D., Trionfini P., Tomasoni S., Rottoli D., Locatelli M., et al. (2017). MicroRNA-184 is a downstream effector of albuminuria driving renal fibrosis in rats with diabetic nephropathy. Diabetologia. 60, 1114–1125. 10.1007/s00125-017-4248-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang X. J., Li L., Du X., Yang B., Mei C. L. (2019). LncRNA TUG1 inhibits the proliferation and fibrosis of mesangial cells in diabetic nephropathy via inhibiting the PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 7519–7525. 10.26355/eurrev_201909_18867 [DOI] [PubMed] [Google Scholar]
- Zha F., Qu X., Tang B., Li J., Wang Y., Zheng P., et al. (2019). Long non-coding RNA MEG3 promotes fibrosis and inflammatory response in diabetic nephropathy via miR-181a/Egr-1/TLR4 axis. Aging (Albany NY). 11, 3716–3730. 10.18632/aging.102011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Freedman B. I., Flekac M., Santos E., Hicks P. J., Bowden D. W., et al. (2009a). Evaluation of genetic association and expression reduction of TRPC1 in the development of diabetic nephropathy. Am. J. Nephrol. 29, 244–251. 10.1159/000157627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Jiang T., Liang X., Shu S., Xiang X., Zhang W., et al. (2019a). lncRNA MALAT1 mediated high glucose-induced HK-2 cell epithelial-to-mesenchymal transition and injury. J. Physiol. Biochem. 75, 443–452. 10.1007/s13105-019-00688-2 [DOI] [PubMed] [Google Scholar]
- Zhang J. Y., Gong Y. L., Li C. J., Qi Q., Zhang Q. M., Yu D. M. (2016). Circulating MiRNA biomarkers serve as a fingerprint for diabetic atherosclerosis. Am J Transl Res. 8, 2650–2658. [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhao S., Zhu Y. (2020). Long noncoding RNA growth arrest-specific transcript 5 alleviates renal fibrosis in diabetic nephropathy by downregulating matrix metalloproteinase 9 through recruitment of enhancer of zeste homolog 2. Faseb. J. 34, 2703–2714. 10.1096/fj.201901380RR [DOI] [PubMed] [Google Scholar]
- Zhang P., Sun Y., Peng R., Chen W., Fu X., Zhang L., et al. (2019b). Long non-coding RNA Rpph1 promotes inflammation and proliferation of mesangial cells in diabetic nephropathy via an interaction with Gal-3. Cell Death & Disease. 10, 526 10.1038/s41419-019-1765-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Z., Qiu X. J., Dong S. S., Zhou L. N., Zhu Y., Wang M. D., et al. (2019c). MicroRNA-770-5p is involved in the development of diabetic nephropathy through regulating podocyte apoptosis by targeting TP53 regulated inhibitor of apoptosis 1. Eur. Rev. Med. Pharmacol. Sci. 23, 1248–1256. 10.26355/eurrev_201902_17018 [DOI] [PubMed] [Google Scholar]
- Zhang Y. Y., Tang P. M., Tang P. C., Xiao J., Huang X. R., Yu C., et al. (2019d). LRNA9884, a novel Smad3-dependent long noncoding RNA, promotes diabetic kidney injury in db/db mice via enhancing MCP-1-dependent renal inflammation. Diabetes. 68, 1485–1498. 10.2337/db18-1075 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhao S., Wu D., Liu X., Shi M., Wang Y., et al. (2018). MicroRNA-22 promotes renal tubulointerstitial fibrosis by targeting PTEN and suppressing autophagy in diabetic nephropathy. J Diabetes Res. 2018, 4728645 10.1155/2018/4728645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Peng H., Chen J., Chen X., Han F., Xu X., et al. (2009b). MicroRNA-21 protects from mesangial cell proliferation induced by diabetic nephropathy in db/db mice. FEBS Lett. 583, 2009–2014. 10.1016/j.febslet.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Zhao C., Hu J., Wang Z., Cao Z. Y., Wang L. (2020). Serum LncRNA PANDAR may act as a novel serum biomarker of diabetic nephropathy in patients with type 2 diabetes. Clin. Lab. 66 10.7754/Clin.Lab.2019.191032 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Yin Z., Li H., Fan J., Yang S., Chen C., et al. (2017). MiR-30c protects diabetic nephropathy by suppressing epithelial-to-mesenchymal transition in db/db mice. Aging Cell. 16, 387–400. 10.1111/acel.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Zeng J., Xue J., Du A., Xu Y. (2020). Knockdown of lncRNA PVT1 alleviates high glucose-induced proliferation and fibrosis in human mesangial cells by miR-23b-3p/WT1 axis. Diabetol. Metab. Syndrome. 12, 33 10.1186/s13098-020-00539-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Chung A. C., Chen H. Y., Dong Y., Meng X. M., Li R., et al. (2013). miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 56, 663–674. 10.1007/s00125-012-2804-x [DOI] [PubMed] [Google Scholar]
- Zhong X., Chung A. C., Chen H. Y., Meng X. M., Lan H. Y. (2011). Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J. Am. Soc. Nephrol. 22, 1668–1681. 10.1681/ASN.2010111168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Chen W., Yu X. Q. (2019). Long non-coding RNAs as novel diagnostic and therapeutic targets in kidney disease. Chronic Dis. Transl. Med. 5, 252–257. 10.1016/j.cdtm.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Huang X. R., Yu J., Yu X., Lan H. Y. (2015). Long noncoding RNA arid2-IR is a novel therapeutic target for renal inflammation. Mol. Ther. 23, 1034–1043. 10.1038/mt.2015.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. J., Zhu M., Xu X. X., Meng X. M., Wu Y. G. (2019a). Exosomes from high glucose-treated macrophages activate glomerular mesangial cells via TGF-β1/Smad3 pathway in vivo and in vitro . Faseb. J. 33, 9279–9290. 10.1096/fj.201802427rrr [DOI] [PubMed] [Google Scholar]
- Zhu X. J., Gong Z., Li S. J., Jia H. P., Li D. L. (2019b). Long non-coding RNA Hottip modulates high-glucose-induced inflammation and ECM accumulation through miR-455-3p/WNT2B in mouse mesangial cells. Int. J. Clin. Exp. Pathol. 12, 2435–2445. 10.21037/ncri.2019.02.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Xu J., Liang W., Li J., Feng L., Zheng P., et al. (2019c). miR-98-5p alleviated epithelial-to-mesenchymal transition and renal fibrosis via targeting Hmga2 in diabetic nephropathy. Internet J. Endocrinol. 7, 4946181 10.1155/2019/4946181 [DOI] [PMC free article] [PubMed] [Google Scholar]