Dear Editor,
The selective degradation of specific pathogenic proteins provides exciting strategies for drug discovery. Emerging new concepts such as proteolysis-targeting chimeras and autophagosome-tethering compounds (ATTECs) are based on the design of “molecular glues” or bifunctional chimeric compounds that tether the target protein (protein of interest, POI) to a specific component of the protein-degradation machinery (PDM) [1–3]. The formed trimer (POI–compound–PDM) then accelerates degradation of the POI, leading its selective reduction. Interestingly, the dose-dependence curves of the POI-compound relationship are U-shaped, with an optimal compound concentration for maximum lowering of the POI and “hook” effects at higher compound concentrations [1], different from traditional Boltzmann dose-dependent curves [4]. This is explained by the logic that when the compound concentration is too high, each molecule may interact with the POI and PDM separately, without tethering them together. Meanwhile, there has been a lack of mathematic modeling describing such effects. While modeling of the trimer formation has been published in a top-tier journal [5], the degradation of the POI was not considered at all. Such modeling has been challenging, because incorporating the degradation greatly increases the modeling calculation. In addition, most of the existing degrader technologies are based on ubiquitination [1], which is a complicated enzymatic reaction that is highly challenging to model [6].
We have proposed and demonstrated a new degrader technology by harnessing autophagy for selective degradation using ATTECs [2]. We demonstrated that compounds that interact with both the POI and the autophagosome protein LC3 tether POI to autophagosomes for subsequent autophagic degradation [2]. Since ATTECs tether POI directly to the PDM without involving complicated enzymatic reactions, they may provide an ideal scenario for mathematical modeling of the degrader’s effects [2]. Here, we describe a simplified model of the degradation effects of an ATTEC, providing possible insights for understanding the dose-dependence data and potential clues for inventing better compounds.
Many parameters may influence the degradation kinetics and the dose-dependence effects [7, 8], and considering all of them as variables may make the model highly complicated and difficult to resolve. Thus, we focused on the relationship between the degradation of the POI and the ATTEC’s affinity to the POI or LC3, and considered them as the only variables. Meanwhile, we estimated the values of all other parameters and considered them as constants to simplify the model as much as possible. Based on these assumptions, we performed the modeling based on the kinetic and steady-state equations of all the relevant chemical reactions (see Modeling Methods in Supplementary Materials).
Kinetic Modeling of Each Species Involved in an ATTEC
We first solved kinetic equations [Eq. (5) to Eq. (10) in Modeling Methods of the Supplementary Materials] using codes written with MatLab and simulated the kinetic changes of different species in the system. With the indicated kinetic parameters, the kinetic changes of each species were calculated (Fig. S1). The kinetic parameters were estimated based on our previous experimental measurements on the GW5074 compound [2].
Based on the kinetic simulation, the free mutant HTT protein (mHTT, the POI) and free LC3 protein had the fastest decay rates and reached steady-state within a few hours (Fig. S1, mHTT and LC3 panels) due to the rapid formation of the ATTEC·mHTT and ATTEC·LC3 binary complexes, respectively. The free ATTEC molecule had a rapid decay but bounced back a little at a much slower rate (Fig. S1, ATTEC panel). The first decay phase was also due to the rapid formation of the ATTEC·LC3 and ATTEC·mHTT binary complexes. The bounce-back illustrated the recycling of ATTEC after degradation of the mHTT·ATTEC·LC3 proteins. The LC3·ATTEC complex increased rapidly due to the formation of this binary complex (Fig. S1, LC3·ATTEC panel). Both the ATTEC·mHTT binary complex and the mHTT·ATTEC·LC3 ternary complex increased rapidly and then decayed slowly to reach a steady-state level (Fig. S1, ATTEC·mHTT and mHTT·ATTEC·LC3 panels). The phase of rapid increase was due to the formation of these complexes, and the slow decay phase of ATTEC·mHTT was due to the degradation of this complex as well as its consumption due to the formation of the ternary complex. The slow decay phase of the mHTT·ATTEC·LC3 complex was due to the degradation of this complex, and was balanced by its formation.
The major purpose of the model was to understand the kinetics of the total mHTT level, which is the sum of free mHTT, the binary ATTEC·mHTT complex, and the ternary mHTT·ATTEC·LC3 complex. Thus, we calculated the sum of the levels of these three species, and simulated the kinetic curves of total mHTT (Fig. S2). The total mHTT level decayed over time, reaching a steady-state after ~30 to ~150 h depending on the kinetic parameters (Fig. S2). We then investigated how the kinetic parameters influence the kinetic curve of the total mHTT level. The Kon1 and Koff1 values determined the kinetics of the formation of the ATTEC·LC3 and mHTT·ATTEC·LC3 complexes. A larger Kon1 or smaller Koff1 led to faster ATTEC·LC3 and mHTT·ATTEC·LC3 formation. Consistent with this, the total mHTT level decayed faster and reached a lower steady-state level with a larger Kon1 and smaller Koff1 (Fig. S2). Notably, these curves were more sensitive to Kon1 changes in a lower value range [between 0.000223 and 0.00223 (h·nmol/L)-1] and to Koff1 changes in a higher value range [between 10.4 and 1.04 (h)-1]. Similar findings were obtained on the influences of Kon2 and Koff2, which determine the kinetics of the formation of the ATTEC·mHTT and mHTT·ATTEC·LC3 complexes (Fig. S2). Under all conditions, the total mHTT level at 48 h was very close to the steady-state level, suggesting that this compound treatment time used in previously published experiments is reasonable [2].
Modeling of the Dose-Dependent Curves for ATTEC
We then solved the steady-state equations [Eq. (11) to Eq. (16) in Modeling Methods of the Supplementary Materials] and simulated the dose-dependence curves of ATTEC. All the ATTEC curves were U-shaped (Fig. S3), consistent with the autophagosome-tethering mechanism in which a sufficient concentration of ATTEC is needed, whereas excessively high concentrations may lead to separate binding of ATTEC to LC3 and mHTT, without tethering the two together. Since at the same Kd values (KdH for the equilibrium dissociation constant of the ATTEC to mHTT, and KdL for the equilibrium dissociation constant of the ATTEC to LC3), the dose-dependence curves changed very little with Kon and Koff varying two orders of magnitude (Fig. S3), we focused on analyzing the effect of Kd on the degradation efficiency of the ATTEC.
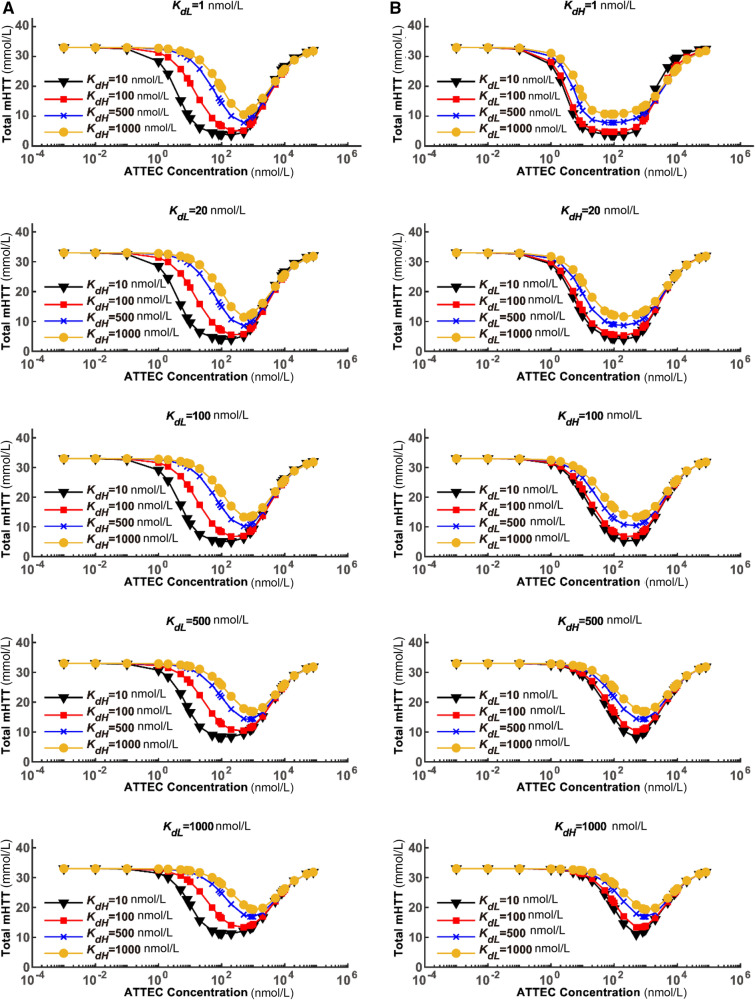
We simulated the dose-dependence curves with different sets of compound–protein affinity parameters (Fig. 1). The shape of the dose-dependence curve closely resembled the experimental data that we published previously [2]. The target protein mHTT was only lowered to a certain degree; the extent of lowering is referred to as maximum degradation (Dmax). Dmax was reached only at optimal concentrations of the ATTEC. It was clear that both Dmax and the optimal concentrations for degradation were influenced by the Kd values (Fig. 1), which are further analyzed below.
Fig. 1.
Modeling of the dose-dependence curves for the ATTEC with varying Kd values. The relationship between the steady-state concentrations of total mHTT and the concentrations of ATTEC molecules are shown. A The ATTEC dose-dependence curves with a set of KdH values and a fixed KdL in each panel. B Similar to A, but with a set of KdL values and a fixed KdH in each panel. The KdL and KdH values are set to a series of representative numbers that are easy for calculation and covering the range of our previously-discovered ATTECs.
Relationship Between Affinity (Kd) and Maximum Degradation (Dmax)
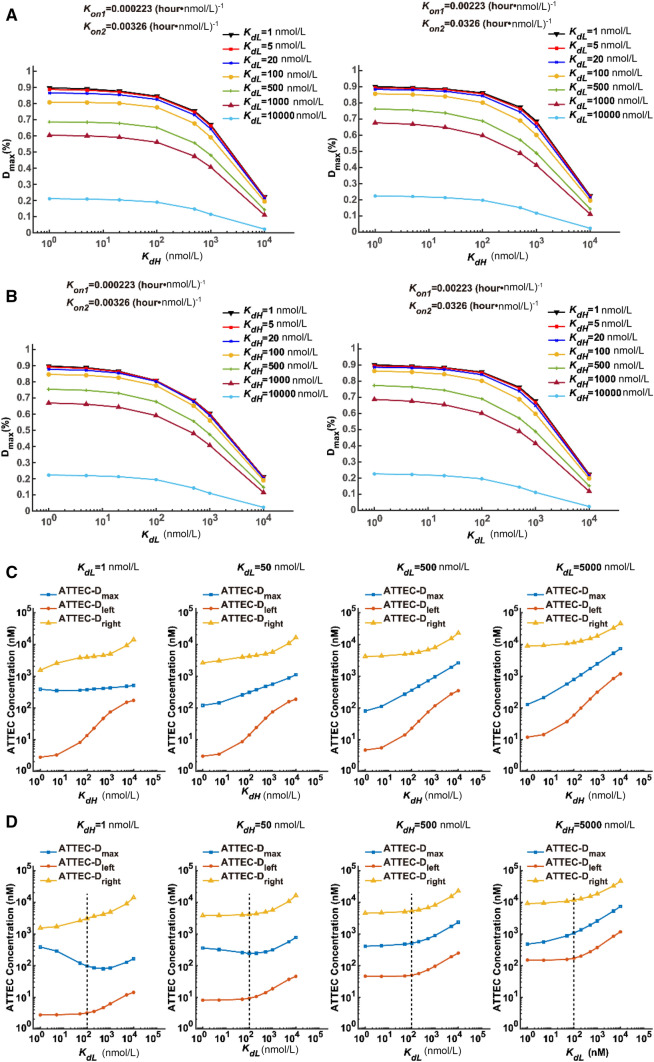
In order to visualize the relationship between Dmax and Kd values, we plotted the Dmax–KdH relationship assuming several different KdL values (Fig. 2A). Since a Kon change between 0.00023 and 0.0023 (hour·nmol/L)-1 led to subtle differences in Dmax (Fig. S3), we simulated based on both of these Kon values and obtained very similar results (Fig. 2A). A KdH increase led to decreased Dmax values, consistent with the prediction that lowering affinity (higher Kd) to the POI leads to less efficient degradation. Meanwhile, the curves were relatively flat within the range 1–100 nmol/L KdH, and dropped rapidly within the range 100–10000 nmol/L KdH (Fig. 2A), suggesting that Dmax is sensitive to KdH only in the low-affinity range (high KdH values at 100–10000 nmol/L). The Dmax–KdL relationship was very similar to the Dmax–KdH relationship (Fig. 2B), suggesting that Dmax is also sensitive to KdL only in the low-affinity range. Thus, while a relatively high affinity of ATTEC to POI (mHTT) and LC3 is desired to achieve a higher Dmax, it may not be necessary to make huge efforts to achieve extremely high affinity (such as ~1 nmol/L Kd), because Kd values within 1–100 nmol/L lead to a similar Dmax.
Fig. 2.
The relationship between the degradation effect (Dmax and the effective concentration range) and the compound–protein affinity (KdL and KdH). A Simulated Dmax–KdH relationship assuming the indicated KdL values. B Simulated Dmax–KdL relationship assuming the indicated KdH values. C, D Plots of concentrations needed to achieve maximum degradation ATTEC–Dmax or half-maximum degradation ATTEC–Dleft and ATTEC–Dright at different sets of KdL and KdH values. C ATTEC–Dmax, ATTEC–Dleft, and ATTEC–Dright concentrations assuming different KdH values at a fixed KdL in each panel. D Similar to C, but assuming different KdL values at a fixed KdH in each panel. Dashed lines indicate 100 nmol/L, below which KdL has little influence on the effective concentration range. The KdL and KdH values were set to a series of representative numbers that are easy for calculation and covering the range of our previously-discovered ATTECs.
Relationship Between Affinity (Kd) and Effective Compound Concentrations
As noted above, the dose-dependence curve of the ATTEC is unlike the common Boltzmann curves that fit most chemical responses. Instead, the ATTEC curve was U-shaped, and an optimal concentration range is desired for efficient degradation. This optimal range could be influenced by the ATTEC’s affinity for both the POI and LC3, as illustrated by our modeling data assuming several different sets of Kd values (Fig. 1). Resolving the relationship between the optimal concentration range and the Kd values may guide compound optimization.
In order to visualize this relationship, we plotted three different ATTEC concentrations against the Kd values. The ATTEC–Dmax value indicates the ATTEC concentration needed to reach the maximum degradation effect. The ATTEC–Dleft and ATTEC–Dright indicate the lower and higher concentrations needed to reach half of the maximum degradation effect, respectively.
We first plotted these concentrations against KdH, assuming several different KdL values (Fig. 2C). The general trend was that ATTEC–Dmax, ATTEC–Dleft, and ATTEC–Dright all increased with KdH (Fig. 2C), suggesting that higher ATTEC concentrations are needed to achieve efficient degradation when the affinity of the ATTEC for mHTT decreases, represented by increased KdH values. In addition, the size of the effective concentration window (Fig. 2C, distance between yellow and red curves, log scale) also decreased as KdH increased, especially at a larger KdH range, suggesting that the effective concentration range is larger when the affinity of the ATTEC for the POI is high. Finally, the ATTEC–Dleft curves generally had a larger slope, suggesting that the lower concentration required to achieve half of the maximum degradation effect is more sensitive to the affinity of the ATTEC for the POI.
We then plotted ATTEC–Dmax, ATTEC–Dleft, and ATTEC–Dright against KdL. Interestingly, the curves differed from those plotted against KdH. The major difference was that the size of the effective concentration window (Fig. 2D, distance between yellow and red curves) was insensitive to KdL values, suggesting that the ATTEC’s affinity for LC3 is not a major factor contributing to the size of the effective concentration range. Second, the ATTEC–Dmax curve (Fig. 2D, blue) did not always increase monotonically, and an optimal KdL between 100 and 1000 nmol/L was required to reach the lowest ATTEC–Dmax, at least when KdH is low (Fig. 2D, KdH = 1 nmol/L or 50 nmol/L). Thus, it seems that a high affinity of ATTEC for LC3 is not always desired to reach maximum degradation effects at low ATTEC concentrations. Finally, for KdL <100 nmol/L (Fig. 2D, left to the dashed lines in each panel), the ATTEC–Dleft and ATTEC–Dright were largely unaffected, whereas they did increase when KdL was >100 nmol/L. Thus, in order to induce efficient degradation at lower ATTEC concentrations, a higher affinity for LC3 (lower KdL) is desired only when KdL is >100 nmol/L. Further increasing the affinity for LC3 may not be necessary, at least in the conditions assumed in our modeling.
In this study, we presented the first simplified model of ATTEC-induced lowering of mHTT. Although we used mHTT for our simulation, the modeling is applicable to other POIs. While the model was over-simplified, the shape of the dose-dependence curves largely fit the experimental data [2]. We further studied the influence of ATTEC’s affinity for the POI and LC3, and found some interesting properties. In general, the Dmax was negatively correlated with both KdH and KdL, but was not so sensitive when the Kd values were <100 nmol/L (Fig. 2A, B). Lower KdH values were also required to achieve the desired effective concentration range (Fig. 2C), but lower KdL values may not be necessary, especially when KdL is <100 nmol/L (Fig. 2D). Thus, optimizing the compounds to reach a higher affinity for the POI is more important, and an attempt to reach extremely high affinity for LC3 (<100 nmol/L) may not be necessary. As we chose mHTT as the POI, it is worth mentioning that the ATTECs that we discovered in a previous study have been confirmed to interact specifically with an expanded stretch of polyQ, without influencing other polyQ protein levels, based on proteomics analysis [2]. These predictions are based on many assumptions, including the estimation of the starting concentrations of the mHTT and LC3 proteins, and certainly need further confirmation by experiments. For example, the starting concentration may be different for other POIs, and this may influence the dose-dependence curve, especially in the lower concentration range (Fig. S4). Meanwhile, as the first model for ATTEC-induced degradation, and even for ternary complex-induced degradation, it may still provide insights into the properties of such degradation and possible factors to consider for compound optimization. Besides neurodegeneration, many other neurological diseases are also associated with protein abnormalities in the brain [9, 10]. ATTEC molecules may selectively degrade these pathogenic proteins and provide entry points to treat these diseases. Understanding how it works by mathematical modeling may provide insights into optimizing such compounds, and thus benefit drug discovery for neurodegeneration and other neurological diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81925012, 81870990, and 31961130379) and a Newton Advanced Fellowship (NAF_R1_191045).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Hang Zhang and Ping An contributed equally to this work.
Contributor Information
Yiyan Fei, Email: fyy@fudan.edu.cn.
Boxun Lu, Email: luboxun@fudan.edu.cn.
References
- 1.Bondeson DP, Mares A, Smith IE, Ko E, Campos S, Miah AH, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z, Wang C, Wang Z, Zhu C, Li J, Sha T, et al. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature. 2019;575:203–209. doi: 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- 3.Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, et al. Drug Development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015, 348: 1376–1381. [DOI] [PMC free article] [PubMed]
- 4.Dubois JM, Ouanounou G, Rouzaire-Dubois B. The Boltzmann equation in molecular biology. Prog Biophys Mol Biol. 2009;99:87–93. doi: 10.1016/j.pbiomolbio.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Douglass EF, Jr, Miller CJ, Sparer G, Shapiro H, Spiegel DA. A comprehensive mathematical model for three-body binding equilibria. J Am Chem Soc. 2013;135:6092–6099. doi: 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schapira M, Calabrese MF, Bullock AN, Crews CM. Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov. 2019;18:949–963. doi: 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Loh C, Chen J, Mainolfi N. Targeted protein degradation mechanisms. Drug Discov Today Technol. 2019;31:53–60. doi: 10.1016/j.ddtec.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Crisp MJ, Mawuenyega KG, Patterson BW, Reddy NC, Chott R, Self WK, et al. In vivo kinetic approach reveals slow SOD1 turnover in the CNS. J Clin Invest. 2015;125:2772–2780. doi: 10.1172/JCI80705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Wang L, Ma Y, Yue W, Zhang D, Li J. P-Rex1 overexpression results in aberrant neuronal polarity and psychosis-related behaviors. Neurosci Bull. 2019;35:1011–1023. doi: 10.1007/s12264-019-00408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Q, Yang H, Wang M, Wei H, Hu F. Role of microtubule-associated protein in autism spectrum disorder. Neurosci Bull. 2018;34:1119–1126. doi: 10.1007/s12264-018-0246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.