Abstract
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides but not translated into proteins. LncRNAs regulate gene expressions at multiple levels, such as chromatin, transcription, and post-transcription. Further, lncRNAs participate in various biological processes such as cell differentiation, cell cycle regulation, and maintenance of stem cell pluripotency. We have previously reported that lncRNAs are closely related to programmed cell death (PCD), which includes apoptosis, autophagy, necroptosis, and ferroptosis. Overexpression of lncRNA can suppress the extrinsic apoptosis pathway by downregulating of membrane receptors and protect tumor cells by inhibiting the expression of necroptosis-related proteins. Some lncRNAs can also act as competitive endogenous RNA to prevent oxidation, thereby inhibiting ferroptosis, while some are known to activate autophagy. The relationship between lncRNA and PCD has promising implications in clinical research, and reports have highlighted this relationship in various cancers such as non-small cell lung cancer and gastric cancer. This review systematically summarizes the advances in the understanding of the molecular mechanisms through which lncRNAs impact PCD.
Subject terms: Cell biology, Molecular biology
Facts
Programmed cell death refers to controlled cell death to maintain the stability of the internal environment.
LncRNAs are a class of RNA molecules that are longer than 200 nucleotides that control gene expression at the epigenetic, transcription, RNA splicing, and post-transcriptional levels.
LncRNA can regulate programmed cell death.
Targeting lncRNA may reduce or reinforce cellular damage caused by lncRNA.
Open questions
What is the mechanism of action of lncRNA on different types of programmed cell death?
How is lncRNA involved in the progression of programmed cell death?
How can we target lncRNAs to modulate programmed cell death mechanisms?
Background
Programmed cell death (PCD) is mediated by several cellular gene expression events, which are mainly divided into apoptosis, autophagy, necroptosis, and ferroptosis1. Caspase is a key factor mediating apoptosis, which can mediate exogenous and intrinsic pathways and thus lead to apoptosis2,3. When caspase activity is inhibited, death receptors (such as Tumor Necrosis Factor [TNF] Receptors [TNFRs]) are activated, followed by activation of receptor-interacting protein kinase 1 (RIPK1), RIPK3, and the mixed lineage kinase domain-like (MLKL) to form necrotic bodies that finally mediate necroptosis1,4,5. The main cause of ferroptosis is the accumulation of iron-dependent reactive oxygen species (ROS) and the consumption of plasma membrane polyunsaturated fatty acids6,7. Among them, major autophagy is the most important type of autophagy8. Under conditions such as starvation, hypoxia, and hormone signaling, cells form autophagosomes that combine with lysosomes, thereby degrading macromolecular substances and causing macroautophagy9,10.
PCD plays an important role in human disease and aging11,12. There are many factors that affect PCD processes, including altered protein levels and other macromolecular substance dysfunction, abnormal glucose levels, mitochondrial dysfunction, and oxidative stress13,14. Recent studies have found that many small molecules, such as lncRNAs, can also regulate PCD in addition to the aforementioned protein factors.
LncRNA is a special RNA type defined as transcripts over 200 nucleotides long, but is not translated into proteins15. LncRNAs are numerous and abundant, and were originally considered to be biologically non-functional transcriptional noise. Although lncRNA is not translated into protein, it is still considered a functional molecule that regulates gene expression at multiple levels, including the chromatin, transcriptional, and post-transcriptional levels. Indeed, lncRNA is involved in cell differentiation, cell cycle regulation, stem cell pluripotency, and maintenance of various biological processes16–19. LncRNAs can affect different apoptotic pathways. For instance, lncRNA overexpression can reduce the expression of membrane surface receptors by affecting extrinsic apoptosis pathway20,21. Autophagy not only promotes cell survival but also causes cell death in some conditions. LncRNA can activate autophagy by activating related enzymes22. In addition, lncRNAs can protect tumor cells from necroptosis by inhibiting the expression of some related proteins20. Some lncRNA types also act as competitive endogenous RNAs to prevent oxidation and inhibit ferroptosis20. Increasing evidence shows that lncRNAs are closely related to PCD, and the relationship between lncRNA and PCD is associated with the occurrence of heart disease, cancer cell apoptosis, and cell survival23,24. In this article, we summarize the role of lncRNAs in PCD to determine the relationship between lncRNAs and PCD.
The relationship between lncRNA and apoptosis
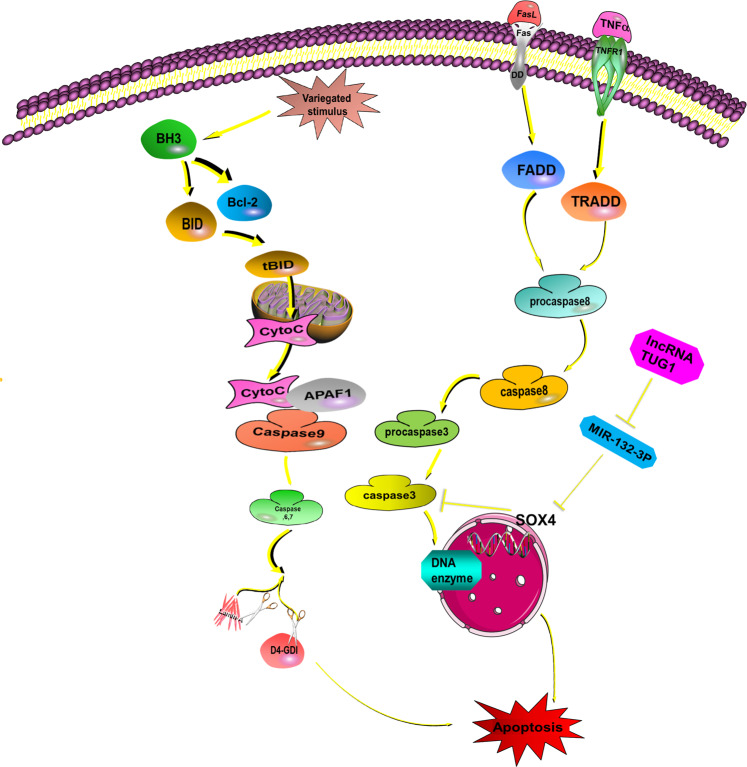
Apoptosis is initiated by many ligands, which regulate specific membrane death receptors (the extrinsic pathway) and/or the Bcl-2 protein family in mitochondria (the intrinsic pathway)25. The extrinsic pathway passes signals from outside the cell to the inside and is regulated by the death receptor TNFR, Fas/CD95, and TNF-related apoptosis-inducing ligand (TRAIL)26. Membrane death receptor activation causes procaspase-8 hydrolysis and activation to form activated caspase-8, which then activates downstream caspase-3. Subsequently, DNA fragmentation factor (DFF) subunit/CAD (DNase) is activated, leading to apoptosis27,28. In the intrinsic pathway, cytotoxic stimuli such as carcinogenic stress, chemotherapeutic agents, and developmental cues activate BH3-only family members and inhibit Bcl-2 proteins before survival, thus activating the pro-apoptotic effectors BAX and BAK, which finally destroy the outer mitochondrial membrane29. After the outer membrane of mitochondria is damaged, cytochrome C and ATF endonuclease are released30,31. Cytochrome C engages the apoptotic protease activating factor-1 (APAF1) and forms apoptosomes, which activate caspase-932. Afterward, caspase-9 activates caspase-3, caspase-6, and caspase-7, which contribute to the cleavage of other proteins and finally result in apoptosis1 (Fig. 1).
Fig. 1. Activation of the apoptotic pathway.
Apoptosis can be divided into the extrinsic pathway and intrinsic pathway. In the extrinsic pathway, the death receptor TNFR, Fas/CD95, and TRAIL combine with the relevant factors from extracellular, such as TNF, FasL. Then, the death receptor, procaspase-8, caspase-8, and caspase-3 are sequentially activated and regulate DNA fragmentation factor (DFF)/caspase-activated DNase (CAD) and cause apoptosis26,27,36. In the intrinsic pathway, cytotoxic stimuli such as carcinogenic stress, chemotherapeutic agents, and developmental cues activate BH3-only family members and inhibit Bcl-2 proteins, thus activating the pro-apoptotic effectors BAX and BAK, which finally destroy the outer mitochondrial membrane. This releases cytochrome C and ATF endonuclease. Cytochrome C engages APAF1 and forms the apoptosome, which activates caspase-9. Subsequently, caspase-9 activates caspase-3, caspase-6, and caspase-7, which contribute to the cleavage of other proteins and eventually promote apoptosis. Various lncRNAs are involved in this process. For instance, TUG1 can inhibit miR-132-3p expression, thereby increasing SOX4 expression. This decreases caspase-3 activity, eventually inhibiting osteosarcoma (OS) cell apoptosis. This indicates that knockdown of TUG1 could regulate the miR-132-3p–SOX4 axis in OS cells and promote apoptosis.
LncRNA can influence apoptosis in several ways. First, lncRNAs can directly or indirectly act on death receptors. We previously reported trophoblast invasion and trophoblast‐mediated VSMC loss in uterine spiral artery remodeling. In that study, we found that extravillous trophoblasts (EVTs) can generate TNF-α, Fas ligand (FasL), and TRAIL. These are important factors mediating EVT-induced VSMC loss, and the combination with VSMC membrane receptors results in VSMC apoptosis. We used lncRNA MEG3-overexpressing plasmid to transfect trophoblast HTR‐8/SVneo cells to increase MEG3 expression. qRT-PCR and Western blotting showed that MEG3 overexpression reduced TNF-α, FasL, and TRAIL expression. Next, VSMCs and transfected HTR-8/SVneo cells were co-cultured for 48 h, and the apoptosis rate of VSMCs was detected by flow cytometry. The results showed that compared with the negative control group, the apoptosis rate of VSMCs in the H‐MEG3 group was significantly reduced, indicating that MEG3 upregulation in HTR‐8/SVneo cells inhibited VSMC apoptosis. These results indicate that MEG3 might be a negative regulator of spiral artery remodeling by acting on the apoptosis-related death receptor, thereby suppressing EVT-mediated VSMC apoptosis20.
Second, lncRNA can act as competitive endogenous RNA (ceRNA) for miRNA by binding with a sequence at the miRNA 5′ end, thereby decreasing target mRNA expression and finally influencing cell apoptosis. A study of osteogenic sarcoma (OS) showed that the lncRNA, Taurine upregulates gene1 (TUG1), regulates disease development. Indeed, TUG1 is overexpressed in OS, potentially targeting miR-132-3p. Luciferase reporter gene analysis and qRT-PCR analysis showed that TUG1 directly binds at its 3′-terminus “-GACUGUUG-” sequence with the miR-132-3p 5′-terminus “-CUGACAA-” sequence, thereby inhibiting miR-132-3p expression, decreasing caspase-3 activity, and eventually inhibiting OS cell apoptosis. Moreover, miR-132-3p inhibitors significantly decreased the apoptosis rate and caspase-3 activity, indicating that miR-132-3p induces apoptosis. The sex-determined region Y-frame 4 (SOX4) is a known oncogene33,34 and a target of miR-132-3p in OS cells. Introducing SOX4 partially reverses the apoptosis-promoting effect of miR-132-3p. This indicates that TUG1 knockdown could regulate the miR-132-3p–SOX4 axis in OS cells and promote apoptosis21.
These studies indicate that lncRNAs can regulate apoptosis via various mechanisms35,36. For instance, Zhang et al. demonstrated that lncRNA TUG1 inhibits apoptosis by downregulating miR-29b37. After LPS treatment, the expression of apoptosis-inducing caspase 3 and cytochrome C in H9c2 cells increased. In addition, qRT-PCR and ELISA showed that the expression of inflammatory factors such as IL-6, IL-8, and TNF-α increased after LPS treatment. These results show that exposure to LPS induces H9c2 cells apoptosis and induces inflammation. Flow cytometry analysis showed that the apoptosis rate in PEx-Tug1-transfected H9c2 cells was significantly reduced following LPS treatment, while TUG1 inhibition significantly increased H9c2 cell apoptosis. With the decrease in apoptosis rate, TUG1 overexpression downregulated caspase 3 and cytochrome C expression, and inhibited IL-6, IL-8, and TNF-α expression and secretion, while TUG1 inhibition enhanced the levels of these cytokines. Overall, these results demonstrate that TUG1 has a protective effect against LPS-induced damage in H9c2 cells. In addition, TUG1 overexpression significantly downregulates miR-29b, while TUG1 inhibition upregulates miR-29b. Based on these results, there may be binding sites between TUG1 and miR-29b: the 5′end “-AACACTGA-” of TUG1-wt could bind to the 3′end “-UUGUGACU-” of miR-29b.
Furthermore, TUG1 also inhibits NF-κB signaling and the JAK/STAT pathway by downregulating miR-29b to induce inflammatory responses and apoptosis. CCK-8 assays showed that LPS exposure in H9c2 cells increases p65 and IκBα phosphorylation, while TUG1 overexpression inhibits NF-κB pathway activation. However, miR-29b mimics reverse TUG1 overexpression by reducing p-p65 and p-IκBα expression. Similar changes were observed in the JAK/STAT pathway. LPS stimulation increase s p-JAK and p-STAT3 expression, while TUG1 overexpression reduces their expression. In addition, miR-29b mimics block TUG1-mediated inhibition of the JAK/STAT pathway. These findings suggest that TUG1 overexpression inactivates the NF-κB and JAK/STAT pathways by downregulating miR-29b.
Similarly, a study of the lncRNA Highly Upregulated in Liver Cancer (HULC) showed that restoring HULC expression could rescue TNF-induced apoptosis, and that HULC regulates TNF-induced apoptosis by regulating miR-9 expression38. Moreover, A study of osteoarthritis demonstrated that knocking down the lncRNA reprogramming (ROR) can inhibit chondrocyte apoptosis through HIF1 (Hypoxia-inducible factors 1) and p5339.
In summary, lncRNAs can affect apoptosis in various ways, such as by acting on apoptosis-related receptors or by acting as ceRNAs.
The relationship between lncRNA and autophagy
Autophagy promotes cell survival, but can also cause cell death. Autophagy usually occurs in three forms: macroautophagy, microautophagy, and CMA8.
Macroautophagy
Macroautophagy is a conserved process in eukaryotic cells, and is the most studied form of autophagy. In this process, the phagocyte membrane envelops the cytoplasmic contents, building a double membrane structure called an autophagosome40. The autophagosome then fuses with a lysosome, which exposes the contents to lysosomal hydrolases. Next, the autophagosome endomembrane and enclosed macromolecular substances degrade and the substances are released into the cytoplasm via the lysosomal membrane osmotic enzyme and are used to synthesize new macromolecules.
In yeast cells, for example, autophagosomes are constructed at the phage assembly site (PAS) and are necessary for the whole process. The ATG (Autophagy Related) gene is involved in autophagosome formation, and when macroautophagy is induced, Atg proteins are recruited to promote autophagosome biogenesis41. The Atg1 protein complex plays an important role in recruiting other Atg proteins to the PAS. Several subunits of the complex are phosphoproteins, including Atg1 protein kinase, Atg13, Atg11, and Atg17–Atg31–Atg29 (sub) complexes. The integration process is as follows: First, by binding with Atg31, Atg29 correctly integrates into the Atg17–Atg31–Atg29 complex. Atg29 phosphorylation plays a key role in binding with other phosphoproteins42,43. Moreover, Atg29 directly interacts with Atg11. Eventually, Atg1 combines with Atg13 and Atg11, and then associates directly with Atg17 and Atg19. Thus, the Atg1 protein complex forms44. Then, the Atg1 protein complex combines and phosphorylates Atg9 on vesicles and then recruits PI3KC3–C1 (ATG34:ATG15:ATG6:ATG14) to the PAS (Fig. 2). Additionally, when the Atg12–atg5–atg16 complex moves to the PAS, Atg8 combines with phosphatidylethanolamine (PE) to form the Atg8–PE complex, which promotes autophagosome membrane elongation, autophagosome formation, and autolysosome formation45. Kotani et al. explain the initiation of membrane expansion in the formation of autophagosomes with the following model: amphiphilic spirals in the Atg2 and Atg18 C-terminal regions cooperate to bind Atg9 vesicles on the PAS, and the Atg2 N-terminal region binds to the endoplasmic reticulum (ER)46. These interactions lead to PAS–ER binding, resulting in membrane expansion and autophagosome formation.
Fig. 2. Autophagosome integration in yeast cells.
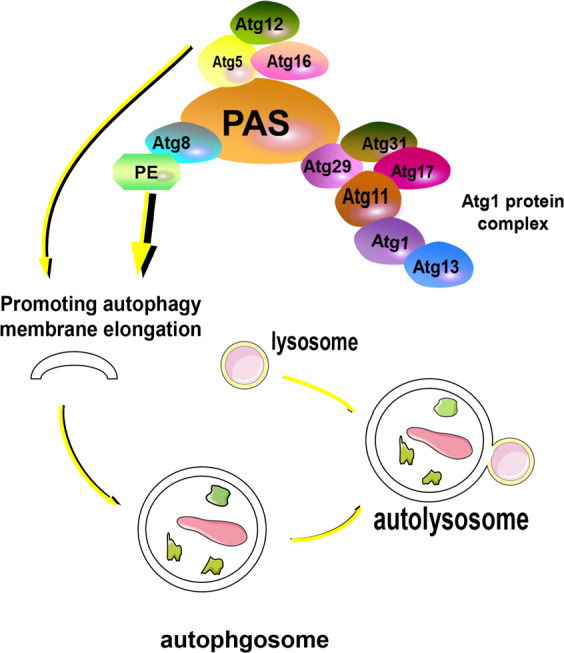
First, binding with Atg31 causes Atg29 to integrate into the Atg17–Atg31–Atg29 complex. Atg29 phosphorylation plays a key role in binding with other phosphoproteins. Moreover, Atg29 and Atg11 directly interact. Eventually, Atg1 combines with Atg13 and Atg11, with the latter directly associating with Atg17 and Atg19. In this way, the Atg1 protein complex takes shape. Moreover, the atg12–atg5–atg16 complex moves to the PAS and makes Atg8 combine with phosphatidylethanolamine (PE) to form the Atg8–PE complex, which promotes the elongation of the autophagosome membrane and forms autophagosomes and autolysosomes.
Microautophagy
Microautophagy is another type of autophagy that occurs via vacuolar phagocytosis, incorporation of organelle components (including organelles), and transport into the lysosome or vacuole system47,48. In yeast cells, microautophagy degrades the proteins and organelles in cells contained within lysosomes or vacuoles. That this degradation has a discriminant capacity, and even portions of the nucleus can be degraded in this way47,49.
Chaperone-mediated autophagy (CMA)
In contrast to other processes, in CMA, the substrate enters the lysosome lumen via the protein transport complex on the lysosome membrane. Additionally, this process directs targeted cytoplasmic proteins to the lysosome50. Moreover, CMA is selective, and the organelles, lipids, nucleic acids, or integral membrane proteins are not degraded51. CMA responds to stress responses such as hunger, lipid toxicity, protein toxicity, hypoxia, oxidative stress, or DNA damage52. By degrading specifically targeted cell proteins, CMA can regulate glucose and lipid metabolism, DNA repair, cellular reprogramming, and cellular responses to stress53.
There are few studies on microautophagy and CMA. Therefore, we will mainly discuss how lncRNA is involved in macroautophagy (hereinafter referred to as autophagy).
lncRNA MALAT1
A 2019 study showed that MALAT1 overexpression recruits Enhancer of Zeste 2 Polycomb repressive complex 2 subunit (EZH2) to the Tuberous sclerosis 2 (TSC2) promoter region, which upregulates H3K27me3 histones, resulting in the inhibition of autophagy54. Contrastingly, lncRNAs also promote the occurrence of autophagy in tumor cells55.
Another study showed that MALAT1 overexpression activates autophagy in colorectal cancer (CRC). The results showed that MALAT1 expression in CRC was very high compared to expression in adjacent normal tissues. MALAT1 expression was greatly reduced by si-MALAT1 transfection, while MALAT1 expression was increased by pcDNA-MALAT1. Western blotting showed that MALAT1 downregulation reduced LC3-II/LC3-I (light chain) expression and increased p62 (a protein provided by SQSTM1 gene) expression. However, MALAT1 upregulation has the opposite effect. This indicates that MALAT1 is related to autophagy activation. In the cells inactivated by MALAT1, caspase-3 expression was increased significantly. Similarly, MALAT1 upregulation resulted in decreased caspase-3 cleavage compared with the pcDNA transduction group. There was no difference in caspase-9 expression between the two groups. Flow cytometry showed that the apoptosis rate of the si-MALAT1 group was significantly higher than in the siRNA group. At the same time, MALAT1 upregulation reduced the apoptosis rate. In addition, MALAT1 upregulation was reduced by the autophagy inhibitor 3-mA. These data show that MALAT1 can increase cell proliferation and inhibit CRC cell apoptosis by activating autophagy.
Importantly, miR-101 and MALAT1 have complementary sequences, suggesting that miR-101 is a target of MALAT1. Compared with adjacent normal tissues, miR-101 expression in CRC was very high, and MALAT1 and miR-101 expression were negatively correlated. The results show that miR-101 overexpression inhibits the transformation of LC3-I to LC3-II and reduces cell proliferation, but increases p62/sqstm1 expression, apoptosis rate, and caspase-3 cleavage. However, co-expressing MALAT1 and miR-101 can eliminate the effect of miR-101 overexpression. This study confirmed that MALAT1 is overexpressed in CRC tissues and cell lines, and is positively correlated with LC3-II expression in CRC. However, introduction of miR-101 into CRC cells promoted cell proliferation and MALAT1-mediated autophagy and inhibited apoptosis56.
Autophagy activation of MALAT1 has been observed in tumors and in other diseases. For example, hypoxia can upregulate MALAT1 levels in endometrial cells in an HIF-1α-dependent manner during endometriosis, thereby activating autophagy. In addition, autophagy has a protective effect and can reduce apoptosis in endometriosis57.
lncRNA H19
The lncRNA H19 gene is located on human chromosome 11 and is a maternally-expressed gene. LncRNA H19 activates autophagy by regulating the dual-specificity phosphatase 5 (DUSP5)-extracellular signal-regulated kinases1/2 (ERK1/2) axis in neuroblastoma. First, a rat model of middle cerebral artery occlusion (MCAO) was established. TTC(Triphenyltetrazolium chloride)-stained brain sections showed that H19 expression during brain ischemia/reperfusion (I/R) is about 35 times higher than expression in control samples. To investigate the role of H19 in neurons, the participants established an oxygen-glucose deprivation/reoxygenation (OGD/R) model of cells simulating I/R injury using SH-SY5Y neuroblastoma cells. The results showed that 8/24 h OGD induced significantly higher lncRNA H19 expression compared to the normal control group. In contrast, lncRNA H19 expression was not significantly changed during 12 and 24 h OGD reperfusion. The expression of lncRNA H19 was upregulated by cerebral I/R in rats and by OGD/R in vitro. Decreased lncRNA H19 expression or autophagy activation could protect cell damage induced by OGD/R. LncRNA H19 upregulation can activate autophagy after OGD/R injury, while transfecting H19 siRNA can inhibit autophagy. However, when rapamycin (RAP), an autophagy inducer, was added, autophagy inhibition by H19 siRNA was activated. These data show that lncRNA H19 activates autophagy during cerebral I/R injury58.
Similarly, Cui et al. found that H19 can induce hypoxia-reoxygenation injury by regulating autophagy in liver cancer cells59. Additional studies show that lncRNA H19 overexpression promotes invasion and autophagy via the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) pathway in trophoblast cells60. Further, H19 decreases DNA methylation at the Beclin1 promoter region via the H19 S-adenosylhomocysteine hydrolase (SAHH)-DNA methyltransferase 3B (DNMT3B) axis, thereby enhancing autophagy61.
lncRNA MEG3
In a study in 2019, Gao et al. explored the regulation of MEG3 on morphine-induced autophagy in HT22 mouse hippocampal neuronal cells. First, HT22 cells were treated with 10 µM morphine, which significantly increased c-fos, p/t-ERK, and p/t-protein kinase C (PKC) protein expression. These results suggest that morphine can enhance the expression of orexinergic receptor 1 (OX1R) and c-fos in HT22 cells and activate the ERK and PKC pathways. In addition, MEG3 expression significantly increased after treating HT22 cells with 10 µM morphine. To investigate the effect of MEG3 upregulation on morphine-induced autophagy in HT22 cells, the cells were transfected with si-MEG3, which sharply decreased MEG3 expression. Compared with the morphine + si-negative control (NC) group, Beclin-1 and LC3-II/LC3-I levels in the morphine + si-MEG3 group decreased. These results suggest that MEG3 upregulation is involved in morphine-induced autophagy in HT22 cells. Subsequently, HT22 cells were incubated with SCH772984, an ERK pathway inhibitor, which significantly reduced morphine-induced Beclin-1 and LC3-II/LC3-I expression, indicating that the ERK pathway plays an important role in HT22 cell autophagy. In addition, compared with the morphine + si-MEG3 group, Beclin-1 and LC3-II/LC3-I expression in HT22 cells in the morphine + si-MEG3 + SCH772984 group were further reduced. These experiments indicate that MEG3 upregulation may be involved in the effect of morphine on autophagy in HT22 cells by promoting ERK pathway activation62.
To study the role of MEG3 in glioma, Zhao et al. used a pcDNA-MEG3 lentiviral vector transfected into U251 cells. qRT-PCR analysis illustrated that compared with the negative control group, MEG3 expression was significantly higher. Flow cytometry analysis illustrated that the cell apoptosis rate was significantly increased after MEG3 overexpression. Meanwhile, immunofluorescence LC3 autophagy assays showed that LC3 was significantly increased in the MEG3 expression group. These data indicate that MEG3 expression can activate glioma cell autophagy63. Other researchers also found that MEG3 expression significantly inhibits glioma cell proliferation and promotes apoptosis and autophagy in vitro. Furthermore, MEG3 levels are negatively correlated with glioma malignancy, and can be used as an independent prognostic factor for glioma63.
Other lncRNAs
In addition to the lncRNAs mentioned above, other lncRNAs play an important role in autophagy regulation. Many studies show that lncRNA plays a key role in regulating autophagy. lncRNA, as a ceRNA for miRNA, participates in the regulation of autophagy. For example, a previous study illustrated that downregulation of growth arrest-specific 5 (GAS5) regulates miR-23a expression, thereby influencing ATG3 and resulting in the inhibition of autophagy. Indeed, GAS5 overexpression inhibited mature miR-23a, but had no influence on pri-miR-23a (primary miR-23a) and pre-miR-23a (miR-23a precursor). Additionally, miR-23a can directly inhibit ATG3 expression in cells. Downregulating GAS5 inhibits the formation of LC3-II, ATG3, and the ATG5-ATG12 complex, thereby inhibiting autophagy. These data indicate that the GAS5/miR-23a/ATG3 axis may be a new regulatory network that could be helpful for understanding autophagy64.
A study of the lncRNA PVT1, in hepatic cellular cancer (HCC), showed that PVT1 lowers miR-365 and relieves miR-365-mediated ATG3 inhibition, resulting in autophagy65. In addition, long intergenic non-protein-coding RNA 665 (LINC00665) also plays a critical role in the development of liver cancer. Shan et al. found that LINC00665 regulates cell viability and autophagy through the miR-186-5p/mitogen-activated protein kinase kinase kinase kinase 3 (MAP4K3) axis in HCC66.
The relationship between lncRNA and necroptosis
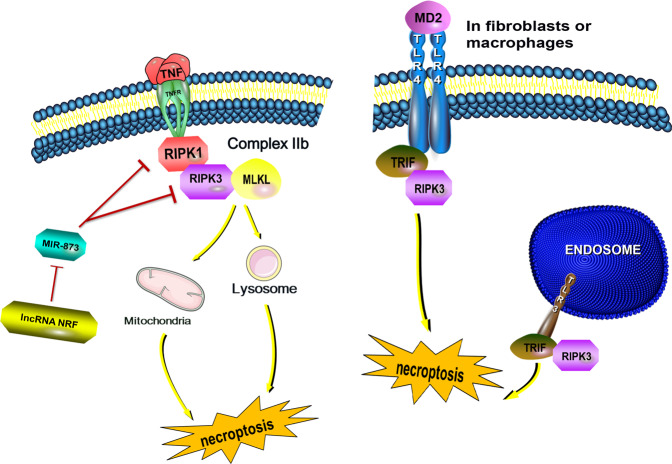
Necroptosis is a newly identified form of PCD, and is mediated by several cell signaling pathways. These reactions activate mixed lineage kinase-like (MLKL) and lead to necroptosis67. Necroptosis has morphological characteristics such as cell swelling, mitochondrial dysfunction, increased membrane permeability, and cytoplasmic content release. Activating TNF and toll-like receptor (TLR) 3/TLR4 can lead to necroptosis. The most widely studied activation mechanism is the TNF-inducible cell signal, which relies on RIPK1, RIPK3, and MLKL1,4,5. In TNF-activated cells, RIPK1 is activated, which causes complex IIb binding with RIPK3. MLKL is then phosphorylated by RIPK3 and is recruited into the necrosome through interaction with RIPK35,68,69. The necrosome targets organelles such as mitochondria and/or lysosomes, destroys the cell membrane, and eventually leads to necroptosis by cell lysis5,70. In fibroblasts and macrophages, TLR3 or TLR4 interacts with RIPK3 through TRIF, an integrin containing a TIR domain that induces interferon β interaction, to directly activate necroptosis. Importantly, this pathway is independent of the RIPK1 signaling pathway71 (Fig. 3).
Fig. 3. TNF-induced necroptosis.
Necroptosis is a newly-described form of programmed cell death. Activating MLKL plays an important role in this process. The most widely studied subtype involves the TNF-inducible cell signal. In TNF-activated cells, RIPK1 is activated, and then activated RIPK1 forms complex IIb with RIPK3. MLKL is then phosphorylated by RIPK3 and recruited into the necrosome through interaction with RIPK3. By targeting organelles such as mitochondria and/or lysosomes, the cell membrane is destroyed, eventually leading to necrosis by cell lysis. LncRNA can regulate necroptosis by influencing this pathway. For example, the lncRNA NRF can directly downregulate miR-873, thereby reducing RIPK1/RIPK3 inhibition by miR-837, finally promoting necroptosis. In fibroblasts and macrophages, TLR3 or TLR4 interacts with RIPK3 through Total Recordable Incident Frequency (TRIF) to directly activate necrosis, independent from the RIPK1 signaling pathway. Cell swelling, mitochondrial dysfunction, membrane permeability, and cytoplasmic content release are observed.
Few studies describe the role of lncRNA in necroptosis. Two studies mentioned a type of lncRNA called TRINGS (Tp53-regulated inhibitor of necrosis under glucose starvation) in human tumor cells and suggested that p53 directly upregulates TRINGS in the absence of glucose72,73. TRINGS binds STRAP (serine–threonine kinase receptor-associated protein) and inhibits STRAP-GSK3β-NF-κB necrotic signaling to protect tumor cells from cell death. This is a novel necroptosis pathway that is different from the traditional necrotic pathway (RIPK1-RIPK3-MLKL). Another study showed that lncRNA Linc00176 affects the cell cycle and liver cancer cell survival by regulating the expression of more than 200 genes, such as miR-9 and miR-185. In HCCs that express Linc00176, these miRNAs are released from Linc00176 and downregulate their target mRNAs, which induce necroptosis of liver cancer cells74.
In addition, lncRNA interacts with miRNA and affects the expression of other miRNA species. Experiments have confirmed that the lncRNA necrosis-related factor (NRF) targets miR-873 and RIPK1/RIPK3 to regulate cardiomyocyte necroptosis75. After RIPK1 and RIPK3 were knocked down in cardiomyocytes, the number of H2O2-induced necrotic cells decreased significantly. This indicates that RIPK1 and RIPK3 are related to H2O2-induced cardiomyocyte necroptosis. After transfecting cardiomyocytes with miR-873 or negative control, RIPK1 and RIPK3 expression were analyzed by Western blotting, which confirmed that miR-873 inhibits RIPK1 and RIPK3 expression in cardiomyocytes. Furthermore, a miR-873 binding site in the RIPK1 or RIPK3 3’ UTR region (-UUCCUG-) is identified, and there is no binding site in mutant miR-873. Luciferase reporter gene assays showed that miR-873 overexpression resulted in a decrease in luciferase activity. The introduction of mutations in miR-873 greatly reduced the inhibitory effect of miR-873. Thus, miR-873 can directly bind to RIPK1 and RIPK3, thereby inhibiting H2O2-induced necroptosis. Bioinformatic analysis showed that lncRNA NRF contains a large number of miR-873 binding sites. RNA pull-down experiments showed that NRF was pulled down by biotinylated wild-type miR-873, but mutations that disrupt the base pairing between NRF and miR-873 inhibited NRF pull-down, indicating that the recognition sequence is specific. In addition, we used a biotin-labeled NRF probe to detect the location of the interaction between NRF and miR-873. The results showed that miR-873 can be co-precipitated by NRF in the cytoplasm. These results show that NRF can directly bind to miR-873 and downregulate the level of miR-873. Further, miR-873 inhibits the translation of RIPK1/RIPK3 in cardiomyocytes and inhibits RIPK1/RIPK3-mediated necroptosis. These results show that lncRNA can be combined with related proteins in the PCD pathway or as ceRNA combined with related miRNAs to regulate necroptosis. Compared with autophagy and apoptosis, the interaction between lncRNAs and necroptosis remains to be further explored.
The relationship between lncRNA and ferroptosis
It is now recognized that ferroptosis is a non-apoptotic form of cell death. The main reason for death is the accumulation of iron-dependent ROS and the consumption of plasma membrane polyunsaturated fatty acids. The morphological characteristics of cellular ferroptosis show that the nucleus is normal, but the mitochondria are smaller than normal, the membrane density is increased, the outer membrane is ruptured, and mitochondria is shrunken or absent76,77.
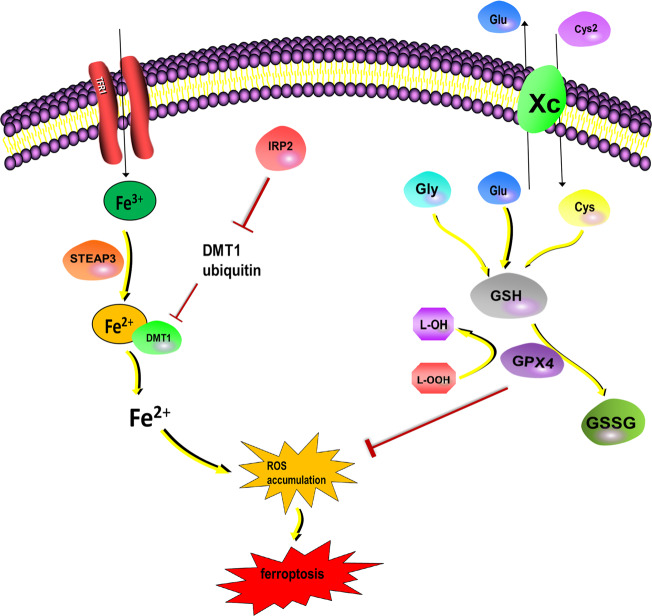
Cellular ferroptosis occurs roughly as follows: System Xc—is a cystine/glutamate antiporter, which transports intracellular glutamate (Glu) to the outside of the cell and transfers extracellular cystine (Cys2) into the cell. Cys2 is converted into cysteine (Cys) for the synthesis of glutathione (GSH)78. GSH peroxidase 4 (GPX4) converts GSH into oxidized glutathione (GSSG) and reduces cytotoxic lipid peroxide (L-OOH) to the corresponding alcohol (L-OH), which ultimately reduces the accumulation of ROS79. In addition, some studies show that p53 is an important tumor suppressor, exerting tumor-suppressive effects by inhibiting ferroptosis80 (Fig. 4).
Fig. 4. Cellular ferroptosis.
The related lipid peroxidation pathway mediated by GPX4 plays an important role in inhibiting ferroptosis. System Xc—is a cystine/glutamate antiporter that transports intracellular Glu to the outside of the cell and transfers extracellular Cys2 into the cell. Then, Cys2 is converted into Cys for the synthesis of GSH. GPX4 converts GSH into oxidized GSSG and reduces cytotoxic lipid peroxide (L-OOH) to the corresponding alcohol (L-OH), which ultimately reduces ROS accumulation. On the other hand, Fe3+ enters the cell and is reduced to Fe2+ by the iron oxidoreductase STEAP3. Then, DMT1 causes Fe2+ release from the endosome, leading to ROS accumulation, which in turn leads to lipid peroxidation and ferroptosis. IRP2 (IREB2) can inhibit the ubiquitination of TfR1 and divalent metal transporter 1 (DMT1) to increase cellular iron uptake and promote ferroptosis.
Circulating iron (Fe3+) enters the cell and is reduced to Fe2+ by the iron oxidoreductase STEAP3 (six-transmembrane epithelial antigen of the prostate 3). Next, Fe2+ is released from the endosome by divalent metal transporter 1 (DMT1) and is transported into the unstable iron pool in the cytoplasm, leading to ROS accumulation, which leads to lipid peroxidation and ferroptosis. Iron regulatory protein 2 (IRP2, also known as IREB2) can inhibit the ubiquitination of transferrin receptor 1 (TfR1) and DMT1 to upregulate cellular iron uptake and promote ferroptosis81. Recent studies showed that ferritin inhibitor 1 (FSP1) (previously known as flavoprotein apoptosis-inducing factor mitochondrial-related 2, AIFM2) is a non-GPX4-dependent ferroptosis inhibitor82–84.
Wang et al. found that the lncRNA LINC00336 inhibits ferroptosis in lung cancer by acting as a ceRNA85. First, the researchers used RSL3 (a ferroptosis activator) to treat A549 and SPC-A-1 lung cancer cells and observed that LINC00336 overexpression limits RSL3-induced cellular ferroptosis. Further, LINC00336 overexpression in A549 or SPC-A cells can resist ferroptosis caused by protein kinases in a dose- and time-dependent manner. In addition, LINC00336 overexpression significantly reduced intracellular Fe2+, lipid ROS, and intracellular mitochondrial superoxide concentrations. These results indicate that LINC00336 overexpression downregulates the occurrence of ferroptosis.
Regarding ferroptosis, several studies show that lncRNA LINC00336 inhibits ferroptosis of lung cancer cells by acting as a ceRNA. Additional experiments confirmed that LINC00336 overexpression significantly enhanced the expression of cystathionine β synthase (CBS), a ferroptosis marker, in A549 and SPC-A-1 cells, while knocking down LINC00336 significantly slowed down the growth of PC9 cell (a lung cancer cell line)85. The expression of CBS in cells indicates that LINC00336 can positively regulate CBS levels, thereby affecting ferroptosis. In addition, transfected MIR6852 can directly bind to LINC00336, and among the binding sites, the 3′-MIR6852 binding site on LINC00336 has a higher binding activity than the 5′-MIR6852 binding site. Finally, a lentivirus was used to introduce MIR6852 and spongy MIR6852 into SPC-A-1 cells. MIR6852 overexpression increased RSL3-induced ferroptosis, while spongy MIR6852 reduced RSL3-induce ferroptosis. These results show that MIR6852 increases cellular ferroptosis and increases the intracellular concentration of lipid ROS, iron, Fe2+, and mitochondrial superoxide. Additionally, MIR6852 directly binds to LINC0033 and inhibits CBS-mediated inhibition of ferroptosis, ultimately increasing overall cellular ferroptosis.
Regulation of the p53 tumor suppressor pathway by lncRNA is a topic of great interest. Mao et al. showed that the p53-related lncRNA P53RRA can directly interact with the functional domain of the signal protein in the cytoplasm by activating the p53 pathway to promote ferroptosis and act as a cancer suppressor86. In more detail, P53RRA combines with Ras GTPase-activating protein-binding protein 1 (G3BP1), using nucleotides 1 and 871 of P53RRA and the RRM interaction domain of G3BP1 (aa177-466). Another study showed that p53 plays a negative role in the regulation of iron oxidation. Spermidine/spermidine N1-acetyltransferase 1 (SAT1) is a transcriptional target of p5387. The expression of SAT1 induces lipid peroxidation and ferroptosis during ROS-induced stress.
In addition, the lncRNA P53RRA can act on the G3BP1 protein complex to promote p53 removal and regulate ferroptosis.
Conclusion
Recent studies on lncRNAs have shown that lncRNAs play an important role in regulating gene expression, and they also participate in the regulation of PCD, including autophagy, apoptosis, necroptosis, and ferroptosis. LncRNA can inhibit or promote PCD by directly or indirectly regulating protein complexes and microRNAs. Indeed, ceRNAs of mRNA are important regulatory pathways. In terms of clinical significance, lncRNAs can regulate PCD in cancer cells88–90.
Several studies have confirmed that lncRNAs affect cancer cell growth in cancer patients, which important for clinical treatment and cancer prognosis. LncRNA plays an important role in drug resistance in breast, gastric, and lung cancer. For instance, lncRNA MALAT1 can inhibit breast cancer metastasis90. MALAT1 overexpression is positively correlated with tumor progression and metastasis of various tumor types, including breast cancer. In addition, lncRNA LINC00673 regulates the proliferation, migration, invasion, and epithelial-mesenchymal transition of non-small cell lung cancer by inhibiting miR-150-5p91. In digestive system cancer, the lncRNA MIF-AS1 promotes cancer cell proliferation and reduces apoptosis92. The lncRNA UICLM (upregulated in colorectal cancer liver metastasis) regulates zinc finger E-box binding homeobox 2 (ZEB2) expression by acting as a ceRNA of microRNA-215, thereby promoting liver metastasis of colorectal cancer93. Cancer is an important public health concern. An in-depth understanding of lncRNAs will provide new targets for cancer treatment and improve clinical treatment of the disease.
Acknowledgements
This work was supported by grants from the Jiangxi Provincial Natural Science Foundation (No. 20202BABL206061), the Cultivation scientific research fund for the junior teachers of medicine in the NanChang University (PY201826), the National Natural Science Foundation of China (No. 81900861).
Conflict of interest
The authors declare no competing interests.
Footnotes
Edited by: Ana Janic.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nunes T, Bernardazzi C, de Souza HS. Cell death and inflammatory bowel diseases: apoptosis, necrosis, and autophagy in the intestinal epithelium. Biomed. Res. Int. 2014;2014:218493. doi: 10.1155/2014/218493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maes ME, Schlamp CL, Nickells RW. BAX to basics: How the BCL2 gene family controls the death of retinal ganglion cells. Prog. Retin. Eye Res. 2017;57:1. doi: 10.1016/j.preteyeres.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong RS. Apoptosis in cancer: from pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao M, et al. c-Jun N-terminal kinases differentially regulate TNF- and TLRs-mediated necroptosis through their kinase-dependent and -independent activities. Cell Death Dis. 2018;9:1140. doi: 10.1038/s41419-018-1189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan J, Amin P, Ofengeim D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 2019;20:19. doi: 10.1038/s41583-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell. Mol. Life Sci. 2016;73:2195. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Y, et al. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 8.Oshima M, Seki T, Kurauchi Y, Hisatsune A, Katsuki H. Reciprocal regulation of chaperone-mediated autophagy/microautophagy and exosome release. Biol. Pharm. Bull. 2019;42:1394. doi: 10.1248/bpb.b19-00316. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura S, Yoshimori T. New insights into autophagosome-lysosome fusion. J. Cell Sci. 2017;130:1209. doi: 10.1242/jcs.196352. [DOI] [PubMed] [Google Scholar]
- 10.Kriegenburg F, Ungermann C, Reggiori F. Coordination of autophagosome-lysosome fusion by Atg8 family members. Curr. Biol. 2018;28:R512. doi: 10.1016/j.cub.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 11.Daneva A, Gao Z, Van Durme M, Nowack MK. Functions and regulation of programmed cell death in plant development. Annu. Rev. Cell Dev. Biol. 2016;32:441. doi: 10.1146/annurev-cellbio-111315-124915. [DOI] [PubMed] [Google Scholar]
- 12.Godlewski M, Kobyliska A. Programmed cell death - strategy for maintenance cellular organisms homeostasis. Postepy Hig. Med. Dosw. 2016;70:1229. [PubMed] [Google Scholar]
- 13.Jeong SY, Seol DW. The role of mitochondria in apoptosis. BMB Rep. 2008;41:11. doi: 10.5483/BMBRep.2008.41.1.011. [DOI] [PubMed] [Google Scholar]
- 14.Garten A, et al. Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat. Rev. Endocrinol. 2015;11:535. doi: 10.1038/nrendo.2015.117. [DOI] [PubMed] [Google Scholar]
- 15.Qiao HP, Gao WS, Huo JX, Yang ZS. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac. J. Cancer Prev. 2013;14:1077. doi: 10.7314/APJCP.2013.14.2.1077. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin. Chim. Acta. 2018;485:229. doi: 10.1016/j.cca.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, et al. LncRNA: shedding light on mechanisms and opportunities in fibrosis and aging. Ageing Res. Rev. 2019;52:17. doi: 10.1016/j.arr.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Liao K, et al. The research progress of LncRNA involved in the regulation of inflammatory diseases. Mol. Immunol. 2018;101:182. doi: 10.1016/j.molimm.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 19.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao H, et al. LncRNA MEG3 inhibits trophoblast invasion and trophoblast-mediated VSMC loss in uterine spiral artery remodeling. Mol. Reprod. Dev. 2019;86:686. doi: 10.1002/mrd.23147. [DOI] [PubMed] [Google Scholar]
- 21.Li G, Liu K, Du X. Long non-coding RNA TUG1 promotes proliferation and inhibits apoptosis of osteosarcoma cells by sponging miR-132-3p and upregulating SOX4 expression. Yonsei Med. J. 2018;59:226. doi: 10.3349/ymj.2018.59.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong H, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, et al. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 2019;20:5758. doi: 10.3390/ijms20225758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhen Q, et al. LncRNA DANCR promotes lung cancer by sequestering miR-216a. Cancer Control. 2018;25:1146205113. doi: 10.1177/1073274818769849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coultas L, Strasser A. The role of the Bcl-2 protein family in cancer. Semin. Cancer Biol. 2003;13:115. doi: 10.1016/S1044-579X(02)00129-3. [DOI] [PubMed] [Google Scholar]
- 26.Dalleau S, Baradat M, Guraud F, Huc L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013;20:1615. doi: 10.1038/cdd.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan TJ, Han LH, Cong RS, Liang J. Caspase family proteases and apoptosis. Acta Biochim. Biophys. Sin. 2005;37:719. doi: 10.1111/j.1745-7270.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- 28.Fulda S. Caspase-8 in cancer biology and therapy. Cancer Lett. 2009;281:128. doi: 10.1016/j.canlet.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 29.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 30.Zhang M, Zheng J, Nussinov R, Ma B. Release of cytochrome C from Bax pores at the mitochondrial membrane. Sci. Rep. 2017;7:2635. doi: 10.1038/s41598-017-02825-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su CH, et al. Fisetin-induced apoptosis of human oral cancer SCC-4 cells through reactive oxygen species production, endoplasmic reticulum stress, caspase-, and mitochondria-dependent signaling pathways. Environ. Toxicol. 2017;32:1725. doi: 10.1002/tox.22396. [DOI] [PubMed] [Google Scholar]
- 32.Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 2008;9:532. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Yan L, Lu F. miR-363-3p inhibits osteosarcoma cell proliferation and invasion via targeting SOX4. Oncol. Res. 2019;27:157. doi: 10.3727/096504018X15190861873459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei X, Li L, Duan X. Long non-coding RNA ABHD11-AS1 promotes colorectal cancer development through regulation of miR-133a/SOX4 axis. Biosci. Rep. 2018;38:BSR20181386. doi: 10.1042/BSR20181386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pal S, Garg M, Pandey AK. Deciphering the mounting complexity of the p53 regulatory network in correlation to long non-coding RNAs (lncRNAs) in ovarian cancer. Cells. 2020;9:527. doi: 10.3390/cells9030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, et al. Long non-coding RNA FOXP4-AS1 is an unfavourable prognostic factor and regulates proliferation and apoptosis in colorectal cancer. Cell Prolif. 2017;50:e12312. doi: 10.1111/cpr.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, et al. Long non-coding RNA TUG1 inhibits apoptosis and inflammatory response in LPS-treated H9c2 cells by down-regulation of miR-29b. Biomed. Pharmacother. 2018;101:663. doi: 10.1016/j.biopha.2018.02.129. [DOI] [PubMed] [Google Scholar]
- 38.Ma Y, et al. Long noncoding RNA highly upregulated in liver cancer regulates the tumor necrosis factor-α-induced apoptosis in human vascular endothelial cells. DNA Cell Biol. 2016;35:296. doi: 10.1089/dna.2015.3203. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, et al. Long non-coding RNA reprogramming (lncRNA-ROR) regulates cell apoptosis and autophagy in chondrocytes. J. Cell. Biochem. 2018;119:8432. doi: 10.1002/jcb.27057. [DOI] [PubMed] [Google Scholar]
- 40.Yang S, Rosenwald A. Small GTPase proteins in macroautophagy. Small GTPases. 2018;9:409. doi: 10.1080/21541248.2016.1246280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Y, et al. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy. 2015;11:28. doi: 10.4161/15548627.2014.984267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Klionsky DJ. The Atg17-Atg31-Atg29 complex and Atg11 regulate autophagosome-vacuole fusion. Autophagy. 2016;12:894. doi: 10.1080/15548627.2016.1162364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao K, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc. Natl Acad. Sci. USA. 2013;110:E2875. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao K, Chew LH, Yip CK, Kionsky DJ. The role of Atg29 phosphorylation in PAS assembly. Autophagy. 2013;9:2178. doi: 10.4161/auto.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, Zhang L. Regulation of ATG and autophagy initiation. Adv. Exp. Med. Biol. 2019;1206:41. doi: 10.1007/978-981-15-0602-4_2. [DOI] [PubMed] [Google Scholar]
- 46.Kotani T, Kirisako H, Koizumi M, Ohsumi Y, Nakatogawa H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl Acad. Sci. USA. 2018;115:10363. doi: 10.1073/pnas.1806727115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oku M, et al. Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 2017;216:3263. doi: 10.1083/jcb.201611029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuck S. Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J. Cell Sci. 2020;133:jcs246322. doi: 10.1242/jcs.246322. [DOI] [PubMed] [Google Scholar]
- 49.Tekirdag K, Cuervo AM. Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone. J. Biol. Chem. 2018;293:5414. doi: 10.1074/jbc.R117.818237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galluzzi L, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirchner P, et al. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 2019;17:e3000301. doi: 10.1371/journal.pbio.3000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:365. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu H, et al. Long non-coding RNA MALAT1 enhances the apoptosis of cardiomyocytes through autophagy inhibition by regulating TSC2-mTOR signaling. Biol. Res. 2019;52:58. doi: 10.1186/s40659-019-0265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu J, Lu AD, Zhang LP, Zuo YX, Jia YP. [Study of clinical outcome and prognosis in pediatric core binding factor-acute myeloid leukemia] Zhonghua Xue Ye Xue Za Zhi. 2019;40:52. doi: 10.3760/cma.j.issn.0253-2727.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Si Y, et al. Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell. Mol. Biol. Lett. 2019;24:50. doi: 10.1186/s11658-019-0175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu H, et al. Long non-coding RNA MALAT1 mediates hypoxia-induced pro-survival autophagy of endometrial stromal cells in endometriosis. J. Cell. Mol. Med. 2019;23:439. doi: 10.1111/jcmm.13947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Cao B, Han D, Sun M, Feng J. Long non-coding RNA H19 induces cerebral ischemia reperfusion injury via activation of autophagy. Aging Dis. 2017;8:71. doi: 10.14336/AD.2016.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui C, Li Z, Wu D. The long non-coding RNA H19 induces hypoxia/reoxygenation injury by up-regulating autophagy in the hepatoma carcinoma cells. Biol. Res. 2019;52:32. doi: 10.1186/s40659-019-0239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu J, et al. Overexpression of long non-coding RNA H19 promotes invasion and autophagy via the PI3K/AKT/mTOR pathways in trophoblast cells. Biomed. Pharmacother. 2018;101:691. doi: 10.1016/j.biopha.2018.02.134. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 2019;12:81. doi: 10.1186/s13045-019-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao S, Li E, Gao H. Long non-coding RNA MEG3 attends to morphine-mediated autophagy of HT22 cells through modulating ERK pathway. Pharm. Biol. 2019;57:536. doi: 10.1080/13880209.2019.1651343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao H, et al. Long non-coding RNA MEG3 regulates proliferation, apoptosis, and autophagy and is associated with prognosis in glioma. J. Neurooncol. 2018;140:281. doi: 10.1007/s11060-018-2874-9. [DOI] [PubMed] [Google Scholar]
- 64.Li L, et al. Knockdown of long non-coding RNA GAS5 increases miR-23a by targeting ATG3 involved in autophagy and cell viability. Cell. Physiol. Biochem. 2018;48:1723. doi: 10.1159/000492300. [DOI] [PubMed] [Google Scholar]
- 65.Yang L, Peng X, Jin H, Liu J. Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene. 2019;697:94. doi: 10.1016/j.gene.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 66.Shan Y, Li P. Long intergenic non-protein coding RNA 665 regulates viability, apoptosis, and autophagy via the MiR-186-5p/MAP4K3 axis in hepatocellular carcinoma. Yonsei Med. J. 2019;60:842. doi: 10.3349/ymj.2019.60.9.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Z, Silke J, Hildebrand JM. Methods for studying TNF-mediated necroptosis in cultured cells. Methods Mol. Biol. 2018;1857:53. doi: 10.1007/978-1-4939-8754-2_5. [DOI] [PubMed] [Google Scholar]
- 68.Zhou W, Yuan J. Necroptosis in health and diseases. Semin. Cell Dev. Biol. 2014;35:14. doi: 10.1016/j.semcdb.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 69.Cai Z, et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 2014;16:55. doi: 10.1038/ncb2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giampietri C, Starace D, Petrungaro S, Filippini A, Ziparo E. Necroptosis: molecular signalling and translational implications. Int. J. Cell Biol. 2014;2014:490275. doi: 10.1155/2014/490275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaiser WJ, et al. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J. Biol. Chem. 2013;288:31268. doi: 10.1074/jbc.M113.462341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khan MR, Xiang S, Song Z, Wu M. The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. EMBO J. 2017;36:3483. doi: 10.15252/embj.201696239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan MR, Wu M, Liu G. Tumor-suppressive or tumor-supportive: for p53, that is the question. Mol. Cell. Oncol. 2018;5:e1408537. doi: 10.1080/23723556.2017.1408537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tran D, et al. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene. 2018;37:75. doi: 10.1038/onc.2017.312. [DOI] [PubMed] [Google Scholar]
- 75.Wang K, et al. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016;23:1394. doi: 10.1038/cdd.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doll S, Conrad M. Iron and ferroptosis: A still ill-defined liaison. IUBMB Life. 2017;69:423. doi: 10.1002/iub.1616. [DOI] [PubMed] [Google Scholar]
- 77.Imai H, Matsuoka M, Kumagai T, Sakamoto T, Koumura T. Lipid peroxidation-dependent cell death regulated by GPx4 and ferroptosis. Curr. Top. Microbiol. Immunol. 2017;403:143. doi: 10.1007/82_2016_508. [DOI] [PubMed] [Google Scholar]
- 78.Lu B, et al. The role of ferroptosis in cancer development and treatment response. Front. Pharmacol. 2017;8:992. doi: 10.3389/fphar.2017.00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li J, et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mou Y, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J. Hematol. Oncol. 2019;12:34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doll S, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 83.Bersuker K, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dai E, et al. AIFM2 blocks ferroptosis independent of ubiquinol metabolism. Biochem. Biophys. Res. Commun. 2020;523:966. doi: 10.1016/j.bbrc.2020.01.066. [DOI] [PubMed] [Google Scholar]
- 85.Wang M, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26:2329. doi: 10.1038/s41418-019-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mao C, et al. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78:3484. doi: 10.1158/0008-5472.CAN-17-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Song X, Long D. Nrf2 and ferroptosis: a new research direction for neurodegenerative diseases. Front. Neurosci. 2020;14:267. doi: 10.3389/fnins.2020.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.shen X, et al. Long non-coding RNA activated by TGF-α expression in cancer prognosis: a meta-analysis. Int. J. Surg. 2018;58:37. doi: 10.1016/j.ijsu.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 89.DAI SP, JIN J, LI WM. Diagnostic efficacy of long non-coding RNA in lung cancer: a systematic review and meta-analysis. Postgrad. Med. J. 2018;94:578. doi: 10.1136/postgradmedj-2018-135862. [DOI] [PubMed] [Google Scholar]
- 90.Li N, Yang M, Shi K, Li W. Long non-coding RNA HOXA11-AS in human cancer: a meta-analysis. Clin. Chim. Acta. 2017;474:165. doi: 10.1016/j.cca.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 91.Lu W, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol. Cancer. 2017;16:118. doi: 10.1186/s12943-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li L, et al. Long non-coding RNA MIF-AS1 promotes gastric cancer cell proliferation and reduces apoptosis to upregulate NDUFA4. Cancer Sci. 2018;109:3714. doi: 10.1111/cas.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen DL, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836. doi: 10.7150/thno.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]