Abstract
IL-36, which belongs to the IL-1 superfamily, is increasingly linked to neutrophilic inflammation. Here, we combined in vivo and in vitro approaches using primary mouse and human cells, as well as, acute and chronic mouse models of lung inflammation to provide mechanistic insight into the intercellular signaling pathways and mechanisms through which IL-36 promotes lung inflammation. IL-36 receptor deficient mice exposed to cigarette smoke or cigarette smoke and H1N1 influenza virus had attenuated lung inflammation compared with wild-type controls. We identified neutrophils as a source of IL-36 and show that IL-36 is a key upstream amplifier of lung inflammation by promoting activation of neutrophils, macrophages and fibroblasts through cooperation with GM-CSF and the viral mimic poly(I:C). Our data implicate IL-36, independent of other IL-1 family members, as a key upstream amplifier of neutrophilic lung inflammation, providing a rationale for targeting IL-36 to improve treatment of a variety of neutrophilic lung diseases.
Subject terms: Immunology, Chronic inflammation
Carolin Koss et al. show that IL-36 produced from neutrophils is a key amplifier of lung inflammation in mice by activating neutrophils, macrophages and fibroblasts in cooperation with GM-CSF and the viral mimic poly(I:C). This study suggests that targeting IL-36 may improve the treatment of neutrophilic lung diseases.
Introduction
The recently described interleukin (IL)-1 family cytokines IL-36α, IL-36β, and IL-36γ1–3 are emerging as contributors to acute and chronic tissue inflammation in human disease, particularly in the neutrophilic skin disease psoriasis4–6. In addition, experimental animal models of skin inflammation, arthritis, and intestinal inflammation further implicate IL-36 cytokines as important inflammatory mediators7–11. In the lung, IL-36 has been suggested to be important in the pathogenesis of experimental bacterial and viral pneumonia in mice7,12. IL-36 cytokines signal via a heterodimeric receptor comprised of an IL-36 receptor (IL-36R) chain and the IL-1 receptor accessory protein (IL-1RAP) that is also shared with the IL-1 and IL-33 receptors13,14. IL-36 cytokines can be expressed by skin epithelial cells9,11, which correlates with increased mRNA and protein concentrations of IL-36 cytokines in human psoriatic skin lesions and experimental models of psoriasis-like diseases in mice, including acanthosis and hyperkeratosis4,5,8–11,15. A common finding in diseases where IL-36 cytokines play a role is the presence of neutrophils8. Like other IL-1 family cytokines, IL-36 cytokines are produced as precursor proteins whose bioactivity is 1000-fold increased after proteolytic processing16,17. Typically, activation of IL-36 cytokines is associated with increased expression of multiple proteases released by neutrophils, such as neutrophil-derived cathepsin G, elastase, and proteinase-318. Consistent with the link between IL-36 cytokines and neutrophils, IL-36 cytokines have been strongly linked to the pathogenesis of generalized pustular psoriasis (GPP) and hidradenitis suppurativa, diseases in which neutrophils are a hallmark feature19. Importantly, blocking IL-36 receptor with a monoclonal antibody markedly reversed the skin lesions in human subjects with GPP20. IL-36 signaling is also activated in intestinal inflammation such as inflammatory bowel disease and experimental colitis21,22. Furthermore, IL-36 also plays an important role in the joint synovium of patients with rheumatoid arthritis23,24. These studies have extended IL-36 and IL-36R expression to include fibroblasts and macrophages25–28, and some reports have also suggested IL-36 expression by lung epithelial cells29,30.
Thus, there is a rationale for considering IL-36 cytokines as important contributors to the pathogenesis of lung diseases, specifically those that are characterized by the accumulation of neutrophils, such as severe non-T2 asthma, chronic obstructive pulmonary disease (COPD), acute respiratory distress syndrome, and cystic fibrosis31. Associative data have linked IL-36 cytokine(s) to neutrophilic inflammation in multiple diseased tissues, yet the mechanisms by which IL-36 drives pathology remain elusive. Here, we used both wild-type (WT) and knockout (KO) in vitro and in vivo murine experimental approaches together with primary human cells to deconvolute the network of stimuli and responses that orchestrate neutrophilic lung disease. Here, we provide the mechanistic rationale by which targeting IL-36 may alleviate our most burdensome respiratory diseases.
Results
IL-36 in neutrophilic lung inflammation
We exposed WT and IL-36 receptor KO mice (Il36r−/−) to cigarette smoke (CS) for 3 weeks. Exposure of WT mice to CS resulted in significantly increased bronchoalveolar lavage (BAL) neutrophil numbers (4.5 × 105 neutrophils/mL). In contrast, alveolar macrophage (AM) numbers did not change after exposure to CS. In Il36r−/− CS-exposed mice (1.7 × 105 neutrophils/mL), we observed a 62% reduction in BAL neutrophil numbers relative to WT mice. In addition, myeloperoxidase (as an indicator of neutrophil activation) concentrations in the BAL of Il36r−/− CS-exposed mice were significantly reduced (to concentrations observed in mice exposed to room air (RA)) relative to CS-exposed WT mice (Fig. 1a). These experiments indicated that IL-36 receptor (encoded by Il1rl2, herein designated as Il36r) signaling was an upstream driver of neutrophil recruitment and activation in mice exposed to CS smoke.
Fig. 1. IL-36γ is an upstream inflammatory driver in mouse neutrophils and alveolar macrophages.
a Neutrophil and macrophage counts and myeloperoxidase concentration in the bronchoalveolar lavage fluid (BALF) from room air (RA) exposed (WT n = 9; Il36r−/− n = 6) and 3-week cigarette smoke (CS) exposed mice (WT n = 10; Il36r−/− n = 10). b–d Neutrophil and macrophage counts, CXCL1, IL-1α, IL-1β, and GM-CSF protein concentrations in BALF from untreated (n = 6) and IL-36γ-exposed mice (intratracheal instillation) (n = 7). e, f Cxcl1, Il1a, Il1b, and Il36g mRNA expression in either e naive mouse alveolar macrophages (pooled n = 15 mice) and stimulated in vitro with no cytokines (−), IL-36γ, or IL-1α/IL-1β or f in mouse bone marrow-derived neutrophils (from n = 4 mice) in vitro stimulated with no cytokines (−), IL-36γ, GM-CSF, or IL-36γ+GM-CSF. g Il36r and Il1r mRNA expression in mouse bone marrow-derived neutrophils (from n = 4 mice) in vitro stimulated with no cytokines (−), IL-36γ, GM-CSF, and IL-36γ+GM-CSF. a, e, f *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs all other groups by one-way ANOVA and Tukey’s correction. b, c, d, g **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs untreated by t test. e AMs were pooled from 15 mice, data are presented as (mean ± SEM) of technical triplicates.
To further determine whether IL-36 cytokines acted as upstream drivers of neutrophil recruitment in the lung, we instilled IL-36γ (as one representative IL36 family cytokine previously described in the literature as a potent pro-inflammatory stimulus)32 intratracheally (i.t.) into naive mice for 4 h (in order to examine early upstream events before the emergence of secondary effects). In contrast to AM numbers, IL-36γ significantly increased BAL neutrophil numbers (Fig. 1b) to amounts comparable to those observed in CS-exposed mice (Fig. 1a). Additionally, C-X-C chemokine ligand 1 (CXCL1; a key neutrophil chemoattractant protein) concentrations were increased in the BAL relative to phosphate-buffered saline (PBS)-exposed mice (Fig. 1c). IL-36γ also significantly increased concentrations of IL-1α, IL-1β, and granulocyte macrophages colony-stimulating factor (GM-CSF; cytokines typically generated by neutrophils, macrophages, or the epithelium33) in the BAL relative to PBS-exposed mice (Fig. 1d). In addition, relative Cxcl1 and Il6 mRNA amounts were increased within the cell pellet recovered from the BAL relative to PBS exposure (Suppl. Fig. 1a).
We next determined whether IL-36 cytokines directly promoted pro-inflammatory activation of AMs and neutrophils. Murine AMs obtained from BAL of naive mice were cultured in the presence of GM-CSF (to maintain normal AM function34,35) and exposed to IL-36 cytokines for 24 h, after which Il1a and Il1b mRNA expression was determined. Both Il1a (19-fold) and Il1b (2-fold) expression was significantly increased relative to that in unstimulated AMs (Fig. 1e). Moreover, relative to unstimulated AMs, IL-36 cytokine-stimulated AMs also had significantly increased mRNA expression for Il36g (4-fold) and Cxcl1 (30-fold), indicating AMs as a source of CXCL1 and IL-36γ in the alveolar compartment. Importantly, a combination of IL-1α and IL-1β did not induce transcription of Il1a, Il1b, or Cxcl1 and only mildly increased Il-36g in AMs (Fig. 1e). We also examined the effect of other lung-associated inflammatory mediators by exposing AMs to lipopolysaccharide (LPS) or its canonical downstream cytokine tumor necrosis factor (TNF)-α. Exposing AMs to TNF-α did not stimulate mRNA expression for Il36g or Cxcl1 in AMs (Suppl. Fig. 1b), while the fold induction of mRNA encoding Cxcl1, Il36g, Il1b, Il1a, and Il6 after stimulating AMs with LPS was similar to that observed in response to IL-36 cytokines, indicating IL-36 cytokines as a potent pro-inflammatory stimulus on AMs (Suppl. Fig. 1c).
Considering that AMs were cultured in GM-CSF, we stimulated bone marrow-derived neutrophils from naive mice in vitro with IL-36γ alone or in combination with GM-CSF. Combining GM-CSF with IL-36γ resulted in significantly increased mRNA expression for Il36g (4-fold), Cxcl1 (10-fold), Il1a (600-fold), and Il1b (24-fold) relative to IL-36γ alone or GM-CSF alone when compared to unstimulated neutrophils (Fig. 1f). GM-CSF was capable of stimulating Il36g expression in mouse neutrophils, suggesting a further mechanism by which IL-36γ can be induced. As was observed for AMs, IL-1α and IL-1β stimulation of neutrophils failed to induce gene expression for these cytokines, even in combination with GM-CSF (Suppl. Fig. 1d). These findings prompted us to determine whether GM-CSF stimulation increased mRNA expression of the Il36r in neutrophils. Indeed, GM-CSF exposure resulted in an ~10-fold increase in Il36r mRNA expression in neutrophils, while no change in the expression of Il1r1 was observed (Fig. 1g). GM-CSF also significantly induced the mRNA expression of the IL-1 receptor antagonist (Il1rn), whereas the mRNA expression of the IL-36 receptor antagonist (Il36rn) remained below the detection limit (Suppl. Fig. 1d). Thus, GM-CSF might skew the responsiveness of neutrophils toward IL-36 relative to IL-1 through upregulation of the IL-36R.
We also determined whether IL-36γ alone or in combination with GM-CSF would increase the transcription of CXCL1, IL1A, IL1B, and IL36G in human neutrophils. In contrast to mouse neutrophils, GM-CSF and not IL-36γ induced mRNA expression for CXCL1 (2-fold), IL1A (69-fold), IL1B (9-fold), and IL-36G (4-fold) relative to unstimulated neutrophils (Fig. 2a). Combining IL-36γ stimulation with GM-CSF did not result in any further increase in the mRNA expression for these cytokines (Fig. 2a). Furthermore, IL-36 did not increase IL36R gene expression in human neutrophils relative to untreated cells (Fig. 2b). However, exposing human neutrophils to GM-CSF resulted in a significant increase of IL36R (encoded by IL1RL2 herein designated as Il36R) mRNA (Fig. 2b). IL1R mRNA was not induced after any of these stimulations (Fig. 2b). Finally, we exposed human lung epithelial cells to IL-36γ and found significantly increased protein secretion and mRNA expression of CXCL1, IL36G, GM-CSF, IL1A, and IL1B (Fig. 2c, d).
Fig. 2. IL-36γ is an upstream inflammatory driver in human neutrophils and small airway epithelial cells.
a, b CXCL1, IL1A, IL1B, and IL36G mRNA expression in human peripheral blood-derived neutrophils (from n = 4 donors) in vitro stimulated with no cytokines (−), IL-36γ, GM-CSF, or IL-36γ+GM-CSF. b IL36R and IL1R mRNA expression in human peripheral blood-derived neutrophils (from n = 4 donors) neutrophils in vitro stimulated with no cytokines (−), IL-36γ, GM-CSF, and IL-36γ+GM-CSF. c, d CXCL1, GM-CSF, and IL1α protein concentrations and CXCL1, IL36G, GM-CSF, IL1A, and IL1B mRNA expression in small airway epithelial cells (SAEC) stimulated with either (−) or IL-36αβγ. a, b *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, vs all other groups by one-way ANOVA and Tukey’s correction. c, d *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs untreated by t test.
Together, these findings indicated that in the alveolar compartment IL-36γ can act as an upstream inflammatory driver to promote neutrophil recruitment and production of pro-inflammatory IL-1 family cytokines in neutrophils and macrophages either alone or in combination with GM-CSF.
IL-36 acts as an upstream inflammatory driver of macrophages and fibroblasts
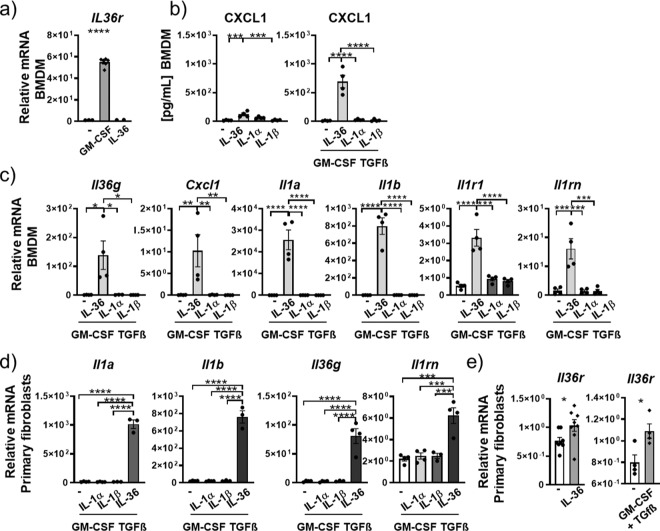
We conditioned bone marrow-derived macrophages (BMDMs) from naive mice toward a putative lung interstitial phenotype by exposing them to transforming growth factor (TGF)-β36,37 and GM-CSF34,35,38 in combination with IL-36 cytokines or IL-1α or IL-1β for 24 h. GM-CSF induced a 55-fold increase in mRNA for Il36r in BMDMs (Fig. 3a) and CXCL1 protein release after IL-36 cytokine stimulation (77-fold) was about 10-fold higher in GMCSF/TGF-β conditioned macrophages relative to that observed in unconditioned BMDMs (Fig. 3b). We therefore continued using GM-CSF/TGF-β conditioned BMDMs. We detected a 140-fold increase in Il36g mRNA expression, a 10-fold increase in Cxcl1 mRNA expression, 4495-fold increase for Il1a mRNA expression, and a 235-fold increase for Il1b mRNA expression in TGF-β/GM-CSF conditioned BMDMs stimulated with IL-36 relative to unstimulated BMDMs (Fig. 3c). In contrast, BMDMs not conditioned in GM-CSF/TGF-β had a reduced responsiveness to IL-36 cytokines with Il36g (6-fold), Cxcl1 (1.4-fold), Il1a (1.7-fold), and Il1b (3.4-fold) only slightly increased after IL-36 cytokine stimulation (Suppl. Fig. 2a). As we observed in AMs and neutrophils in Fig. 1, stimulation with a combination of IL-1α and IL-1β did not affect mRNA expression for Cxcl1, Il1b, and Il1a relative to untreated BMDMs (Fig. 3c and Suppl. Fig. 2a). Additionally, mouse BMDMs upregulated mRNA expression of Il1r1 and Il1rn in response to IL-36γ stimulation, while Il36rn mRNA expression was undetected (Fig. 3c and Suppl. Fig. 2a).
Fig. 3. IL-36γ is an upstream amplifier in mouse macrophages and fibroblasts.
a Relative mRNA amounts of Il36r in naive mouse bone marrow-derived macrophages (BMDMs, n = 6) in response to no stimulation (−) or stimulation with GM-CSF and IL-36γ. b CXCL1 protein concentrations in supernatant of BMDMs (n = 4) after no stimulation (−), or stimulation with IL-36αβγ. IL-1α, IL-1β alone, or in combination with GM-CSF and TGFβ c, d Relative mRNA amounts of Il36g, Cxcl1, Il1a, Il1b, Il1r1, Il1rn, and Il36rn in BMDMs (n = 4) (c) and primary mouse fibroblasts (n = 4) (d) after no stimulation (−) or stimulation with IL-36αβγ, IL-1α, IL-1β alone, or in combination with GM-CSF and TGF-β. e Relative mRNA amounts in primary mouse fibroblasts (n = 4) of Il36r after no stimulation (−) or stimulation with IL-36αβγ or GM-CSF and TGF-β. Shown are the mean values ± SEM of biological replicates. a–d *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs all other groups by one-way ANOVA and Tukey’s correction. e *P ≤ 0.05, vs untreated by t test.
In human monocyte-derived macrophages (MDMs) from healthy volunteer donors, IL36R mRNA was not induced by IL-36 cytokines or by GM-CSF stimulation (Fig. 4a). Stimulation with IL-36 in combination with GM-CSF significantly increased relative mRNA amounts of IL1A (53-fold) and IL1B (30-fold) (Fig. 4a). Stimulation of human MDMs with IL-36γ also promoted mRNA expression for CXCL1 (Suppl. Fig. 3a). However, MDMs did not exhibit a significant increase in IL36G mRNA expression after IL-36 stimulation relative to untreated MDMs, which was only slightly increased (1.3-fold) after stimulation with GM-CSF. The combination of IL-36 cytokines and GM-CSF significantly increased the IL36G expression relative to GM-CSF alone (Fig. 4a).
Fig. 4. IL-36γ is an upstream amplifier in human macrophages and fibroblasts.
a Relative mRNA amounts in human monocyte-derived macrophages (MDM) (depicted are mean values ± SME of technical triplicates from one representative of four experiments) of IL-36G, IL-36R, IL1A, and IL1B and b IL-36γ, IL-36R, IL1A, IL1B, GMCSF, and CXCL1 in primary human lung fibroblasts (n = 4) after no stimulation (−) or stimulation with IL-36αβγ, GM-CSF alone, or with the combination of IL-36αβγ and GM-CSF. Shown are the mean values ± SEM of biological replicates. a, b *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs all other groups by one-way ANOVA and Tukey’s correction.
When primary mouse fibroblasts obtained from the lungs of naive mice were exposed in vitro to IL-36γ or IL-1α and IL-1β stimulation for 24 h, we observed significantly increased mRNA expression for Il1a (284-fold), Il1b (337-fold), Cxcl1 (151-fold), Il36g (50-fold), Il1r1 (1.3-fold), Il1Rn (5-fold), and Il36a (22-fold) in response to IL-36γ relative to unexposed cells (Suppl. Fig. 2b). Stimulation with IL-1α and IL-1β did not result in any detectable increases in mRNA expression for these genes and Il36b and Il36rn were also not expressed in response to any of the stimulation conditions (Suppl. Fig. 2b). Conditioning fibroblasts with GM-CSF and TGF-β resulted in an additional increase in mRNA for Il1a (1015-fold), Il1b (761-fold), Il36g (81-fold), and Il1rn (6.2-fold) when exposed to IL-36 cytokines (Fig. 3d). Analogous to the findings in mouse neutrophils and mouse BMDMs, GM-CSF also increased mRNA expression for the Il36r in primary lung fibroblasts, albeit with a lower fold induction. IL-36 also slightly increased the Il36r in primary lung fibroblasts (Fig. 3e).
When primary human lung fibroblasts (from otherwise healthy donors) were stimulated with IL-36 cytokines, GM-CSF, or the combination thereof, we observed increased mRNA expression for IL36G, IL36R, IL1A, IL1B, GM-CSF, and CXCL1 after IL-36 stimulation relative to untreated fibroblasts (Fig. 4b).
Inflammatory conditions in the lung also promote tissue remodeling39, in which matrix metalloproteinase 9 (MMP9) has been shown to be an important mediator40. MMP9 protein and mRNA expression were increased in both primary lung fibroblasts and BMDMs after stimulation with IL-36 cytokines (Suppl. Fig. 2c).
In summary, these findings highlight the difference between IL-1α/β and IL-36 activation and pinpoint IL-36 as an upstream amplifier of human and mouse macrophages and fibroblasts.
IL-36 is important in neutrophilic lung inflammation in vivo
Thus far, the data suggested that IL-36 signaling was important for neutrophil recruitment and activation both in vivo (Fig. 1a–c) and in vitro (Figs. 1 and 3). We therefore predicted that IL-36 would play an important role in a lung inflammatory condition associated with high neutrophil numbers. To test this hypothesis, we advanced our CS smoke model depicted in Fig. 1 to either CS for 1, 2, or 3 weeks or to CS (for 2 weeks) followed by exposure to H1N1 influenza virus for 48 h (CS + H1N1). Neutrophil numbers in recovered BAL were 1.8 × 105/mL after 1 week CS exposure, 8 × 104/mL after 2 weeks of CS exposure, 3.6 × 105/mL after 3 weeks of CS exposure, 8.2 × 105/mL after H1N1, and 1.05 × 106/ mL after CS + H1N1 (Fig. 5a, c). Importantly, consistent with the higher numbers of neutrophils in CS + H1N1 mice relative to CS mice, CS + H1N1-exposed mice exhibited significantly higher increases in Il36g mRNA in the BAL pellet (1759-fold increased relative to RA) compared to CS-exposed mice (40-fold relative to RA) (Fig. 5b). In addition, as indicators of lung inflammation, we found a significant increase of IL-1β, CXCL1, and IL-6 protein in lung tissue of CS + H1N1-exposed mice relative to CS-exposed mice (Fig. 5d). These findings showed that this model was suitable to generate lung inflammatory conditions with high neutrophil recruitment to the alveolar space, which also correlated with marked Il36g expression.
Fig. 5. IL-36γ is critical in neutrophilic lung inflammation.
a Neutrophil counts in BALF from room air (RA) (n = 18) exposed or from 1-week (n = 9), 2-week (n = 9), and 3-week (n = 8) cigarette smoke (CS) exposed mice and from H1N1(4 days post treatment) exposed mice (n = 8) and from mice exposed to 2 weeks CS followed by 48 h of H1N1 exposure (n = 8). b Relative mRNA amounts of IL-36g in the cellular BAL pellet from room air (RA) and 2 week cigarette smoke (CS)-exposed mice (n—4). c, d Neutrophil numbers in BALF and IL1β, CXCL1, and IL-6 protein concentrations in lung homogenate of room air-exposed mice (RA; n = 4), 2-week CS-exposed mice (n = 4–8), and 2-week CS-exposed mice challenged with H1N1 for 48 h (CS + H1N1) (n = 4–8). e Neutrophil numbers in BALF and relative mRNA amounts of IL-36g and Cxcl1 in lung homogenate from room air-exposed (RA, n = 5) and 2-week CS-exposed mice challenged with H1N1 for 48 h (n = 7–8) exposed WT and Il36r−/− mice. f, g IL1β, CXCL1, IL6, and TNF protein concentrations in BALF and in lung homogenate from room air-exposed (RA, n = 5) and 2-week CS-exposed mice challenged with H1N1 for 48 h (n = 7–8) exposed WT and Il36r−/− mice. Depicted are mean values ± SEM of biological replicates. a, c–g *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs all other groups by one-way ANOVA and Tukey’s correction. b *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs untreated by t test.
We therefore utilized this model to determine the contribution of IL-36 cytokine signaling to lung inflammation in vivo. Il36r−/− mice exposed to CS + H1N1 exhibited a significant decrease (~72%) of BAL neutrophil numbers relative to that observed in CS + H1N1-exposed WT mice (Fig. 5e). Il36r−/− mice exposed to CS + H1N1 also exhibited significantly decreased mRNA expression for Cxcl1 in the lung homogenate relative to CS + H1N1-exposed WT mice (Fig. 5e). Furthermore, Il36r−/− mice exposed to CS + H1N1 also exhibited significantly decreased mRNA expression for Il36g in the lung homogenate (Fig. 5e). Finally, IL-1β, CXCL1, IL6, and TNF-α protein concentrations were also decreased in CS + H1N1-exposed Il36r−/− mice in both BAL fluid (Fig. 5f) and lung homogenate (Fig. 5g and Suppl. Fig. 4a) relative to CS + H1N1-exposed WT.
We next used mice with genetic deficiency in IL-1RAP (the common receptor chain for the receptors for IL-1α, IL1-β, IL-33, and IL-362) to define the effect of IL-36R signaling (reflecting the contribution of IL-36) within the IL-1 family in this model. CS + H1N1-exposed Il1rap−/− exhibited a significant reduction in the expression of markers for lung inflammation (Suppl. Fig. 4b). Specifically, we observed a 61% reduction in BAL neutrophil numbers relative to WT mice exposed to CS + H1N1. In addition, IL-1β, CXCL1, and IL6 protein concentrations in lung homogenate were significantly reduced in CS + H1N1-exposed Il1rap−/− mice relative to CS + H1N1-exposed WT mice (Suppl. Fig. 4b). Importantly, the percentage of reduction in BAL neutrophil numbers was comparable between Il1rap−/− and Il36r−/− CS + H1N1-exposed mice (Suppl. Fig. 4c). While Il1rap−/− CS + H1N1-exposed mice exhibited an 81% reduction for CXCL1 and an 80% reduction for IL-1β protein concentrations in lung homogenate, Il36r−/− CS + H1N1-exposed mice exhibited a 47% reduction for CXCL1 and 44% reduction for IL-1β. Notably, the percentage of reduction in IL-6 protein amounts in the lung homogenate was equal between Il1rap−/− (45%) and Il36r−/− (48%) CS + H1N1-exposed mice (Suppl. Fig. 4c). These findings indicated that, within the IL-1 family of cytokines that signal through IL-1RAP, IL-36 cytokines contribute to a large degree to the lung inflammatory response in CS + H1N1 mice.
IL-36 cooperates with polyinosinic-polycytidylic acid (poly(I:C)) on macrophages and fibroblasts
Based on the observed attenuated inflammation in Il36r−/− mice in response to CS + H1N1, we next employed in vivo approaches in which we used the Toll-like receptor 3 (TLR3) agonist poly(I:C) as a viral mimetic and focused on measuring neutrophils, GM-CSF, CXCL1, IL-36, and MMP9 cytokine expression in the BAL of mice. Mice exposed to i.t. poly(I:C) had increased neutrophils (but no increases in macrophages) in the BAL, 12 and 24 h post poly(I:C) exposure relative to PBS-exposed mice (Suppl. Fig. 5a). In addition, we found significantly increased protein concentrations for CXCL1 (at 4 and 24 h), GM-CSF (at 24 h), and MMP9 (at 24 h) in BAL, as well as increased Il36g mRNA in the BAL pellet (at 12 h) relative to PBS-exposed mice (Suppl. Fig. 5a, b).
We next exposed AMs obtained by BAL and bone marrow-derived neutrophils from naive mice to poly(I:C) in vitro. Poly(I:C) induced small but significant increases in mRNA expression for Il36g (2-fold), Cxcl1 (2.3-fold), Il1a (1.16–fold), Il1b (2.17-fold), and Il36r (1.11-fold) in AMs relative to untreated controls (Suppl. Fig. 5c). In mouse bone marrow-derived neutrophils, poly(I:C) alone failed to induce increased mRNA expression for Il36g, Cxcl1 Il1a, and Il1b (Fig. 6a). Combining poly(I:C) stimulation with IL-36γ stimulation resulted in numerical increased mRNA expression for Il1b but had similar effects on Cxcl1 and trended toward increased expression of Il1a (Fig. 6a).
Fig. 6. IL-36γ cooperates with poly(I:C) on mouse macrophages and fibroblasts.
a Relative mRNA amounts of Il36g, Cxcl1, Il1a, and Il1b in mouse neutrophils (n = 2–6) unstimulated with (−) or stimulated with IL-36αβγ, poly(I:C), or the combination of IL-36αβγ and poly(I:C) (depicted are mean values ± SEM of biological replicates). b Relative mRNA amounts of Il36g, Cxcl1, Il1a, Il1b, and Gmcsf in BMDMs (n = 4) and primary mouse fibroblasts (n = 4) (depicted are mean values ± SEM of biological replicates) unstimulated (−) or stimulated with IL-36αβγ, poly(I:C), or the combination of IL-36αβγ and poly(I:C). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs all other groups by one-way ANOVA and Tukey’s correction.
Combining IL-36 cytokines with poly(I:C) resulted in significantly increased relative mRNA amounts in BMDMs and primary mouse fibroblast for Il36g, Cxcl1, Il1a, Il1b, and Gmcsf (Fig. 6b) and increased protein concentrations for CXCL1, IL-36γ (pro-form), GM-CSF, and MMP9 relative to the individual stimulations compared to untreated cells (Suppl. Fig. 5d, f). Interestingly, as observed for AMs (Suppl. Fig. 5c), poly(I:C) promoted mRNA expression for the Il36r on BMDMs (Suppl. Fig. 5e). Primary mouse lung fibroblasts were comparable to BMDMs (Fig. 6b). Of note, poly(I:C) alone failed to induce Il36g, Cxcl1, Il1a, Il1b, Gmcsf, and Il36r mRNA in primary lung fibroblasts, highlighting the requirement for cooperating with IL-36 in activating fibroblasts (Fig. 6b and Suppl. Fig. 5e, f). The combination of poly(I:C) and IL-36 (7.5-fold) resulted in a 50% reduced Ifnb1 mRNA expression in BMDMs relative to poly(I:C) (15-fold) stimulation alone. IL-36 alone did not induce Infb1 expression in BMDMs (Suppl. Fig. 5g). Neither poly(I:C) nor IL-36 stimulation resulted in increased Infb1 mRNA expression in fibroblasts (Suppl. Fig. 5g).
Similar to mouse fibroblasts, human lung fibroblasts stimulated with the combination of poly(I:C) and IL-36 cytokines exhibited a significant increase in CXCL1, IL1A, IL1B, and GM-CSF mRNA relative to untreated controls (Fig. 7a). Neither poly(I:C) nor IL-36 cytokine exposure promoted IL36G mRNA expression in human fibroblasts (Fig. 7a).
Fig. 7. IL-36γ cooperates with poly(I:C) on human macrophages and fibroblasts.
a, b Relative mRNA amounts of IL-36g, CXCLl1, IL1A, IL1B, and GM-CSF in human fibroblasts and human MDMs (depicted are mean values ± SME of technical triplicates from one representative of three to four experiments) unstimulated (−) or stimulated with IL-36αβγ, poly(I:C), or the combination of IL-36αβγ, and poly(I:C). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs all other groups by one-way ANOVA and Tukey’s correction.
In human MDMs, IL-36 cytokine stimulation resulted in significant increase of CXCLl1, IL1A, and IL1B mRNA expression relative to untreated or poly(I:C) alone and was not changed by combining IL-36 cytokines with poly(I:C) (Fig. 7b). Similar to the fibroblast data, IL36G mRNA expression in MDMs was not increased after IL-36 cytokine, poly(I:C), or IL-36 cytokine + poly(I:C) stimulation (Fig. 7b).
Human basal lung epithelial cells exposed to poly(I:C) exhibited significant and marked increases in protein concentrations of GM-CSF (63-fold), CXCL1 (2.1-fold), and IL-36γ (71-fold) and mRNA expression of GM-CSF (7-fold), CXCL1 (4-fold), IL36G (364-fold), IL1A (186-fold), and IL1B (49-fold) (Fig. 8a, b). LPS, used as an additional TLR agonist did not induce mRNA increases of these cytokines. (Fig. 8a, b). IL-1α protein concentration was not increased after poly(I:C) or LPS stimulation relative to untreated control and IL-1β was only detected after poly(I:C) stimulation (Fig. 8a).
Fig. 8. Small airway epithelial cells as the main cellular source of GMCSF.
a GM-CSF, CXCL1, IL-36γ, IL-1β, and IL-1α protein concentrations and GM-CSF, CXCL1, IL36G, IL1A, and IL1B mRNA expression of lung epithelial basal cells from healthy donor stimulated with poly(I:C) and LPS (depicted are mean values ± SEM of technical triplicates from one representative of three experiments). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 vs all other groups by one-way ANOVA and Tukey’s correction.
These data indicated that poyl(I:C) increased the production of pro-inflammatory cytokines in lung epithelial cells and cooperated with IL-36 to further amplify pro-inflammatory pathways.
Neutrophils are a source of IL-36γ in acute lung injury
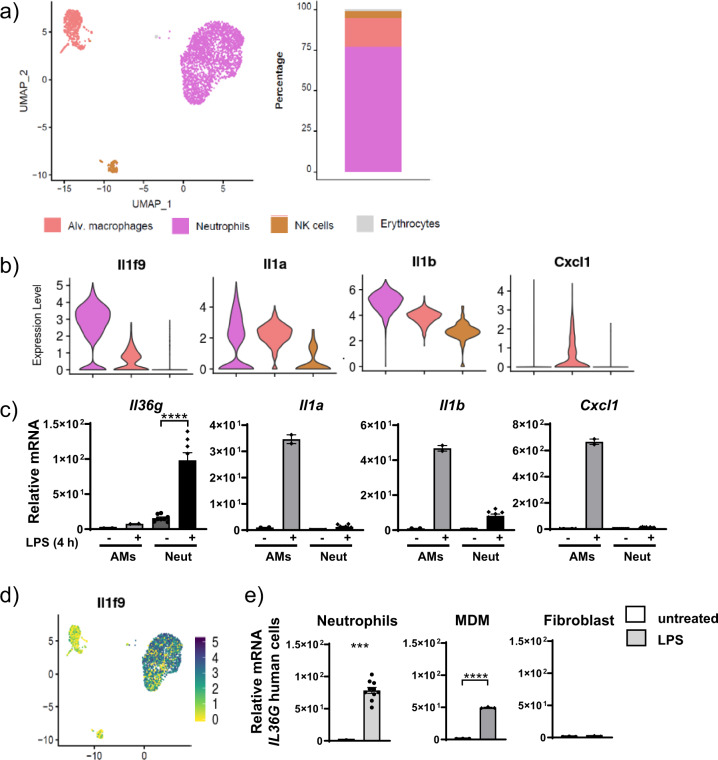
To determine neutrophils as a source of IL-36 not only in a lung inflammation model generated by CS or CS and virus, we also examined a lung inflammation model simulating exposure to bacterial stimuli. We therefore exposed mice to aerosolized LPS for 4 h before proceeding with single-cell RNA sequencing (scRNA-seq) analysis of the BAL cells. We chose 4 h exposure as it was determined to be the earliest time point with maximal neutrophil numbers in the BAL fluid (2.12 × 106 neutrophils/mL) in a pilot experiment (Suppl. Fig. 6a). After LPS challenge, the majority of the sequenced BAL cells represented neutrophils, which accounted for 77% of all BAL cells, while AMs represented only 18% of the BAL cell population (Fig. 9a). Additionally, we identified natural killer cells (4%) and few erythrocytes (1%) based on their reported characteristic gene expression profiles41 (Fig. 9a and Suppl. Fig. 6b, c). We then determined the relative transcriptional expression of Il36a, Il36b, and Il36g in comparison to Il1a and Il1b in BAL neutrophils and macrophages. High-resolution scRNA-seq analysis allowed us to identify neutrophils as the main source of Il36g. Il36g was detected in 84% of the neutrophils and its transcript levels were 10.5-fold increased relative to those detected in AMs (Fig. 9b, d). In contrast, Il1b was strongly expressed by both neutrophils and AMs. Nevertheless, Il1b was still increased 3.6-fold in neutrophils relative to AM while Il1a was detected in both populations, whereas the overall expression was significantly (1.5-fold) increased in neutrophils relative to AMs (Fig. 9b). In contrast to Il36g, Il36a was only detected in very few single cells (while Il36b was not detected in any cell type (Suppl. Fig. 6d)). Next, we investigated the expression of Cxcl1 across the different cell types. Cxcl1 expression was predominantly restricted to AMs and increased 1.9-fold compared to neutrophils (Fig. 9b). We next determined mRNA expression for Il36a,b,g and Il1a,b as well as Cxcl1 by quantitative polymerase chain reaction (qPCR) in mouse bone marrow-derived neutrophils and mouse AMs from naive mice in response to in vitro stimulation with LPS for 4 h to closely reflect the in vivo stimulation protocol. Consistent with the scRNA-seq data, we found that neutrophils were more responsive to LPS stimulation in terms of Il36g expression (48-fold increased) compared to the expression in AMs (8-fold increased) (Fig. 9c). In addition, induction of Cxcl1 mRNA expression upon LPS stimulation was restricted to AM and not induced in stimulated neutrophils (Fig. 9c). Il1a and Il1b were induced in both cell types after LPS stimulation but more predominantly in AMs. AMs had increased Il36a (690-fold) mRNA expression after LPS stimulation and Il36b was undetectable (Suppl. Fig. 6e). Furthermore, IL-36α and IL-36β mRNA expression remained below the detection limit for neutrophils.
Fig. 9. Neutrophils are a source of IL-36γ in acute lung injury.
a UMAP representation of the cell populations identified in BAL from LPS-exposed mice. Cell identity is indicated by the color code. Relative frequency of the cell types is shown in the bar plot using the same color code. b Violin plots display normalized expression levels of Il36g (Il1f9), Il1a, Il1b, and Cxcl1 across the different cell populations. c Relative mRNA amounts of the same genes used in b after 4 h in vitro LPS stimulation of naive mouse alveolar macrophages (AM) and naive mouse bone marrow-derived neutrophils; depicted are mean values ± SEM of biological duplicates (each biological duplicate represents pooled AMs from 4 mice) and n = 8 from the neutrophils analyzed by one-way ANOVA. d Visualization of normalized IL-36g (Il1f9) expression levels per single cell. e mRNA expression of IL-36g after LPS stimulation in vitro of human neutrophils, MDMs (depicted are mean values ± SME of technical triplicates from one representative of three to four experiments) and human fibroblasts n = 4. Data represents the depicted mean ± SEM of biological replicates; ***P ≤ 0.001, ****P ≤ 0.0001 vs untreated by t test.
Human neutrophils and human MDMs exhibited a 100- and 200-fold increase in IL36G mRNA expression after LPS stimulation relative to unstimulated cells. Human fibroblasts exhibited a 2.5-fold IL36G mRNA induction after LPS stimulation relative to untreated controls (Fig. 9e).
A more detailed analysis of the neutrophil clusters in the scRNA-seq analysis revealed two subpopulations that we designated N1 and N2 (Suppl. Fig. 6f). To differentiate the subclusters, we did a pathway analysis. Subcluster N2 showed increased IL-1 family and IFN pathways, whereas subcluster N1 exhibited increased remodeling pathways (Suppl. Fig. 6g). Finally, increased differential expression of MMP9 was observed to be restricted to subcluster N1, suggesting that these neutrophils could promote tissue remodeling (Suppl. Fig. 6h).
A hypothetical model of how cells and mediators interplay with IL-36 to promote neutrophilic lung inflammation is depicted in Fig. 10.
Fig. 10. Proposed signaling pathways involving IL-36 as an upstream amplifier in neutrophilic lung inflammation.
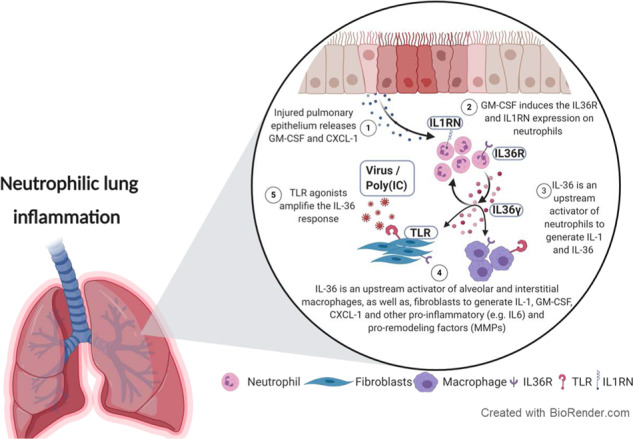
1 Chronic (e.g., cigarette smoke) or acute (H1N1 or poly(I:C) injury to the lung epithelium promotes release of CXCL1 and GMCSF. 2 CXCL1 promotes neutrophil recruitment while GM-CSF upregulates the IL-36R and IL1RN by neutrophils and promotes IL-36γ expression. 3 IL-36γ activates neutrophils to generate IL1 and IL36. 4 IL-36γ subsequently activates alveolar and interstitial macrophages and fibroblasts to generate IL-1, CXCL1, GM-CSF, MMPs, and more IL-36. GM-CSF also upregulates the lL36R on macrophages and on fibroblasts and sensitizes cells to IL-36. 5 Poly(I:C) as a TLR agonist cooperates with IL-36γ in amplifying activation of macrophages and fibroblasts. Note that IL-36RN is not induced while IL1RN is induced, shifting the balance toward heightened responsiveness to IL-36.
Discussion
Here, we combined chronic and acute lung inflammation mouse models induced by CS, viral (poly(I:C)), and bacterial (LPS) components with additional in vivo and in vitro approaches using primary mouse and human cells to provide mechanistic insight that identifies IL-36 within the IL-1 family of cytokines as an early upstream innate immune driver and amplifier of acute and chronic lung inflammation, particularly when associated with neutrophils. Mechanistically, lung neutrophils were identified as both a cellular source and target of IL-36 and IL-36 promoted the pro-inflammatory activation of lung macrophages, fibroblasts, and epithelial cells. IL-36 stimulated feed-forward signaling that resulted in the generation of IL-36 and IL-1 in fibroblasts and macrophages. Moreover, IL-36 cooperated with viral analog poly(I:C) and with GM-CSF to enhance the pro-inflammatory responses of macrophages and fibroblasts.
There is substantial evidence associating the pro-inflammatory role of IL-36 with the presence of neutrophils in mouse models of skin inflammation and also in human skin diseases8,42, such as GPP or hidradenitis suppurativa, where blockade of IL-36 signaling can greatly reduce disease symptoms20. In addition, IL-36 has been described to be increased in patients with COPD43, and in COPD, neutrophils are correlated with the severity of the symptoms and with microbial exacerbation, while high neutrophil numbers are correlated with poor prognosis and disease progression44,45. Previous reports also showed GM-CSF release after pulmonary infection and the importance of GM-CSF in inflammatory processes in the lung has been reported, including in COPD46,47. Finally, previous reports had suggested an important role for IL-36 in mouse models of bacterial and viral pneumonia7,12. Yet, no studies had investigated the interplay between IL-36, GM-CSF, and neutrophils in chronic lung inflammation or in lung inflammation in which acute virus challenge is superimposed on chronic injury, a reductionist mechanistic model for human lung disease secondary to smoking and virus infection.
Using a genetic approach employing mice with Il36r deficiency, we showed that IL-36 signaling was a critical component of the pro-inflammatory response in both CS- and CS + HN1N-exposed mice, demonstrating a role for IL-36 in both low-grade chronic lung inflammation (CS model) as well as high-grade acute microbial lung inflammation super-imposed on pre-existing chronic injury, such as during acute viral exacerbations in COPD (CS + H1N1 model). Specifically, Il36r−/− mice had significantly attenuated lung neutrophil influx and reduced alveolar IL-1β, CXCL1, IL-6, and TNF-α concentrations. Making use of IL1rap−/− mice, we isolated the contribution of IL-36 cytokines within the IL-1 family of cytokines in this model and showed that the phenotype of the Il1rap−/− mice was largely replicated in Il36r−/− mice. These data solidify IL-36 as a key upstream pro-inflammatory driver among the IL-1 family of cytokines that signal through IL-1RAP. Importantly, genetic ablation of IL-36 signaling was sufficient to attenuate recruitment of neutrophils to the same degree as seen in Il1rap−/− mice, while the protein expression of IL-1β and CXCL1 was more attenuated in the Il1rap−/− mice. These findings can be interpreted that blocking IL-36R allows for maintaining at least part of the IL-1 innate-mediated immune response necessary to maintain some level of immune defense, while it abrogates the acute inflammatory and tissue destructive effects of IL-36, which superimpose on the underlying inflammation and exacerbate immune pathology and substantially compromise organ function in the face of acute pathogen challenge. The findings that Il36g expression within the alveolar immune cell compartment was restricted to neutrophils in LPS-challenged mice and that stimulation of mouse AMs, mouse neutrophils, and human neutrophils with LPS in vitro increased IL-36γ production demonstrate that neutrophils can also be an important source of IL-36γ in humans and mice in neutrophilic inflammation associated with bacterial infection. Thus, we propose that IL-36R blockade in patients with chronic lung disease prone to microbial exacerbations will alleviate symptoms while preserving sufficient innate immune signaling mediated by IL-1α and IL-1β to conserve host defense.
We also demonstrate that IL-36γ promoted generation of IL-36γ itself and was upstream of both IL-1α and IL-1β in neutrophils, fibroblasts, and macrophages. IL-36γ was also capable of inducing CXCL1 in vivo and in a variety of cell types in vitro, including lung macrophages, lung fibroblasts, neutrophils, and lung epithelial cells. Notably, CXCL1, as well as IL-1α and IL-1β, expression in macrophages was restricted to IL-36γ stimulation and not observed with the other IL-1 family cytokines IL-1α and IL-1β, placing an important role on IL-36 in an upstream amplification loop for the production of CXCL1 and IL-1 family cytokines in a variety of innate immune cells. Neutrophils themselves also responded to IL-36 stimulation with CXLC1 production, highlighting a feed-forward loop between IL-36, CXCL1, and neutrophils. Thus, the IL-36γ-triggered pro-inflammatory mediator output broadly facilitates feed-forward amplification of the inflammatory response by engaging a variety of cells to increase neutrophil recruitment (CXCL1) and sensitization to IL-36γ or IL-1.
Our findings furthermore demonstrate that, downstream of neutrophils, activated IL-36 cytokines subsequently cooperate with additional pro-inflammatory signals, such as GM-CSF or poly(I:C)/viruses as an upstream driver of IL-1, GM-CSF, and CXCL1, further amplifying the pro-inflammatory interplay between neutrophils, IL-36, IL-1, and GM-CSF. Moreover, mice exposed to poly(I:C) had increased neutrophils and protein concentrations of GM-CSF and CXCL1 in the BAL. Notably, poly(IC) did not induce GM-CSF or CXCL1 in macrophages or neutrophils, making the lung epithelium the most likely source of GM-CSF and CXCL1 in vivo in response to poly(I:C). Therefore, poly(I:C) can promote IL-36 generation indirectly through epithelial-derived cytokines such as GM-CSF, which acts on neutrophils to produce IL-36. Another important finding of our study was that IL-36 also cooperated with poly(I:C) to directly increase the expression of Il36g, Cxcl1, Il1a, Il1b, and Gmcsf in macrophages and fibroblasts. Thus poly(I:C) can also promote IL-36 signaling. Previous reports had suggested the cooperation between TLR signaling and cytokine signaling with IL-36 in promoting inflammation32 (Fig. 6).
We also identified human lung epithelial cells as a source of IL-36γ in response to poly(I:C) but not LPS exposure (Fig. 8). There is literature evidence for IL-36γ expression in the lung epithelium in patients with lung disease31,48. Furthermore, IL-36γ is prominently expressed in keratinocytes of the skin9,11, particularly in response to bacterial challenge or the presence of neutrophils8,49. Therefore, the lung epithelium might be an additional source of IL-36γ in the face of viral exacerbation, while the LPS-mediated IL-36 generation in neutrophils described here suggests neutrophils as a major source of IL-36 in the face of bacterial exacerbation.
In human neutrophils, IL36G mRNA was increased in response to IL-1 rather than IL-36γ. Therefore, it is tempting to speculate that in humans with underlying chronic lung inflammation the increased levels of IL-150 induce IL-36 signaling to promote tissue inflammation. The finding that IL-36R expression in murine neutrophils and human neutrophils was increased by GM-CSF and the observation that combining GM-CSF with IL-36 (mouse) or IL-1 (human) amplified the production of IL-36 suggests that GM-CSF tailors responsiveness of mouse and human cells toward IL-36 and IL-1, respectively. This suggests a tightly regulated and context-dependent restriction of IL-1 family cytokine generation and signaling. Context-dependent activity of IL-36 has been described previously by showing that the activity of IL-36 cytokines is ~1000-fold increased after extracellular processing by neutrophil-derived proteases, enabling IL-36 activity in the presence of neutrophils16. Furthermore, it was recently reported that neutrophil extracellular traps facilitate generation of IL-1 family members, particularly IL-3651. Thus neutrophils are important for both generating and activating IL-36, placing neutrophils as an early upstream pro-inflammatory event in the IL-36 signaling cascade.
Here, we made an additional observation that GM-CSF was an inducer of IL-36R in mouse and human neutrophils. Intriguingly, GM-CSF promoted the expression of IL-36R but not the IL-1 receptor on macrophages and fibroblasts. There is evidence for a relationship between GM-CSF and neutrophils by the fact that humans treated with GM-CSF had increased numbers of neutrophils52. Another study demonstrated that LPS-exposed GM-CSF−/− mice had reduced infiltration of neutrophils compared to the WT control53.
We also observed that IL-36γ- or IL-1-stimulated cells increased the expression of the IL-1RN but not the IL-36RN, suggesting unrestrained IL-36γ signaling while limiting IL-1 signaling in these conditions. Intriguingly, severe neutrophilic skin diseases in humans are associated with polymorphisms in IL-36RN genes54,55.
Finally, inflammatory conditions in the lung are associated with tissue remodeling. Therefore, we used fibroblasts as structural target cells reported to produce MMP9 and examined MMP9 production as a surrogate for an important remodeling factor after IL-36 cytokine stimulation56,57. Consistent with previous reports, IL-36 was indeed capable of inducing MMP9 expression in primary lung fibroblasts57, involving IL-36 signaling in pro-fibrotic fibroblast activity21,56.
In conclusion, we have used several mouse models to reflect important triggers of lung inflammation that bear physiological relevance in humans with COPD, such as CS, CS + H1N1, poly(I:C), and LPS, allowing us to mechanistically investigate the interplay between IL-36 and neutrophilic lung inflammation. We have shown that injured epithelium released GM-CSF, which increased IL-36 expression in the alveolar compartment. IL-36 subsequently activated alveolar and interstitial macrophages, as well as fibroblasts, to generate IL-1, CXCL1, GM-CSF, MMPs, and IL-36 (Fig. 10). Our study thus pinpoints IL-36 as a key upstream pro-inflammatory driver and amplifier of neutrophilic lung inflammation and provides a mechanistic explanation for the link between IL-36 and neutrophilic inflammation that had been observed previously in human diseases and in animal models19,29,57–59 (Fig. 10). Our data therefore provide an experimental rationale for exploring the therapeutic potential of IL-36 signaling blockade to attenuate pro-inflammatory events in human lung disease with a significant contribution of neutrophils, such as asthma and COPD.
Methods
Animals
Female and male WT C57BL/6J mice and Il1rap−/− mice on the C57BL/6 background or Il36r−/− mice or WT Balb/cAnCrl mice, all 8–13 weeks of age, were purchased from Charles River (Sulzfeld, Germany or the US). Animals were housed in groups of 5 mice per cage under specific pathogen-free conditions in isolated ventilated cages at 20–25 °C and a humidity of 46–65% with a dark/night cycle of 12 h. Mice had free access to water and chow. All experiments were approved by the animal welfare officers within Boehringer Ingelheim Pharma GmbH and Co KG, as well as by the local authorities for the care and use of experimental animals (Regierungspräsidium Tübingen; TVV 12-009-G and 14-016-G; 35/9185.81-8). Experiments with influenza virus were performed under biosafety level 2 conditions and were in accordance with German national guidelines and legal regulations.
Reagents
Human and mouse primary cells in this study were stimulated with the cytokines and the concentrations depicted in Table 1. Unless otherwise described, the cells were stimulated with macrophage media containing Dulbecco’s modified Eagle’s medium (DMEM) (1×)+GlutaMAXTM-I (GIBCO #31966-021); 10% Spezial-HI fetal calf serum (FCS; GIBCO #16140-071); 1%NEAA (100x GIBCO #11140-035); 1% P/S (10,000 U/mL Penicillin, 10,000 µg/mL Streptomycin GIBCO #15140-122) and recombinant human MCSF.
Table 1.
Cytokines used in this study.
| Species | Cytokine | Concentration | Catalog number | Manufacturer |
|---|---|---|---|---|
| Human/mouse | rhMCSF |
100 ng/mL (macrophage differentiation) 10 ng/mL (macrophage stimulation) |
216-MCC/CF | R&D Systems |
| Human/mouse | Salmonella LPS | 100 ng/mL | L6143-1MG | Sigma |
| Mouse | rmIL-36α | 33 ng/mL | 7059-ML/CF | R&D Systems |
| Mouse | rmIL-36β | 33 ng/mL | 7060-ML/CF | R&D Systems |
| Mouse | rmIL-36γ | 33 ng/mL | 6996-IL/CF | R&D Systems |
| Mouse | rmIL-1α | 10 ng/mL | 400-ML/CF | R&D Systems |
| Mouse | rmIL-1α | 50 ng/mL (Suppl. Fig. 1c) | 400-ML/CF | R&D Systems |
| Mouse | rmIL-1β | 10 ng/mL | 401-ML/CF | R&D Systems |
| Mouse | rmIL-1β | 50 ng/mL (Suppl. Fig. 1c) | 401-ML/CF | R&D Systems |
| Mouse | rmTNFα | 10 ng/mL | 410-MT/CF | R&D Systems |
| Mouse | rmGM-CSF | 10 ng/mL | 415-ML/CF | R&D Systems |
| Human/mouse | Poly(I:C) (LMW) | 1 µg/mL | tlrl-picw | InvivoGen |
| Human | rhTGFβ | 10 ng/mL | 7666-M/CF | R&D Systems |
| Human | rhIL-36α | 33 ng/mL | 6995-IL-010/CF | R&D Systems |
| Human | rhIL-36β | 33 ng/mL | 6834-ILB-025/CF | R&D Systems |
| Human | rhIL-36γ | 33 ng/mL | 2320-IL-025/CF | R&D Systems |
| Human | rhIL-1α | 10 ng/mL | 200-LA-010/CF | R&D Systems |
| Human | rhIL-1β | 10 ng/mL | 201-LB-010/CF | R&D Systems |
| Human | rhTNFα | 10 ng/mL | 410-MT/CF | R&D Systems |
| Human | rhGMCSF | 10 ng/mL | 7954-GM-010/CF | R&D Systems |
Poly(I:C) administration
For i.t. administration of poly(I:C), female C57BL/6 mice, 8–10 weeks old (Javier Laboratories), were anesthetized with isoflurane for 3 min before instilling 2 mg/kg poly(I:C) (LMW) (#tlrl-picw/tlrl-picw-250, InvivoGen, Lot: PIW-40-03) diluted in NaCl (0.9%)) in a total volume of 50 µL/animal with a 1-mL syringe during inspiration. BAL was performed 4, 12, 24, and 48 h after the poly(I:C) administration.
IL-36 i.t. administration
For i.t. administration of IL-36γ, female Balb/c mice were anesthetized with isoflurane for 3 min and 1 µg IL-36γ (6996-IL/CF, Lot DAQQ041703A, R&D) that was diluted in PBS in a total volume of 50 µL/animal was administered with a 1-mL syringe during inspiration. BAL was performed 4 h after the IL-36γ administration. For time course analysis, mice were sacrificed 10, 20, or 30 min after the IL-36gγ administration (BAL was pooled from 2 to 3 animals from each group).
LPS aerosol exposure
Male Balb/c mice were exposed once to a single dose of aerosolized Escherichia coli LPS (Serotyp 055:B5, Sigma Aldrich) in PBS (1 mg/mL) for 30 min. LPS was administered by a nebulizer (Parimaster®) connected to a self-made Plexiglas box. After pre-flooding the box and all tubes for 30 min with the LPS aerosol, mice were transferred into the box and were exposed to a continuous flow of LPS aerosol for 25 min and left to remain for another 5 min after the aerosol was discontinued60–62. Mice were sacrificed 2, 4, 6, 8, 12, 18, 24, 36, 48, and 60 h after LPS exposure and BAL was performed as described in the section “Bronchoalveolar lavage.”
CS exposure model
Female and male C57BL/6J mice were either exposed to RA or CS in a heated (38 °C) perspex box (homemade, whole-body exposure chamber) for 4 days (4–5 cigarettes/day), with the experimental readout on day 5 (1-week CS exposure model) or for 5 days/week (4–5 cigarettes/day) with the experimental readout on day 11 (2-week CS exposure model)63. For the 3-week CS exposure model, Il36r−/− mice and littermate WT control C57BL/6J mice were CS exposed 5 days per week with the experimental readout on day 19. Mice were exposed to five cigarettes (Roth-Händle without filters, tar 10 mg, nicotine 1 mg, carbon monoxide 6 mg, Imperial Tobacco) per day. A semi-automatic cigarette lighter and smoke generator with an electronic timer was used to control CS exposure (Boehringer Ingelheim Pharma GmbH & Co. KG, Biberach, Germany) as previously described64. Eighteen hours after the last CS exposure, mice were euthanized on either day 5 or day 19 with an overdose of pentobarbital (400 mg/kg intraperitoneal (i.p.)) and BAL was performed.
CS exposure and H1N1 infection model
Il36r−/− and Il1rap−/− mice and littermate WT control C57BL/6J mice received RA or were infected with Influenza virus in combination with CS exposure for 2 weeks as described in the 2-week CS exposure model in the section “CS exposure model.” For viral infection, mice were anesthetized with 3% isoflurane and infected 2 h after smoke exposure by administering 3 Infections Units (IU) of H1N1 for IL1rap−/− and WT controls and 30 IU of H1N1 for Il36r−/− mice and WT controls in PBS in a total volume of 50 µL intranasally, 25 µL per nostril on day 7. Influenza virus A/PR/8/34 (H1N1) was obtained from Boehringer Ingelheim in Laval, Canada.
Lung homogenate
Lungs were removed and washed with PBS and snap frozen in liquid nitrogen immediately after extraction and weighing, and they were stored at −80 °C. For homogenization, lungs were thawed and a FastPrep-24 Sample Preparation System following the manufacturer’s instructions (MP Biomedicals, Irvine, CA, USA) was used. Homogenates were then centrifuged for 10 min at 300 × g and 4 °C and the supernatant was stored at −20 °C. Cytokine amounts in lung homogenate were measured by using MSD multiplex technology (Meso Scale Discovery, Gaithersburg, MD, USA) according to the manufacturer’s instructions.
Murine bone marrow-derived neutrophil isolation and stimulation
Neutrophils were isolated from the suspension of bone marrow of male C57BL/6J mice (8–13 weeks) according to the manufacturer’s instructions (Miltenyi Biotec, Neutrophil Isolation Kit mouse; #130-097-658). The neutrophils were resuspended and plated at 5 × 105 cells/mL/well in a 24-well plate in macrophage media without MCSF and stimulated with Salmonella LPS, IL-36α, IL-36β, IL-36γ, IL-1α, IL-1β, GM-CSF, and poly(I:C) (LMW) in vitro for 4 h. Cells were cultured in an incubator at 37 °C at 5% CO2.
Bronchoalveolar lavage
Mice were euthanized by i.p. administration of an overdose of pentobarbital (400–800 mg/kg). The trachea was cannulated and BAL performed by flushing lungs twice with 0.8 mL of lavage buffer (Hanks’ balanced salt solution (HBSS) containing 0.6 mM EDTA). Cell counts per mL BAL fluid (neutrophils, AMs) were measured and differentiated using the Sysmex XT-1800i automated hematology analyzer as described previously63,64. The BAL fluid was centrifuged for 5 min at 200 × g at 4 °C. Supernatant was decanted and cytokines were measured using MSD (Meso Scale Discovery, Gaithersburg, MD, USA) or enzyme-linked immunosorbent assays (ELISAs) following the manufacturers’ instructions. BAL pellet was lysed using RLT buffer (350 µL, #1053393, Qiagen) to isolate RNA for downstream cDNA generation and gene expression analyses by qPCR.
AM isolation and stimulation
BAL pellet was isolated as described in the section “Bronchoalveolar lavage” and resuspended and 2.5 × 105 cells/0.5 mL/well were plated in a 24-well plate in macrophage media with GM-CSF and stimulated with Salmonella LPS (4 h), IL-36α, IL-36β, IL-36γ, IL-1α, IL-1β, TNF-α, and poly(I:C) (LMW) in vitro. Cells were cultured in an incubator at 37 °C at 5% CO2.
Bone marrow-derived macrophages
C57BL/6J mice (male, 8–13 weeks) were sacrificed by i.p. administration of an overdose of pentobarbital (400–800 mg/kg), and bone marrow cells were harvested from femurs and tibiae by flushing the respective bone with PBS. Isolated cells were centrifuged at 200 × g for 5 min, supernatant was decanted, and pellets were resuspended in 100 mL of macrophage media. Cells were differentiated to macrophages in an incubator at 37 °C at 5% CO2 for 6–7 days before scraping, counting, and plating for downstream experiments.
Stimulation of BMDMs
In all experiments, 4 × 105 macrophages/mL were seeded in a 12-well plate in macrophage media. Macrophages were allowed to adhere for 16–24 h before stimulating for 24 h with Salmonella LPS (4 h), IL-36α, IL-36β, IL-36γ, IL-1α, IL-1β, TNF-α, and poly(I:C) (LMW) in vitro. Cells were cultured in an incubator at 37 °C at 5% CO2. To mimic the lung milieu, BMDMs were stimulated with TGF-β and GM-CSF in combination with IL-36α, IL-36β, IL-36γ, IL-1α, and IL-1β for 24 h.
Fibroblast isolation and stimulation
C57BL/6J mice (male, 8–13 weeks) were sacrificed by i.p. administration of an overdose of pentobarbital (400–800 mg/kg), and the lungs were removed and washed by dipping into ice-cold PBS. The lungs were minced and transferred into a 50-mL tube containing 10 mL DMEM (DMEM (1×)+GlutaMAXTM-I (GIBCO #31966-021), 10% FCS (GIBCO #16140-071), 1% Antibiotic Antimycotic solution (Life Technologies, cat.no. 15240062), and 100 µL of liberase solution (26 U/5 mg/mL; Roche, cat.no. 05401119001). After 2 h shaking at 37 °C, the lung pieces were strained (70 µM, BD, #352350) and flushed twice with 5 mL PBS. The tissue solution was then centrifuged for 5 min at 190 × g. The pellet was resuspended and washed twice in 10 mL PBS (centrifugation for 5 min at 190 × g). The pellet was then resuspended in DMEM (DMEM (1×)+GlutaMAXTM-I (GIBCO #31966-021); 10% FCS (GIBCO #16140-071); 1% Antibiotic Antimycotic solution (Life Technologies, cat.no. 15240062), and transferred into a 75-cm2 flask and cultured in an incubator at 37 °C at 5% CO2. After the first 24 h, medium was changed. Every 2 days, the fibroblasts were washed with PBS and medium was changed until a confluence of 70% was reached to transfer the fibroblasts into a 175-cm2 flask. In all, 5 × 104 primary mouse fibroblasts were seeded in a 12-well plate in macrophage media without MCSF. After 16–24 h, fibroblasts were simulated for 24 h with IL-36α, IL-36β, IL-36γ, IL-1α, IL-1β, or poly(I:C) (LMW) in vitro. Cells were cultured in an incubator at 37 °C at 5% CO2.
Stimulation of human epithelial basal cells
In all experiments, 3 × 104 small airway epithelial cells from a human donor obtained from Lonza were seeded in a 96-well plate in 200 µL PneumaCult™-Ex Plus Medium (Stem cell Technologies, # 05040 Kit, 05041 Basal Medium). After 24 h, epithelial cells were simulated for 24 h with Salmonella LPS, IL-36α, IL-36β, IL-36γ, GMCSF, and poly(I:C) (LMW) IL-1α and IL-1β. Cells were cultured in an incubator at 37 °C at 5% CO2.
Generation of human MDMs
Monocytes were isolated from peripheral blood mononuclear cells from human volunteer donors by negative magnetic separation using the Monocyte Isolation Kit II (MACS Miltenyi #130-091-153, Miltenyi Biotech, Bergisch Gladbach, Germany) and the AutoMACS pro system (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. In order to generate MDMs, monocytes were resuspended in 100 mL of macrophage media. Cells were cultured in an incubator at 37 °C at 5% CO2 for 6–7 days before using for experiments. All experiments with blood from human volunteer donors were approved by the blood donation service from Boehringer Ingelheim Pharma GmbH and Co. KG following ethical standards and local regulation.
Stimulation of human MDMs
In all experiments, 5 × 105 macrophages were seeded in a 12-well plate in macrophage media. After 16–24 h macrophages were simulated for 24 h with Salmonella LPS, IL-36α, IL-36β, IL-36γ, GM-CSF, and poly(I:C) (LMW). Cells were cultured in an incubator at 37 °C at 5% CO2.
Neutrophil isolation and stimulation
Neutrophils were isolated from whole blood from volunteer human donors by negative magnetic separation using the MACSexpess Whole Blood Neutrophil Isolation Kit (#130-104-434, Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. In all, 3–5 × 105 neutrophils were seeded in a 24-well plate in macrophage media without MCSF. All experiments with blood from human volunteer donors were approved by the blood donation service from Boehringer Ingelheim Pharma GmbH and Co KG following ethical standards and local regulation.
Stimulation of primary human fibroblasts
In all experiments, 2 × 104 normal human lung fibroblasts obtained from Lonza were seeded in a 12-well plate in macrophage media without MCFS and simulated for 24 h with Salmonella LPS, IL-36α, IL-36β, IL-36γ, GM-CSF, and poly(I:C) (LMW). Cells were cultured in an incubator at 37 °C at 5% CO2.
Single-cell RNA sequencing
The 10× Genomics Chromium system and Single Cell 3’ Reagent v2 Kits (10× Genomics, Pleasanton, CA) were used to generate scRNA-seq libraries according to the manufacturer’s instructions. The reverse transcribed cDNA was first amplified by 14 PCR cycles and then used to generate the sequencing library, which was finally amplified by 14 PCR cycles. The sequencing library was purified as described by the manufacturer and then subjected to another cleanup step using one volume of SPRISelect Beads (Beckman Coulter, Brea, CA, USA) in order to remove leftover primers and dimers. The average length of the purified library was 494 bp. A HiSeq 4000 and two 50-cycle SBS kits (Illumina, San Diego, CA, USA) were used for sequencing and clusters were generated on a cBot using the HiSeq 3000/4000 PE Cluster Kit. The paired-end sequencing run comprised a 26-bp read 1 (16 bp cell barcode and 10 bp UMI) and 98-bp read 2 (transcript sequence read) as well as a 8-bp index read. On average, approximately 50,000 reads were sequenced per cell.
Data processing and analysis
Raw data were processed using the Cell Ranger v2.1.165 pipeline to finally obtain a count matrix that was used as input for downstream analysis by the Seurat R package v3.0.0.966. During data processing, reads were aligned to the GRCm38 reference genome and annotated according to Ensembl release 8667.
For downstream analysis, low quality cells with <300 detected genes and/or >10% of mitochondrial transcripts were removed from the data set. Additionally, cells with >5000 detected genes and genes that were detected in less than three cells were removed from the data set, which left 3075 cells and 14,728 genes for further analysis.
Data were then normalized and log-transformed per cell using the “NormalizeData” function. In order to identify cell clusters within the data set, the highly variable genes were first calculated by the “FindVariableFeatures” function and “mean.var.plot” method. Data were then scaled and centered and principal component (PC) analysis was computed on the variable genes using the “RunPCA” function. Cell clusters were then calculated using the first 13 PCs as input and a resolution of 0.2 and visualized by UMAP68 representation that was also calculated on the first 13 PCs.
Cell clusters were annotated based on the expression of characteristic marker genes. A small cluster that represented 3.4% of the cells was characterized by the expression of both macrophage and neutrophil marker genes (Suppl. Fig. 6b, c) as well as a low UMI count that was 12- and 2.3-fold lower than for macrophages and neutrophils, respectively. Therefore, this cluster was identified as a low-quality cluster and removed for further analysis. Subcluster analysis of the neutrophils was performed as described for the entire data set using only the neutrophils. Neutrophil clusters were computed on the first 10 PCs and a resolution of 0.2. The first ten PCs were also used for UMAP dimensional reduction.
Differential expression analysis and gene set enrichment analysis
Differential expression analysis was calculated by the MAST R package v1.6.169 via Seurat’s “FindMarkers” function. Genes were kept for differential expression analysis if they were detected in at least 10% of the cells of the compared groups and considered to be significantly differentially expressed if they were at least 1.5-fold differentially expressed with a P value adjusted for multiple testing <0.05 (Bonferroni correction). Gene set enrichment analysis was performed for the significantly increased genes per neutrophil cluster using the g:Profiler webtool70 and the default settings data sources.
LPS exposure for scRNA-seq study
Female C57BL/6JCR mice (18–20 g, Charles River Research Models and Services Germany GmbH, Sulzfeld, Germany) were used for the acute LPS model that was used for the scRNA-seq study. Mice were exposed to LPS as described in the section “LPS aerosol exposure” and euthanized 4 h post LPS inhalation by i.p. pentobarbital injection (Narcoren, Merial GmbH, Halbermoos, Germany). Mice were given ad libitum access to water and food during the experiment.
BAL and cell purification for single-cell study
BAL was performed four times per animal using 0.8 mL of HBSS per lavage per animal. For scRNA-seq analysis, BAL fluids of eight animals were equally pooled to obtain a single BAL pool that contained the same cell count per animal. BAL cells were centrifuged at 400 × g and 4 °C for 5 min and the supernatant was removed. The cell pellet was washed once using 10 mL of HBSS/0.04% bovine serum albumin (BSA) buffer and finally resuspended in 500 µL of HBSS/0.04% BSA buffer and filtered through a 40-µm cell strainer.
RNA isolation and qPCR
Cells were washed with PBS and lysed using 350 µL RLT buffer (350 µL, Qiagen #1053393). RNA was purified using MagMax (Biosystems #AM1830) following the manufacturer’s instructions. RNA was transcribed to cDNA using the High Capacity cDNA Reverse Transcription Kit (Biosystems #4368813) according to the manufacturer’s instructions. Taqman master mix (Applied Biosystems #4352042) was used to prepare the RT-qPCR reaction mix. Taqman gene expression assays were obtained from ThermoFischer Scientific: Housekeeper Hprt1: Mm03024075_m1 or 18S: Mm03928990_g1; IL1F9: Mm00463327_m1; MMP-9: Mm00463327_m1; CXCL1: Mm04207460_m1; IL1β: Mm00434228_m1; IL1F6: Mm00457645_m1; IL1F8: Mm01337546_g1;Il1a: Mm00439620_m1; Il6: Mm00446190_m1; IL1RL2: Mm00519245_m1. Plates were prepared following the manufacturer’s instructions and analyzed on an Applied Biosystems QuantStudio 6 Flex Real-Time PCR System. Gene expression was normalized to the housekeeper (dCt) and depicted as 2(ddCt).
Cytokine measurements
To measure the protein concentrations in supernatant, MSDs (Meso Scale Discovery, Gaithersburg, MD, USA; V-Plex Proinflammatory Panel 1 mouse Kit; K15048D-1; and ELISAs such as the CXCL1/KC DuoSet ELISA (R&D Systems, #DY453-05) and the mouse IL-36γ ELISA (Aviva Systems Biology, #OKEH03002) were used according to the manufacturer’s instructions.
Statistical analyses and reproducibility
Data were analyzed by using the Prism 8 software package (GraphPad Software, San Diego, CA). All data were expressed as mean and standard error of the mean (SEM) and were analyzed using Student’s t test or one-way analysis of variance with a value of <0.05 considered to be statistically significance. The reproducibility was determined by using several biological replicates and repeating the experiment several times as indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We are grateful to Dr. Peter Murray for his critical appraisal of the manuscript. We acknowledge Dr. David Lamb for his in vivo model expertise. We thank Jochen Blender for help with the technical in vivo aspects.
Author contributions
K.C.E.K.: conceived of the study, designed the study, interpreted the data, and wrote the manuscript; C.K.K.: conceived and designed the research study, conducted the bulk of the experiments, interpreted the data, and wrote the manuscript; P.J.B., L.E.D., J.R.B.: contributed to interpreting the data; C.T.W.: conducted the bulk of the single cell experiment; C.T., C.L., M.P., S.F., M.K.: contributed to the experimental execution of the experiments; C.M.M.W., D.P., M.R., J.F., F.G., M.T.: contributed to the conception and design of the study; all authors contributed to scientific discussions, revision of the manuscript, had full access to all the data, and agreed to submit for publication.
Data availability
All data are available from the corresponding author upon request and the data related to the RNA-seq experiment are deposited in GEO reference GSE159161. Source data underlying the main and supplementary figures are available in Supplementary Data 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-01703-3.
References
- 1.Rivers-Auty J, Daniels MJD, Colliver I, Robertson DL, Brough D. Redefining the ancestral origins of the interleukin-1 superfamily. Nat. Commun. 2018;9:1156. doi: 10.1038/s41467-018-03362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinarello C, et al. IL-1 family nomenclature. Nat. Immunol. 2010;11:973. doi: 10.1038/ni1110-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg H, et al. IL-1RL2 and its ligands contribute to the cytokine network in psoriasis. J. Immunol. 2010;185:4354–4362. doi: 10.4049/jimmunol.1000313. [DOI] [PubMed] [Google Scholar]
- 5.Tortola L, et al. Psoriasiform dermatitis is driven by IL-36-mediated DC-keratinocyte crosstalk. J. Clin. Investig. 2012;122:3965–3976. doi: 10.1172/JCI63451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marrakchi S, et al. Interleukin-36-receptor antagonist deficiency and generalized pustular psoriasis. N. Engl. J. Med. 2011;365:620–628. doi: 10.1056/NEJMoa1013068. [DOI] [PubMed] [Google Scholar]
- 7.Kovach MA, et al. IL-36gamma is a crucial proximal component of protective type-1-mediated lung mucosal immunity in Gram-positive and -negative bacterial pneumonia. Mucosal Immunol. 2017;10:1320–1334. doi: 10.1038/mi.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumberg H, et al. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J. Exp. Med. 2007;204:2603–2614. doi: 10.1084/jem.20070157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debets R, et al. Two novel IL-1 family members, IL-1 delta and IL-1 epsilon, function as an antagonist and agonist of NF-kappa B activation through the orphan IL-1 receptor-related protein 2. J. Immunol. 2001;167:1440–1446. doi: 10.4049/jimmunol.167.3.1440. [DOI] [PubMed] [Google Scholar]
- 10.Carrier Y, et al. Inter-regulation of Th17 cytokines and the IL-36 cytokines in vitro and in vivo: implications in psoriasis pathogenesis. J. Invest. Dermatol. 2011;131:2428–2437. doi: 10.1038/jid.2011.234. [DOI] [PubMed] [Google Scholar]
- 11.Johnston A, et al. IL-1F5, -F6, -F8, and -F9: a novel IL-1 family signaling system that is active in psoriasis and promotes keratinocyte antimicrobial peptide expression. J. Immunol. 2011;186:2613–2622. doi: 10.4049/jimmunol.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wein AN, et al. IL-36gamma protects against severe influenza infection by promoting lung alveolar macrophage survival and limiting viral replication. J. Immunol. 2018;201:573–582. doi: 10.4049/jimmunol.1701796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towne JE, Garka KE, Renshaw BR, Virca GD, Sims JE. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J. Biol. Chem. 2004;279:13677–13688. doi: 10.1074/jbc.M400117200. [DOI] [PubMed] [Google Scholar]
- 14.Barksby HE, Lea SR, Preshaw PM, Taylor JJ. The expanding family of interleukin-1 cytokines and their role in destructive inflammatory disorders. Clin. Exp. Immunol. 2007;149:217–225. doi: 10.1111/j.1365-2249.2007.03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vigne S, et al. IL-36R ligands are potent regulators of dendritic and T cells. Blood. 2011;118:5813–5823. doi: 10.1182/blood-2011-05-356873. [DOI] [PubMed] [Google Scholar]
- 16.Towne JE, et al. Interleukin-36 (IL-36) ligands require processing for full agonist (IL-36alpha, IL-36beta, and IL-36gamma) or antagonist (IL-36Ra) activity. J. Biol. Chem. 2011;286:42594–42602. doi: 10.1074/jbc.M111.267922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry CM, et al. Neutrophil-derived proteases escalate inflammation through activation of IL-36 family cytokines. Cell Rep. 2016;14:708–722. doi: 10.1016/j.celrep.2015.12.072. [DOI] [PubMed] [Google Scholar]
- 18.Vergnolle N. Protease inhibition as new therapeutic strategy for GI diseases. Gut. 2016;65:1215–1224. doi: 10.1136/gutjnl-2015-309147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston A, et al. IL-1 and IL-36 are dominant cytokines in generalized pustular psoriasis. J. Allergy Clin. Immunol. 2017;140:109–120. doi: 10.1016/j.jaci.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachelez H, et al. Inhibition of the interleukin-36 pathway for the treatment of generalized pustular psoriasis. N. Engl. J. Med. 2019;380:981–983. doi: 10.1056/NEJMc1811317. [DOI] [PubMed] [Google Scholar]
- 21.Scheibe K, et al. IL-36R signalling activates intestinal epithelial cells and fibroblasts and promotes mucosal healing in vivo. Gut. 2017;66:823–838. doi: 10.1136/gutjnl-2015-310374. [DOI] [PubMed] [Google Scholar]
- 22.Russell SE, et al. IL-36alpha expression is elevated in ulcerative colitis and promotes colonic inflammation. Mucosal Immunol. 2016;9:1193–1204. doi: 10.1038/mi.2015.134. [DOI] [PubMed] [Google Scholar]
- 23.Boutet MA, et al. Distinct expression of interleukin (IL)-36alpha, beta and gamma, their antagonist IL-36Ra and IL-38 in psoriasis, rheumatoid arthritis and Crohn’s disease. Clin. Exp. Immunol. 2016;184:159–173. doi: 10.1111/cei.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding L, Wang X, Hong X, Lu L, Liu D. IL-36 cytokines in autoimmunity and inflammatory disease. Oncotarget. 2018;9:2895–2901. doi: 10.18632/oncotarget.22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nerlich A, et al. C/EBPbeta is a transcriptional key regulator of IL-36alpha in murine macrophages. Biochim. Biophys. Acta. 2015;1849:966–978. doi: 10.1016/j.bbagrm.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Bachmann M, Scheiermann P, Hardle L, Pfeilschifter J, Muhl H. IL-36gamma/IL-1F9, an innate T-bet target in myeloid cells. J. Biol. Chem. 2012;287:41684–41696. doi: 10.1074/jbc.M112.385443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magne D, et al. The new IL-1 family member IL-1F8 stimulates production of inflammatory mediators by synovial fibroblasts and articular chondrocytes. Arthritis Res. Ther. 2006;8:R80. doi: 10.1186/ar1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietrich D, et al. Interleukin-36 potently stimulates human M2 macrophages, Langerhans cells and keratinocytes to produce pro-inflammatory cytokines. Cytokine. 2016;84:88–98. doi: 10.1016/j.cyto.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Ramadas RA, Ewart SL, Medoff BD, LeVine AM. Interleukin-1 family member 9 stimulates chemokine production and neutrophil influx in mouse lungs. Am. J. Respir. Cell Mol. Biol. 2011;44:134–145. doi: 10.1165/rcmb.2009-0315OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramadas RA, Ewart SL, Iwakura Y, Medoff BD, LeVine AM. IL-36alpha exerts pro-inflammatory effects in the lungs of mice. PLoS ONE. 2012;7:e45784. doi: 10.1371/journal.pone.0045784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsanejad R, Fields WR, Steichen TJ, Bombick BR, Doolittle DJ. Distinct regulatory profiles of interleukins and chemokines in response to cigarette smoke condensate in normal human bronchial epithelial (NHBE) cells. J. Interferon Cytokine Res. 2008;28:703–712. doi: 10.1089/jir.2008.0139. [DOI] [PubMed] [Google Scholar]
- 32.Chustz RT, et al. Regulation and function of the IL-1 family cytokine IL-1F9 in human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 2011;45:145–153. doi: 10.1165/rcmb.2010-0075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 34.Akagawa KS, Kamoshita K, Tokunaga T. Effects of granulocyte-macrophage colony-stimulating factor and colony-stimulating factor-1 on the proliferation and differentiation of murine alveolar macrophages. J. Immunol. 1988;141:3383–3390. [PubMed] [Google Scholar]
- 35.Chen BD, Mueller M, Chou TH. Role of granulocyte/macrophage colony-stimulating factor in the regulation of murine alveolar macrophage proliferation and differentiation. J. Immunol. 1988;141:139–144. [PubMed] [Google Scholar]
- 36.Yu X, et al. The cytokine TGF-beta promotes the development and homeostasis of alveolar macrophages. Immunity. 2017;47:903.e4–912.e4. doi: 10.1016/j.immuni.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Lambrecht BN. TGF-beta gives an air of exclusivity to alveolar macrophages. Immunity. 2017;47:807–809. doi: 10.1016/j.immuni.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Dranoff G, et al. Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science. 1994;264:713–716. doi: 10.1126/science.8171324. [DOI] [PubMed] [Google Scholar]
- 39.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 40.Legrand C, et al. Airway epithelial cell migration dynamics. MMP-9 role in cell-extracellular matrix remodeling. J. Cell Biol. 1999;146:517–529. doi: 10.1083/jcb.146.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franzen, O., Gan, L.-M. & Bjorkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, baz046 (2019). [DOI] [PMC free article] [PubMed]
- 42.Terui T, Ozawa M, Tagami H. Role of neutrophils in induction of acute inflammation in T-cell-mediated immune dermatosis, psoriasis: a neutrophil-associated inflammation-boosting loop. Exp. Dermatol. 2000;9:1–10. doi: 10.1034/j.1600-0625.2000.009001001.x. [DOI] [PubMed] [Google Scholar]
- 43.Tolle, L. B., Newstead, M., Kovach, M. A., Zeng, X. & Standiford, T. The role of IL-36γ in experimental lung injury and in patients with acute lung injury. Am. J. Respir. Crit. Care Med.187, A3857 (2013).
- 44.Meijer M, Rijkers GT, van Overveld FJ. Neutrophils and emerging targets for treatment in chronic obstructive pulmonary disease. Expert Rev. Clin. Immunol. 2013;9:1055–1068. doi: 10.1586/1744666X.2013.851347. [DOI] [PubMed] [Google Scholar]
- 45.Guillon A, et al. Neutrophil proteases alter the interleukin-22-receptor-dependent lung antimicrobial defence. Eur. Respir. J. 2015;46:771–782. doi: 10.1183/09031936.00215114. [DOI] [PubMed] [Google Scholar]
- 46.Cakarova L, et al. Macrophage tumor necrosis factor-alpha induces epithelial expression of granulocyte-macrophage colony-stimulating factor: impact on alveolar epithelial repair. Am. J. Respir. Crit. Care Med. 2009;180:521–532. doi: 10.1164/rccm.200812-1837OC. [DOI] [PubMed] [Google Scholar]
- 47.Rosler B, Herold S. Lung epithelial GM-CSF improves host defense function and epithelial repair in influenza virus pneumonia-a new therapeutic strategy? Mol. Cell. Pediatr. 2016;3:29. doi: 10.1186/s40348-016-0055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bochkov YA, et al. Rhinovirus-induced modulation of gene expression in bronchial epithelial cells from subjects with asthma. Mucosal Immunol. 2010;3:69–80. doi: 10.1038/mi.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakagawa S, et al. Staphylococcus aureus virulent PSMalpha peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe. 2017;22:667–677. doi: 10.1016/j.chom.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pauwels NS, et al. Role of IL-1alpha and the Nlrp3/caspase-1/IL-1beta axis in cigarette smoke-induced pulmonary inflammation and COPD. Eur. Respir. J. 2011;38:1019–1028. doi: 10.1183/09031936.00158110. [DOI] [PubMed] [Google Scholar]
- 51.Hudock KM, et al. Neutrophil extracellular traps activate IL-8 and IL-1 expression in human bronchial epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020;319:L137–L147. doi: 10.1152/ajplung.00144.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnberg H, Letocha H, Nou F, Westlin JF, Nilsson S. GM-CSF in chemotherapy-induced febrile neutropenia–a double-blind randomized study. Anticancer Res. 1998;18:1255–1260. [PubMed] [Google Scholar]
- 53.Choi JC, et al. Granulocyte macrophage-colony stimulating factor (GM-CSF) augments acute lung injury via its neutrophil priming effects. J. Korean Med. Sci. 2008;23:288–295. doi: 10.3346/jkms.2008.23.2.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mossner R, et al. The genetic basis for most patients with pustular skin disease remains elusive. Br. J. Dermatol. 2018;178:740–748. doi: 10.1111/bjd.15867. [DOI] [PubMed] [Google Scholar]
- 55.Sugiura K, et al. The majority of generalized pustular psoriasis without psoriasis vulgaris is caused by deficiency of interleukin-36 receptor antagonist. J. Invest. Dermatol. 2013;133:2514–2521. doi: 10.1038/jid.2013.230. [DOI] [PubMed] [Google Scholar]
- 56.Scheibe K, et al. Inhibiting interleukin 36 receptor signaling reduces fibrosis in mice with chronic intestinal inflammation. Gastroenterology. 2019;156:1082–1097. doi: 10.1053/j.gastro.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 57.Heath JE, Scholz GM, Veith PD, Reynolds EC. IL-36gamma regulates mediators of tissue homeostasis in epithelial cells. Cytokine. 2019;119:24–31. doi: 10.1016/j.cyto.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Milora KA, Uppalapati SR, Sanmiguel JC, Zou W, Jensen LE. Interleukin-36beta provides protection against HSV-1 infection, but does not modulate initiation of adaptive immune responses. Sci. Rep. 2017;7:5799. doi: 10.1038/s41598-017-05363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gabay C, Towne JE. Regulation and function of interleukin-36 cytokines in homeostasis and pathological conditions. J. Leukoc. Biol. 2015;97:645–652. doi: 10.1189/jlb.3RI1014-495R. [DOI] [PubMed] [Google Scholar]
- 60.Chignard M, Balloy V. Neutrophil recruitment and increased permeability during acute lung injury induced by lipopolysaccharide. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000;279:L1083–L1090. doi: 10.1152/ajplung.2000.279.6.L1083. [DOI] [PubMed] [Google Scholar]
- 61.Weathington NM, et al. A novel peptide CXCR ligand derived from extracellular matrix degradation during airway inflammation. Nat. Med. 2006;12:317–323. doi: 10.1038/nm1361. [DOI] [PubMed] [Google Scholar]
- 62.Mould, K. J., Jackson, N. D., Henson, P. M., Seibold, M. & Janssen, W. J. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight4, e126556 (2019). [DOI] [PMC free article] [PubMed]
- 63.Bucher H, Duechs MJ, Tilp C, Jung B, Erb KJ. Tiotropium attenuates virus-induced pulmonary inflammation in cigarette smoke-exposed mice. J. Pharmacol. Exp. Ther. 2016;357:606–618. doi: 10.1124/jpet.116.232009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tilp C, et al. Effects of conventional tobacco smoke and nicotine-free cigarette smoke on airway inflammation, airway remodelling and lung function in a triple allergen model of severe asthma. Clin. Exp. Allergy. 2016;46:957–972. doi: 10.1111/cea.12665. [DOI] [PubMed] [Google Scholar]
- 65.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888.e21–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yates AD, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diaz-Papkovich A, Anderson-Trocme L, Ben-Eghan C, Gravel S. UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts. PLoS Genet. 2019;15:e1008432. doi: 10.1371/journal.pgen.1008432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raudvere U, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data are available from the corresponding author upon request and the data related to the RNA-seq experiment are deposited in GEO reference GSE159161. Source data underlying the main and supplementary figures are available in Supplementary Data 1.