Highlights
-
•
VSG mRNA copy number varies with the identity of the VSG.
-
•
Early premature termination codons result in degradation of VSG mRNA.
-
•
Late premature termination codons translated to produce incomplete VSG result in an increase in VSG mRNA.
-
•
There is a feedback pathway between non-functional VSG production and VSG mRNA levels.
Keywords: Trypanosoma brucei, VSG mRNA
Abstract
The bloodstream form of Trypanosoma brucei persists in mammalian hosts through a population survival strategy depending on antigenic variation of a cell surface coat composed of the variant surface glycoprotein (VSG). The integrity of the VSG coat is essential and blocking its synthesis results in a cell division cycle arrest just prior to cytokinesis. This observation indicates that VSG levels are monitored and that the cell has mechanisms to respond to a disruption of synthesis. Here, the regulation of VSG mRNA levels has been investigated by first measuring VSG mRNA copy number, and second using ectopic expression of VSG transgenes containing premature termination codons. The findings are that (i) VSG mRNA copy number varies with the identity of the VSG and (ii) a pathway detects synthesis of non-functional VSG protein and results in an increase in VSG mRNA levels.
1. Introduction
Trypanosoma brucei can sustain long-term infections in mammalian hosts by antigenic variation of the variant surface glycoprotein (VSG) that forms a coat covering the entire surface of the cell [1]. The active VSG gene is located near a telomere at the 3′ end of a transcription unit known as a bloodstream expression site (BES). There are 10–20 BESs in T. brucei genomes but only one BES is transcribed at any one time [2,3]. Antigenic variation occurs when a low frequency event, either gene conversion in the active BES or a less well characterised epigenetic switch to an alternative BES, results in transcription of a different VSG gene [4]. The processes involved in antigenic variation and monoallelic VSG expression have been studied in some detail, the active BES is transcribed by RNA polymerase I (RNAPI) [5] and is located in the nuclear expression site body (ESB), a compartment containing RNAPI lying outside the nucleolus [6]. Expression of protein coding genes by RNAPI evolved in a cell in which the maturation of every cytoplasmic mRNA included trans-splicing of a capped 39 nucleotide exon to the 5′ end [7], thus the first 39 bases of a VSG mRNA are transcribed by RNAPII and the remainder by RNAPI. Presumably, RNAPI transcription allows the exceptionally high levels of VSG mRNA in the cell to be transcribed from a single gene. The precise details of VSG monoallelic expression, both the default silencing and the escape from silencing by the active BES, are less well understood but involve a VEX/CAF complex identified in a whole genome RNAi screen for loss of monoallelic VSG expression [3,8].
There is strong evidence that mechanisms have evolved to ensure that the supply of newly synthesised VSG is adequate to maintain the densely packed coat as new plasma membrane is added during cellular growth. Blocking VSG synthesis by RNAi knockdown of the mRNA or by blocking translation using a morpholino oligonucleotide results in cell division cycle arrest at the onset of cytokinesis [9,10]. It is assumed that cytokinesis is the point in the cell cycle where there is the highest demand for new VSG coated membrane. Trypanosomes with a common genetic background but expressing different VSGs proliferate at different rates in identical culture conditions, providing evidence that the identity of the expressed VSG can affect cellular growth rates with some VSGs effectively constraining proliferation [11]. As VSGs constitute 10–20 % of total protein and VSGs have very different amino acid sequences, the rate of successful folding and export from the endoplasmic reticulum may be limiting, thus affecting growth rates. Studies have reached mixed conclusions, either that VSG protein is synthesised in amounts greater than is needed and the excess degraded [12,13] whereas another report found that just sufficient is synthesised [14].
As might be expected for an mRNA encoding a highly abundant protein, VSG mRNA has a long half-life in proliferating cells with measurements ranging from 4.5 h [15] down to 1 h [16]. Its levels can be modulated by the cell as expression of a second VSG from a transgene reduces steady state mRNA levels from the endogenous VSG gene. Inducible expression of a T7 RNA polymerase-driven VSG6 (also known as VSG121) transgene located in the non-transcribed spacer of a ribosomal RNA gene cluster was shown to cause a rapid decrease in endogenous VSG2 (also known as VSG221) mRNA levels and a slower transcriptional attenuation of the active BES dependent on the activity of DOT1B (disruptor of telomeric silencing-1B) [17]. It is not clear whether attenuation of the active VSG expression site is an entirely intranuclear event or if there is a cytoplasmic contribution. In a second set of experiments, insertion of a VSG117 gene in various locations in the genome of cells expressing VSG2 resulted in a reduction of VSG2 mRNA proportional to the amount of VSG117 mRNA expressed [16]. These observations imply that the total VSG mRNA level is regulated and that the mechanism can recognise different VSG mRNAs despite their diverse sequences. The only obvious conserved motif in VSG mRNAs is a 16 nucleotide sequence (16-mer) in the 3′UTR close to the poly(A) tail. Although the 16-mer is necessary for the very high levels of cytoplasmic VSG mRNA [16], no mechanism has been characterised for how it is involved in positive regulation of high levels of VSG mRNA and it is unknown whether it is also involved in down regulation when a second VSG mRNA is expressed.
Here, expression of a second VSG from transgenes containing a premature termination codon (PTC) at various locations in the open reading frame was used to produce cytoplasmic degradation of VSG mRNA [18,19] and the consequent effect on VSG mRNA measured. In yeast and metazoa, recognition of a PTC during translation triggers a reduction in the half-life of an mRNA via the nonsense-mediated decay (NMD) pathway mediated by a complex containing the RNA helicase UPF1 (reviewed in [20,21]). In yeast, NMD is not a binary process and only rarely is a PTC-containing mRNA completely absent from a cell (for example [22]). The degree of reduction in the PTC-containing mRNAs is affected by the location of the PTC within the open reading frame, the nearer to the initiation codon the more probable that NMD will be triggered on any round of translation [23]. In trypanosomes, investigation of PTC-containing mRNAs showed a similar relationship between PTC location in an ORF and reduction in mRNA levels but any role for the UPF1 homologue remained ambiguous [24].
Expression of PTC-containing VSG transgenes and measurement of transgene and endogenous VSG mRNA levels confirmed the presence of a pathway that reduced PTC-containing VSG mRNAs, with the decrease being proportional to the proximity of the PTC to the initiation codon. Measurements of the VSG mRNA in wild type cells showed that VSG6 expressing cells have a higher copy number than VSG2 expressers and that double expressers had intermediate levels. An unexpected finding was that expression of VSG transgenes with PTCs close to the C-terminus resulted in increases in both transgenic and endogenous VSG mRNAs. Together, these experiments provide further evidence for a pathway that decreases PTC-containing mRNAs, and a second pathway regulating VSG mRNA levels that is linked to production of functional VSG protein.
2. Materials and methods
2.1. Plasmids
The sequence of the VSG6 BES was taken from Genbank entry FM162569 and VSG2 BES from FM162566. Genomic DNAs from yeast containing these cloned telomeres (a kind gift of Gloria Rudenko) were used as templates for PCR reactions to recover the fragments described below. Constructs to introduce a VSG2 transgene were based on the plasmid p3952. Digestion of p3952 with restriction enzymes Acc65I and SacI released a 4.6 kbp fragment (Supp. Fig. 1A) that contained, in order: bases −800 to −300 upstream of the VSG6 initiation codon (5′ targeting); XhoI site; bases -182 to -1 upstream of the VSG2 initiation codon (VSG2 splice acceptor and 5′UTR); an SphI site; VSG2 ORF; PacI site; bases 1–615 downstream of the VSG2 termination codon (VSG2 3′UTR and polyadenylation); XmaI site; a tubulin/blasticidin resistance cassette (alpha to beta tubulin inter-ORF, blasticidin-S-deaminase ORF, beta to alpha tubulin inter-ORF); SpeI site; bases −300 upstream to +210 downstream of the VSG6 initiation codon (3′ targeting) (see Supp. Fig. 1B for sequence). The plasmid was constructed by joining fragments using the restriction enzyme sites listed above. Subsequent changes to the VSG2 ORF used SphI and PacI. NEB Phusion DNA polymerase was used for site-directed mutagenesis according to the manufacturer’s instructions, oligonucleotides are listed in Supp. Table 1.
2.2. Cell culture and sample preparation
Trypanosoma brucei Lister 427 bloodstream form trypanosomes were used as parental cell lines throughout this study. HMI-11 [25] was used to culture bloodstream form trypanosomes with 5 μg/ml blasticidin to maintain genetically modified cell lines. All experiments were performed using logarithmically growing cells at a density of less than 1 × 106 cells/ml. Preparation of whole cell lysates, western blotting, preparation of total RNA and northern blotting all used standard techniques [26] with the following details. The primary antibodies used for Western blotting were: (i) Rabbit anti-VSG2 N-terminal peptide [27] (a kind gift of Peter Overath), (ii) mouse monoclonal anti-VSG6 (a kind gift of Miguel Navarro), and (iii) mouse monoclonal L13D6 anti-paraflagellar rod proteins (PFR) 1 and 2 (a kind gift from Keith Gull). Secondary antibodies were: (i) goat anti‐Rabbit AlexaFluor 680, and (ii) goat anti‐Mouse IRDye800. The LI-COR Odyssey infrared imaging system was used to visualise western blots. For northern blots, the relevant whole VSG ORF was used as a probe. Phosphorimaging was used to quantitate RNA from northern blots, background from the blot was subtracted from the band-of-interest, and then normalised against the rRNA loading control signal.
2.3. RNAseq for transcript abundances
Total RNA was prepared as above for single sample RNAseq to estimate VSG mRNA abundance. The cDNA libraries were prepared and sequenced at the Beijing Genomics Institute (Shenzhen, China) [28] In brief, polyadenylated RNA was purified from total RNA, converted to cDNA using random hexamer primers sheared and size selected for fragments ∼200 bp in length using the Illumina TruSeq RNA Sample Preparation Kit v2. RNAseq of the resulting libraries was used for the determination of transcript abundances. Sequencing was performed on an Illumina Hiseq 2000 (Illumina, CA) platform. Paired end reads were subject to quality trimming and adaptor filtering using Trimmomatic [29] using the settings “LEADING:10 TRAILING:10 SLIDINGWINDOW:5:15 MINLEN:50”. The quality filtered paired-end reads were then mapped to the complete set of CDS from version 6 of T. brucei genome annotation using bowtie2 [30] and transcript abundances were estimated using eXpress [31]. The sequence reads are in EBI ArrayExpress accession E-MTAB-9122.
3. Results
3.1. VSG2 transgenes were expressed and PTCs were effective in terminating translation
Expression of a VSG protein is essential for the proliferation of bloodstream form trypanosomes [9]. To investigate any cytoplasmic control of VSG mRNA levels independently of cell viability, trypanosomes were made to express two VSG mRNAs from a single bloodstream form expression site (BES), generating double expressers, using a similar construct design to others [32] (Fig. 1A). This approach allowed the manipulation of one VSG mRNA whilst maintaining viability through expression of the endogenous VSG. In the experiments here, the parental cell line was Trypanosoma brucei Lister 427 expressing VSG6 and the transgene encoded VSG2 including the native 5′ and 3′UTRs (Supplementary Fig. 1).
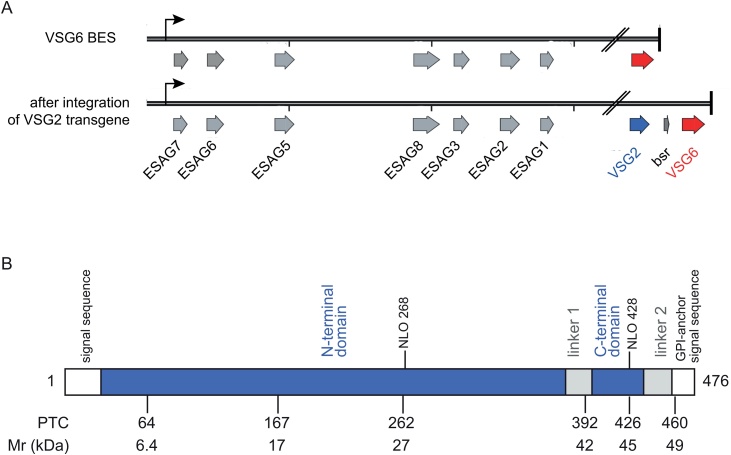
Fig. 1.
Integration of the VSG2 transgenes into the VSG6 bloodstream expression site (BES) and consequent effect on growth of transgene containing cell lines.
A. Site of integration of transgenes. Electroporation of Acc65I and SacI digested targeting construct (p3952; Supp. Fig. 1) containing either the wild-type or a mutant VSG2 ORF into bloodstream form trypanosomes expressing VSG6 and selection with blasticidin resulted in stable cell lines expressing either wild-type or mutant VSG2 in addition to wild-type VSG6. Sequence of the BES3 containing VSG6 is from Genbank FM162569; intact ORFs are indicated and the vertical ticks are spaced at 10 kbp.
B. Map of VSG2 showing the location of PTCs, at codons 64, 167, 262, 392, 426 and 460. The signal peptide, the N-terminal domain, linkers (1 and 2), the C-terminal domain, and the GPI-anchor signal sequence, and the two N-linked oligosaccharides (NLO and residue number) are shown. The approximate molecular weights (kDa) of wild-type or each of the PTC-containing VSG monomers are shown.
In addition to a construct designed to express wildtype VSG2 after insertion into the active VSG6 expression site, a series of VSG2 mutants were made by introducing premature termination codons (PTCs) (Fig. 1B). VSG2 is encoded by a 476 codon open reading frame (ORF) and the mature VSG contains 433 amino acids after processing which removes a 26 residue N-terminal signal sequence and a 17 residue C-terminal GPI-anchor addition sequence [33]. Mature VSG2 has two folded domains, the N-terminal domain (residues 27–377, all numbering from the nascent polypeptide), an unstructured linker (378–400), the C-terminal domain (401–442) and a second linker (443–459) with the GPI-anchor attached. The PTCs in the VSG2 open reading frame were located at codons 64, 167 and 262 in the N-terminal domain, 392 in the interdomain linker, 426 in the C-terminal domain and 460, the first residue of the GPI-addition sequence designed to produce a mature VSG without a GPI-anchor (Fig. 1B).
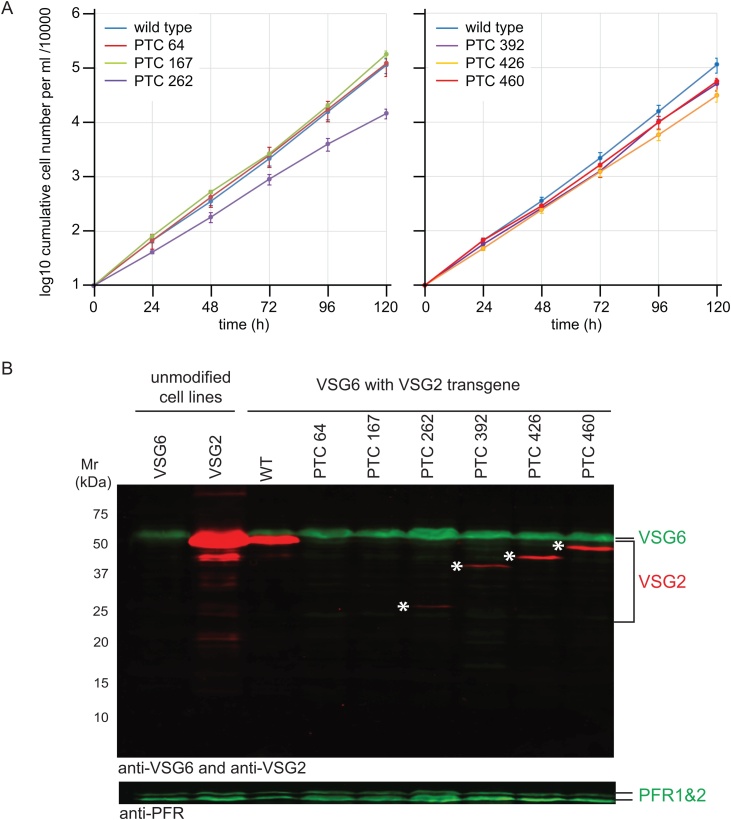
Three independent clones of each transgenic cell line were selected and any effect on proliferation was measured in continuous culture with daily passage over 5 days (Fig. 2A and Supp. Table 2). On averaging the three clones for any one cell line we observed that: (i) expression of a wild type VSG2 transgene had little effect on proliferation when compared to the VSG6 expressing parental cell line, (ii) cell lines that contained the transgene with a PTC at codon 262 grew more slowly with a ∼10-fold reduction in population growth after 5 days as measured by cumulative cell number, (iii) the three cell lines with PTCs closest to the wild type stop codon (codons 392, 426 and 460) all had slightly reduced proliferation over 5 days, with 2- to 3-fold reduction in population growth as measured by cumulative cell number, (iv) PTCs at other locations had no effect.
Fig. 2.
VSG2 transgenes were expressed and PTCs were effective in terminating translation.
A. Measurement of cell number over a five day time course for three independent clones of each transgenic cell line. Proliferation is expressed as cumulative cell number/10,000 and the standard error is shown (Supp. Table 2). Two panels are used for clarity.
B. Western blot of whole cell lysates for the series of cell lines expressing a VSG2 transgene in a VSG6 background. One clone of each cell line is shown: VSG6, the parental cell line expressing VSG6. VSG2, a distinct cell line expressing VSG2 from its endogenous locus. The remaining cell lines express a VSG2 transgene in a VSG6 background: VSG6 VSG2 WT, or WT VSG6 with PTCs at one of the following codons within VSG2: 64, 167, 262, 392, 426 or 460 were loaded on the gel. Whole cell lysates of 2 × 106 cell equivalents were loaded per lane and the white asterix (*) indicates truncated VSG2 polypeptides of the expected predicted molecular weights.
Expression of VSGs in the cell lines was tested by western blotting for both the endogenous VSG6 and any product from the VSG2 transgene (Fig. 2B). VSG2 was detected using an antiserum raised against a peptide corresponding to the N-terminal 15 residues of mature VSG2 [27], these residues were present in all VSG2s encoded by the PTC-containing transgenes. VSG2 polypeptides of the expected molecular weight were detected in cell lines containing PTCs at codons 262, 392, 426 and 460 as well as more abundant expression of the wild type VSG2 transgene. No polypeptides were detected from VSG2 transgenes with PTCs at 67 and 167 (Fig. 2B), these were assumed to be below the limit of detection in this experiment. These findings indicated that the VSG2 transgenes were expressed as expected and that the PTCs were effective in terminating translation.
3.2. PTC-containing VSG2 transgenes are subject to NMD and an additional pathway that detects non-functional VSG resulting in increased VSG mRNA
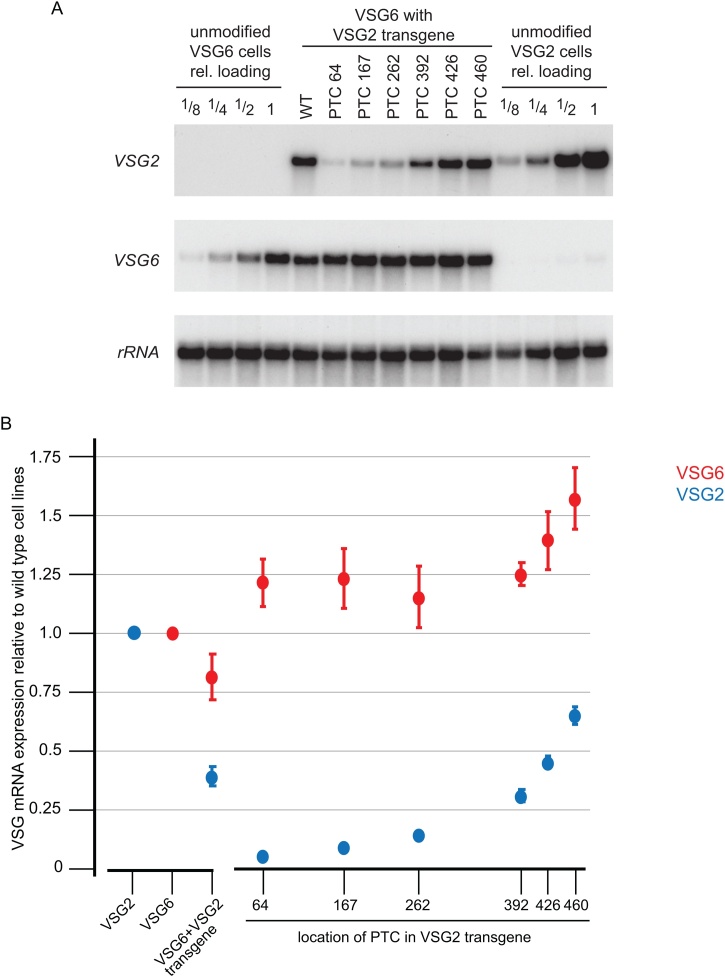
The expression of VSG mRNAs in cell lines containing each transgene was measured using quantitative northern blotting (Fig. 3A). mRNA levels were estimated as the average of three independent clones and expressed relative to the VSG mRNA from wild type cell lines expressing either VSG2 or VSG6 (Supp. Table 3), for example a value of 0.60 represents 60 % of the VSG mRNA present in the wild type cell line expressing that specific VSG. The values were then plotted against the location of the stop codon in the VSG2 transgene (Fig. 3B).
Fig. 3.
Expression of PTC-containing VSG2 transgenes and measurement of transgene and endogenous VSG mRNA levels.
A. Northern blot analysis of VSG2 and VSG6 mRNA levels in one set of cell lines expressing a VSG2 transgene in a VSG6 background. To assist with quantitation, mRNA from unmanipulated VSG6 and VSG2 expressing cell lines was also analysed as a titration of the amount of total RNA loaded. A relative loading of 1 indicated 1 μg total RNA. The blot was probed for rRNA to adjust quantitation for precise loading.
B. VSG mRNA levels determined from quantitative northern blots as above, expressed relative to the VSG mRNA level measured in unmodified cell lines VSG2 (blue) and VSG6 (red). First, an average value for cell lines expressing a wild type VSG2 transgene in a VSG6 background (VSG6 + VSG2 transgene) is shown followed by a plot of VSG mRNA expression against the location of the stop codon in the VSG2 transgene. The error bars are standard error of the mean from measurements of three independent clones for each transgenic cell line. The measurements of VSG6 mRNA in all cell lines with a PTC-containing VSG2 transgene were significantly different (p < 0.05 by Student’s unpaired t-test) to VSG6 mRNA in cell lines with a wild type VSG2 transgene (Supp. Table 3).
The first observation from these measurements was that the insertion of the wild type VSG2 transgene resulted in the expression of two VSG mRNAs as expected, but the levels of the VSG2 and VSG6 mRNAs were not the same, VSG2 mRNA expression was 0.39 and VSG6 mRNA was 0.81.
This observation was investigated further by using RNAseq to estimate VSG mRNA abundance as transcripts per million transcripts (TPM). RNA from wild type cell lines expressing either VSG2 or VSG6 was used along with a cell line expressing wild type VSG2 in a VSG6 background at two time points after electroporation of the transgene construct. RNAseq data was processed to produce estimates of mRNA abundance as transcripts per million transcripts (Table 1). In the wild type cell lines, VSG mRNA levels varied: VSG2 mRNA represented 15 % and VSG6 mRNA 22 % of total mRNA. In the cell line expressing transgenic VSG2, total VSG mRNA was intermediate with 18 % and 20 % of total mRNA respectively, and the ratio of VSG mRNAs was 0.26:0.74 (VSG2:VSG6) (Table 2).
Table 1.
Measurement of VSG mRNA abundance by RNAseq. Two moderately abundant mRNAs (DHH1 and NOT1) an an abundant mRNA (RPL4) are shown for comparison.
| mRNA expression in transcripts per million transcripts (TPM) | |||||
|---|---|---|---|---|---|
| Gene | Accession number | T. brucei L427 VSG2 | T. brucei L427 VSG6 parental | T. brucei L427 VSG6:VSG2 d28 | T. brucei L427 VSG6:VSG2 d44 |
| VSG6 | Tb427.BES15.12 | 25 | 219777 | 136573 | 147716 |
| VSG2 | Tb427.BES40.22 | 151633 | 5 | 47619 | 51168 |
| total VSG | 151658 | 219782 | 184192 | 198884 | |
| DHH1 | Tb427.10.3990 | 294 | 259 | 292 | 306 |
| NOT1 | Tb427.10.1510 | 197 | 161 | 135 | 155 |
| RPL4 | Tb427.03.5050 | 2214 | 1891 | 2202 | 2060 |
Table 2.
The relative abundance of VSG mRNA in a transgenic cell line expressing wild type VSG2 compared to wild type cell lines.
| Fractional VSG mRNA expression | |||||
|---|---|---|---|---|---|
| Gene | Accession number | T. brucei Lister 427 VSG6 parental | T. brucei Lister 427 VSG2 | T. brucei Lister 427 VSG6 p3952 VSG6:VSG2 double expresser d28 | T. brucei Lister 427 VSG6 p3952 VSG6:VSG2 double expresser d44 |
| VSG6 | Tb427.BES15.12 | 1 | 0 | 0.74 | 0.74 |
| VSG2 | Tb427.BES40.22 | 0 | 1 | 0.26 | 0.26 |
The RNAseq estimates of the abundance of VSG2 and VSG6 mRNAs were in agreement with the northern blot data. When the northern blot measurements were converted to abundance estimates, the ratio of mRNAs was 0.25:0.75, similar to that estimated by RNAseq (calculation in Supp. Fig. 2). Thus, the relative abundance of VSG mRNA can vary with the identity of the VSG and the decrease in VSG6 mRNA on introduction of a VSG2 transgene indicated that there is a regulatory system able to adjust the total VSG mRNA concentration. The adjustment could be occurring at the transcriptional and/or post-transcriptional level.
Next, the expression of VSG2 mRNAs in cell lines with PTC-containing transgenes was measured by northern blotting (Fig. 3 and Supp. Table 3). The expression showed a position-dependent effect, with PTCs at codons 64, 167, 262, 392, 426 and 460 resulting in relative levels of 0.05, 0.09, 0.14, 0.31, 0.45 and 0.65 (Fig. 3B). The measurements of VSG2 mRNA for the first four PTCs was as expected for an NMD pathway and provides further evidence that such a pathway is present in trypanosomes. However, an unexpected observation was that in the cell lines with PTCs at codons 426 and 460, the level of VSG2 mRNA, 0.45 and 0.65 respectively, was greater than the wild type VSG2 transgene, 0.39.
When expression of the endogenous VSG6 mRNA was measured, two observations were made: first, expression of all the PTC-containing VSG2 transgenes resulted in an increase in VSG6 mRNA from 0.81 for the wild type to at least 1.15, more than the amount present in the parental cell line (Fig. 3B). Second, for VSG2 transgenes with PTCs close to the C-terminus, there was a larger increase to 1.39 and 1.57 for PTCs at 426 and 460 respectively (Fig. 3B). Thus, synthesis of a faulty VSG protein triggers a general increase in VSG mRNA.
4. Discussion
Trypanosomes tolerate the expression of two VSGs with little effect on growth in culture [32] and, as VSG expression is essential, investigations have used ectopic expression of a second VSG to investigate regulation. Here, transgenes encoding VSG mRNAs containing PTCs were used to trigger VSG mRNA decay in the cytoplasm and the effects on both transgene and endogenous VSG mRNA were measured. The insertion of a VSG2 transgene upstream of the active VSG6 gene (Fig. 1A) followed the original approach to successfully force the simultaneous expression of two VSGs [32], and was used here to ensure that both transgenic and endogenous VSGs were co-transcribed. In previous work [32], three different VSG transgenes were inserted upstream of the endogenous VSG2 gene and, in each case, high levels of expression of VSG2 and the transgenic VSG were detected by immunofluorescence and western blotting [32]. Here, a VSG2 transgene was inserted upstream of the endogenous VSG6 gene and measurements of wild type transgenic and endogenous VSG expression by western blotting were similar to earlier work (Fig. 2B).
When the VSG mRNA levels were measured, the findings were: first, wild type cells expressing either VSG2 or VSG6 contained different amounts of VSG mRNA as a fraction of total mRNA: VSG2 mRNA was 15 %, and VSG6 mRNA was 22 %. Cells expressing a wild type VSG2 transgene contained intermediate levels of VSG mRNA (Fig. 3 and Table 1). These measurements could arise from a genuine difference in VSG mRNA levels and/or result from differences in all other mRNAs, that is the amount of VSG mRNA remains constant but all other mRNAs change. It is not possible to rule out the latter but the former is more likely given probable folding efficiency variation in different VSG proteins discussed below.
Second, cell lines expressing a wild type VSG2 transgene in a VSG6 background contained a decreased amount of VSG6 mRNA. However, the cells did not contain equal numbers of the two VSG mRNAs, the ratio was approximately 1:3 (VSG2:VSG6) measured by both RNAseq and northern blotting (Fig. 3 and Table 1).
Third, there was the expected position dependent response of VSG2 mRNA levels to the inclusion of a PTC, the nearer the PTC to the N-terminus the lower the steady state levels of VSG2 mRNA (Fig. 3). However, reduction in VSG2 mRNA only occurred with PTCs located in the N-terminal domain or the inter-domain linker. This decrease in VSG2 mRNA resulted in an increase in VSG6 mRNA.
Fourth, VSG2 transgenes containing a PTC in the C-terminal domain or at codon 460, which directed synthesis VSG2 without a GPI-anchor, resulted in both VSG2 and VSG6 mRNAs increasing 1.6 to 2-fold compared to the levels in cells expressing a wild type VSG2 transgene (Fig. 3). These variations in copy number indicate that it is unlikely that there is a direct counting mechanism for VSG mRNA, but rather a feedback from the production of functional VSG protein.
The first two findings concern the difference in VSG mRNA copy number in cells expressing different VSGs. Since the variation is present in the cell line expressing two VSGs from the same BES, it does not result from differential transcription and must be explained by post-transcriptional process(es). The major determinant of stability for most mRNAs in trypanosomes is codon use, expressed numerically as a gene expression codon adaptation index (geCAI) [26]. In this case, the geCAI value for VSG2 is 0.33 and VSG6 is 0.35, and this difference might contribute towards the higher levels of VSG6 mRNA but is unlikely to be the sole determinant. The second observation that expression of a second VSG results in a reduction of the endogenous VSG mRNA has been reported before [16,17].
The expression of VSG2 with a PTC in the N-terminal domain or inter-domain linker resulted in the reduction in VSG2 mRNA levels, inversely proportional to the length of the residual open reading frame (Fig. 3). This is a typical characteristic of an NMD pathway [23] and has been reported before in trypanosomes [24]. The decrease in VSG2 mRNA resulted in an increase in VSG6 mRNA to ∼1.5 fold more than cells expressing a full length VSG2 transgene and ∼1.2 fold more than parental cells expressing VSG6. This increase to levels above that present in wild type cells indicated that the trypanosome has spare capacity for the production of VSG mRNA and is emphasised in cells expressing VSG2 transgenes with PTCs in the C-terminal domain or immediately after the mature C-terminus, so that a complete but not GPI-anchored VSG was synthesised. In the latter case, both VSG2 and VSG6 mRNAs increased 1.6 to 2-fold.
One model arising from the measurements above is that VSG mRNA was subject to two pathways. First, an NMD pathway that decreased the amount of PTC-containing mRNA but became less effective when the PTC was close to the C-terminus. Second, a pathway that detected unsuccessful production of membrane-anchored VSG and triggered an increase in total VSG mRNA. The second pathway is apparent as an increase in VSG6 mRNA in cells expressing VSG2 with an early PTC and an increase in both VSG mRNAs in cell expressing VSG2 with a late PTC, when the NMD pathway is less potent.
The kinetics and route of VSG synthesis are well characterised (reviewed in [34,35]). As VSGs are translated by ER-associated ribosomes, the PTC-containing VSG mRNAs would have directed translocation of a nascent VSG into the ER. It is likely that several could not fold correctly and were potential substrates for an unfolded protein response (UPR) pathway. However, there is evidence that the UPR is absent in bloodstream form trypanosomes [36] and that GPI-anchorless VSG is selectively retained and degraded in the lysosome [37,38]. Our favoured model for the function of the pathway that increases VSG mRNA in response to incorrect VSG protein is that it evolved to compensate for variation in the rates of successful folding of different VSGs. If only a small fraction of VSG protein folds successfully, the pathway increases VSG mRNA and if a larger fraction of the VSG folds correctly, then less VSG mRNA is produced. This model predicts that VSG2 folds more efficiently than VSG6. The model is also consistent with the observation that bloodstream form trypanosomes seem to have a capacity for degrading a large amount of non-functional VSG without any great effect on proliferation.
As VSG mRNAs are highly variable in sequence, how is the increased mRNA that occurred in response to the anchorless VSG specific to VSG mRNA? Different VSG mRNAs have one specific conserved sequence feature, the ‘16-mer’ element in the 3′ UTR [16] and this is an obvious candidate for mediating a specific response but the experiments here do not distinguish whether this happens in the cytoplasm via mRNA half-life and/or in the nucleus via transcription rate or more efficient mRNA maturation.
The strict selection pressure imposed by the mammalian immune response on bloodstream trypanosomes makes it likely that VSG synthesis is tightly regulated. Here, evidence is provided for sensors that detect the production of functional VSG coated membranes and a response that alters VSG mRNA levels. The regulation of VSG synthesis is central to the survival of the African trypanosome in a mammalian host and as such its regulation will contain further complexities.
Author contributions
I.E.M. generated cell lines, performed proliferation experiments, performed sample preparation and subsequent western and northern blotting and analyses. S.K. performed RNA sequencing data analysis. A.S generated cell lines and prepared RNA for RNAseq. M.C. and A.S. devised and supervised the study. M.C. analysed data and wrote the manuscript. The final version of the manuscript was approved by all authors.
Acknowledgements
This work was funded by the Wellcome Trust award 093008/Z/10/Z. MC is a Wellcome Trust Investigator217138/Z/19/Z. We would like to thank Sarah Marsden, Markus Engstler, Mark Field, Jay Bangs and Nancy Standart for reading and commenting on the manuscript.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.molbiopara.2020.111348.
Contributor Information
Angela Schwede, Email: angela.schwede@gmx.de.
Mark Carrington, Email: mc115@cam.ac.uk.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Mugnier M.R., Stebbins C.E., Papavasiliou F.N. Masters of disguise: antigenic variation and the VSG coat in Trypanosoma brucei. PLoS Pathog. 2016;12(9) doi: 10.1371/journal.ppat.1005784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cestari I., Stuart K. Transcriptional regulation of telomeric expression sites and antigenic variation in trypanosomes. Curr. Genomics. 2018;19(2):119–132. doi: 10.2174/1389202918666170911161831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faria J., Glover L., Hutchinson S., Boehm C., Field M.C., Horn D. Monoallelic expression and epigenetic inheritance sustained by a Trypanosoma brucei variant surface glycoprotein exclusion complex. Nat. Commun. 2019;10(1):3023. doi: 10.1038/s41467-019-10823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sima N., McLaughlin E.J., Hutchinson S., Glover L. Escaping the immune system by DNA repair and recombination in African trypanosomes. Open Biol. 2019;9(11) doi: 10.1098/rsob.190182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L.C., Chung H.M., Lee P.T., Lee M.G. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell. 2003;2(3):542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navarro M., Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414(6865):759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 7.Clayton C.E. Gene expression in Kinetoplastids. Curr. Opin. Microbiol. 2016;32:46–51. doi: 10.1016/j.mib.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Glover L., Hutchinson S., Alsford S., Horn D. VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 2016;113(26):7225–7230. doi: 10.1073/pnas.1600344113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheader K., Vaughan S., Minchin J., Hughes K., Gull K., Rudenko G. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 2005;102(24):8716–8721. doi: 10.1073/pnas.0501886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ooi C.P., Smith T.K., Gluenz E., Wand N.V., Vaughan S., Rudenko G. Blocking variant surface glycoprotein synthesis alters endoplasmic reticulum exit sites/Golgi homeostasis in Trypanosoma brucei. Traffic. 2018;19(6):391–405. doi: 10.1111/tra.12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D., Albergante L., Newman T.J., Horn D. Faster growth with shorter antigens can explain a VSG hierarchy during African trypanosome infections: a feint attack by parasites. Sci. Rep. 2018;8(1):10922. doi: 10.1038/s41598-018-29296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field M.C., Sergeenko T., Wang Y.N., Bohm S., Carrington M. Chaperone requirements for biosynthesis of the trypanosome variant surface glycoprotein. PLoS One. 2010;5(1):e8468. doi: 10.1371/journal.pone.0008468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y.N., Wang M., Field M.C. Trypanosoma brucei: trypanosome-specific endoplasmic reticulum proteins involved in variant surface glycoprotein expression. Exp. Parasitol. 2010;125(3):208–221. doi: 10.1016/j.exppara.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiengwe C., Muratore K.A., Bangs J.D. Surface proteins, ERAD and antigenic variation in Trypanosoma brucei. Cell. Microbiol. 2016;18(11):1673–1688. doi: 10.1111/cmi.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehlers B., Czichos J., Overath P. RNA turnover in Trypanosoma brucei. Mol. Cell. Biol. 1987;7(3):1242–1249. doi: 10.1128/mcb.7.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ridewood S., Ooi C.P., Hall B., Trenaman A., Wand N.V., Sioutas G., Scherwitzl I., Rudenko G. The role of genomic location and flanking 3’UTR in the generation of functional levels of variant surface glycoprotein in Trypanosoma brucei. Mol. Microbiol. 2017;106(4):614–634. doi: 10.1111/mmi.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batram C., Jones N.G., Janzen C.J., Markert S.M., Engstler M. Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. Elife. 2014;3 doi: 10.7554/eLife.02324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand S., Lykke-Andersen J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat. Struct. Mol. Biol. 2013;20(6):702–709. doi: 10.1038/nsmb.2575. [DOI] [PubMed] [Google Scholar]
- 19.Rufener S.C., Muhlemann O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2013;20(6):710–717. doi: 10.1038/nsmb.2576. [DOI] [PubMed] [Google Scholar]
- 20.Wen J., Brogna S. Nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2008;36(Pt 3):514–516. doi: 10.1042/BST0360514. [DOI] [PubMed] [Google Scholar]
- 21.He F., Jacobson A. Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 2015;49:339–366. doi: 10.1146/annurev-genet-112414-054639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen J., Brogna S. Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J. 2010;29(9):1537–1551. doi: 10.1038/emboj.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc. Natl. Acad. Sci. U. S. A. 1979;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delhi P., Queiroz R., Inchaustegui D., Carrington M., Clayton C. Is there a classical nonsense-mediated decay pathway in trypanosomes? PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirumi H., Hirumi K. Axenic culture of African trypanosome bloodstream forms. Parasitol. Today. 1994;10(2):80–84. doi: 10.1016/0169-4758(94)90402-2. [DOI] [PubMed] [Google Scholar]
- 26.de Freitas Nascimento J., Kelly S., Sunter J., Carrington M. Codon choice directs constitutive mRNA levels in trypanosomes. Elife. 2018;7 doi: 10.7554/eLife.32467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grunfelder C.G., Engstler M., Weise F., Schwarz H., Stierhof Y.D., Boshart M., Overath P. Accumulation of a GPI-anchored protein at the cell surface requires sorting at multiple intracellular levels. Traffic. 2002;3(8):547–559. doi: 10.1034/j.1600-0854.2002.30805.x. [DOI] [PubMed] [Google Scholar]
- 28.Fiebig M., Kelly S., Gluenz E. Comparative life cycle transcriptomics revises Leishmania Mexicana genome annotation and links a chromosome duplication with parasitism of vertebrates. PLoS Pathog. 2015;11(10) doi: 10.1371/journal.ppat.1005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts A., Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10(1):71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz-Jordan J.L., Davies K.P., Cross G.A. Stable expression of mosaic coats of variant surface glycoproteins in Trypanosoma brucei. Science. 1996;272(5269):1795–1797. doi: 10.1126/science.272.5269.1795. [DOI] [PubMed] [Google Scholar]
- 33.Carrington M., Miller N., Blum M., Roditi I., Wiley D., Turner M. Variant specific glycoprotein of Trypanosoma brucei consists of two domains each having an independently conserved pattern of cysteine residues. J. Mol. Biol. 1991;221(3):823–835. doi: 10.1016/0022-2836(91)80178-w. [DOI] [PubMed] [Google Scholar]
- 34.Silverman J.S., Bangs J.D. Form and function in the trypanosomal secretory pathway. Curr. Opin. Microbiol. 2012;15(4):463–468. doi: 10.1016/j.mib.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manna P.T., Boehm C., Leung K.F., Natesan S.K., Field M.C. Life and times: synthesis, trafficking, and evolution of VSG. Trends Parasitol. 2014;30(5):251–258. doi: 10.1016/j.pt.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiengwe C., Brown A.E., Bangs J.D. Unfolded protein response pathways in bloodstream-form Trypanosoma brucei? Eukaryot. Cell. 2015;14(11):1094–1101. doi: 10.1128/EC.00118-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Triggs V.P., Bangs J.D. Glycosylphosphatidylinositol-dependent protein trafficking in bloodstream stage Trypanosoma brucei. Eukaryot. Cell. 2003;2(1):76–83. doi: 10.1128/EC.2.1.76-83.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohme U., Cross G.A. Mutational analysis of the variant surface glycoprotein GPI-anchor signal sequence in Trypanosoma brucei. J. Cell. Sci. 2002;115(Pt 4):805–816. doi: 10.1242/jcs.115.4.805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.