Abstract
Objective
Accumulating evidence illustrates that sirtuins (SIRTs) regulate autophagy and apoptosis in cancer cells; however, the role of SIRT5 in gastric cancer (GC) cells remains unknown. In this study, we examined the role of SIRT5 in GC cells.
Methods
We detected SIRT5 protein levels in freshly collected samples from patients with GC. Next, we studied the function of SIRT5 in autophagy. Furthermore, the signaling pathway through which SIRT5 enhanced autophagy in GC cells was detected. In addition, we established a GC cell apoptosis model to analyze the role of SIRT5 in apoptosis.
Results
SIRT5 expression was downregulated in GC tissues. We discovered that SIRT5 promoted autophagy in GC cells. We demonstrated that SIRT5 enhanced autophagy in GC cells via the AMP-activated protein kinase–mammalian target of rapamycin signaling pathway. In addition, SIRT5 was degraded during apoptosis in GC cells. Meanwhile, we observed that calpains and caspase-related proteins were associated with SIRT5-related GC cell apoptosis.
Conclusions
SIRT5 is a crucial regulator of autophagy and apoptosis in GC cell lines that can maintain the balance of autophagy and apoptosis.
Keywords: Sirtuin 5, autophagy, apoptosis, gastric cancer, calpain, caspase, AMP-activated protein kinase, mammalian target of rapamycin
Introduction
Gastric cancer (GC) is one of the most common cancers and the third leading cause of cancer-related mortality worldwide,1 accounting for 723,000 mortalities in 2013.2 To our knowledge, many factors can contribute to its development and progression, including exogenous and endogenous factors. The therapeutic effect of surgery on GC is remarkable; however, the median overall survival (OS) rate of patients with GC remains poor.3 Thus, it is necessary to identify an effective and efficient therapy for GC.
In recent years, increasing evidence has revealed that changes in autophagy are associated with the genesis and development of tumors.4–6 In cells, autophagy is responsible for the degradation of excessive nutrient deposits.7 Autophagy is also extremely vital for cells to maintain their basic functions because it eliminates aged organelles and accumulated toxic material. Autophagy is induced in response to cellular stress to promote survival. However, excessive autophagy will lead to cell death.
Sirtuins (SIRTs) comprise a family of nicotinamide adenine dinucleotide-dependent lysine deacetylases that are characterized by a highly conserved SIRT catalytic core domain.8 Humans express seven SIRTs that are located in different parts of the cell. SIRT1, SIRT6, and SIRT7 are localized to the nucleus. SIRT2 is present primarily in the cytoplasm, whereas SIRT3, SIRT4, and SIRT5 are located in mitochondria.9 In addition to regulating multiple aspects of physiological responses, including cell stress and metabolism,10,11 SIRTs are also believed to participate in the development of various cancers.12–14
SIRT5 is believed to regulate cytosolic and mitochondrial protein malonylation, with glycolysis as its major target (i.e., TCA cycle, urea cycle, glycolysis, fatty acid oxidation).9,15,16 Increasing evidence indicates that the SIRT family, including SIRT5, regulates both autophagy and apoptosis.17–19 Among the SIRTs family members, SIRT5 is the least known. In 2019, an interesting study proved that SIRT5 plays an important role in autophagy in colorectal cancer.20 However, the functions of SIRT5 in both autophagy and apoptosis in GC cells remain largely obscure.
Methods and materials
Cells and culture conditions
Human GC cells (MKN7, AGS, SUN1, and HCG27) were purchased from the American Type Culture Collection (Manassas, VA, USA). GC cells were cultured in Leibovitz’s L-15 medium (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) at 37°C in a humidified atmosphere of 5% CO2. The apoptosis-inducing agent etoposide (Sigma-Aldrich, St. Louis, MO, USA) was used at a concentration of 5 µg/mL for 4 or 6 hours. The cells were treated with 100 nM rapamycin (Invitrogen, Thermo Fisher Scientific) for 24 hours to induce autophagy. All agents were dissolved in dimethyl sulfoxide.
Transfection
All GC cells (2 × 105) were seeded into six-well plates, cultured for 24 hours, and then transfected with 100 pmol small-interfering RNA (siRNA) using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) in serum-free medium according to the manufacturer’s protocol. After 4 to 6 hours, the medium was replaced with complete culture medium. The transfected cells were harvested for protein extraction or used for other experiments after 2 days. SIRT5 siRNAs and negative-control siRNA were purchased from GenePharma (Shanghai, China). The three SIRT5 siRNA sequences were as follows: human SIRT5 siRNA #1, 5′-GCCAAGUUCAAGUAUGGCATT-3′; siRNA #2, 5′-GCUGGUGUUAGUGCAGAAATT-3′; and siRNA #3, 5′-GCAUCCCAGUUGAGAAACU-3′. The SIRT5-overexpressing (OE) lentivirus and control lentivirus were both purchased from GenePharma. In total, 1 × 106 lentiviral particles were added to each well with gentle mixing, and puromycin was used to select the SIRT5-OE cell line.
Patients and tissue specimens
Fifteen fresh GC tissue samples and paired noncancerous tissue samples were collected from patients with GC during surgery between January 2017 and May 2017 at the Department of Pathology at the First Affiliated Hospital of Soochow University (Soochow, China). None of the patients had received radiotherapy or chemotherapy before surgery.
All patients were diagnosed with GC according to the sixth edition of the American Joint Committee on Cancer staging manual. Patients with GC who underwent curative surgery between January 2010 and September 2010, who were diagnosed with gastric adenocarcinoma, and who had adequate tumor samples were enrolled in this study. Immunohistochemistry was used to determine the level of SIRT5 expression in this cohort. Basic patient data were collected, including age, sex, tumor location, gross appearance, histologic analysis, lymph node metastasis status, and pathologic stage. Surgical specimens were obtained after gastrectomy. The tumor tissues were fixed and wax-embedded. After surgery, patients were followed up at our outpatient clinic every month. Recurrence was defined as the first imaging evidence of tumor relapse, cytologic analysis of ascites from abdominal tapering, endoscopic findings of tumor recurrence, and/or metastases observed on a bone scan. The pattern of tumor recurrence was recorded in detail. The follow-up data were prospectively collected and regularly updated. OS was defined as the time from the date of surgery to that of death or the latest follow-up. Disease-free survival (DFS) was defined as the period from the date of surgery for GC to that of recurrence.
All research involving human participants was approved by the Ethics Committee of the First Affiliated Hospital of Soochow University (Soochow, China). Informed consent was obtained from all patients.
Immunohistochemistry
Tissues were fixed in formalin, embedded in paraffin, and sectioned before being mounted on slides, which were then subjected to deparaffinization and rehydration. Then, the slides were microwaved for 30 minutes in 0.01 mol/L sodium citrate buffer (pH 6.0). After antigen retrieval and pre-incubation with 10% normal goat serum, anti-SIRT5 (1: 400; Cell Signaling Technology, Danvers, MA, USA) was employed at 4°C overnight. These slides were stained using a VECTSDTSIN Elite ABC Kit (Vector Laboratories, Maravai LifeSciences, San Diego, CA, USA) and counterstained with hematoxylin. The intensity of staining was scored as follows: 0, no staining; 1, weak, light yellow; 2, moderate, yellow-brown; and 3, strong, brown. In addition, the proportion of positive cells was categorized as follows: 0, <10%; 1, 10% to 49%; 2, 50% to 74%; and 3, >75%. Then, the staining score was calculated by multiplying the staining intensity score by the proportion of positive cells score. A staining score of 0 or 1 indicated negative SIRT5 expression, whereas higher scores indicated positive expression.
Western blot analysis
All antibodies were purchased from Cell Signaling Technology. Phosphate-buffered saline (PBS) supplemented with 1 mmol/L phenylmethylsulfonyl fluoride (PMSF) was used to wash cells twice. Cells were scraped off the dishes, pelleted at 500 × g for 10 minutes, lysed in cold lysis buffer (20 mmol/L Tris-HCl, 1 mmol/L EDTA, 150 mmol/L NaCl, 1 mmol/L EGTA, 1% Triton X-100, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, 1 µg/mL leupeptin, and 1 mmol/L PMSF), and sonicated for 5 s. The lysates were clarified by centrifugation at 12,000 × g for 30 minutes at 4°C. Identical amounts of cell lysates were resolved via 8% or 15% SDS-PAGE. Membranes were incubated in blocking solution consisting of 5% powdered milk in Tris-buffered saline with Tween-20 (10 mmol/L Tris-HCl, 150 mmol/L NaCl, and 1% Tween-20) for 1 hour and then immunoblotted with the appropriate antibodies. Then, the polyvinylidene fluoride membranes were treated with a horseradish peroxide-conjugated anti-rabbit IgG antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 hour at room temperature. All primary antibodies were used at 1:1000, and all secondary antibodies were used at 1:2000.
Specific bands were detected using an enhanced chemiluminescence system (Pierce, Thermo Fisher Scientific). The band intensity was semiquantitated using BandScan software (Bio-Rad, Hercules, CA, USA) after digitizing using a V300 scanner (Epson, Tokyo, Japan).
Immunofluorescence staining
The cells were fixed in 4% paraformaldehyde for 10 minutes and then washed with PBS for 5 minutes. The slides were immersed in 100 mg/mL digitonin for 15 minutes at room temperature and then washed. Subsequently, the cells were incubated with an anti-microtubule-associated protein 1 light chain 3 (LC3) antibody (MBL, Nagoya, Japan) and washed three times. A FITC-conjugated anti-rabbit IgG antibody (MBL) was added to the cells, and the cells were washed after the incubation.
Apoptosis analysis
An Annexin-V-PE Apoptosis Detection kit (Invitrogen, Thermo Fisher Scientific) was used to measure apoptosis. Cells were washed with PBS and resuspended in 1× binding buffer at a concentration of 1 × 106 cells/mL. Subsequently, 5 mL of annexin V–phycoerythrin and 5 mL of propidium iodide were added to 100 mL of the cell suspension, and the mixture was incubated for 15 minutes in the dark. After incubation, 400 mL of 1× binding buffer were added. The analyses were performed using a FACScan flow cytometer (Beckman Coulter, Fullerton, CA, USA).
Electron microscopy
HCG27 cells were treated with 100 nM rapamycin for 24 hours to induce autophagy. Then, the cells were fixed with 2% paraformaldehyde–2% glutaraldehyde in 0.1 mL/L phosphate buffer (pH 7.4), followed by 1% osmium tetroxide. After dehydration, thin sections were stained with uranyl acetate and lead citrate for observation using a JEM 100 CX electron microscope (JEOL, Tokyo, Japan).
Statistical analysis
Statistical analysis was performed using SPSS version 22.0 (IBM, Armonk, NY, USA). All experiments were repeated at least three times. Statistical significance was determined using Student’s t-test. P ≤ 0.05 indicated a significant difference.
Results
SIRT5 is downregulated in GC tissues
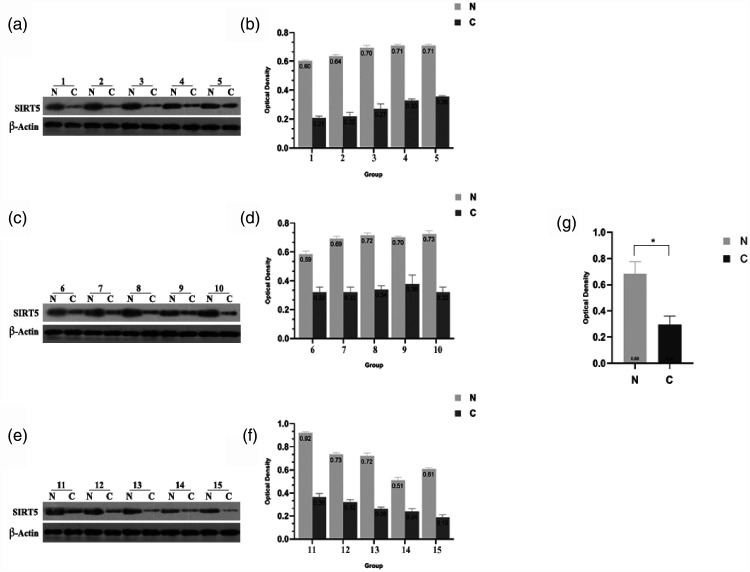
Western blotting was performed to confirm the expression of SIRT5 in GC tissues. We collected 15 pairs of gastric tumor tissues and adjacent tissues from patients. As presented in Figure 1a, c, and e, SIRT5 expression was substantially downregulated in most GC tissues compared with its levels in the paired adjacent noncancerous tissues. Figure 1b, d, and f reveals the relative optical density of SIRT5, illustrating the higher expression of SIRT5 in normal gastric tissues. After incorporating the relative optical density data, tissues were divided into two groups (tumor and adjacent noncancerous tissues). Figure 1g reveals that SIRT5 expression is much higher in adjacent noncancerous tissues than in tumor tissues (P < 0.05). Thus, we concluded that SIRT5 is downregulated in GC tissues. To further study whether the expression of SIRT5 was correlated with tumor progress, we determined the associations of SIRT5 with several clinicopathological factors. Notably, SIRT5 expression was correlated with nodal involvement, lymphovascular invasion, 5-year OS, and 5-year DFS (all P < 0.05, Table 1).
Figure 1.
SIRT5 protein expression in GC samples. (a–b) Western blotting and quantification of SIRT5 expression in paired fresh tissues from patients with GC (groups 1–5). (c–d) Western blotting and quantification of SIRT5 expression in paired fresh tissues from patients with GC (groups 6–10). (e–f) Western blotting and quantification of SIRT5 expression in paired fresh tissues from patients with GC (groups 11–15). Western blotting and quantification in 15 paired fresh tissues from patients with GC revealed decreased SIRT5 expression in tumor tissues. Data represent the average and SD of three independent experiments (*P < 0.05).
SIRT5, sirtuin 5; GC, gastric cancer.
Table 1.
Associations of SIRT5 expression with clinicopathological variables in patients with gastric cancer (n = 176).
| No expression | Expression | ||
|---|---|---|---|
| Characteristic | (n = 112) | (n = 64) | P |
| Age | 65.89 | 67.88 | 0.38 |
| Tumor size (cm) | |||
| <4 | 40 | 28 | |
| 4–8 | 44 | 24 | |
| >8 | 28 | 12 | 0.49 |
| Tumor location | |||
| Upper stomach | 28 | 18 | |
| Middle stomach | 34 | 18 | |
| Lower stomach | 46 | 26 | |
| Whole stomach | 4 | 2 | 0.97 |
| Cell grade | |||
| Poorly differentiated | 90 | 40 | |
| Moderately differentiated | 20 | 22 | |
| Well-differentiated | 2 | 2 | 0.35 |
| Gross appearance | |||
| Superficial type | 30 | 16 | |
| Borrmann types I and II | 38 | 24 | |
| Borrmann types III and IV | 44 | 28 | 0.89 |
| Lauren’s histology | |||
| Intestinal type | 54 | 28 | |
| Diffuse type | 58 | 36 | 0.56 |
| Nodal involvement | |||
| Negative | 18 | 20 | |
| Positive | 94 | 44 | 0.02* |
| Lymphovascular invasion | |||
| No | 20 | 28 | |
| Yes | 92 | 36 | 0.0002* |
| Depth of cancer invasion | |||
| Mucosa | 10 | 4 | |
| Submucosa | 14 | 6 | |
| Proper muscle | 48 | 24 | |
| Serosa | 40 | 30 | 0.52 |
| TNM stage | |||
| I | 10 | 4 | |
| II | 70 | 38 | |
| III | 32 | 22 | 0.64 |
| 5-Year OS | 39.10% | 51.20% | 0.005* |
| 5-Year DFS | 38.00% | 49.60% | 0.010* |
P < 0.05.
SIRT5, sirtuin 5; OS, overall survival; DFS, disease-free survival.
SIRT5 plays an important role in inducing GC cell autophagy
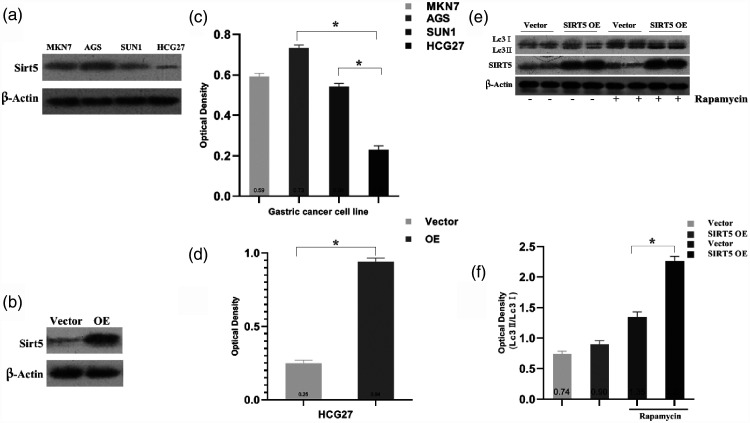
Previous work revealed SIRT5 expression in human GC tissue. In addition, there is evidence that SIRT family members can regulate autophagy.21 Therefore, we investigated the role of SIRT5 in autophagy in GC cells. First, four gastric cell lines, namely MKN7, AGS, SUN1, and HCG27 cells, were used to detect SIRT5 levels via western blotting. We found that SIRT5 expression was highest in AGS cells and lowest in HCG27 cells, as presented in Figure 2a. Figure 2c demonstrates the relative optical density in GC cell lines. The average relative optical density was highest in AGS cells and lowest in HGC27 cells (0.73 vs. 0.23, P < 0.05). Therefore, in our subsequent study, AGS cells were used to examine the downregulation of SIRT5, and HCG27 cells were used to examine the upregulation of SIRT5. As illustrated in Figure 2b and d, HCG27 cells were transfected with vector or SIRT5 lentivirus plasmids, and SIRT5 expression was detected using western blotting. The transfection effect was obvious, and the expression of SIRT5 was upregulated in HCG27 cells (P < 0.05).
Figure 2.
SIRT5 was detected in GC cell lines. SIRT5 overexpression promoted autophagy in HCG27 cells treated with 100 nM rapamycin. (a, c) Western blotting and quantification of SIRT5 expression in GC cell lines (MKN7, AGS, SUN1, and HCG27). (b, d) Western blotting and optical density quantification of SIRT5 levels in HCG27 cells after transfection with an SIRT5 lentivirus or control lentivirus. (e) Western blotting of SIRT5 expression and the LC3-II/LC3-I ratio in SIRT5-OE or vector cells incubated with or without rapamycin. (f) Relative optical density quantification of the LC3-II/LC3-I ratio. Data represent the average and SD of three or six independent experiments (*P < 0.05).
SIRT5, sirtuin 5; GC, gastric cancer; LC3-II, autophagosome-associated form of microtubule-associated protein 1 light chain 3; LC3-I, cytosolic form of microtubule-associated protein 1 light chain 3; SIRT5-OE, sirtuin 5-overexpressing lentivirus; Vector, control lentivirus.
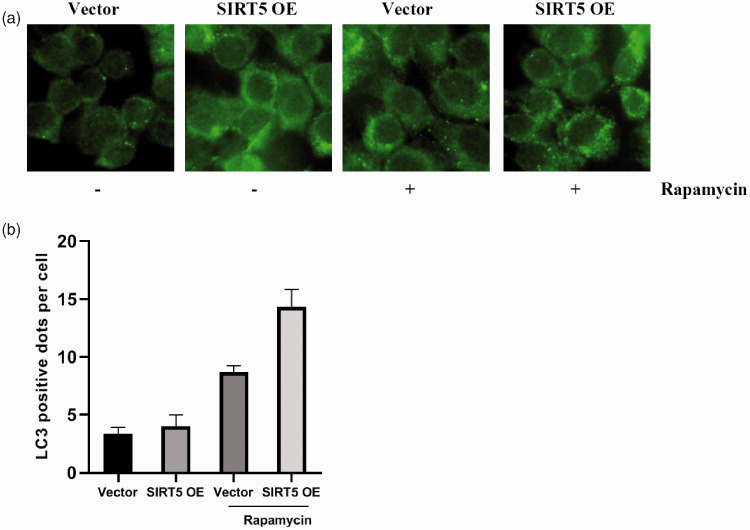
LC3 is a homolog of Apg8p for autophagy in yeast and a marker of autophagy formation. The C-terminal fragment of LC3 is cleaved immediately to yield a cytosolic form called LC3-I. A subpopulation of LC3-I is further converted to an autophagosome-associated form named LC3-II that can be detected by western blotting.22 Rapamycin was applied to induce autophagy in HCG27 cells. As highlighted in Figure 2e and f, we detected an elevated ratio of LC3-II/LC3-I in SIRT5-OE HCG27 cells when autophagy was induced by rapamycin (P < 0.05). Furthermore, we analyzed autophagosome formation by fluorescence microscopy (Figure 3a and b).
Figure 3.
Autophagy is induced by rapamycin in HCG27 cells. (a) Immunofluorescent images of HCG27 cells after transfection with an SIRT5 lentivirus or control lentivirus and incubation with or without rapamycin. (b) Quantification of autophagy in HCG27 cells after transfection with an SIRT5 lentivirus or control lentivirus and incubation with or without rapamycin. Data represent the average and SD of three independent experiments (*P < 0.05).
SIRT5, sirtuin 5; SIRT5-OE, sirtuin 5-overexpressing lentivirus; Vector, control lentivirus.
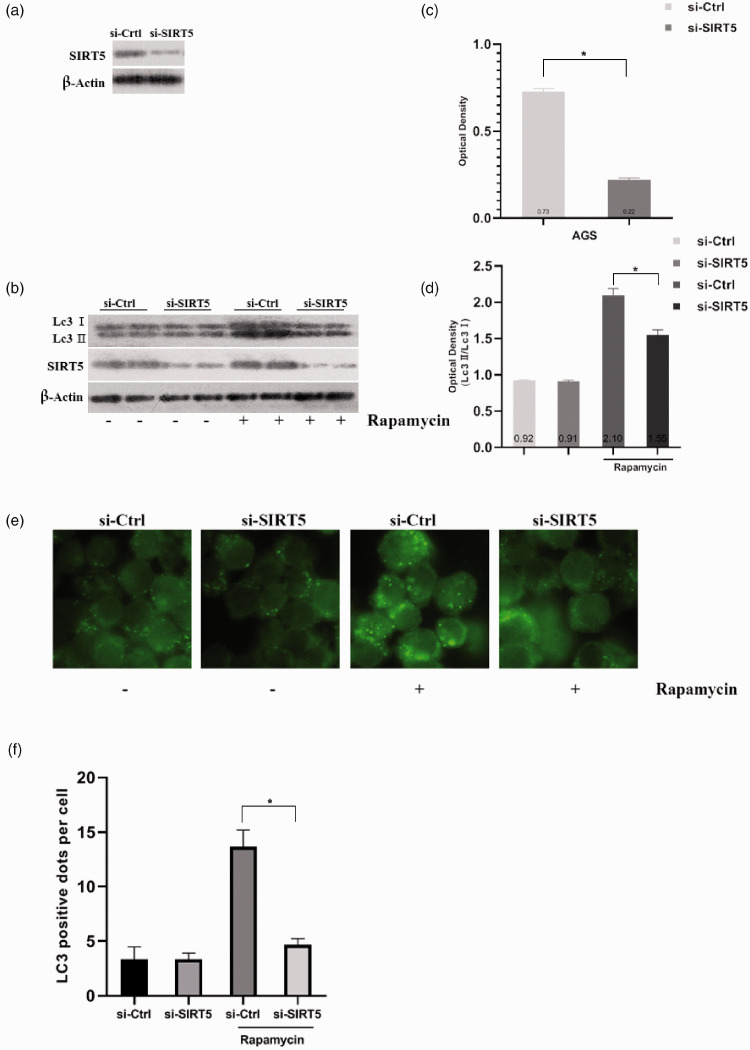
On the contrary, we depleted SIRT5 using siRNA in AGS cells. The interference efficiency was confirmed, as displayed in Figure 4a and c (P < 0.05). Then, we observed a lower LC3-II/LC3-I ratio in SIRT5-depleted cells when autophagy was induced (P < 0.05, Figure 4b and d). We also used fluorescent microscopy to observe autophagosomes (Figure 4e and f). All of these results were similar, and they all preliminarily proved that SIRT can promote autophagy.
Figure 4.
Autophagy is suppressed in AGS cells when SIRT5 was depleted using siRNA and SIRT5-OE HCG27 cells were observed using electron micrographs after treated with rapamycin. (a, c) Western blot analysis and corresponding optical density quantification of SIRT5 levels in AGS cells after transient transfection with negative-control and SIRT5 siRNA oligonucleotides. (b, d) Western blot analysis and optical density quantification of SIRT5 and LC3-II/LC3-I levels in AGS cells after transient transfection with negative-control and SIRT5 siRNA oligonucleotides. (e) Immunofluorescent images of AGS cells after transient transfection with negative-control and SIRT5 siRNA oligonucleotides and incubation with or without 100 nM rapamycin. (f) Quantification of autophagy in AGS cells after transient transfection with negative-control and SIRT5 siRNA oligonucleotides and incubation with or without rapamycin. Data represent the average and SD of three or six independent experiments (*P < 0.05).
SIRT5, sirtuin 5; SIRT5-OE, sirtuin 5-overexpressing lentivirus; Vector, control lentivirus; siRNA, small-interfering RNA; LC3-II, autophagosome-associated form of microtubule-associated protein 1 light chain 3; LC3-I, cytosolic form of microtubule-associated protein 1 light chain 3.
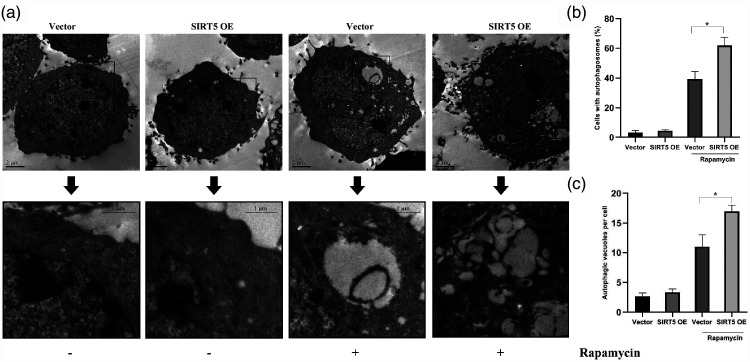
Finally, transmission electron microscopy was used to inspect autophagosomes in HCG27 cells, as presented in Figure 5a. We discovered changes between vector-transfected and SIRT5-OE cells. We detected more autophagosomes in SIRT5-OE cells than in vector-transfected cells when autophagy was induced by rapamycin (P < 0.05, Figure 5b and c).
Figure 5.
Rapamycin induced the formation of autophagic vacuoles in HCG27 cells. (a) Electron micrographs of autophagy in vector-transfected or SIRT5-OE HCG27 cells with or without 100 nM rapamycin. (b) The percentage of HCG27 cells with autophagosomes. (c) The average number of autophagic vacuoles per cell. Data represent the average and SD of three independent experiments (*P < 0.05).
SIRT5, sirtuin 5; SIRT5-OE, sirtuin 5-overexpressing lentivirus; Vector, control lentivirus.
These data revealed that SIRT5 promotes autophagy in GC cells.
SIRT5 promotes autophagy in GC cells via the AMP-activated protein kinase (AMPK)–mammalian target of rapamycin (mTOR) pathway
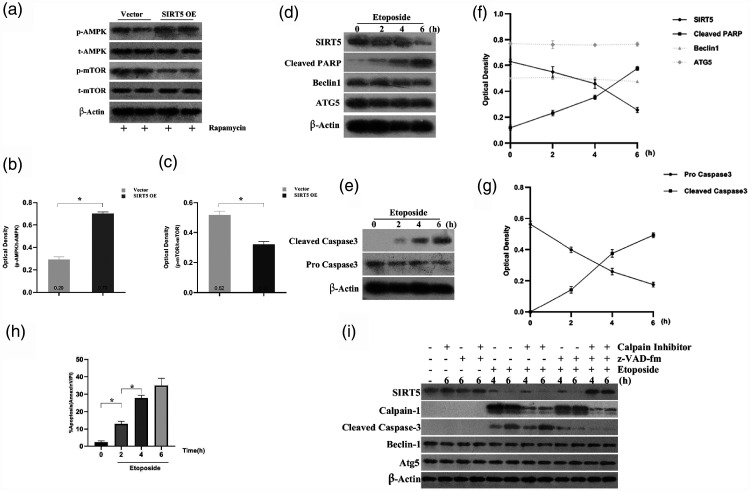
In our previous study, we discussed the function of SIRT5 in autophagy in GC cells. However, the mechanism by which SIRT5 modulates autophagy remains unknown, and we further investigated the mechanism by assessing the levels of the autophagy regulators AMPK and mTOR in HCG27 cells treated with rapamycin. Western blotting and quantification of the results indicated that the ratio of phosphorylated AMPK (p-AMPK) to total AMPK (t-AMPK) was increased in the SIRT5 group, whereas that of phosphorylated mTOR (p-mTOR) to total mTOR (t-mTOR) was decreased (both P < 0.05, Figure 6a–c).
Figure 6.
SIRT5 promoted autophagy in HCG cells via AMPK–mTOR-regulated autophagy, SIRT5 is degraded during etoposide-induced apoptosis, and calpains and caspases are responsible for the cleavage of SIRT5. (a) Western blot analysis of p-AMPK, t-AMPK, p-mTOR, and t-mTOR in vector-transfected or SIRT5-OE HCG27 cells induced by 100 nM rapamycin. (b–c) Relative protein levels were determined after normalization to t-AMPK or t-mTOR. (d) AGS cells were treated with 1 mg/mL etoposide for the indicated times. SIRT5, Beclin 1, cleaved PARP, and ATG5 expression was detected via western blotting at various time points. (e) AGS cells were treated with 1 mg/mL etoposide for the indicated times, and pro-caspase-3 and cleaved caspase-3 expression was analyzed via western blotting. (f) Normalized quantitation of the optical density of SIRT5, Beclin 1, cleaved PARP, and ATG5 in AGS cells treated with etoposide for the indicated times. (g) Normalized quantitation of the optical density of pro-caspase-3 and cleaved caspase-3 in AGS cells treated with etoposide for the indicated times. (h) AGS cells treated with 1 mg/mL etoposide for 6 hours in the presence or absence of 20 mM calpain inhibitor (CL) and/or 10 mM caspase inhibitor (z-VAD-fmk). SIRT5 expression was evaluated via western blotting. All data represent the average and SD of three or six independent experiments (*P < 0.05).
SIRT5, sirtuin 5; AMPK, AMP-activated protein kinase, mTOR, mammalian target of rapamycin; p-AMPK, phosphorylated AMPK; t-AMPK, total AMPK; p-mTOR, phosphorylated mTOR; t-mTOR; total mTOR; SIRT5-OE, sirtuin 5-overexpressing lentivirus; Vector, control lentivirus; PARP, poly(ADP-ribose) polymerase; ATG5, autophagy-related 5.
These data indicate that SIRT5 regulates HCG27 cell autophagy through the AMPK/mTOR/autophagy pathway.
SIRT5 is associated with apoptosis in GC cells
Autophagy and apoptosis have many similarities: both can lead to cell death, and many autophagy-related proteins can also regulate apoptosis.23–25 Some SIRTs can regulate both apoptosis and autophagy;19,26 therefore, we investigated the function of SIRT5 in apoptosis in GC cells, as the aspect of autophagy was examined in our previous study. Thus, we assessed the function of SIRT5 in apoptosis in GC cells. AGS cells were treated with etoposide to induce apoptosis. A decrease in the protein levels of SIRT5 was observed in the presence of etoposide, whereas cleaved caspase-3 and cleaved poly(ADP-ribose) polymerase, inducers of apoptosis, were activated after treatment with etoposide for 0–6 hours. However, the levels of Beclin 1 or autophagy-related 5 remained unchanged (Figure 6d–g). Flow cytometry revealed that increasing exposure to etoposide resulted in a higher rate of apoptosis (P < 0.05, Figure 6h). Thus, these findings suggest that SIRT5 may regulate the apoptotic process in GC cells.
Interestingly, we discovered that the variation in SIRT5 and cleaved caspase 3 expression was different during apoptosis in GC cells, and prior research revealed that caspases and calpains were responsible for cleavage of the autophagy-related protein Ambra1.27 Further experiments were performed to examine whether SIRT5 is a target of caspases and calpains during apoptosis in AGS cells. AGS cells were treated with etoposide in the presence or absence of a calpain inhibitor (CL) or caspase inhibitor (z-VAD-fmk). Figure 4i reveals that etoposide-induced SIRT5 degradation was stopped only when the cells were treated with both the calpain and caspase inhibitors, proving that SIRT5 cleavage in apoptosis may be mediated by both calpains and caspases in GC cells.
These results demonstrated that SIRT5 expression was downregulated and that SIRT5 was a target of caspases and calpains during apoptosis in GC cells.
Discussion
Aberrant SIRT5 protein expression has been reported in a variety of human cancers.28 SIRT5 expression obviously differs in different types of tumors. SIRT5 was reported to be increasingly upregulated in breast and lung cancers.29,30 This finding indicates that SIRT5 may exert a cancer-promoting effect in some cancers. However, in other cancers, such as liver cancer, SIRT5 expression was found to significantly decreased.31 Whether SIRT5 acts as a tumor suppressor was unclear until a recent report by Lu et al.32 demonstrated that oxoglutarate dehydrogenase mediates the inhibitory effects of SIRT5 on the proliferation and migration of GC cells. In this study, we examined the expression of SIRT5 in GC patient specimens and found that tumor tissues had lower SIRT5 expression than normal tissues. This laid the foundation of our study: SIRT5 expression is abnormal in GC.
Autophagy is a tightly regulated process that removes cytosolic components or damaged organelles, and the process begins with the formation of double-membrane vesicles called autophagosomes.33 Subsequently, autophagosomes fuse and deliver their cargo to lysosomes for degradation and recycling. Initially considered a random process, autophagy is now regarded as an adaptive response specifically activated by stress or environmental changes.34
Autophagy is controlled by specific genes known as autophagy-related genes. In addition, many other genes can also affect autophagy.35–37 Accumulating evidence has proven that SIRTs have a regulatory function in autophagy.21,38 The role of autophagy differs by tumor type.39–41
In a previous study, He et al.42 found that SIRTs, including SIRT5, are involved in autophagy-mediated cell death in BGC-823 human gastric carcinoma cells. However, the role of SIRT5 in GC cell autophagy was not deeply revealed before this study. We detected SIRT5 expression in four GC cell lines: MKN7, AGS, SUN1, and HCG27. Interestingly, HCG27 cells were believed to be the most malignant of the examined cell lines, and SIRT5 expression was lowest in these cells. The protein level of SIRT5 decreased as the degree of malignancy decreased, indicating that SIRT5 may have an anticancer effect in GC cells. Therefore, in the subsequent experiment, SIRT5 was depleted in AGS cells and overexpressed in HCG27 cells to clarify the function of the protein in autophagy. When autophagy was induced by rapamycin, we found that the ratio of LC3-II/LC3-I was decreased when SIRT5 was inhibited in AGS cells, whereas the ratio was increased when SIRT5 was upregulated in HCG27 cells, indicating that SIRT5 can promote autophagy. Based on these findings, electron microscopy was used to determine the changes in autophagosome quantity in HCG27 cells, and the results were consistent with those of a previous study. In summary, we demonstrated that SIRT5 could induce autophagy in GC cells.
Autophagy is controlled by two main nutrient-sensing pathways: mTOR and AMPK signaling.43 Once activated, AMPK regulates many mitochondrial functions through the phosphorylation of downstream effectors. We believe that the same pathways are involved in SIRT5-related autophagy. The mechanism underlying the protective effect of SIRT5 through the modulation of autophagy was further investigated by assessing the expression of the autophagy regulators AMPK and mTOR in SIRT5-OE HCG27 cells. Western blotting and quantification of the results indicated that when HCG27 cells were treated with rapamycin, the p-AMPK/t-AMPK ratio was higher in SIRT5-OE cells than in vector-transfected cells, whereas that of p-mTOR/t-mTOR was decreased. This finding suggests that SIRT5 induces autophagy in GC cells through the AMPK–mTOR pathway.
Autophagy and apoptosis constitute distinct cellular processes, often with opposing outcomes, and their signaling pathways are extensively interconnected through various mechanisms of crosstalk. Many related proteins can regulate both autophagy and apoptosis.44 With evidence of the downregulation of SIRT5 during apoptosis, we speculated that SIRT5 might negatively contribute to apoptosis in GC cells. However, the specific mechanism by which SIRT5 contributes to apoptosis was unknown. We preliminarily confirmed that the apoptotic function of SIRT5 is correlated with calpain-1 and cleaved caspase-3, evidenced only in the presence of both calpain and caspase inhibition, and SIRT5 degradation was stopped.
In summary, our results suggest that SIRT5 regulates the crosstalk between autophagy and apoptosis in GC cells. SIRT5 has the potential to be used as a prognostic or diagnostic marker, and we would like to extend this finding to other cancers and explore the regulatory role of SIRT5.
Footnotes
Availability of data and materials: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Project of Nature Science Foundation of China (81672348), National Science Foundation of Jiangsu Province of China (BK20191172), and Project of Gusu Medical Key Talent of Suzhou City of China (2020–2024).
ORCID iD: Dechun Li https://orcid.org/0000-0002-0419-8144
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Kassebaum NJ, Lopez AD, Murray CJ, et al. A comparison of maternal mortality estimates from GBD 2013 and WHO. Lancet 2014; 384: 2209–2210. doi: 10.1016/S0140-6736(14)62421-1. [DOI] [PubMed] [Google Scholar]
- 3.Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet 2012; 44: 570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Yin X, Yang Y. Rasal2 suppresses breast cancer cell proliferation modulated by secretory autophagy. Mol Cell Biochem 2019; 462: 115–122. doi: 10.1007/s11010-019-03615-7. [DOI] [PubMed] [Google Scholar]
- 5.Xu H, Zhang L, Qian X, et al. GSK343 induces autophagy and downregulates the AKT/mTOR signaling pathway in pancreatic cancer cells. Exp Ther Med 2019; 18: 2608–2616. doi: 10.3892/etm.2019.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li HF, Huang LF, Chen LH. Chitooligosaccharides inhibit A549 lung cancer cell line proliferation by regulating cell autophagy. J Biol Regul Homeost Agents 2019; 33: 1527–1532. [PubMed] [Google Scholar]
- 7.Madrigal-Matute J, Cuervo AM. Regulation of Liver Metabolism by Autophagy. Gastroenterology 2016; 150: 328–339. doi: 10.1053/j.gastro.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J 2007; 404: 1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michishita E, Park JY, Burneskis JM, et al. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 2005; 16: 4623–4635. doi: 10.1091/mbc.e05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature 2009; 460: 587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell 2006; 126: 257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Garrean S, Muhs A, Bui JT, et al. Complete eradication of hepatic metastasis from colorectal cancer by Yttrium-90 SIRT. World J Gastroenterol 2007; 13: 3016–3019. doi: 10.3748/wjg.v13.i21.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JE, Lou Z, Chen J. Interactions between DBC1 and SIRT 1 are deregulated in breast cancer cells. Cell Cycle 2009; 8: 3784–3785. doi: 10.4161/cc.8.22.10055. [DOI] [PubMed] [Google Scholar]
- 14.Pant K, Yadav AK, Gupta P, et al. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biol 2017; 12: 340–349. doi: 10.1016/j.redox.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida Y, Rardin MJ, Carrico C, et al. SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol Cell 2015; 59: 321–332. doi: 10.1016/j.molcel.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rardin MJ, He W, Nishida Y, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab 2013; 18: 920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu X, Han D, Chen W, et al. SIRT1-mediated FoxOs pathways protect against apoptosis by promoting autophagy in osteoblast-like MC3T3-E1 cells exposed to sodium fluoride. Oncotarget 2016; 7: 65218–65230. doi: 10.18632/oncotarget.11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Jiang S, Wang H, et al. SIRT6 protects against hepatic ischemia/reperfusion injury by inhibiting apoptosis and autophagy related cell death. Free Radic Biol Med 2018; 115: 18–30. doi: 10.1016/j.freeradbiomed.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Xu D, Jiang X, He H, et al. SIRT2 functions in aging, autophagy, and apoptosis in post-maturation bovine oocytes. Life Sci 2019; 232: 116639. doi: 10.1016/j.lfs.2019.116639. [DOI] [PubMed] [Google Scholar]
- 20.Shi L, Yan H, An S, et al. SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Mol Oncol 2019; 13: 358–375. doi: 10.1002/1878-0261.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo G, Jian Z, Zhu Y, et al. Sirt1 promotes autophagy and inhibits apoptosis to protect cardiomyocytes from hypoxic stress. Int J Mol Med. 2019; 43: 2033–2043. doi: 10.3892/ijmm.2019.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol 2004; 36: 2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Ye LX, Yu J, Liang YX, et al. Beclin 1 knockdown retards re-endothelialization and exacerbates neointimal formation via a crosstalk between autophagy and apoptosis. Atherosclerosis 2014; 237: 146–154. doi: 10.1016/j.atherosclerosis.2014.08.052. [DOI] [PubMed] [Google Scholar]
- 24.Mo ZT, Li WN, Zhai YR, et al. Icariin Attenuates OGD/R-Induced Autophagy via Bcl-2-Dependent Cross Talk between Apoptosis and Autophagy in PC12 Cells. Evid Based Complement Alternat Med 2016; 2016: 4343084. doi: 10.1155/2016/4343084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010; 29: 1717–1719. doi: 10.1038/onc.2009.519. [DOI] [PubMed] [Google Scholar]
- 26. Retracted: Sirt3 activation attenuated oxidized low-density lipoprotein-induced human umbilical vein endothelial cells' apoptosis by sustaining autophagy by Luo, X, Yang, Z, Zheng, S, Cao, Y and Wu, Y. Cell Biol Int 2017; 41: 932. doi: 10.1002/cbin.10291. [DOI] [PubMed] [Google Scholar]
- 27.Pagliarini V, Wirawan E, Romagnoli A, et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ 2012; 19: 1495–1504. doi: 10.1038/cdd.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahlknecht U, Ho AD, Letzel S, et al. Assignment of the NAD-dependent deacetylase sirtuin 5 gene (SIRT5) to human chromosome band 6p23 by in situ hybridization. Cytogenet Genome Res 2006; 112: 208–212. doi: 10.1159/000089872. [DOI] [PubMed] [Google Scholar]
- 29.Lu W, Zuo Y, Feng Y, et al. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol 2014; 35: 10699–10705. doi: 10.1007/s13277-014-2372-4. [DOI] [PubMed] [Google Scholar]
- 30.Igci M, Kalender ME, Borazan E, et al. High-throughput screening of Sirtuin family of genes in breast cancer. Gene 2016; 586: 123–128. doi: 10.1016/j.gene.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 31.Chen XF, Tian MX, Sun RQ, et al. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep 2018; 19: e45124. doi: 10.15252/embr.201745124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X, Yang P, Zhao X, et al. OGDH mediates the inhibition of SIRT5 on cell proliferation and migration of gastric cancer. Exp Cell Res 2019; 382: 111483. doi: 10.1016/j.yexcr.2019.06.028. [DOI] [PubMed] [Google Scholar]
- 33.Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature 2008; 451: 1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakahira K, Cloonan SM, Mizumura K, et al. Autophagy: a crucial moderator of redox balance, inflammation, and apoptosis in lung disease. Antioxid Redox Signal 2014; 20: 474–494. doi: 10.1089/ars.2013.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang R, Zeh HJ, Lotze MT, et al. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18: 571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molejon MI, Ropolo A, Re AL, et al. The VMP1-Beclin 1 interaction regulates autophagy induction. Sci Rep 2013; 3: 1055. doi: 10.1038/srep01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 2007; 9: 1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu N, Lei Y, Wang Y, et al. Inhibition of SIRT1/2 upregulates HSPA5 acetylation and induces pro-survival autophagy via ATF4-DDIT4-mTORC1 axis in human lung cancer cells. Apoptosis 2019; 24: 798–811. doi: 10.1007/s10495-019-01559-3. [DOI] [PubMed] [Google Scholar]
- 39.Jain K, Paranandi KS, Sridharan S, et al. Autophagy in breast cancer and its implications for therapy. Am J Cancer Res 2013; 3: 251–265. [PMC free article] [PubMed] [Google Scholar]
- 40.Jaboin JJ, Hwang M, Lu B. Autophagy in lung cancer. Methods Enzymol 2009; 453: 287–304. doi: 10.1016/S0076-6879(08)04014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui J, Gong Z, Shen HM. The role of autophagy in liver cancer: molecular mechanisms and potential therapeutic targets. Biochim Biophys Acta 2013; 1836: 15–26. doi: 10.1016/j.bbcan.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 42.He XX, Huang CK, Xie BS. Autophagy inhibition enhanced 5-FU-induced cell death in human gastric carcinoma BGC-823 cells. Mol Med Rep 2018; 17: 6768–6776. doi: 10.3892/mmr.2018.8661. [DOI] [PubMed] [Google Scholar]
- 43.Cao Y, Luo Y, Zou J, et al. Autophagy and its role in gastric cancer. Clin Chim Acta 2019; 489: 10–20. doi: 10.1016/j.cca.2018.11.028. [DOI] [PubMed] [Google Scholar]
- 44.Rubinstein AD, Kimchi A. Life in the balance - a mechanistic view of the crosstalk between autophagy and apoptosis. J Cell Sci 2012; 125: 5259–5268. doi: 10.1242/jcs.115865. [DOI] [PubMed] [Google Scholar]