Summary
Cell entry of the pandemic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is mediated by its spike protein S. As a main antigenic determinant, S protein is in focus of various therapeutic strategies. Besides particle-cell fusion, S mediates fusion between infected and uninfected cells resulting in syncytia formation. Here, we present sensitive assay systems with a high dynamic range and high signal-to-noise ratios covering not only particle-cell and cell-cell fusion but also fusion from without (FFWO). In FFWO, S-containing viral particles induce syncytia independently of de novo synthesis of S. Neutralizing antibodies, as well as sera from convalescent patients, inhibited particle-cell fusion with high efficiency. Cell-cell fusion, in contrast, was only moderately inhibited despite requiring levels of S protein below the detection limit of flow cytometry and Western blot. The data indicate that syncytia formation as pathological consequence during coronavirus disease 2019 (COVID-19) can proceed at low levels of S protein and may not be effectively prevented by antibodies.
Subject areas: Membrane Architecture, Cell Biology, Biochemical Assay
Graphical abstract
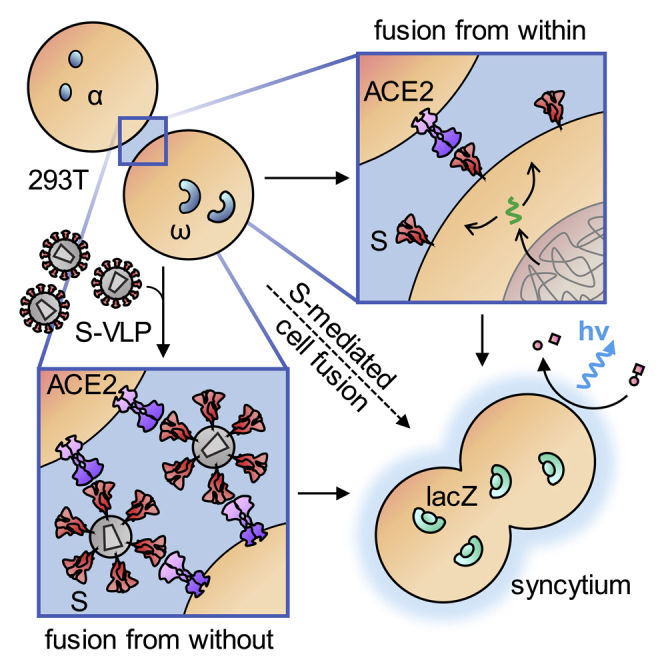
Highlights
-
•
Minimal levels of SARS-CoV-2 spike protein can cause cell fusion
-
•
Spike protein displayed on virus-like particles induces fusion from without
-
•
Particle-cell fusion is more sensitive toward neutralization than cell-cell fusion
-
•
Highly sensitive and scalable membrane fusion assays are applicable at BSL-1
Membrane Architecture; Cell Biology; Biochemical Assay
Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is met with an unprecedented global scientific effort. As the causative agent of the pandemic and the associated coronavirus disease 2019 (COVID-19), SARS-CoV-2 is a typical coronavirus, with a large spike protein (S) inserted in the membrane of the enveloped particle. Being the main surface-exposed antigen and the mediator of cell entry, S protein forms the most important target of current efforts to develop therapeutic drugs and prophylactic vaccines (Gioia et al., 2020; Tang et al., 2020).
As a typical class I viral fusion protein, S protein forms trimers with a globular head domain containing the receptor-binding site which contacts angiotensin-converting enzyme 2 (ACE2) as its entry receptor. Proteolytic cleavage separates the globular head (S1 domain) and the stalk domain (S2), which contains the hydrophobic fusion peptide at its N-terminus and the transmembrane domain toward the C-terminus. Priming of the fusion-competent state requires processing of two cleavage sites localized in close N-terminal proximity of the fusion peptide. These cleavage sites can be recognized by alternative proteases, thus explaining the high flexibility of the virus to adapt to various tissues. In particular, the S1/S2 site is cleaved by proprotein convertases like furin localized in the trans-Golgi network during trafficking of the newly synthesized S protein to the cell surface. The S2′ site can be cleaved by the serine protease TMPRSS2 which is exposed on the surface of target cells and contacted when the virus binds to ACE2. In absence of TMPRSS2, virus particles are endocytosed and cleaved by cathepsins (Hasan et al., 2020; Hoffmann et al., 2020a, 2020b; Shang et al., 2020a; Walls et al., 2020). In cell culture, trypsin was described as further alternative protease able to activate membrane fusion (Ou et al., 2020; Xia et al., 2020; Zhang and Kutateladze, 2020). Thus, two alternative entry routes exist for SARS-CoV-2, notably determined by the availability of activating proteases (Hoffmann et al., 2020b; Tang et al., 2020). For both routes, membrane fusion is pH independent.
Like other fusion proteins that are active pH independently, S protein mediates not only fusion between the viral and the cellular membranes during particle entry but also fusion of infected cells with uninfected cells (Buchrieser et al., 2020). This process is mediated by newly synthesized S protein accumulating at the cell surface. The resulting syncytia are giant cells containing at least three, often many more nuclei. Cell-cell fusion is used by viruses such as human immunodeficiency virus (HIV) (Compton and Schwartz, 2017; Bracq et al., 2018), measles virus (MV) (Griffin, 2020), or herpesvirus (Cole and Grose, 2003) to spread in a particle-independent way. The resulting syncytia are documented as pathological consequence detectable in various tissues such as the lung (MV), skin (herpesvirus), or lymphoid tissues (HIV). In the brain, cell-to-cell transmission via hyperfusogenic F proteins constitutes a hallmark of MV-caused encephalitis as a fatal consequence of acute MV infections manifesting years later (Ferren et al., 2019).
Besides particle-cell and cell-cell fusion, a third membrane fusion process mediated by viral fusion proteins has been designated fusion from without (FFWO) (Roller et al., 2008). FFWO results in syncytia in absence of newly expressed fusion protein. In presence of a sufficient concentration of particle-associated fusion protein, adjacent cells are fused, e.g., by bound HIV or herpesvirus particles either directly or after uptake of fusion protein and its presentation at the cell surface (Clavel and Charneau, 1994; Melikyan, 2014).
Here, we investigated the competence of SARS-CoV-2 S protein for these three membrane fusion processes. For each of them, we established quantitative assays relying on expressed S protein, thereby avoiding work with infectious virus at biosafety level 3 (BSL-3). The data reveal a strong membrane fusion activity of the S protein and demonstrate syncytia formation even at undetectable levels of S protein and FFWO. Examination of sera from convalescent patients with COVID-19 revealed potent neutralizing capacity against particle-cell fusion but only moderate or low activity against cell-cell fusion.
Results
Spike-protein-mediated particle entry
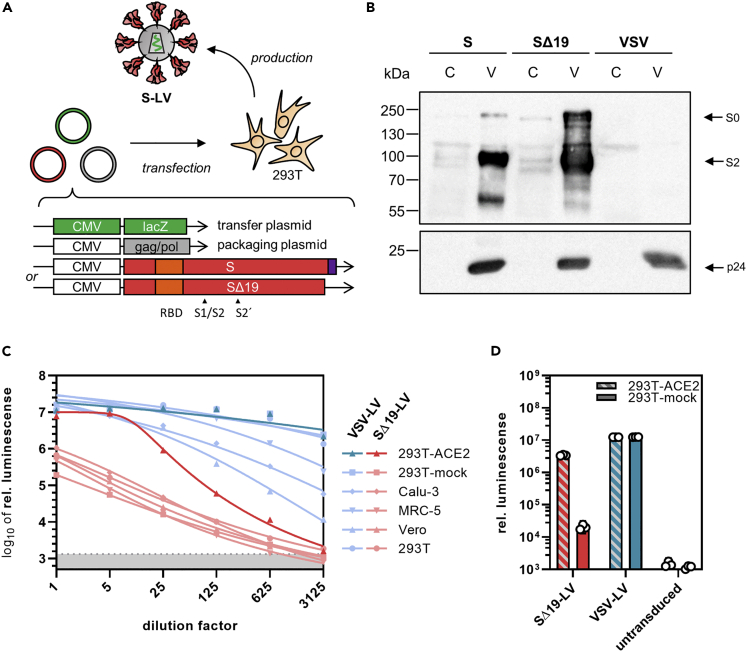
To follow particle entry mediated by S protein, we set out to generate lentiviral vectors (LVs) pseudotyped with S protein. S protein was codon optimized for expression in human cells, and either the full-length protein or the previously described C-terminal truncation variant SΔ19 (Ou et al., 2020) was used for pseudotyping. LVs transferring the β-galactosidase gene lacZ were generated by transient transfection of HEK-293T cells and subsequently purified and concentrated (Figure 1A) (see Transparent Methods for details). Western blot analysis of LV batches revealed a stronger spike signal relative to p24 for SΔ19, demonstrating its better incorporation into LV particles (Figure 1B). SΔ19-LVs were titrated on cell lines frequently used in coronavirus research, i.e., Vero E6, MRC-5, Calu-3, HEK-293T, and HEK-293T overexpressing ACE2 (293T-ACE2). On all cell lines included, the luminescence signal decreased linearly with increasing dilution of the vector (Figure 1C). In contrast to LVs pseudotyped with the G protein of vesicular stomatitis virus (VSV-LV), SΔ19-LV did not reach a signal plateau, indicating that only a subsaturating fraction of the cells was transduced. However, transduction rates with SΔ19-LV increased more than 100-fold upon overexpression of hACE2 on HEK-293T cells, reaching a signal-to-noise ratio of more than 2000. VSVG-LV-mediated gene delivery was not affected by overexpression of hACE2 (Figure 1D). Remarkably, with a saturating dose of SΔ19-LV, a similar luminescence signal was reached on 293T-ACE2 cells as with VSV-LV (Figure 1C).
Figure 1.
Spike-mediated particle entry
(A) Generation of pseudotyped lentiviral vectors. Second-generation LVs pseudotyped with S protein were generated by transfection of HEK-293T cells with a packaging plasmid encoding HIV-1 gag/pol, a transfer vector plasmid with a lacZ reporter gene and one of two envelope plasmids encoding codon-optimized SARS-CoV-2 S with or without (SΔ19) the 19 C-terminal amino acids. The C-terminal endoplasmic reticulum retention signal (purple) and the receptor-binding domain (RBD, orange) are indicated.
(B) Incorporation of S protein into LVs determined by Western blotting. S-LV and SΔ19-LV particles (V) and lysates of their producer cells (C) were stained for the presence of S protein (top) and p24 as particle loading control. Top blot was exposed for 30 s and bottom blot for 5 s. Image contrast was adjusted, retaining relative signal strengths.
(C) Gene transfer activities on the indicated cell lines. The indicated dilutions of 5 μL vector stock of SΔ19-LV or VSV-LV were added to the cells. Cell lysates were prepared three days after vector addition, and lacZ reporter activity was quantified as a luminescence readout. Symbols represent means of technical triplicates. Gray shaded area indicates 95% confidence interval (CI) of signals from untransduced cells (blanks).
(D) Effect of ACE2 overexpression on reporter transfer. 293T cells transfected with ACE2 expression plasmid or mock plasmid were incubated with 0.2 μL of SΔ19-LV or VSV-LV. Cell lysates were prepared three days after vector addition, and reporter activity was quantified as a luminescence readout. Bars represent geometric means of technical triplicates ±95% CIs.
Quantifying cell fusion mediated by SARS-CoV-2 S protein
Upon transfection, SARS-CoV-2 S protein showed a remarkable fusogenic activity. When transfected HEK-293T cells producing SΔ19-LV were detached and cocultured overnight in a 1:1 ratio with Vero E6 cells, plates were covered by large syncytia, each containing at least 10 (and up to 100) nuclei (Figure S1A). To examine this further, the full-length and truncated forms of S were overexpressed in HEK-293T, and cocultures with Vero E6 target cells were imaged by confocal laser scanning microscopy (Figure S1B). Both, S and SΔ19 protein, induced many large syncytia characterized by cytoskeletal rearrangement, clustering of more than five nuclei and colocalization of the red and green fluorescent protein (RFP and GFP) reporter dye signals (Figure S1C).
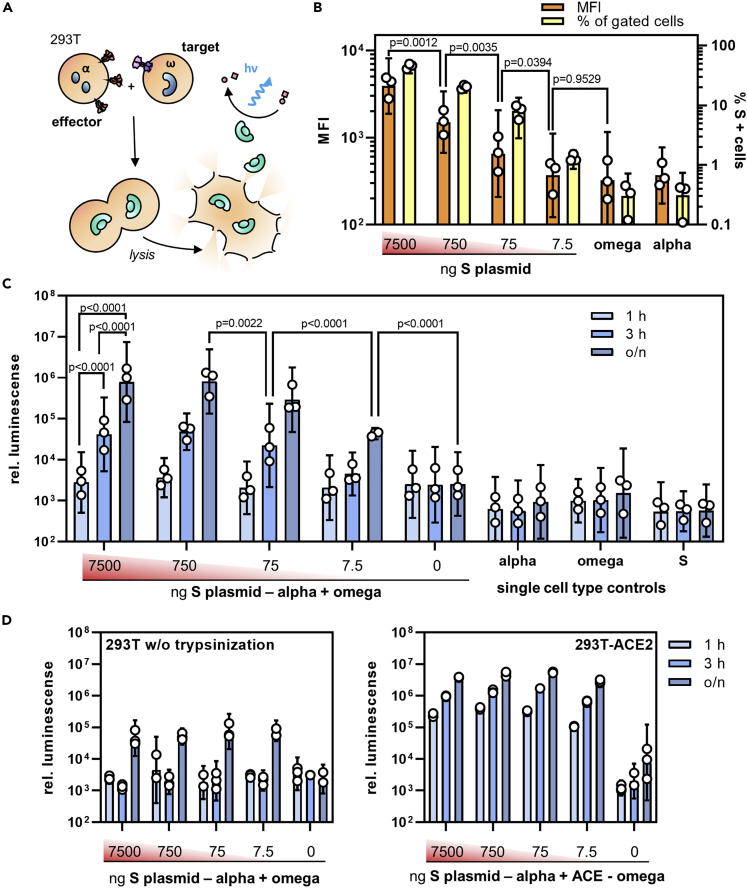
To quantify the cell-cell fusion mediated by S protein, an α-complementation assay based on β-galactosidase previously used to evaluate HIV-mediated fusion (Holland et al., 2004) was adapted to the S protein (see Transparent Methods for details): Upon cell-cell fusion, complementation of ω enzyme fragment with the α fragment inside the syncytium results in the formation of active β-galactosidase that is then quantified by a luminescence reaction (Figure 2A). In a first step, the minimal amount of S protein required to result in a detectable signal was determined. Effector cells were transfected with varying quantities of S protein-encoding plasmid (ranging from 7.5 μg to 7.5 ng per T75 flask) and S protein surface expression was followed by flow cytometry. S protein expression was still clearly detectable with 75 ng of plasmid, while the mean fluorescence intensity (MFI) at 7.5 ng plasmid was indistinguishable from background (Figure 2B). Notably, this low level of S protein was still sufficient to induce significant cell-cell fusion when the assay was allowed to proceed overnight, highlighting the potent fusogenic activity of S protein (Figure 2C). With higher amounts of plasmid, three hours of incubation were sufficient to obtain signals more than 10-fold above background. Interestingly, when cells were detached prior to coculture with ethylenediaminetetraacetic acid (EDTA) only instead of trypsin-EDTA, the cell fusion activity decreased by at least one order of magnitude, and only overnight incubation yielded signals significantly over background (Figure 2D). When target cells overexpressed ACE2 and were detached with trypsin, the maximum extent of the fusion signal increased by another order of magnitude (Figure 2D). Now, the signal-to-noise ratio reached two orders of magnitude already after one-hour incubation including the samples expressing the lowest amounts of S protein. This highlights the dependence of S protein activity on the presence of ACE2. Under optimal conditions and with ACE2 overexpressing cells, a signal-to-noise ratio of 2.9 orders of magnitude was reached.
Figure 2.
Quantifying S protein-mediated cell-cell fusion
(A) Experimental setup for the quantification of cell-cell fusion. Active β-galactosidase is formed when effector cells expressing S protein and the α-fragment fuse with target cells expressing ACE2 and the ω-fragment.
(B–D) Effector cells transfected with different amounts of S-protein expression plasmid (ranging from 7500 ng to 7.5 ng per T75 flask) were assessed for S protein expression by flow cytometry (B) and then used in the fusion assay (C-D). (B) Mean fluorescence intensity (MFI, orange bars) and the percentage of S-positive cells (yellow bars) were determined in flow cytometry. Bars represent (geometric) means ±95% confidence intervals (CIs), n = 3. p values are from one-way analysis of variance (ANOVA).
(C) Activities of β-galactosidase in cocultures of effector and target cells in absence of ACE2 overexpression. Effector cells were transfected with the indicated amounts of S protein encoding plasmid, detached with trypsin and cultivated for the indicated time periods (o/n: overnight) with target cells which were transfected with the ω-fragment encoding plasmid only. Bars represent geometric means ±95% CIs, n = 3. p values are from two-way ANOVA.
(D) Influence of proteolytic processing and ACE2 overexpression on cell fusion activity. Left panel: Effector and target cells were detached without trypsin using 5 mM EDTA in PBS. Right panel: Target cells were co-transfected with the ω-fragment and ACE2 encoding plasmids. Bars represent geometric means of technical triplicates ±95% CIs.
See also Figure S1.
Fusion from without mediated by S protein
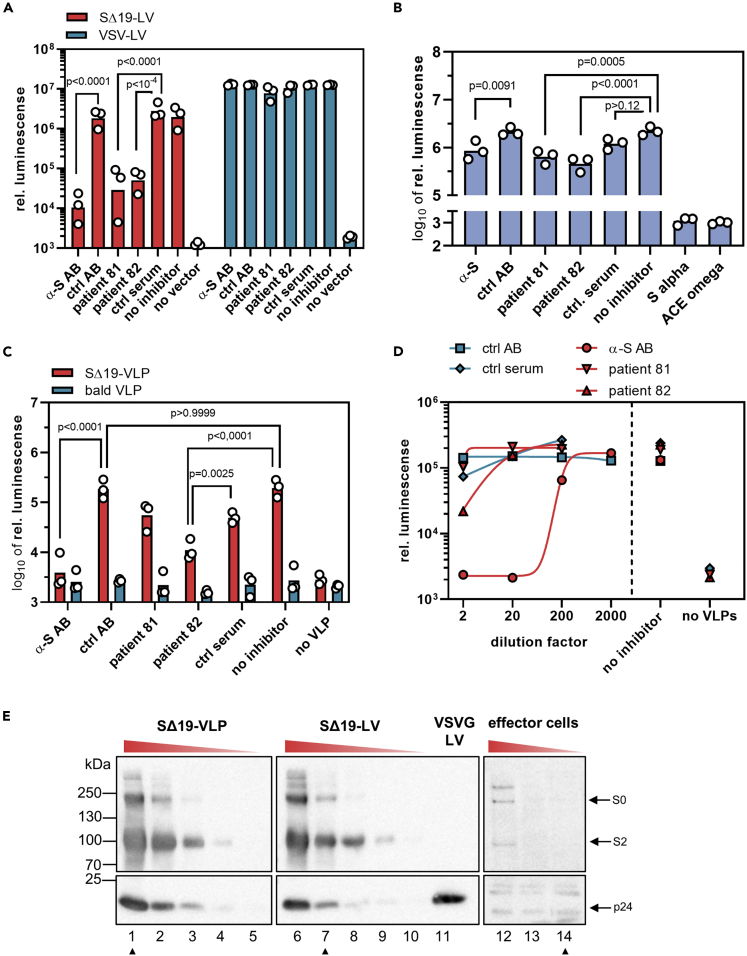
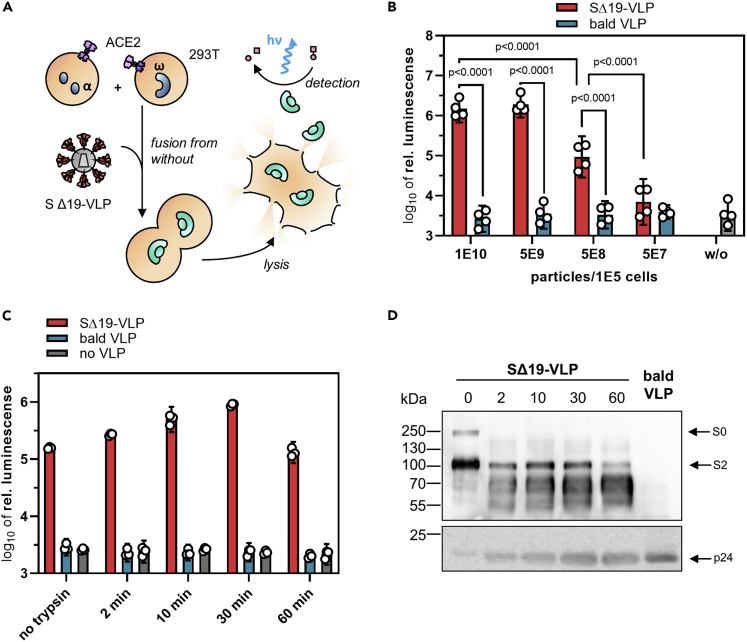
In light of its substantial fusogenicity, we explored whether the spike protein present on virus particles is sufficient to cause cell fusion. In contrast to the previous experiments, where one cell type in the α-complementation assay had served as a spike-bearing effector and the other as a target cell, both α- and ω-expressing cells served as target cells in the FFWO assay. Spike protein was introduced into the system by means of LV-based virus-like particles (VLPs) having the SΔ19 protein incorporated but devoid of any packaged transgene. Luminescence signal was a readout for the activity of the split β-galactosidase reporter, which was restored by cell fusion-caused combination of the α and ω fragments. This way, luminogenic activity is directly linked to cell fusion mediated by the VLPs (see Transparent Methods for details). Western blot analysis confirmed that similar levels of SΔ19 protein were present in VLPs and SΔ19-LV particles (Figure 4E). To quantify FFWO, cocultures of 293T-ACE2 cells transfected with the plasmids encoding the α or ω fragment were incubated with different amounts of VLPs to induce cell fusion and thus complementation (Figure 3A). At a dose of 5000 VLPs per cell, a substantial fusion signal more than one order of magnitude over background was obtained (Figure 3B). This signal increased further with increasing particle numbers, reaching a plateau at 5 x 104 particles per cell, with a signal-to-noise ratio of 2.7 orders of magnitude (Figure 3B). This FFWO activity of the S protein was again strongly dependent on ACE2 overexpression since no luminescence activity significantly above that of background (as determined with bald VLPs devoid of spike protein) was observed in absence of ACE2 (Figure S2). When the VLPs were treated with 2 mg/mL trypsin, the fusion signal increased by approximately 5.7-fold after 30 min of incubation. The chosen incubation period was critical for this increase since trypsin digestion for 60 min was not beneficial (Figure 3C). This correlated well with processing of the S0 protein into S2 upon trypsin exposure (Figure 3D). Prolonged digestion for 60 min resulted in loss of S2, while p24 levels did not decrease (Figure 3D). Finally, VLPs displaying full-length S protein were generated and tested for FFWO. Being slightly less active than SΔ19-VLPs, ACE2-expressing cells incubated with S-VLPs showed significant levels of luminescence activity well over background (Figure S2C). Therefore, we can conclude that FFWO is also mediated by full-length S protein and not a particular property of the SΔ19 spike protein variant.
Figure 4.
Antibody-mediated neutralization of membrane fusion
The neutralizing activities of an S-protein specific antibody and sera from two convalescent patients with COVID-19 were determined against S-protein-mediated particle entry (A), cell-cell fusion (B), and FFWO (C-D). Bars represent (geometric) means, n = 3. p values are from two-way (A, C) or one-way (B) analysis of variance (ANOVA).
(A) 0.2 μL of SΔ19-LV or VSV-LV was incubated for 30 min with the indicated antibodies or sera before being added to HEK-293T cells transfected with ACE2 expression plasmid. Cell lysates were prepared 3 days after vector addition, and reporter activity was quantified as luminescence readout.
(B) Effector cells coexpressing S protein (7.5 ng S plasmid per T75) and the α-fragment were incubated for 30 min with antibodies or sera before being added to target cells cotransfected with ω-fragment and ACE2 expression plasmids. Bars represent means, n = 3.
(C) 5 x 108 SΔ19-VLPs or bald VLPs were incubated for 30 min with antibodies or sera before being added to the cocultures of α/ACE2 and ω/ACE2 cells. Luminescence was quantified after overnight incubation. Select multiplicity-adjusted p values are reported.
(D) Neutralizing capacity of the indicated dilutions of antibodies and sera on FFWO. Dilution factors apply to pure serum or 80 μg/mL for the antibodies.
(E) Semi-quantitative Western blots determining S protein levels present in the three fusion assays. Lysates of 2.5 x 105 effector cells transfected with 750 ng (lane 12), 75 ng (lane 13), and 7.5 ng (lane 14) of S protein-encoding plasmid per T75 flask, respectively, were analyzed for S protein and p24 protein levels along with three-fold dilutions of 2.5 x 109 SΔ19-VLPs (lane 1–5) or 3 μL of SΔ19-LVs (lane 6–11). The blots showing spike and p24 levels of LVs and VLPs were exposed for 3 s whereas the blot for effector cells was exposed for 800 s. Lanes were rearranged, but no lanes were omitted. Samples corresponding to the material used in (A-D) are marked with arrowheads.
See also Figure S3 for alternative representation of (A-D).
Figure 3.
S protein-mediated fusion from without
(A) Experimental setup. HIV-1-derived SΔ19-VLPs were added as effector triggering fusion from without (FFWO) of cocultures of ACE2 overexpressing HEK-293T cells transfected with plasmids encoding the α-fragment or the ω-fragment of β-galactosidase. Cocultures were lysed, and galactosidase activity of the lysates was determined in luminescence reactions.
(B) The indicated numbers of SΔ19-VLPs or bald VLP, produced without the S protein, were added to the coculture. Activities were quantified after overnight incubation. Bars represent means ±95% confidence intervals (CIs), n = 4. p values are from two-way analysis of variance (ANOVA).
(C-D) Effect of proteolytic processing on the induction of FFWO. 5 x 108 SΔ19-VLPs, bald VLPs, or no VLPs were incubated in 2 mg/mL trypsin for the indicated time periods before addition to cocultures (105 cells). Luminescence was quantified after overnight cultivation.
(C). Bars represent means of technical triplicates ±95% CIs.
(D) Processing of the S protein after trypsin treatment of the VLPs for indicated times by Western blotting for S and p24 proteins. Blots were cropped to show the relevant signals of the spike protein. Exposure time was set to 600 s for spike detection and 60 s for p24 detection. Image contrast was adjusted, retaining relative signal strengths.
See also Figure S2.
Neutralization of the membrane fusion activities
Having established three different quantitative assay systems with an extraordinary high signal-to-noise ratio, we next assessed the neutralizing capacity of an S protein specific antibody, as well as convalescent sera from two donors (see Transparent Methods for details). The sera were obtained four months after diagnosis of the infection with SARS-CoV-2 by polymerase chain reaction (PCR). Both donors had experienced mild symptoms. The strongest neutralization was observed in the particle entry assay. Addition of a neutralizing monoclonal anti-S antibody reduced the reporter signal by 2.3 orders of magnitude, which corresponded to more than 99% neutralization (Figures 4A and S3). Diluted 1:1 with vector stock, both sera reduced the reporter activity by 2.0 (98.5%) and 1.7 (97.4%) orders of magnitude, respectively, compared to control serum (Figures 4A and S3). In sharp contrast, their effect on cell-cell fusion remained below one order of magnitude for all three inhibitors (α-S AB: 0.4 orders [60.8%]; serum 81: 0.5 orders [71.0%]); serum 82: 0.7 orders [79.1%]). Notably, the signal-to-background ratio was comparable for both assays, and identical amounts of sera had been applied (Figures 4A and S3). For the inhibition of FFWO, a mixed picture was obtained. The monoclonal anti-S antibody decreased FFWO signal by 1.7 orders of magnitude to the background level (97.9% neutralization) (Figure 4C). Titration of the antibody yielded an IC50 of 0.37 μg/mL, well in line with the value determined by the vendor in pseudovirus neutralization assays (0.11 μg/mL) (Figure 4D). The effect of the patient sera was clearly less pronounced than what was observed in particle entry neutralization. The strongest effect was seen with serum 82 which decreased the fusion signal by 1.2 orders of magnitude (94% neutralization). Serum 81, which was highly active in the entry assay, had no stronger effect on the fusion signal than the control serum (0.5 orders of magnitude, 72% neutralization) (Figures 4C and 4D). Notably, antigen amounts on SΔ19-LV particles and the corresponding VLPs were similar, while a higher dose of S was used in the FFWO neutralization experiment than in the particle entry neutralization experiment (Figure 4E). They exceeded the amounts of S protein present in the cell-cell fusion assay by far, thus excluding higher amounts of antigen in the cell-cell fusion assay as causative for this difference.
Discussion
Here, we provide unique assay systems for a systematic side-by-side comparison of the membrane fusion activities of the SARS-CoV-2 spike protein S. Three quantitative assays are presented, covering (i) entry of LV particles pseudotyped with S, (ii) fusion between S and ACE2-expressing cells, and (iii) fusion between ACE2-expressing cells via VLPs displaying S protein. All assays exhibit high signal-to-noise ratios covering two to three orders of magnitude as important prerequisite to identify subtle differences, e.g., when screening for the activities of inhibitors. Dependence on trypsin treatment and ACE2 expression confirmed the validity of the assays. Several studies have recently published S-protein-based particle cell entry assays relying either on replication incompetent VSV particles (Hoffmann et al., 2020b) or LVs similar to those used here in this study. With a maximum signal-to-noise ratio of four orders of magnitude, the system we describe here is well among or even better than the top-performing LV-based assays published so far with second (Ou et al., 2020) or simply first-generation LVs with their significant safety concerns (Zeng et al., 2020; Zhu et al., 2020). It thus offers improved safety, as vectors are produced without the use of helper virus and with separate packaging and transfer plasmids, significantly lowering the risk of replication-competent virus formation. The truncation of the 19 C-terminal amino acids of S suggested by (Ou et al., 2020) indeed improves incorporation of S protein into lentiviral particles and likely contributes to the system's performance. This is expected, as viral envelope proteins are commonly truncated for pseudotyping of lentiviral particles (Funke et al., 2008; Bender et al., 2016). Truncation is thought to optimize spacing and in this case removes an endoplasmic reticulum (ER) retention signal (Ou et al., 2020). Regarding cell fusion assays, previously published studies relied on nucleus counting or reporter complementation (Ou et al., 2020; Zhu et al., 2020). The assay system we describe here improves upon the previously reported maximum signal-to-noise ratios by at least an order of magnitude (Zhu et al., 2020).
The syncytia forming activity of the S protein is remarkable not only with respect to speed and extent but even more so with respect to the low amounts of S protein required, even when ACE2 is not overexpressed on the target cells. The high sensitivity of the assay we established here allowed the detection of cell fusion activity at levels of S protein which were below the level of detection of flow cytometry and Western blot. This remarkable activity is likely enabled by the high affinity of S protein for its receptor ACE2 (Shang et al., 2020b; Walls et al., 2020). Being well in the low nanomolar range, it is even lower than that of the highly fusogenic MV for its receptors (Navaratnarajah et al., 2008; Mühlebach et al., 2011). To our knowledge, we here provide the first demonstration of FFWO not only for the SARS-CoV-2 S protein but also for any coronavirus, while FFWO has been observed for other enveloped viruses entering cells by pH-independent membrane fusion, such as retroviruses (Clavel and Charneau, 1994), paramyxoviruses (Bratt and Gallaher, 1969), and herpesviruses (Falke et al., 1985). FFWO is a relevant addition to syncytia formation between infected and uninfected cells since it means that S protein present on viral particles can trigger fusion of uninfected cells, thus inducing cytopathic effects in absence of a productive infection. Potentially, this may also be caused by defective particles. In cell culture experiments, relatively high particle numbers are required to trigger FFWO since diffusion barriers have to be overcome to allow sufficient particles to contact the cells. Accordingly, much higher amounts of S protein were needed in the FFWO setting than in the conventional syncytia formation assay, where neighboring adherent cells grow in direct contact. In this respect, it appears evident that in vivo, where in organs such as the lung, kidney, or liver, epithelial cells are tightly packed with very limited extracellular space, fewer particles will be necessary to trigger fusion. While it is conceivable that sufficient concentrations of viral particles accumulate in patient epithelia, clear evidence for a pathogenic relevance of FFWO in patients has yet to be provided, although some evidence from animal studies exists for retroviruses (Murphy and Gaulton, 2007).
Independently of that, the FFWO assay described here can become a prime choice for the preclinical evaluation of neutralizing binders for the following reasons: First, it can be performed at BSL-1 conditions, while LV-based assays require BSL-2. Second, it exhibits a high signal-to-noise ratio, even exceeding the maximum ratio recently reported for a benchmark LV-based neutralization assay (Zeng et al., 2020). Third, this assay is unique in combining neutralization of particle-associated S protein with neutralization of cell fusion, thus providing information about two mechanisms of infection in a single readout.
Syncytia formation has recently been described as main and unique lung pathology in patients affected with COVID-19 in an occurrence not seen in other lung infections before. The polynucleated pneumocytes detected by histology and immunostaining were virus positive (Bussani et al., 2020). Especially in persistent virus infections, syncytia formation is an important mechanism of virus spread as described for other important human pathogens like MV (Ferren et al., 2019) or various herpesviruses (Cole and Grose, 2003). The low level of S protein sufficient for cell fusion we determined here appears to be ideal to support persistence of SARS-CoV-2. Evidence for SARS-CoV-2 persistence including neurological complications is recently accumulating, and it is particularly the persistence which appears to result in fatal outcomes (Hamming et al., 2004; Berger, 2020). In this respect, it may be worrying that the antibodies and convalescent sera we tested were less efficient in neutralizing cell-cell fusion than particle entry, even more so since the amounts of S protein present in the latter assay were substantially higher. This suggests that cell fusion is not only proceeding with minimal amounts of S protein but also difficult to access for neutralizing antibodies. Further studies assessing sera from many more patients including severe and fatal cases will have to be performed to investigate this hypothesis further. The biological assay systems required are now available.
Limitations of the study
This manuscript describes quantitative assay systems for the membrane fusion activity of SARS-CoV-2 spike proteins. The assays rely on expression of spike protein from plasmid and on lentiviral particles displaying the spike protein. While not likely, formally, we cannot exclude that the results obtained were influenced by this experimental context and thus they may not be completely transferrable to the biology of actual SARS-CoV-2 infections. Independently from that however, the membrane fusion function of the SARS-CoV-2 spike protein as biochemical function is adequately reflected by the assays. As indicated in the discussion, it remains to be demonstrated if the FFWO activity observed in the assay systems is of relevance for clinical SARS-CoV-2 infections.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Christian J. Buchholz, Molecular Biotechnology and Gene Therapy, Paul-Ehrlich-Institut, Paul-Ehrlich-Straße 51–59, 63,225 Langen, Germany; christian.buchholz@pei.de, phone: +496103774011, fax: +496103771255.
Material availability
Plasmids are available from the lead contact upon completion of a Material Transfer Agreement.
Data and code availability
This study did not use any unpublished custom code, software, or algorithm.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors like to thank Klaus K. Conzelmann (Munich) for providing the plasmid encoding codon-optimized spike protein. Additionally, we like to thank Gundula Braun and Manuela Gallet for excellent technical assistance in generating lentiviral vector stocks and cloning of plasmids.
Author contributions
S.T. designed, performed, and evaluated experiments. A.M. designed experiments, analyzed data, and contributed to writing of the manuscript. V.R. designed and performed experiments and generated unique research material. T.J.M. provided unique research material. E.F. and K.C. contributed to conceiving the study and supervising work. C.J.B. conceived the study, supervised work, acquired grants, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102170.
Supplemental information
References
- Bender R.R., Muth A., Schneider I.C., Friedel T., Hartmann J., Pluckthun A., Maisner A., Buchholz C.J. Receptor-targeted nipah virus glycoproteins improve cell-type selective gene delivery and reveal a preference for membrane-proximal cell attachment. PLoS Pathog. 2016;12:e1005641. doi: 10.1371/journal.ppat.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J.R. COVID-19 and the nervous system. J. Neurovirol. 2020;26:143–148. doi: 10.1007/s13365-020-00840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracq L., Xie M., Benichou S., Bouchet J. Mechanisms for cell-to-cell transmission of HIV-1. Front. Immunol. 2018;9:260. doi: 10.3389/fimmu.2018.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt M.A., Gallaher W.R. Preliminary analysis of the requirements for fusion from within and fusion from without by Newcastle disease virus. Proc. Natl. Acad. Sci. U S A. 1969;64:536–543. doi: 10.1073/pnas.64.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel-Benhassine F., Van der Werf S. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39:e106267. doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussani R., Schneider E., Zentilin L., Collesi C., Ali H., Braga L., Volpe M.C., Colliva A., Zanconati F., Berlot G., Silvestri F. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EbioMedicine. 2020 doi: 10.1016/j.ebiom.2020.103104. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel F., Charneau P. Fusion from without directed by human immunodeficiency virus particles. J. Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole N.L., Grose C. Membrane fusion mediated by herpesvirus glycoproteins: the paradigm of varicella-zoster virus. Rev. Med. Virol. 2003;13:207–222. doi: 10.1002/rmv.377. [DOI] [PubMed] [Google Scholar]
- Compton A.A., Schwartz O. They might Be giants: does syncytium formation sink or spread HIV infection? PLoS Pathog. 2017;13:e1006099. doi: 10.1371/journal.ppat.1006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke D., Knoblich A., Müller S. Fusion from without induced by herpes simplex virus type 1. Intervirology. 1985;24:211–219. doi: 10.1159/000149645. [DOI] [PubMed] [Google Scholar]
- Ferren M., Horvat B., Mathieu C. Measles encephalitis: towards new therapeutics. Viruses. 2019;11:1017. doi: 10.3390/v11111017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke S., Maisner A., Mühlebach M.D., Koehl U., Grez M., Cattaneo R., Cichutek K., Buchholz C.J. Targeted cell entry of lentiviral vectors. Mol. Ther. 2008;16:1427–1436. doi: 10.1038/mt.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia M., Ciaccio C., Calligari P., Simone G.de, Sbardella D., Tundo G., Francesco Fasciglione G., Di Masi A., Di Pierro D., Bocedi A. Role of proteolytic enzymes in the Covid-19 infection and promising therapeutic approaches. Biochem. Pharmacol. 2020;182:114225. doi: 10.1016/j.bcp.2020.114225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.E. Measles virus persistence and its consequences. Curr. Opin. Virol. 2020;41:46–51. doi: 10.1016/j.coviro.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B.A., Hussain A., Qadir F.A., Attar F., Aziz F.M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.U., Munk C., Lucero G.R., Nguyen L.D., Landau N.R. α-Complementation assay for HIV envelope glycoprotein-mediated fusion. Virology. 2004;319(2):343–352. doi: 10.1016/j.virol.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Melikyan G.B. HIV entry: a game of hide-and-fuse? Curr. Opin. Virol. 2014;4:1–7. doi: 10.1016/j.coviro.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlebach M.D., Mateo M., Sinn P.L., Prüfer S., Uhlig K.M., Leonard Vincent H.J., Navaratnarajah C.K., Frenzke M., Wong X.X., Sawatsky B. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480:530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S.L., Gaulton G.N. TR1.3 viral pathogenesis and syncytium formation are linked to Env-Gag cooperation. J. Virol. 2007;81:10777–10785. doi: 10.1128/JVI.00816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnarajah C.K., Vongpunsawad S., Oezguen N., Stehle T., Braun W., Hashiguchi T., Maenaka K., Yanagi Y., Cattaneo R. Dynamic interaction of the measles virus hemagglutinin with its receptor signaling lymphocytic activation molecule (SLAM, CD150) J. Biol. Chem. 2008;283:11763–11771. doi: 10.1074/jbc.M800896200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller D.G., Dollery S.J., Doyle J.L., Nicola A.V. Structure-function analysis of herpes simplex virus glycoprotein B with fusion-from-without activity. Virology. 2008;382:207–216. doi: 10.1016/j.virol.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal. Transduction Targeted Ther. 2020;5:92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Evans J.P., Pearson R., Qu P., Zheng Y.-M., Robinson R.T., Hall-Stoodley L., Yount J.S., Pannu S., Mallampalli R.K. Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers and convalescent plasma donors. JCI Insight. 2020 doi: 10.1172/jci.insight.143213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kutateladze T.G. Molecular structure analyses suggest strategies to therapeutically target SARS-CoV-2. Nat. Commun. 2020;11:2920. doi: 10.1038/s41467-020-16779-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yu D., Yan H., Chong H., He Y. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J. Virol. 2020;94 doi: 10.1128/JVI.00635-20. e00635–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not use any unpublished custom code, software, or algorithm.