Abstract
Background
The emergence and spread of SARS-CoV-2 throughout the world has created an enormous socioeconomic impact. Although there are several promising drug candidates in clinical trials, none is available clinically. Thus, the drug repurposing approach may help to overcome the current pandemic.
Methods
The main protease (Mpro) of SARS-CoV-2 is crucial for cleaving nascent polypeptide chains. Here, FDA-approved antiviral and anti-infection drugs were screened by high-throughput virtual screening (HTVS) followed by re-docking with standard-precision (SP) and extra-precision (XP) molecular docking. The most potent drug's binding was further validated by free energy calculations (Prime/MM-GBSA) and molecular dynamics (MD) simulation.
Results
Out of 1397 potential drugs, 157 showed considerable affinity toward Mpro. After HTVS, SP, and XP molecular docking, four high-affinity lead drugs (Iodixanol, Amikacin, Troxerutin, and Rutin) with docking energies −10.629 to −11.776 kcal/mol range were identified. Among them, Amikacin exhibited the lowest Prime/MM-GBSA energy (−73.800 kcal/mol). It led us to evaluate other aminoglycosides (Neomycin, Paramomycin, Gentamycin, Streptomycin, and Tobramycin) against Mpro. All aminoglycosides were bound to the substrate-binding site of Mpro and interacted with crucial residues. Altogether, Amikacin was found to be the most potent inhibitor of Mpro. MD simulations of the Amikacin-Mpro complex suggested the formation of a complex stabilized by hydrogen bonds, salt bridges, and van der Waals interactions.
Conclusion
Aminoglycosides may serve as a scaffold to design potent drug molecules against COVID-19. However, further validation by in vitro and in vivo studies is required before using aminoglycosides as an anti-COVID-19 agent.
Keywords: COVID-19, SARS-CoV-2, Docking and simulation, Aminoglycosides, Antibiotics
Introduction
At the end of 2019, many patients were diagnosed with a respiratory tract infection exhibiting severe pneumonia in Wuhan, the capital of Hubei province in China [1], [2]. In a short time, the disease spread throughout the world and caused a pandemic. It was revealed that the causal organism is a novel coronavirus closely related to bat Severe Acute Respiratory Syndrome (SARS)-like coronavirus, and thus named as SARS-CoV-2 [3], [4]. The RNA genome of SARS-CoV-2 is about 82% identical to the SARS coronavirus (SARS-CoV), with both viruses belonging to clade b of the genus Betacoronavirus [1], [2]. In general, coronaviruses are enveloped, positive-sense, single-stranded RNA viruses in the genus Coronavirus of the family Coronaviridae that can infect humans and several animals, including mammals and aves [5], [6], [7]. The famous outbreak of SARS-CoV in Guangdong, China [8], and Middle East respiratory syndrome coronavirus (MERS-CoV) in many countries of the Middle East region [9] establish the fact that some coronaviruses can cause life-threatening infection in patients. Likewise, COVID-19 has been confirmed to be transmitted via a human-to-human transmission that quickly spread to the majority of countries worldwide [10], affecting millions (40,657,071 reported cases), with a death toll reaching 1,123,122 till Oct 19, 2020 (https://www.worldometers.info/coronavirus/).
Unfolding COVID-19 pandemics shows a necessity of rapid finding of drug-candidates that could be used immediately in numerous hotspots of virus activity. One of the most attractive drug targets among coronaviruses is the main protease (Mpro), also named chymotrypsin-like protease (3CLpro) [11]. This enzyme plays a crucial role in processing the polyproteins translated from the viral RNA [12]. Inhibiting the activity of this enzyme would severely block viral replication. Since no human proteases with similar cleavage specificity are known, this approach sounds promising, as the prospective drug candidate's toxic manifestations against this enzyme would be negligible.
Several studies have now reported putative inhibitors using bioinformatics studies. For example, Gentile et al. (2020) screened a collection of 14,064 compounds searching for potential SARS-CoV-2 Mpro inhibitors [13]. Similarly, Jin and colleagues identified 30 drugs and compounds as SARS-CoV-2 Mpro inhibitors through protein modeling and virtual screening [14], which represents rapid progress in dealing with the crisis. Virtual screening suggested that ledipasvir or velpatasvir might be particularly attractive as therapeutics to combat the new coronavirus [15]. Moreover, this protease's potential inhibitors were postulated to be alpha-keto amides with modifications added based on the protein–compound interactions [16]. However, to the best of our knowledge, no study to date exploited aminoglycosides as Mpro inhibitors. Therefore, we screened a library of antiviral agents against the Mpro enzyme. Initial results indicate Amikacin's effectiveness (aminoglycoside) in binding at the substrate site of Mpro and making contacts with the catalytic residues. Further, some other aminoglycosides were selected to examine their effect on the Mpro binding. Overall, Amikacin was found to be the best amongst aminoglycosides.
Materials and methods
Retrieval and processing of ligands
The ligands in antiviral (L7000) and anti-infection (L3100) agents libraries available at Selleck Inc (www.selleckchem.com) were processed using “LigPrep-2018 (Schrodinger, LLC, NY, USA)” as described previously [17]. Briefly, the ionization states of ligands were defined at pH 7.0 ± 2.0, and the salt (if any) was removed using “Epik-2018 (Schrodinger, LLC, NY, USA)”. For each ligand, a maximum of 32 conformations was allowed to be generated. The energies of each ligand were minimized by OPLS3e (Optimized Potential for Liquid Simulations). A total of 3809 conformations representing different ionization states of ligands were generated and employed further in the study.
Retrieval and processing of protein (Mpro)
The 3D structural coordinates of the SARS-CoV-2 main protease (Mpro) were retrieved from the PDB databank (www.rcsb.org/structure/6LU7). The X-ray crystal structure of Mpro (PDB Id: 6LU7) was resolved to 2.16 Å and contained a bound peptide-based inhibitor (N3) [18]. Before virtual screening and molecular docking, the structure of Mpro was processed using “Protein Preparation Wizard-2018 (Schrodinger, LLC, NY, USA)” as reported earlier [17]. Briefly, hydrogen atoms were added, bond orders were assigned, any missing loops or side chains were added using “Prime-2018 (Schrodinger, LLC, NY, USA)”. Non-catalytic water molecules and any other heterogeneous atoms were removed. A hydrogen bond network was created and optimized at a physiological pH of 7.4. Finally, the energy of the protein was minimized using OPLS3e forcefield. The grid enclosing substrate-binding site was created using the “Receptor Grid Generation tool (Schrodinger, LLC, NY, USA)” by selecting the bound ligand (N3) as the center of the grid. A grid box of 88 × 88 × 88 Å dimension was generated for virtual screening and molecular docking.
High throughput virtual screening (HTVS) and molecular docking
Screening of ligands toward the substrate-binding site of Mpro was performed using the HTVS module in “Glide-2018 (Schrodinger, LLC, NY, USA)”. The ligands displaying a good affinity for Mpro were again docking at the substrate-binding site using standard precision (SP) docking. Further, the ligands filtered through SP docking were once again docked at the substrate-binding site of Mpro using extra precision (XP) docking of “Glide-2018 (Schrodinger, LLC, NY, USA)” [17]. Analyses of the results were performed using “Maestro-2018 (Schrodinger, LLC, NY, USA)”. The binding energy (ΔG) was used to calculate binding affinity (K d) of ligands using the relation, as described previously [19].
where R and T represent Boltzmann's gas constant (= 1.987 cal (mol K)−1 and temperature (= 298 K), respectively.
Prime/molecular mechanics-generalized born surface area (MM-GBSA) free energy calculations
The solvent effect on the binding of a ligand to protein was evaluated by estimating the MM-GBSA using “Prime-2018 (Schrodinger, LLC, NY, USA)” as reported earlier [17]. Briefly, the optimization feature of Prime-2018 (Schrodinger, LLC, NY, USA) was employed to minimize the docked poses, while the binding free energy of ligands was computed using OPLS3e forcefield, VSGB solvent model, and rotamer search algorithm, i.e., MM-GBSA continuum solvent model [20], [21], [22].
Molecular dynamics (MD) simulation
MD simulation of the most potent protein-ligand complex was performed to evaluate its stability and dynamics behavior. The initial conformation of the selected protein-ligand complex was subjected to an MD simulation of 100 ns using “Desmond-2018 (Schrodinger, LLC, NY, USA)” as described earlier [17]. An orthorhombic box was utilized to submerge protein-ligand complex in TIP3P explicit water solvent, at least 10 Å away from the box's walls. Proper counterions were added to neutralize the system, and physiological conditions were mimicked by adding 150 mM NaCl. Further, the whole system's energy was minimized with a convergence criterion of 1 kcal/mol/Å. MD simulation was performed under NTP conditions (300 K temperature and 1.013 bar pressure) wherein the temperature and pressure were maintained using Nose-Hoover-Chain thermostat and Martyna-Tobias-Klein barostat, respectively [23], [24]. A time-step of 2 fs was fixed, and the energy and structure were recorded and saved in the trajectory every 10 ps. The results were analyzed using “Maestro-2018 (Schrodinger, LLC, NY, USA)”.
Results
HTVS and SP and XP molecular docking analysis
A total of 3809 conformations representing different ionization states of ligands were screened for the substrate-binding site of Mpro using HTVS. The HTVS analysis showed that 1397 ligands could bind Mpro with varying docking energies in the range of −4.001 to −9.742 kcal/mol. These 1397 ligands were again docked to the substrate-binding site of Mpro using SP docking mode. Out of these, only 157 ligands showed considerable affinity toward Mpro with docking energies ≤ −4.245 kcal/mol (Supplementary Table 1). The Glide g-score, Glide e-model and Glide energy of ligands varied between −9.651 to −4.681 kcal/mol, −119.797 to −31.220 kcal/mol, and −76.950 to −12.790 kcal/mol, respectively. Further, XP docking was performed on top scoring ligands obtained by SP docking analysis, i.e., the ligands having ≤ −7.5 kcal/mol docking energy (13 ligands only) were again subjected to XP docking. The analysis of XP docking revealed that the docking energies of Iodixanol, Amikacin, Troxerutin, Rutin, Proanthocyanidins, Lomitapide, Birinapant, Netilmicin, Bortezomib, Lumefantrine, Leuprolide, Cobicistat, and Cangrelor were −11.776, −11.238, −11.075, −10.629, −8.427, −7.644, −7.635, −7.144, −7.078, −7.067, −6.464, −4.167 and −4.112 kcal/mol respectively (Table 1 ). Similarly, the Glide g-score, Glide e-model, and Glide energy of ligands in XP docking mode were −11.776 to −4.254 kcal/mol, −119.344 to −64.286 kcal/mol, and −87.118 to 53.821 kcal/mol, respectively. The most promising ligands (Iodixanol, Amikacin, Troxerutin, and Rutin) with docking score ≤ −10.000 kcal/mol were subjected to free energy calculations by Prime/MM-GBSA. The Prime/MM-GBSA score of Amikacin was lowest (−73.800 kcal/mol), followed by Troxerutin (−58.236 kcal/mol), Rutin (−56.651 kcal/mol) and Iodixanol (−45.963 kcal/mol) (Table 1).
Table 1.
XP docking and Prime/MM-GBSA scores of compounds having a docking score ≤ −7.5 kcal/mol in SP mode.
| S. no. | Name of compounds | Docking score (kcal/mol) | Glide g-score (kcal/mol) | Glide e-model (kcal/mol) | Glide energy (kcal/mol) | Primer/MM-GBSA (kcal/mol) |
|---|---|---|---|---|---|---|
| 1. | Iodixanol | −11.776 | −11.776 | −119.344 | −83.600 | −45.963 |
| 2. | Amikacin | −11.238 | −11.315 | −89.722 | −65.561 | −73.800 |
| 3. | Troxerutin | −11.075 | −11.078 | −100.903 | −69.179 | −58.236 |
| 4. | Rutin | −10.629 | −10.657 | −98.651 | −66.120 | −56.651 |
| 5. | Proanthocyanidins | −8.427 | −8.427 | −89.788 | −64.829 | ND |
| 6. | Lomitapide | −7.644 | −7.647 | −87.585 | −54.886 | ND |
| 7. | Birinapant | −7.635 | −8.556 | −93.992 | −64.414 | ND |
| 8. | Netilmicin | −7.144 | −7.863 | −69.786 | −54.375 | ND |
| 9. | Bortezomib | −7.078 | −7.078 | −68.339 | −55.127 | ND |
| 10. | Lumefantrine | −7.067 | −7.091 | −64.286 | −52.821 | ND |
| 11. | Leuprolide | −6.464 | −6.999 | −77.576 | −63.923 | ND |
| 12. | Cobicistat | −4.167 | −4.697 | −137.01 | −87.118 | ND |
| 13. | Cangrelor | −7.505 | −7.519 | −78.278 | −60.657 | ND |
ND stands for not determined.
Amikacin (shown in bold) is selected for further analysis.
XP docking and Prime/MM-GBSA analysis of aminoglycosides
The initial screening of ligands by HTVS and shortlisting by different docking procedures (SP and XP docking) led to the identification of Amikacin (an aminoglycoside) as the most promising inhibitor of SARS-CoV-2 Mpro. We extended the study by evaluating the binding affinity of other structurally similar aminoglycoside candidates such as Gentamycin, Neomycin, Paramomycin, Streptomycin, and Tobramycin toward the substrate-binding site of Mpro using XP docking and Prime/MM-GBSA (Table 2 ). Amongst aminoglycosides, Amikacin was the most potent inhibitor of Mpro as its XP docking score (−11.238 kcal mol−1), and Prime/MM-GBSA score (−73.800 kcal mol−1) was the lowest. A detailed analysis of Mpro-Amikacin interaction is given below, while the interaction of other aminoglycosides with Mpro is described in supplementary data.
Table 2.
Molecular docking (SP and XP) and Prime/MM-GBSA calculations of selected aminoglycosides toward Mpro of SARS-CoV-2.
| S. no. | Name of antibiotics | XP docking score (kcal/mol) | Binding affinity, Kd (M−1) | Prime/MM-GBSA (kcal/mol) |
|---|---|---|---|---|
| 1. | Amikacin | −11.238 | 1.75 × 108 | −73.800 |
| 2. | Neomycin | −9.114 | 4.84 × 106 | −58.174 |
| 3. | Paramomycin | −8.503 | 1.72 × 106 | −56.228 |
| 4. | Gentamycin | −7.770 | 4.99 × 105 | −53.394 |
| 5. | Streptomycin | −7.533 | 3.35 × 105 | −50.094 |
| 6. | Tobramycin | −5.831 | 1.89 × 104 | −53.582 |
Amikacin (shown in bold) is selected for further analysis.
Mpro-Amikacin interaction
Amikacin-Mpro interaction analysis revealed that it was bound at the substrate-binding site and interacted with key amino acid residues (Fig. 1 and Table 3 ). Amikacin formed seven hydrogen bonds (one hydrogen bond with each of Phe140, Pro168, and Gln189, and two hydrogen bonds with Cys145 and Glu166 each), and two salt bridges with Cys145 and Glu166. It also networked through van der Waals’ interaction with some other amino acid residues such as Hie41, Met49, Tyr54, Leu141, Asn142, Gly143, Ser144, His163, His164, Met165, Leu167, Gly170, Hie172, Asp187, and Arg188. It is worth to note that His41 and Cys145 are catalytic amino acid residues of SARS-CoV-2 Mpro. The docking energy and binding affinity of Amikacin toward Mpro were estimated to be −11.238 kcal/mol and 1.75 × 108 M−1, respectively. The Prime/MM-GBSA score of Amikacin-Mpro interaction was estimated to be −73.800 kcal/mol (Table 2).
Fig. 1.
Molecular docking of Amikacin with SARS-CoV-2 Mpro. (A) 2D view of Amikacin binding at the substrate-binding site of Mpro, (B) 3D view of Amikacin binding at the substrate-binding cavity of Mpro, and (C) Molecular interaction between Amikacin and Mpro, showing various kinds of bonds and amino acid residues responsible for the formation of a stable Amikacin-Mpro complex.
Table 3.
Molecular interaction between SARS-CoV-2 Mpro and different aminoglycosides.
| S. no. | Name of antibiotics | Residues of Mpro involved in various interactions |
||
|---|---|---|---|---|
| Hydrogen bonding | Salt bridges | Other interactions | ||
| 1. | Amikacin | Phe140, Cys145a, Glu166a, Pro168, Gln189 | Cys145, Glu166 | Hie41, Met49, Tyr54, Leu141, Asn142, Gly143, Ser144, His163, His164, Met165, Leu167, Gly170, Hie172, Asp187, Arg188 |
| 2. | Neomycin | Thr26, Ser46, Phe140, Cys145a, Glu166, Gln189 | Cys145, Glu166 | Thr25, Leu27, Hie41, Thr45, Met49, Leu141, Asn142, Gly143, Ser144, His163, His164, Met165, Hie172 |
| 3. | Paramomycin | Phe140a, Cys145, Glu166d, Thr190 | Cys145, Glu166a | Leu141, Asn142, Gly143, Ser144, His163, His164, Met165, Leu167, Pro168, Gly170, Hie172, Arg188, Gln189, Ala191, Gln192 |
| 4. | Gentamycin | Leu141, Asn142, Glu166b,Gln189a, Leu167 | Glu166a | Hie41, Met49, Phe140, Ser144, Cys145, His163, His164, Met165, Pro168, Gly170, Hie172, Arg188, Thr190, Gln192 |
| 5. | Streptomycin | Asn142, Cys145a, Glu166c, Gln189 | Cys145, Glu166 | Thr25, Thr26, Leu27, Hie41, Met49, Phe140, Leu141, Gly143, Ser144, His163, His164, Met165, Leu167, Pro168, Gly170, Hie172, Arg188, Thr190 |
| 6. | Tobramycin | Phe140, Asn142, Cys145, Glu166c, Leu167 | Cys145, Glu166a | Hie41, Met49, Leu141, Gly143, Ser144, His163, His164, Met165, Pro168, Gly170, Hie172, Gln189 |
Two interactions.
Three interactions.
Four interactions.
Five interaction.
Residues in bold are catalytic residues.
Mpro-Amikacin MD simulation analysis
Root mean square deviation (RMSD) analysis
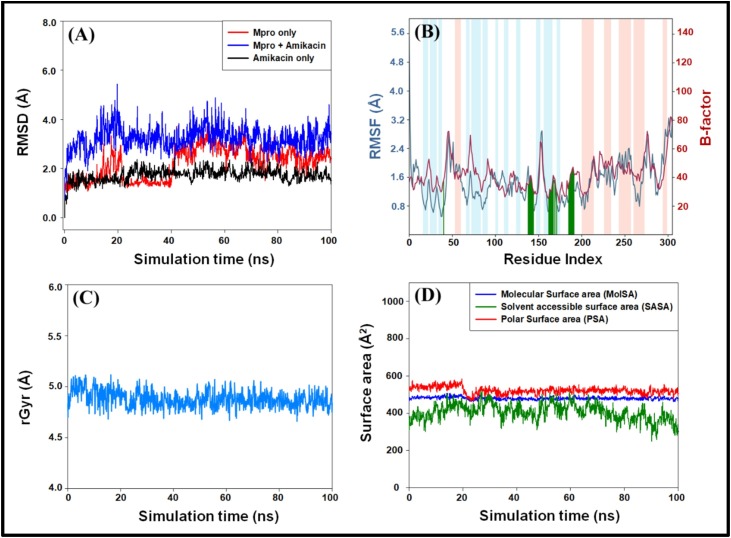
RMSD is a measure of deviation in the position of Cα-atoms compared to the initial frame, as a function of simulation time. An RMSD value of ±2.00 Å suggests that the structure of protein remains stable throughout the simulation. The RMSD plots of Mpro, Amikacin, and Mpro-Amikacin complex is shown in Fig. 2A. A sharp increase in the RMSD value of Mpro was observed during the initial phase of simulation; however, it was stabilized in the latter part of the simulation (40–100 ns). The RMSD values of Amikacin were observed to remain within the limit throughout the simulation. Moreover, the RMSD value of the Mpro-Amikacin complex was increased sharply during 0–15 ns but remained constant within ±2.00 Å for the rest of the simulation time. During the initial phase, the variation in the RMSD value of the Mpro-Amikacin complex was due to the entry of a large ligand (Amikacin) into the substrate-binding site of Mpro. However, once stable interactions were formed between Mpro and Amikacin, the RMSD value became stable (Fig. 2A). The mean ± standard deviations in the RMSD values of Mpro, Amikacin, and Mpro-Amikacin complex during 40–100 ns duration were estimated to be 2.65 ± 0.40 Å, 3.27 ± 0.42 Å, and 1.79 ± 0.21 Å, respectively.
Fig. 2.
Molecular docking simulation of the Amikacin-Mpro complex. (A) Root mean square deviations (RMSDs) in the Cα-atoms of Mpro only (red), Amikacin-Mpro complex (blue) and Amikacin only (black), during simulation, (B) root mean square fluctuations (RMSFs) in the Cα-atoms of Mpro (teal) as compared to experimentally determined B-factor of Mpro (brown). The vertical lines (green) on X-axis represent the amino acid residue with which Amikacin formed a contact. The light brown and light teal vertical bars represent the secondary structures α-helices and β-sheets respectively, (C) variation in the radius of gyration (rGyr) as a function of simulation time, and (D) variations in molecular surface area (MolSA), solvent accessible surface area (SASA) and polar surface area (PSA) as a function of simulation time.
Root mean square fluctuation (RMSF) analysis
RMSF measures the conformation changes in the side chains of individual amino acid residues during a simulation. Fig. 2B represents the RMSF plot of the Mpro-Amikacin complex (blue curve) and the B-factor of Mpro (brown curve) determined experimentally during X-ray crystallography. The RMSF values of the Mpro-Amikacin complex were in agreement with the B-factor of Mpro, i.e., the protein did not undergo any significant conformational changes due to the inhibitor's binding. The vertical green lines on X-axis show the contact between Amikacin and Mpro, while the light brown and teal vertical bars represent the regions of α-helices and β-sheets. The RMSF plot of the Mpro-Amikacin complex also confirmed that all the contacts between Mpro and Amikacin were located in domain II, harboring the substrate-binding site (Fig. 2B).
Analysis of radius of gyration (rGyr) and surface areas
The analysis of rGyr of a ligand demonstrates its compactness as a function of simulation. In this study, the rGyr of Amikacin in the Mpro-Amikacin complex was found to remain constant around 4.88 ± 0.08 Å (Fig. 2C). It indicates the formation of a stable Mpro-Amikacin complex due to the formation of favorable interactions. Further, the variation in molecular surface area (MolSA), solvent accessible surface area (SASA), and polar surface area (PSA) of Amikacin was also determined during the simulation (Fig. 2D). It was found that the MolSA, SASA, and PSA of Amikacin remained constant throughout the simulation. The mean value along with standard deviations of MolSA, SASA, and PSA of Amikacin was estimated to be 480.30 ± 7.71 Å2, 405.55 ± 34.89 Å2, and 522.16 ± 19.24 Å2, respectively. It is evident that MolSA, SASA, and PSA's values were fluctuating within the prescribed limits, thus indicating the formation of a stable Mpro-Amikacin complex.
Analysis of interactions pattern during simulation
The analysis of the interaction between Amikacin and Mpro that occurred during simulation indicates that hydrogen bonds played a significant role in stabilizing the Mpro-Amikacin complex (Fig. 3 ). Amikacin formed various interactions such as hydrogen bonds, hydrophobic interactions, ionic interactions, and water bridges with different substrate binding sites’ residues (Fig. 3A). The total number of contacts formed between Mpro and Amikacin during the simulation was in the 2-17 range, with an average of 8 contacts (Fig. 3B, upper panel). The significant amino acid residues participated in different capacities to stabilize the Mpro-Amikacin complex were His41, Ser139, Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, His163, His164, Met165, Glu166, Leu167, Pro168, Gly170, His172, Val180, Asp187, Arg188, Gln189, Thr190, Ala191, and Gln192. Interestingly, Asn142, Ser144, Cys145, His164, and Glu166 played a significant role in making a stable Mpro-Amikacin complex (Fig. 3B, lower panel). The catalytic residue Cys145 formed a hydrogen bond and a water bridge with Amikacin for 76% and 37% of simulation time, respectively. Moreover, Glu166 formed two hydrogen bonds with Amikacin for 97% and 73% of simulation time, and a water bridge for more than 50% simulation time. His164 also formed a hydrogen bond with Amikacin for a shorter duration of around 37% simulation time (Fig. 3C).
Fig. 3.
Molecular interactions between Amikacin and Mpro during molecular dynamics simulation. (A) Participation of different amino acid residues during simulation. An interaction fraction of >1 shows that the residue was involved in more than one kind of interaction, (B) contribution of amino acid residues in making contacts with Amikacin as a function of simulation time, and (C) percent interaction between different amino acid residues and Amikacin during the simulation. Only residues having interaction with Amikacin for >30% simulation are shown.
Analysis of the secondary structure
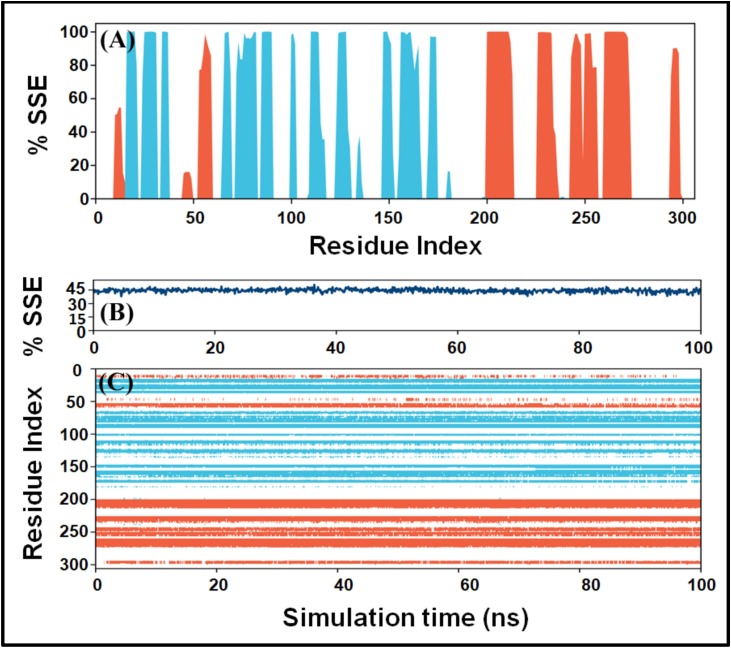
The analysis of variations in the secondary structure of a protein during simulation is crucial to examine protein conformation's stability. Fig. 4A shows the contribution of individual amino acid residues in secondary structure formation, i.e., α-helices (blue bars) and β-sheets brown bars). It is evident that the majority of domain I and domain II amino acid residues, which are part of α-helices and β-sheets in the native structure, continue to contribute nearly 100% of simulation time in maintaining Mpro's secondary structural conformation. Similarly, the amino acid residues of domain III, which are part of β-sheets in the native structure, continue to participate nearly 100% of simulation time in maintaining the protein's conformation (Fig. 4A). Moreover, the secondary structural elements (SSE) of Mpro reported in X-ray crystal structure were 52% (α-helix: 27% + β-sheet: 25%). During the simulation, the average secondary structure of Mpro was estimated to be 48% (α-helix: 24% + β-sheet: 24%), which was in agreement with the reported value (Fig. 4B). The contribution of individual amino acid residues over the simulation duration is represented in Fig. 4C. It is evident that the secondary structure of Mpro remained steady and did not deviate significantly upon Amikacin's binding during the whole simulation duration. The minor local changes might be due to the entry of a big ligand such as Amikacin into the substrate-binding cavity of Mpro.
Fig. 4.
Variation in the secondary structure of Mpro as a function of simulation. (A) Contribution of individual amino acid residues in the formation of protein's secondary structure elements (SSE). Orange and teal bars represent changes in α-helices and β-sheets. (B) Fluctuations in the SSE (%) as a function of simulation, and (C) contribution of individual amino acid residues in the overall secondary structure of Mpro during the simulation.
Discussion
The recent outbreak of COVID-19 caused by SARS-CoV-2 has implicated a substantial socioeconomic burden. The increasing number of cases and high mortality has fueled the academic and pharmaceutical industries to discover effective therapeutics. However, the conventional method of drug discovery is time-consuming and requires massive financial support. An alternative approach to identify lead compounds is the engagement of computational methods such as structure-based drug designing against potential targets of SARS-CoV-2. In this study, we have employed high-throughput virtual screening, molecular docking, and molecular dynamics simulation studies to identify novel inhibitors of SARS-CoV-2. We have applied drug-repurposing strategies to screen FDA-approved antiviral and anti-infective agents’ library available at Selleck Inc. against the main protease (Mpro or 3CLpro) of SARS-CoV-2.
The main protease (Mpro) of SARS-CoV-2 is a suitable target for drug development as Mpro, along with PLpro, are responsible for the cleavage of polypeptides pp1a and pp1ab into 16 fully functional non-structural proteins (nsps). Earlier, Yang et al. [6] have used Mpro of SARS-CoV as the most suitable target and designed an effective peptide-based inhibitor (N3), which binds at the substrate-binding site of Mpro. Recently, Mpro has emerged as the target of choice to design effective therapeutics against SARS-CoV-2. For instance, Jin et al. [14] have reported an X-ray crystal structure of Mpro bound with a mechanism-based inhibitor (N3). Some other computer-based drug designing and drug repurposing approaches have identified potential inhibitors of Mpro, such as Nelfinavir and Lopinavir [25]; Hesperidin and Diosmin [26]; Kaempferol, Quercetin, and Rutin [26]; and Ebselen [14].
This study identified 4 potential lead compounds after screening 3809 different conformational states of compounds using different methods such as HTVS, SP docking, XP docking, and Prime/MM-GBSA. The lead compounds (Amikacin, Iodixanol, Troxerutin, and Rutin) have varied antiviral and anti-infective activities. Amikacin is an aminoglycoside antibiotic widely used as an anti-bacterial agent against various infections such as meningitis, pneumonia, sepsis, abdominal infection, joint infection, urinary tract infection [27], [28]. Amikacin acts by inhibiting the protein synthesis after it binds the 16S rRNA and the RNA-binding S12 protein of the 30S subunit of the prokaryotic ribosome [29]. Iodixanol is an osmomolar, non-ionic radiocontrast agent generally used in computed tomography (CT) during coronary angiography. It has a short half-life of 2 h and excreted unchanged through the body. Troxerutin (a derivative of Rutin) is a naturally occurring flavonoid found in most fruits and vegetables. Troxerutin has hepatoprotective, vasoprotective, anti-erythrocytic, anti-thrombin, fibrinolytic, and edema-protective rheological activities, owing to its antioxidant potential [30], [31], [32], [33]. Moreover, it protects radiation-induced damage to DNA and micro-nuclei formation. Rutin, one of the top-ranked drugs in our study, is a natural flavonoid commonly found in citrus fruits. It has diverse biological activities such as anti-allergic, anti-inflammatory, anti-proliferative, and anti-carcinogenic. Rutin acts as a quercetin's carrier to the large intestine, where it gets metabolized to release the active Quercetin [34].
Among the lead compounds, Amikacin exhibited the lowest Prime/MM-GBSA energy. Other aminoglycosides such as Gentamycin, Neomycin, Paramomycin, Streptomycin, and Tobramycin were also evaluated for their potential to inhibit Mpro. All the aminoglycosides have been found to interact with Mpro at the substrate-binding site, primarily through extensive hydrogen-bonding interactions with the key/catalytic residues. Among aminoglycosides, Amikacin was an effective inhibitor of Mpro due to its lowest Prime/MM-GBSA score. Hence, the stability of Mpro-Amikacin was evaluated by molecular dynamics simulation. Amikacin interacted with Mpro with crucial amino acid residues, Asn142, Ser144, Glu166, Cys145, and His164 in domain II. Among the crucial amino acids at the binding site, Glu166 forms a strong hydrogen bond with the Amikacin terminal –NH3 and –OH atoms. It has been shown that the catalytic dyad (His41 and Cys145) at the junction of domain I and domain II of Mpro is responsible for its catalytic activity. Our analysis revealed that the residues Pro168 and Glu166, adjacent to Cys145, are involved in the interaction with the drug molecule, suggesting their crucial role in inhibiting the protein–protein interaction between the main protease and the human epigenetic regulatory proteins.
In addition to anti-bacterial activity, aminoglycosides have been discovered to possess antiviral properties against herpes simplex virus, influenza virus, and Zika virus [35], [36]. Thus, aminoglycosides could serve as lead molecules to develop potential inhibitors against SARS-CoV-2.
Conclusion
During the COVID-19 pandemic, drug repurposing is being pursued as a fast strategy to develop safe and effective COVID-19 treatments. With some candidates being moved into clinical trials, no drug has shown a beneficial response against COVID-19 infection. In this study, a variety of potential ligands were considered for its affinity toward SARS-CoV-2 protease. Out of four potential lead compounds, Amikacin (an aminoglycoside) emerged as the most potent drug-candidate exhibiting the highest binding affinity toward Mpro. Other aminoglycosides such as Gentamycin, Neomycin, Paramomycin, Streptomycin, and Tobramycin also showed a high affinity toward Mpro. However, before clinical application, detailed studies are needed to establish the antiviral potential of aminoglycosides.
Funding
No funding sources.
Competing interests
None declared.
Ethical approval
Not required.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group no. RG-1441-461.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jiph.2021.01.016.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79 doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. 2005;287 doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H., Bartlam M., Rao Z. Drug design targeting the main protease, the Achilles’ heel of coronaviruses. Curr Pharm Des. 2006;12:4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- 7.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu R.H., He J.F., Evans M.R., Peng G.W., Field H.E., Yu D.W. Epidemiologic clues to SARS origin in China. Emerg Infect Dis. 2004;10 doi: 10.3201/eid1006.030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87:7790–7792. doi: 10.1128/jvi.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 12.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281 doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentile D., Patamia V., Scala A., Sciortino M.T., Piperno A., Rescifina A. Putative inhibitors of SARS-COV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar Drugs. 2020;18 doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y. Structure of M pro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 doi: 10.1101/2020.02.26.964882. [DOI] [Google Scholar]
- 15.Chang Y., Tung Y., Lee K., Chen T., Hsiao Y., Chang C. Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking. Preprints. 2020:1–7. doi: 10.20944/preprints202002.0242.v2. [DOI] [Google Scholar]
- 16.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed M.Z., Muteeb G., Khan S., Alqahtani A.S., Somvanshi P., Alqahtani M.S. Identifying novel inhibitor of quorum sensing transcriptional regulator (SdiA) of Klebsiella pneumoniae through modelling, docking and molecular dynamics simulation. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1767209. [DOI] [PubMed] [Google Scholar]
- 18.Zeng H., Liu Z., Hu G., Qu L., Yang R. Investigation on the binding of aloe-emodin with tyrosinase by spectral analysis and molecular docking. Spectrochim Acta A: Mol Biomol Spectrosc. 2019;211:79–85. doi: 10.1016/j.saa.2018.11.045. [DOI] [PubMed] [Google Scholar]
- 19.Rehman M.T., Ahmed S., Khan A.U. Interaction of meropenem with ‘N’ and ‘B’ isoforms of human serum albumin: a spectroscopic and molecular docking study. J Biomol Struct Dyn. 2016;34:1849–1864. doi: 10.1080/07391102.2015.1094411. [DOI] [PubMed] [Google Scholar]
- 20.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput. 2016;12 doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 21.Li J., Abel R., Zhu K., Cao Y., Zhao S., Friesner R.A. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins Struct Funct Bioinform. 2011;79 doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brańka A.C. Nosé-Hoover chain method for nonequilibrium molecular dynamics simulation. Phys Rev E: Stat Phys Plasmas Fluids Relat Interdiscip Top. 2000;61:4769–4773. doi: 10.1103/PhysRevE.61.4769. [DOI] [PubMed] [Google Scholar]
- 24.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. doi: 10.1063/1.467468. [DOI] [Google Scholar]
- 25.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S. Potential inhibitor of COVID-19 main protease (M pro) from several medicinal plant compounds by molecular docking study. Preprints. 2020:1–14. doi: 10.20944/preprints202003.0226.v1. [DOI] [Google Scholar]
- 26.Adem S., Eyupoglu V., Sarfraz I., Rasul A., Ali M. Identification of potent COVID-19 main protease (Mpro) inhibitors from natural polyphenols: an in silico strategy unveils a hope against CORONA. Preprints. 2020 doi: 10.20944/preprints202003.0333.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmad S., Mokaddas E. Current status and future trends in the diagnosis and treatment of drug-susceptible and multidrug-resistant tuberculosis. J Infect Public Health. 2014;7:75–91. doi: 10.1016/j.jiph.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Caminero J.A., Sotgiu G., Zumla A., Migliori G.B. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis. 2010;10:621–629. doi: 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
- 29.Lambert T. Antibiotics that affect the ribosome. Rev Sci Tech. 2012;31:57–64. doi: 10.20506/rst.31.1.2095. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z.F., Fan S.H., Zheng Y.L., Lu J., Wu D.M., Shan Q. Troxerutin improves hepatic lipid homeostasis by restoring NAD+-depletion-mediated dysfunction of lipin 1 signaling in high-fat diet-treated mice. Biochem Pharmacol. 2014;91:74–86. doi: 10.1016/j.bcp.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Turner R.B., Fowler S.L., Berg K. Treatment of the common cold with troxerutin. APMIS. 2004;112:605–611. doi: 10.1111/j.1600-0463.2004.apm1120908.x. [DOI] [PubMed] [Google Scholar]
- 32.Adam B.S., Pentz R., Siegers C.P., Strubelt O., Tegtmeier M. Troxerutin protects the isolated perfused rat liver from a possible lipid peroxidation by coumarin. Phytomedicine. 2005;12:52–61. doi: 10.1016/j.phymed.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Maurya D.K., Balakrishnan S., Salvi V.P., Nair C.K. Protection of cellular DNA from γ-radiation-induced damages and enhancement in DNA repair by troxerutin. Mol Cell Biochem. 2005;280:57–68. doi: 10.1007/s11010-005-8052-3. [DOI] [PubMed] [Google Scholar]
- 34.Kim H., Kong H., Choi B., Yang Y., Kim Y., Mi J.L. Metabolic and pharmacological properties of rutin, a dietary quercetin glycoside, for treatment of inflammatory bowel disease. Pharm Res. 2005;22:1499–1509. doi: 10.1007/s11095-005-6250-z. [DOI] [PubMed] [Google Scholar]
- 35.Cohen J.I. New activities for old antibiotics. Nat Microbiol. 2018;3:531–532. doi: 10.1038/s41564-018-0152-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gopinath S., Kim M.V., Rakib T., Wong P.W., Van Zandt M., Barry N.A. Topical application of aminoglycoside antibiotics enhances host resistance to viral infections in a microbiota-independent manner. Nat Microbiol. 2018;3:611–621. doi: 10.1038/s41564-018-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.