Abstract
Background
The outbreak of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) that was first detected in the city of Wuhan, China has now spread to every inhabitable continent, but now the attention has shifted from China to other epicentres. This study explored early assessment of the influence of spatial proximities and travel patterns from Italy on the further spread of SARS-CoV-2 worldwide.
Methods
Using data on the number of confirmed cases of COVID-19 and air travel data between countries, we applied a stochastic meta-population model to estimate the global spread of COVID-19. Pearson's correlation, semi-variogram, and Moran's Index were used to examine the association and spatial autocorrelation between the number of COVID-19 cases and travel influx (and arrival time) from the source country.
Results
We found significant negative association between disease arrival time and number of cases imported from Italy (r = −0.43, p = 0.004) and significant positive association between the number of COVID-19 cases and daily travel influx from Italy (r = 0.39, p = 0.011). Using bivariate Moran's Index analysis, we found evidence of spatial interaction between COVID-19 cases and travel influx (Moran's I = 0.340). Asia-Pacific region is at higher/extreme risk of disease importation from the Chinese epicentre, whereas the rest of Europe, South-America and Africa are more at risk from the Italian epicentre.
Conclusion
We showed that as the epicentre changes, the dynamics of SARS-CoV-2 spread change to reflect spatial proximities.
Keywords: COVID-19, Coronavirus, Epicentre, Spatial proximity, Travel, China, Italy, One health, Health security
1. Introduction
As at June 30th 2020, 215 countries and territories have confirmed at least one case of COVID-19 and a total number of 10,185,374 COVID-19 cases have been confirmed globally – mostly in Europe (10.4%) and the Americas (50.4%) [1]. Human population movement plays an important role in the spread of infectious diseases. Although initially China has served as the epicentre of the current SARS-CoV-2 outbreak, real-time phylogenetic analysis suggests that the virus was introduced to most African countries and Latin America from people traveling from Italy [2]. Migration has remained the major source of concern for the current COVID-19 outbreak and most countries have focused on China as the likely source of any importation. While the movement of people between China and sub-Saharan Africa has increased rapidly over the last decade, the spread of COVID-19 to the African continent has been more related to the current COVID-19 outbreak in Italy [[3], [4], [5]]. There has been much attention on the importation of infectious diseases such as Ebola, tuberculosis, malaria or viral hepatitis from Africa to Europe [6,7]. Until now, there are few reports that focus on cases of imported infectious diseases from Europe to Africa and South America. This, perhaps, is why most African and South American countries initially focused their COVID-19 surveillance efforts on travellers from China without much attention paid to the possible importation from other countries.
In addition to phylogenetic analysis [2], a number of other circumstantial factors support the introduction of SARS-CoV-2 to countries other than from China. For example, it appears more likely that the introduction of COVID-19 to Africa and South America occurred from Italy rather than from China. First, since January 23rd 2020, China increased its containment measures and decreased the number of outbound international flights. Second, early febrile airport screening mostly targeted travellers from Asia overlooking those from other parts of the world – including travellers from Europe. However, though, febrile airport screening turned out to have limited effectiveness [8]. As most of the infections by SARS-CoV-2 are asymptomatic and mild, it is more likely that infected carriers could enter a country without being detected by the temperature screening at the airports [[9], [10], [11]]. Lastly, in the case of South America, Brazilians of European origin are the largest group of foreigners with full or partial Italian ancestry outside Italy. Travel by these individuals could partially explain the introduction of SARS-CoV2 to Brazil when Italy (and Europe in general) became the epicentre of the pandemic in March 2020 which is also supported by phylogenetic analysis [12,13]. In this paper, we provided an early assessment of the epidemic in Italy in comparison to the Chinese epicentre. We also investigated the dynamic of the spread of COVID-19 for the two epicentres and how it affect the spread of the pandemic worldwide using measurements of spatial proximity and travel volume between countries.
2. Materials and methods
In this early assessment of the pandemic, we obtained data on the number of confirmed cases of COVID-19 in each country from the World Health Organisation (WHO) situation reports and real-time online COVID-19 monitoring sites until March 10th 2020 [1,14,15]. The air travel data between countries were obtained from the Official Airline Guide (OAG) database [16]. Preliminary analysis includes Pearson's correlation analysis to examine the association between the number of COVID-19 cases and travel influx (and arrival time) from the source country. Spatial data reveal the degree of dependency among observations in geographical space [17,18]; consequently, spatial dependence measures such as Moran's Index to detect spatial patterns in COVID-19 data [19]. Bivariate Moran's Index was used to measure the spatial autocorrelation between the number of confirmed COVID-19 cases and travel influx [20]. The spatial clusters and type of spatial autocorrelation for each country were presented as Local Indicator of Spatial Association (LISA) plot [21]. For the spatial weights, we used Queen-style contiguity 1st order nearest neighbour matrix (i.e., two countries are neighbouring if they share common borders or a point). Empirical semi-variogram plots were also used to graphically visualize the spatial autocorrelation between the number of confirmed COVID-19 in each country and their distances from an epicentre.
Additionally, we developed a stochastic meta-population model of global spread using the flight data of travel volumes between each country. The country-specific compartmental model is an adaptation of the classical SEIR (Susceptible-Exposed-Infectious-Recovered) model based on reported dynamics of the novel coronavirus for global spread of COVID-19 [22], [23], [35]. See the appendix for model details and Bosco et al. [24] for deterministic version. In other to compare the relative risk of importation of COVID-19 from Italy and China to other countries, we assumed the same number of confirmed COVID-19 cases exported from China and Italy to initialise the number of people infected. We classified each country's risk of importation into quartiles, slight risk (<25%), moderate risk (25%–50%), high risk (50%–75%) and extreme risk (>75%).
3. Results and discussion
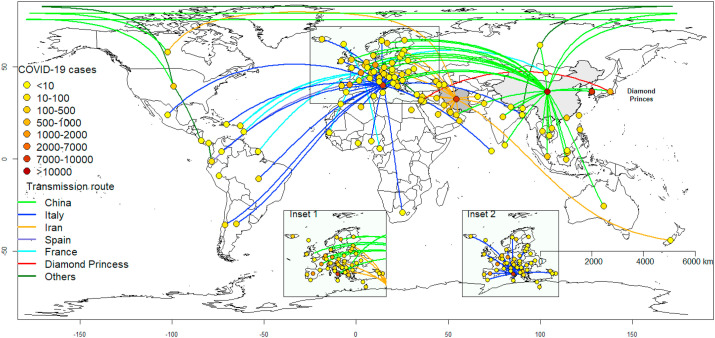
Up until the February 21st 2020 Italian outbreak, all reported European cases were imported from China (Fig. 1 ) [25]. By March 10th 2020 the disease had spread to 46 out of the 53 countries within the WHO European region [1]. At that time, Italy had 9172 confirmed cases and 463 deaths. The European region alone accounted for 46% of the total cases outside of China. There are currently three COVID-19 epicentres –China, Italy, and Iran, which are mostly responsible for spreading the virus globally (Fig. 1).
Fig. 1.
Transmission routes of COVID-19 as at March 7th, 2020. The lines represent transmission routes from the source of COVID-19 into a country. Inset 1: European cases originating from China (green lines) and Iran (orange lines). Inset 2: Cases originating from Italy. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
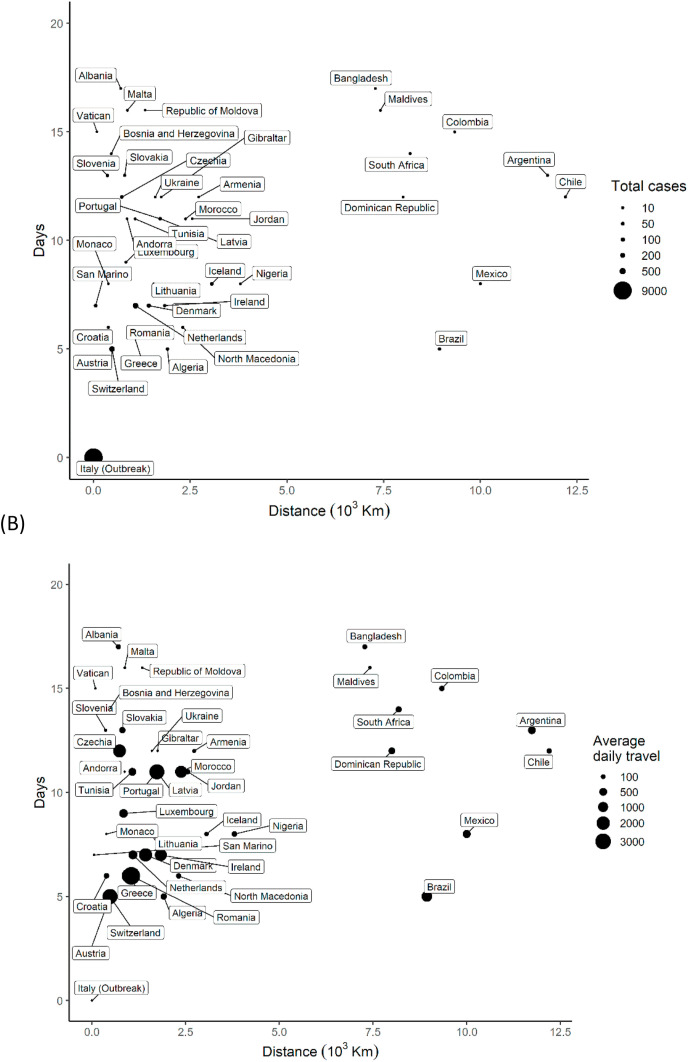
We present in Fig. 2 a, the distribution of time elapsed since February 21st 2020 when the first imported case of COVID-19 from Italy was reported in each country against the geographical distance from Italy. The outbreak in Italy was highly sporadic in the last three weeks. While the shortest arrival time (after the Italy outbreak) was five days to Austria, Switzerland, Brazil and Algeria, the virus spread to 44 countries within 17 days (median, 11 days) (Fig. 2b). The global arrival times for three epicentres from the first reported cases in Wuhan, China are shown in Fig. S1.
Fig. 2.
Distribution of arrival times of COVID-19 cases against geographic distance from the source (Italy). The dots are proportional to the size of, (A) total cases, and (B) travel influx from the source country.
The Pearson's correlation analysis (r = −0.43, p = 0.004) suggests a negative association between disease arrival time and the number of cases imported from Italy. That is, countries with a shorter introduction time have more cases. There was a significant association between the number of cases and daily travel influx from Italy (r = 0.39, p = 0.011). The apparent spatial autocorrelation in the COVID cases within Europe (Moran's Index = 0.310, p=<0.001) is also significant (Fig. S2). Notably, France and Italy are located in the right upper quadrant, while Switzerland, Austria and Slovakia are on the left upper quadrant, indicating a positive and negative spatial autocorrelation patterns respectively. Additionally, semi-variograms indicated spatial autocorrelation of the disease incidence exists up to a distance of 120 decimal degrees for Italy (Fig. S3). Using bivariate LISA analysis, we found evidence of spatial interaction between COVID-19 cases and arrival time as well as travel influx (Moran's I = 0.340). This suggests that spatial variations within the European region were non-random, exhibiting effects of neighbouring interactions and travel influx.
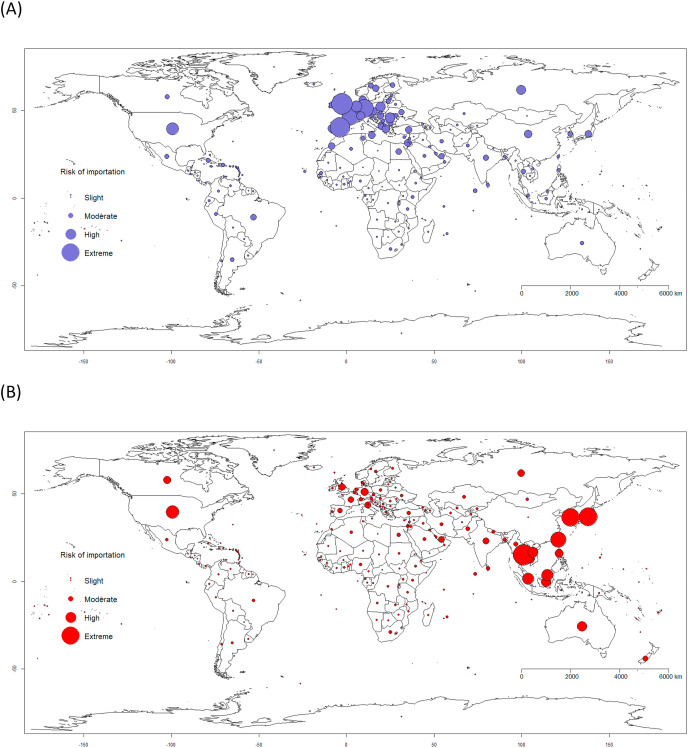
We simulated COVID-19 spread comparing two outbreak epicentres – Italy and China. Fig. 3 shows the relative risk of importation from the two epicentres. The results show that whereas the Asia-Pacific is more at risk (high to extreme risk) from the Chinese epicentre, Europe, South-America and Africa are more at risk from the Italian epicentre. High population density and highly interconnected transportation networks connecting tourism hubs in Northern Italy with major European cities have made it extremely difficult to contain and reduce infections. This aligns well with our prediction of extreme risks of case importations from Italy to other European countries, while Latin American and African countries are at high risk (Fig. 3). In the case of Italy, our modelling results could have provided an argument for the Italian government to institute an early national quarantine and travel restrictions to mitigate the spread of infection. Based on the evidence from the Chinese policies of containment and quarantine that showed considerable effectiveness [26], these interventions would likely to slow the spread of the disease within Europe and abroad.
Fig. 3.
Global risk of importation of COVID-19 from, (A) Italy (purple dot) and (B) China (red dot) based on air travel influx from source country. The dots are proportional to the cumulative relative risk that an infected individual will be arriving at a specific country for an epicenter. Risk of importation: slight risk (<25%), moderate risk (25%–50%), high risk (50%–75%) and extreme risk (>75%). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Although most African countries have focused their COVID-19 surveillance efforts on travellers from China, our analysis shows that this approach may need to be reconsidered as the number of cases in Europe soar. This makes the risk of importation to Africa higher from European countries than from China through stronger transportation connectivity and migration flows (especially, with North African countries). The burden of COVID-19 (cases per 100,000 people) in Africa was highest in Djibouti and South Africa [27]. The preparedness and capacity of African countries to detect, respond and prevent infectious disease have been suggested to be low [28,29]. This may be complicated by the indirect effects associated with disruptions to other critical healthcare services due to concurrent outbreaks in some African countries [30].
4. Conclusion
Using spatial analysis and a web-based mathematical modelling, we estimated the risk of importation from two major epicentres to other countries and showed that spatial proximity and mobility are important factors that fuel disease importation. These non-random spatial variations among neighbouring countries were supported by high quality mobility data in the study. However, the data was limited to air travel passenger flows and train and bus data could further improve the accuracy of our estimates. Similarly, we did not account for underlying health conditions in the meta-population model projections. These limitations in addition to data quality have been a challenge for modellers [31].
This analysis illustrates the potential development of the infection spread from emerging epicentres to other regions and may be useful, especially in the early stages of the pandemic, in countries’ epidemic preparedness including large-scale interventions such as travel restrictions and containment strategies. As at now, Italy has put itself under lockdown and other countries such as Australia and the USA have restricted flights from Italy [[32], [33], [34]]. As new epicentres are emerging, countries must adapt quickly and adjust their containment measures to reduce the spread of infection.
CRediT authorship contribution statement
Oyelola A. Adegboye: Conceptualization, Data curation, Formal analysis, Methodology, Roles, Writing - original draft. Adeshina I. Adekunle: Formal analysis, Methodology, Writing - original draft. Anton Pak: Formal analysis, Methodology, Writing - original draft. Ezra Gayawan: Formal analysis, Methodology, Writing - original draft. Denis HY. Leung: Methodology, Writing - original draft. Diana P. Rojas: Resources, Investigation, Writing - original draft. Faiz Elfaki: Visualization, Writing - review & editing. Emma S. McBryde: Resources, Writing - review & editing, Supervision. Damon P. Eisen: Writing - original draft, Writing - review & editing, Supervision.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tmaid.2021.101988.
Appendix 1.
Table S1.
State variable and parameter descriptions
| Variable | Description | |
|---|---|---|
| Susceptible class | ||
| Latent stage not infectious | ||
| Latent stage infectious | ||
| First stage of symptomatic | ||
| Second stage of symptomatic | ||
| Recovered class | ||
| Total population | ||
|
Parameter |
Description |
Values (references) |
| Transmission rates for classes | (23) | |
| First stage incubation rate | 0.3125 (23) | |
| Second stage incubation rate | 0.5 (23) | |
| First stage of recovery | 0.5 (23) | |
| second stage of recovery | 0.176 (23) | |
| COVID-19 case fatality | 0.018 (35) | |
| Death rate | Not used |
Appendix 2. Mathematical Model details
The infectious classes are late latency and the two stages of symptomatic infectiousness (). Patients either recover and moved to R class or die and are replaced to ensure a constant population. The two-country dynamical model is as shown below:
| (1) |
where the superscript 1 and 2 on the compartmental variable in equation (1) denotes country 1 and country 2, respectively.
This model is extended to all countries and coded in R [36] using the infectious disease node of the Australia Nectar Research cloud (www.nectar.org.au) as an individual-based model with a binomial distribution of the number of people experience an event at a specific time step. The events are infection, migration, emigration, recovery and death due to COVID-19. We neglected natural death, as this does not affect our result. We further assumed that no countries except the epicentres are experiencing an outbreak. Hence the basic reproduction number at the epicentres is . Each country's COVID-19 dynamics follows the schematic representation in Fig. S4.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Distribution of COVID-19 arrival times (days from first reported cases in China) and distance from China. The size of the dot is proportional to the daily air travellers between China and other countries as at March 10th, 2020.
Local indicator of spatial autocorrelation (LISA) plot for COVID-19 cases in Europe.
Semi-variogram showing spatial autocorrelation for COVID-19 data up 0.000 decimal degrees (~1.2 km) for Italy.
Schematic diagram of the transmission dynamics.
References
- 1.World Health Organization . In: Coronavirus disease (covid-2019) situation reports. ˆ, editor. Geneva, Switzerland; 2020. [Google Scholar]
- 2.Nextstrain . In: Genomic epidemiology of novel coronavirus (hcov-19) ˆ, editor. 2020. [Google Scholar]
- 3.Pigato M., Tang W. World Bank Working Paper; 2015. China and africa: expanding economic ties in an evolving global context. Investing in Africa forum. 95161. [Google Scholar]
- 4.Shen X. vol. 20. May; Washington, DC: 2014. Chinese manufacturers moving to Africa—who? What? Where? Does Africa benefit? (Conference “China in the world economy: building a new partnership with Africa” world bank). [Google Scholar]
- 5.Adegboye O.A., Adekunle A.I., Gayawan E. Early transmission dynamics of novel coronavirus (covid-19) in Nigeria. Int J Environ Res Publ Health. 2020;17(9):3054. doi: 10.3390/ijerph17093054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castelli F., Sulis G. Migration and infectious diseases. Clin Microbiol Infect. 2017;23(5):283–289. doi: 10.1016/j.cmi.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Khyatti M., Trimbitas R.-D., Zouheir Y., et al. Infectious diseases in north Africa and north African immigrants to Europe. Eur J Publ Health. 2014;24(suppl_1):47–56. doi: 10.1093/eurpub/cku109. [DOI] [PubMed] [Google Scholar]
- 8.Quilty B.J., Clifford S. Effectiveness of airport screening at detecting travellers infected with novel coronavirus (2019-ncov) Euro Surveill. 2020;25(5) doi: 10.2807/1560-7917.ES.2020.25.5.2000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou L., Ruan F., Huang M., et al. Sars-cov-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothe C., Schunk M., Sothmann P., et al. Transmission of 2019-ncov infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (covid-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. J Am Med Assoc. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 12.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of sars-cov-2 genomes. Proc Natl Acad Sci Unit States Am. 2020;117(17):9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candido D.D.S., Watts A., Abade L., et al. Routes for covid-19 importation in Brazil. J Trav Med. 2020;27(3) doi: 10.1093/jtm/taaa042. taaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu T., Ge X., Yu G. An R package and a website with real-time data on the covid-19 coronavirus outbreak. medRxiv. 2020 [Google Scholar]
- 15.Worldometer Covid-19 coronavirus outbreak. 2020. https://www.worldometers.info/coronavirus/ Assessed on February 5 2020.
- 16.Officoal Aviation Guide. OAG Flight database. Available at www.oag.com.
- 17.Liu W., Yang K., Qi X., et al. Spatial and temporal analysis of human infection with avian influenza a (h7n9) virus in China. Euro Surveill. 2013;18(47) doi: 10.2807/1560-7917.es2013.18.47.20640. 20640. [DOI] [PubMed] [Google Scholar]
- 18.Adegboye O.A., Leung D.H., Wang Y.G. Analysis of spatial data with a nested correlation structure. J Roy Stat Soc: Series C (Applied Statistics) 2018;1(67):329–354. [Google Scholar]
- 19.Moran P.A. A test for the serial independence of residuals. Biometrika. 1950;37(1/2):178–181. [PubMed] [Google Scholar]
- 20.Lee S.-I. Developing a bivariate spatial association measure: an integration of Pearson’s r and Moran’s I. J Geogr Syst. 2001;3(4):369–385. [Google Scholar]
- 21.Anselin L. A local indicator of multivariate spatial association: extending Geary’s C. Geogr Anal. 2019;51(2):133–150. [Google Scholar]
- 22.Lin Q., Chiu A.P., Zhao S., He D. Modeling the spread of middle east respiratory syndrome coronavirus in Saudi Arabia. Stat Methods Med Res. 2018;27(7):1968–1978. doi: 10.1177/0962280217746442. [DOI] [PubMed] [Google Scholar]
- 23.Kucharski A.J., Russell T.W., Diamond C., et al. Early dynamics of transmission and control of covid-19: a mathematical modelling study. Lancet Infect Dis. 2020;20(5):553–558. doi: 10.1016/S1473-3099(20)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho Bosco, Adekunle Adeshina, James Trauer, McBryde E. Global pandemic map. 2019. http://www.pandemic.org.au/v2/#/about Available at.
- 25.Stoecklin S.B., Rolland P., Silue Y., et al. First cases of coronavirus disease 2019 (covid-19) in France: surveillance, investigations and control measures, january 2020. Euro Surveill. 2020;25(6) doi: 10.2807/1560-7917.ES.2020.25.6.2000094. 2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chinazzi M., Davis J.T., Ajelli M., et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (covid-19) outbreak. Science. 2020;368(6489):395–400. doi: 10.1126/science.aba9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gayawan E., Oo Awe, Oseni B.M., et al. The spatio-temporal epidemic dynamics of covid-19 outbreak in Africa. Epidemiol Infect. 2020;148:e212. doi: 10.1017/S0950268820001983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert M., Pullano G., Pinotti F., et al. Preparedness and vulnerability of african countries against importations of covid-19: a modelling study. Lancet. 2020;395(10227):871–877. doi: 10.1016/S0140-6736(20)30411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Xu C., Wong Y.K., et al. Preparedness is essential for malaria-endemic regions during the covid-19 pandemic. Lancet. 2020;395(10230):1094–1096. doi: 10.1016/S0140-6736(20)30561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBryde E.S., Meehan M.T., Adegboye O.A., et al. Role of modelling in covid-19 policy development. Paediatr Respir Rev. 2020;35(2020):57–60. doi: 10.1016/j.prrv.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meehan M.T., Rojas D.P., Adekunle A.I., et al. Modelling insights into the covid-19 pandemic. Paediatr Respir Rev. 2020;35(2020):64–69. doi: 10.1016/j.prrv.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker P. U.S. To suspend most travel from Europe as world scramble to fight pandemic. N Y Times. 2020 https://www.nytimes.com/2020/03/11/us/politics/anthony-fauci-coronavirus.html Assessed on. [Google Scholar]
- 33.Mercer P. Australia extends virus travel ban to Italy. VOA. 2020 https://www.voanews.com/science-health/coronavirus-outbreak/australia-extends-virus-travel-ban-italy Assessed on. [Google Scholar]
- 34.Feuer W. Italy expands its quarantine to the entire country as coronavirus cases and deaths surge. CNBC. 2020 https://www.cnbc.com/2020/03/09/italy-extends-its-quarantine-to-the-entire-country-pm-asks-residents-to-stay-at-home.html Assessed on March 15, 2020. [Google Scholar]
- 35.Riou J., Hauser A., Counotte M.J., Althaus C.L. medRxiv; 2020. Adjusted age-specific case fatality ratio during the covid-19 epidemic in Hubei, China, January and February 2020. [Google Scholar]
- 36.Rc Team R . R foundation for statistical computing; Vienna, Austria: 2020. A language and environment for statistical computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of COVID-19 arrival times (days from first reported cases in China) and distance from China. The size of the dot is proportional to the daily air travellers between China and other countries as at March 10th, 2020.
Local indicator of spatial autocorrelation (LISA) plot for COVID-19 cases in Europe.
Semi-variogram showing spatial autocorrelation for COVID-19 data up 0.000 decimal degrees (~1.2 km) for Italy.
Schematic diagram of the transmission dynamics.