Abstract
Long noncoding RNAs play an important role in the occurrence, invasion, as well as metastasis of various human cancers, including hepatocellular carcinoma. Long noncoding RNAs can affect the biological functions of hepatocellular carcinoma cells by regulating various genes; however, only a small fraction of molecular mechanisms of long noncoding RNAs have been elucidated. In the present study, lnc AC010973.1 (lnc-ATG9B-4) was first identified by microarray analysis from 8 patients with hepatocellular carcinoma and confirmed by quantitative PCR in 176 patients with hepatocellular carcinoma. We demonstrated that lnc-ATG9B-4 was tightly relative to the tumorous size, TNM stages, portal vein tumor thrombus (PVTT), the tumor capsule, metastasis, degree of differentiation, and poor prognosis of hepatocellular carcinoma according to long-term follow-up data. In hepatocellular carcinoma cells, overexpression of lnc-ATG9B-4 promoted proliferation, invasion, as well as migration, while inhibiting lnc-ATG9B-4 by siRNA significantly attenuated the proliferation, invasion, as well as migration. Interestingly, lnc-ATG9B-4 increased the expression of cyclin-dependent kinase 5 (CDK5), which was closely related to the development and chemotherapy sensitivity of hepatocellular carcinoma. In summary, our results revealed that lnc-ATG9B-4 suggests an unfavorable prognosis of hepatocellular carcinoma and facilitates the proliferation, invasion, as well as migration of hepatocellular carcinoma cells by upregulating CDK5. This research suggests that lnc-ATG9B-4 may be a new biomarker for predicting the prognosis of hepatocellular carcinoma; meanwhile, targeting lnc-ATG9B-4 might serve as a potential strategy for the treatment hepatocellular carcinoma.
Keywords: Long noncoding RNA, lnc-ATG9B-4, hepatocellular carcinoma, metastasis, proliferation, CDK5
Impact statement
In this study, we explored the expression profile of lncRNA in HCC tumor tissues and paracancerous tissues using microarray assays. Furthermore, a new lncRNA (lnc-ATG9B-4) was identified, which was about 3.5 times more expressed in tumor tissues than in paracancerous tissues. Through clinicopathological analysis, lnc-ATG9B-4 was determined to be related to the tumorous size, TNM stages, portal vein tumor thrombus (PVTT), the tumor capsule, metastasis, and the degree of differentiation. Lnc-ATG9B-4 promoted the proliferation, invasion, as well as migration of the HCC cells by upregulating the expression of CDK5. Here, we further exploited the molecular mechanisms of lnc-ATG9B-4 to screen new drug intervention targets for recurrence and metastasis of HCC.
Introduction
Hepatocellular carcinoma (HCC) is one of the main malignancies, accounting for 75%–85% of liver malignancies1 and about 12.5% of new malignant tumors per year in China.2 Up to 2018, a total of 841,100 cases of liver cancer have accounted for 4.7% of new cancers, and 786,600 cases of liver cancer-related deaths have accounted for 8.2% of cancer-related deaths in the world,3 of which over 50% of new cases and deaths occur in China each year.2 With the latest advances in new diagnostic and treatment strategies, the morbidity and mortality of liver cancer have decreased.4 To date, the five-year survival rate for liver cancer patients worldwide has increased; unfortunately, it is still less than 30%.1 Due to limitations in radical surgery indications and the quite high recurrence and metastasis rate, the outcome of patients with HCC remains unsatisfactory. The mechanisms of HCC progression are rather complicated. Meanwhile, there is still a lack of effective prognostic biomarkers for the early diagnosis and intervention for the recurrence and metastasis of HCC. There is a pressing need to discern new effective molecular biomarkers for the diagnosis, treatment, as well as prognosis of HCC.
Long noncoding RNAs (lncRNAs), a class of endogenous transcripts longer than 200 nucleotides, contain no open reading frame and have no protein coding potential.5 In mammals, most lncRNAs are transcribed and spliced by RNA polymerase II and are thought to be “noise” and by-products of the transcription process without biological functions.6
LncRNAs is reported to play a vital role in tumorigenesis, invasion, and metastasis of different cancers in humans and are promising potential prognostic biomarkers and possible therapeutic targets.7 It has been reported that the lncRNA HULC is immensely expressed in HCC and promotes the proliferation of HCC cells.8 The expression of the lncRNA HOTAIR has been confirmed to be remarkably increased in HCC and closely related to the metastasis and recurrence of HCC.9 It was demonstrated that the lncRNA MEG3, which is expressed at a low level in HCC, inhibited apoptosis also promoted the proliferation of HCC cells.10 Moreover, the lncRNAs UCID11 and PVT112 promote the proliferation, invasion, as well as metastasis of HCC cells. In summary, lncRNAs can affect the biological functions of HCC cells by regulating various genes; however, the molecular mechanisms of only a small portion of lncRNAs have been illustrated.
In recent years, researchers have paid more attention to the role of cyclin-dependent kinase 5 (CDK5) in tumors, particularly HCC. Some studies have shown that CDK5 mediates the phosphorylation of TPX2 to promote the proliferation and tumorigenicity of hepatocytes,13 and promotes DNA replication resulting in HCC cell proliferation and inhibition of apoptosis.14 Inhibition of CDK5 expression significantly inhibited the proliferation of HCC cells15 and neovascularization,16 and even improved the efficacy of the chemotherapy drug sorafenib in HCC patients and prevented the drug resistance to sorafenib in the treatment of HCC.17 These results showed that CDK5 can participate in the emergence and development of HCC, and that it plays a crucial regulatory role in tumor cell proliferation, invasion, and metastasis.
Some lncRNAs aggravated progress of HCC through promoting the proliferation, invasion, as well as metastasis of HCC cells. Meanwhile, CDK5 was overexpression in the HCC and played an essential role in regulating biological functions of HCC cells. Therefore, we will explore the correlation between lncRNAs and CDK5, which will be beneficial to the exploration of the molecular mechanism underlying the appearance and development of HCC.
Materials and methods
Clinical sample
A total of 176 primary HCC tissues and matched paracancerous tissues were collected at the time of surgical resection at the First People’s Hospital of Huaihua (Huaihua, Human, China) from August 2014 to August 2018. The clinical sample were stored at −80°C. The eight fresh-frozen primary HCC tissues and matched paracancerous tissues were used to microarray assay. Meanwhile, a total of 176 fresh primary HCC and paracancerous tissue were analyzed for quantitative PCR (qPCR) to validate the differential expression of lnc-ATG9B-4.
RNA extraction and lncRNAs microarray assay
The total RNAs of 16 tissues including 8 primary HCC tissues and 8 paracancerous tissues were extracted by using TRIzol reagent (15596026, Invitrogen), and small RNAs were purified in accordance with the manufacturer’s protocol of mirVana miRNA Isolation Kit (AM1561, Ambion). The RNAs were determined by spectrophotometer (NanoDrop ND-2000, Thermo) for concentration and purity. The integrity of RNA was determined by capillary electrophoresis. Further analysis was carried out for the RNAs with RNA integrity number greater than 6. Furthermore, lncRNA microarray assay was carried out by CapitalBio Corporation (CapitalBio, China).
qPCR analysis
To validate the differential expression of lnc-ATG9B-4 from the lncRNA microarray assay and the correlation between expression of CDK5 and lnc-ATG9B-4, the expression levels of lnc-ATG9B-4 and CDK5 were detected by qPCR in HCC patients including primary HCC tissues and paracancerous tissues. The qPCR analysis using the 2−ΔΔCt method18 was performed following our previously published protocol.19
Cells culture
The 293 T and HepG2 cells were cultivated in DMEM (Gibco) with 10% FBS (Gibco) and 1% penicillin-streptomycin (Sigma). All cells were purchased from ATCC and cultured in 5% CO2 at 37°C incubators.
Plasmids constructed and lentivirus packing
To analyze whether the lnc-ATG9B-4 regulated the proliferation, invasion, as well as migration of HCC cells, the plasmids with lnc-ATG9B-4 or shRNA- lnc-ATG9B-4 were constructed and the recombinant lentivirus particles were packed following the methods of previously published.19 The HepG2 cells were infected by the lentivirus particles and identified for further proliferation, invasion, and migration analysis by qPCR. All primers are shown in Table 1.
Table 1.
The primer pairs of lnc-ATG9B-4.
| Names | Nucleotide Sequence (5ʹ-3ʹ) | |
|---|---|---|
| lnc-ATG9B-4 PCR | Forward | ATCGGGATTCCAGGCAGATCACCCGAGG |
| Reverse | ATCGTCTAGAGACACTCCAGTCGCCAGC | |
| lnc-ATG9B-4 qPCR | Forward | ATTGGCTCTTTATCCCTGCTGA |
| Reverse | AGTGGTGCTTCCAGGATTCAA | |
| 18s RNA | Forward | AGAAACGGCTACCACATCCA |
| Reverse | CACCAGACTTGCCCTCCA | |
| SiRNA-control | TTCTCCGAACGTGTCACGTAA | |
| shRNA-control | Top strand | GATCCGTTCTCCGAACGTGTCACGTAATTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTC |
| Bottom strand | AATTGAAAAAATTCTCCGAACGTGTCACGTAATCTCTTGAATTACGTGACACGTTCGGAGAACG | |
| siRNA1 | GAGAATTGCTTGAACCCAGGAGGCAGAGAATTGCTTGAACCCAGGAGGCA | |
| shRNA1-lnc-ATG9B-4 | Top strand | GATCCGAGAATTGCTTGAACCCAGGAGGCATTCAAGAGATGCCTCCTGGGTTCAAGCAATTCTCTTTTTTG |
| Bottom strand | AATTCAAAAAAGAGAATTGCTTGAACCCAGGAGGCATCTCTTGAATGCCTCCTGGGTTCAAGCAATTCTCG | |
| siRNA2 | GGCCTATTAGATTGGCTCTTTATCC | |
| shRNA2-lnc-ATG9B-4 | Top strand | GATCCGGCCTATTAGATTGGCTCTTTATCCTTCAAGAGAGGATAAAGAGCCAATCTAATAGGCCTTTTTTG |
| Bottom strand | AATTCAAAAAAGGCCTATTAGATTGGCTCTTTATCCTCTCTTGAAGGATAAAGAGCCAATCTAATAGGCCG | |
| siRNA3 | TCCAAAGTGCTGGGATTACAGGAGC | |
| shRNA3-lnc-ATG9B-4 | Top strand | GATCCGTCCAAAGTGCTGGGATTACAGGAGCTTCAAGAGAGCTCCTGTAATCCCAGCACTTTGGATTTTTTG |
| Bottom strand | AATTCAAAAAATCCAAAGTGCTGGGATTACAGGAGCTCTCTTGAAGCTCCTGTAATCCCAGCACTTTGGACG | |
Proliferation analysis
The proliferation of HepG2 cells was assessed with the CCK-8 assay (Dojindo) in accordance with the protocol of products following our previously published methods.19
Migration and invasion analysis
The migration of cells was detected using wound healing assay. The culture inser (ibidi, 80241) was placed in a 12-well plate. After all cells were starved for 6 h, the cells were seeded in the culture chambers. The element was not removed until cells were 100% confluence in the chambers, and the 12-well plate was incubated in 5% CO2 incubator at 37°C. Then, the region of wounds was measured at the indicated times, and quantified with Image J software.
The invasion of cells was analyzed using transwell chambers (3422, Corning) treated with Matrigel (1:8, BD). A 12-well plate was filled completely with medium. Cells were suspended in free-serum medium with a final concentration of 5 × 105 cells/mL. A 100 μL cells suspension was seeded in the trasnswell chambers. The chamber was placed in 12-well plates, and incubated in incubator. After 24 h of culture, the transwell chambers were removed, washed by PBS, and fixed by paraformaldehyde. The cells were stained by 0.1% crystal violet. Then, cells in the upper of chambers were swabbed with cotton swab, while cells in the bottom of chambers were imaged and counted under randomly five fields with the microscope.
Western blotting
The HepG2 cells with overexpressed lnc-ATG9B-4 or shRNA-lnc-ATG9B-4 were collected and ultrasonically lysed in RIPA buffer (P0013B, Beyotime) on ice for 30 s. Supernatant of lysates was collected after centrifuging and concentrations of total protein were measured with BCA protein assay kit (PA101-01, Biomed) in accordance with the protocol of manufacturer. About 25 μL of total protein was isolated by 10% SDS-PAGE. Then, the proteins were transferred to a PVDF membrane (IPVH00010, Millipore), obstructed by 5% skim milk (D8340, Solarbio) at 4°C overnight. The membrane was incubated with anti-CDK5 antibody (ab40773, abcam) or anti-actin antibody (ab179467, abcam) at room temperature (RT) for 2 h, and subsequently incubated with the anti-rabbit IgG (ab6721, abcam) at RT for 1–2 h. Finally, the proteins were visualized by chemiluminescence and analyzed by Image J software.
Hematoxylin and eosin stain
The H&E Kit was purchased from abcam (ab245880, abcam). The H&E stain was performed to identify HCC and paracancerous in accordance with the protocol of manufacturer. Briefly, human HCC and paracancerous tissues were fixed by 4% paraformaldehyde for 12 h and embedded in paraffin. Afterwards, sections were cut at 2 μm thickness and dewaxed. Then, the sections were incubated with Hematoxylin for 5 min, and subsequently incubated with Eosin for 2–3 min. Finally, the images were viewed and photographed using microscope (Carl Zeiss, GER).
Fluorescence in situ hybridization
The Cy3 labeled lnc-ATG9B-4 nucleotides fluorescent probe was designed and synthesized by Servicebio (Wuhan, China). The FISH had been performed to detect the expression of lnc-ATG9B-4 in the human HCC tissue samples as previously reported.20 Briefly, the sections were cut to a thickness of 5 μm and dewaxed. Next, sections were retrieved at 100°C in 10 mM sodium citrate buffer (pH 6.0) for 15 min and digested by protease K (20 μg/mL) at 37°C for 20–30 min. The sections were prehybridized in a prehybridization solution at 37°C for 1 h. Afterwards, the sections were incubated with fluorescent probe in the dark at 37°C overnight. After washing using 2× saline sodium citrate (S6639, Sigma), the sections were incubated with DAPI (D9542, Sigma) in the dark at RT for 8 min. Finally, images of FISH were observed at wave of 510–560 nm using fluorescent microscope (Carl Zeiss, GER).
Statistical analysis
All the data from three independent trials were expressed as the mean ± standard deviation. SPSS 23.0 and GraphPad Prism 8.0 software were used for Student t-test to analyze the differences between the two groups. Statistical comparisons of the HCC patients’ overall survival (OS) were operated by the Kaplan–Meier (K-M) analysis with SPSS23.0. The correlation between expression of CDK5 and lnc-ATG9B-4 was analyzed suing the Spearman correlation analysis with SPSS23.0. The P value <0.05 was regarded as statistically significant.
Results
High expression of lnc-ATG9B-4 in HCC
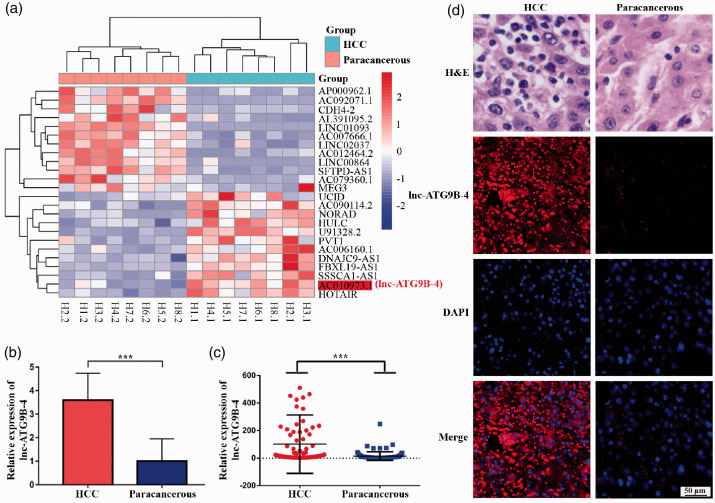
The total RNAs of HCC and matched paracancerous tissues were extracted from eight patients with HCC. Then, an lncRNA microarray assay was performed. The results are shown in Figure 1(a). There were multiple differentially expressed lncRNAs between HCC and paracancerous tissues. Among them, lncRNA AC010973.1 (lnc-ATG9B-4) was expressed approximately 3.5 times higher than that in paracancerous tissues (Figure 1(b) and Table 2). To verify its differential expression, the expression of lnc-ATG9B-4 was analyzed by qPCR in 176 patients with HCC. The expression of lnc-ATG9B-4 in HCC was higher than in paracancerous tissues (Figure 1(c)), consistent with the microarray assay results. To verify whether lnc-ATG9B-4 was expressed in HCC or paracancerous tissues, the FISH for lnc-ATG9B-4 had been performed in the human HCC tissue samples. Red fluorescence in paracancerous tissues was hardly showed; however, the intensity of red fluorescence in the HCC tissues increased significantly, which indicated that the lnc-ATG9B-4 was mainly expressed in the HCC tissue (Figure 1(d)). The results suggested that the expression of lnc-ATG9B-4 in HCC tissues was markedly greater than that in paracancerous tissues.
Figure 1.
High expression of lnc-ATG9B-4 in HCC. (a) Heat map of lncRNAs expression in eight patients with HCC. The red box represents lnc-ATG9B-4. (b) The relative expression of lnc-ATG9B-4 in eight patients with HCC by lncRNA microarray. (c) Scattergram of the relative lnc-ATG9B-4 expression level. The expression level of lnc-ATG9B-4 was verified by qPCR in 176 HCC tissue samples. The plots represent the average level of lnc-ATG9B-4 from three independent trials (Student’s t-test, ***P < 0.001). (d) Representative images of the lnc-ATG9B-4 FISH in HCC. HCC and paracancerous tissues were showed by H&E stains (top). The expression of lnc-ATG9B-4 was detected by FISH (bottom). (A color version of this figure is available in the online journal.)
Table 2.
The lnc-ATG9B-4 of lncRNA microarray from eight HCC patients.
| ID | Cancer | Paracancerous | |
|---|---|---|---|
| H1 | R1713 | 1.296895593 | 0.707635 |
| H2 | R1759 | 1.403965938 | 0.213507 |
| H3 | R1765 | 0.922186001 | 0.586083 |
| H4 | R1773 | 0.914181525 | 0.10441 |
| H5 | R1819 | 0.666374719 | 0.106621 |
| H6 | R1847 | 1.105416648 | 0.242376 |
| H6 | R1853 | 0.717246559 | 0.147415 |
| H8 | R1881 | 0.561007062 | 0 |
Lnc-ATG9B-4 is closely associated with the pathological characteristics of HCC patients
The 176 HCC patients were split into two groups based on the expression of lnc-ATG9B-4. The expression of lnc-ATG9B-4 in HCC tissues was subtracted from that in paracancerous tissues. Then, the expression of lnc-ATG9B-4 in patients was listed in descending order, and the patients in whom lnc-ATG9B-4 expression was higher than the median of lnc-ATG9B-4 expression were defined as the lnc-ATG9B-4 high expression group; otherwise, the patients were defined as the lnc-ATG9B-4 low expression group. In order to confirm the role of lnc-ATG9B-4 in the occurrence and development of HCC, a correlation analysis between the expression of lnc-ATG9B-4 and the clinicopathological features of HCC patients was conducted. As illustrated in Table 3, lnc-ATG9B-4 was closely associated with tumor size, TNM stages, PVTT, the tumor capsule, metastasis, and the degree of differentiation, which indicated that lnc-ATG9B-4 was associated with the pathological characteristics of patients with HCC.
Table 3.
Relationship between lnc-ATG9B-4 expression and various clinicopathological variables.
| Variables | Total |
lnc-ATG9B-4 expression |
χ2 | P value | |
|---|---|---|---|---|---|
| Low (n = 88) | High (n = 88) | ||||
| Gender | 3.610 | 0.0574 | |||
| Male | 156 | 74 | 82 | ||
| Female | 20 | 14 | 6 | ||
| Age | 0.8186 | 0.3656 | |||
| ≥55 | 90 | 48 | 42 | ||
| <55 | 86 | 40 | 46 | ||
| Size of tumor (cm) | 6.155 | 0.0461 | |||
| <3 | 84 | 38 | 46 | ||
| 3–5 | 28 | 20 | 8 | ||
| >5 | 64 | 30 | 34 | ||
| No. of tumorous | 0.5176 | 0.4718 | |||
| Solitary | 40 | 22 | 18 | ||
| Multiple | 136 | 66 | 70 | ||
| Depth of invasion | 3.4493 | 0.3275 | |||
| T1 | 80 | 40 | 40 | ||
| T2 | 76 | 40 | 36 | ||
| T3 | 14 | 4 | 10 | ||
| T4 | 6 | 4 | 2 | ||
| TNM stage | 9.7953 | 0.0204 | |||
| I | 70 | 44 | 26 | ||
| II | 96 | 42 | 54 | ||
| III | 6 | 1 | 5 | ||
| IV | 4 | 1 | 3 | ||
| PVTT | 29.701 | <0.0001 | |||
| Yes | 80 | 22 | 58 | ||
| No | 96 | 66 | 30 | ||
| AFP (ng/mL) | 1.2081 | 0.2717 | |||
| ≥400 | 38 | 16 | 22 | ||
| <400 | 138 | 72 | 66 | ||
| HBV-DNA (IU/mL) | 0.0949 | 0.7581 | |||
| ≥1000 | 70 | 34 | 36 | ||
| <1000 | 106 | 54 | 52 | ||
| Capsule | 20.551 | <0.0001 | |||
| Yes | 82 | 56 | 26 | ||
| No | 94 | 32 | 62 | ||
| Portal hypertension | 0.9258 | 0.3360 | |||
| Yes | 58 | 32 | 26 | ||
| No | 118 | 56 | 62 | ||
| Ascites | 1.1001 | 0.2943 | |||
| Yes | 16 | 6 | 10 | ||
| No | 160 | 82 | 78 | ||
| Cirrhosis | 1.4471 | 0.2291 | |||
| Yes | 146 | 70 | 76 | ||
| No | 30 | 18 | 12 | ||
| Metastasis | 4.4001 | 0.0359 | |||
| Yes | 16 | 4 | 12 | ||
| No | 160 | 84 | 76 | ||
| Degree of differentiation | 6.9881 | 0.0082 | |||
| Well/moderate | 52 | 34 | 18 | ||
| Poor and not | 124 | 54 | 70 | ||
| Recurrence | 0.8539 | 0.3554 | |||
| Yes | 106 | 50 | 56 | ||
| No | 70 | 38 | 32 | ||
High expression of lnc-ATG9B-4 indicates the poor outcomes of HCC
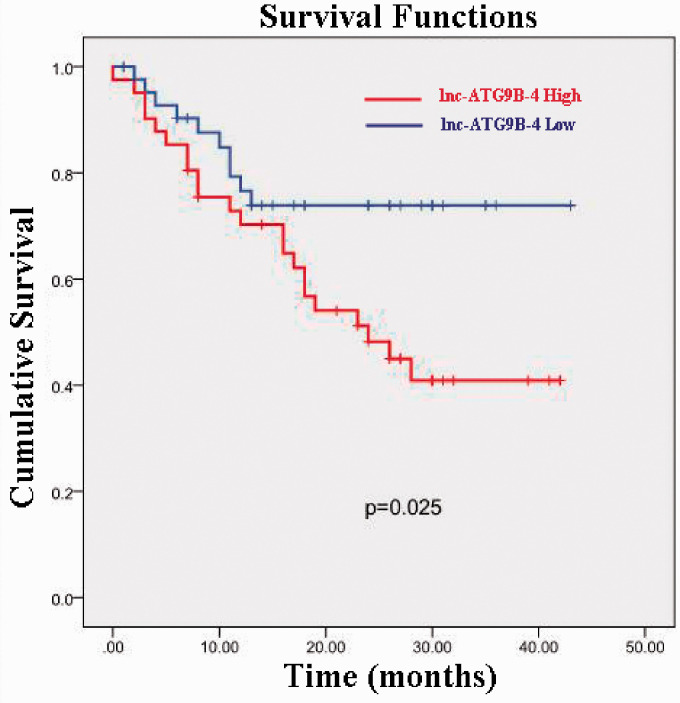
Based on the above results, lnc-ATG9B-4 is associated with the pathological characteristics of patients with HCC. To further investigate whether lnc-ATG9B-4 has any effects on OS, K-M analysis was performed based on long-term follow-up data. As shown in Figure 2, the cumulative survival rate of HCC patients with high lnc-ATG9B-4 expression was approximately 40% at 2.5 years. Nevertheless, the cumulative survival rate of patients with low expression of lnc-ATG9B-4 was up to 75%, which was prominently higher than that of patients with high lnc-ATG9B-4 expression (P = 0.025). In summary, these data manifest that lnc-ATG9B-4 is highly correlated with the poor prognosis of HCC patients
Figure 2.
The association between the lnc-ATG9B-4 expression level and OS for HCC patients. The OS of the patients with high lnc-ATG9B-4 expression was apparently shorter than that of patients with low lnc-ATG9B-4 expression (K-M, P = 0.025). (A color version of this figure is available in the online journal.)
Lnc-ATG9B-4 facilitated the proliferation, invasion, as well as migration of HepG2 cells
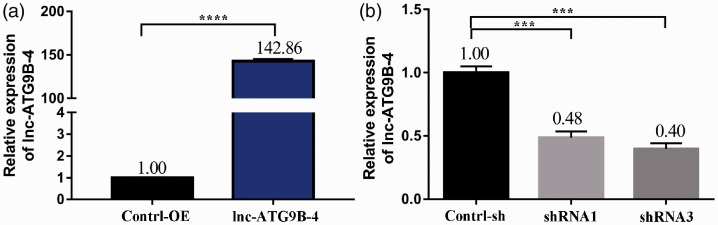
In order to analyze the influence of lnc-ATG9B-4 on the biological function of HCC cells, the expression of lnc-ATG9B-4 was detected by qPCR. The expression of lnc-ATG9B-4 was distinctly increased in the HepG2 cells overexpressing lnc-ATG9B-4 (HepG2-ATG9B-4) (P < 0.05) (Figure 3(a)). The lnc-ATG9B-4 expression was approximately 0.48 times higher in the HepG2 cells overexpressing shRNA1-lnc-ATG9B-4 (HepG2-shRNA1) and 0.40 times higher in the HepG2 cells overexpressing shRNA3-lnc-ATG9B-4 (HepG2-shRNA3) (Figure 3(b)). The HepG2-ATG9B-4 and HepG2-shRNA3 (HepG2-shRNA-ATG9B-4) cells were used for further experiments.
Figure 3.
Generation of stable HepG2 cells. The expression of lnc-ATG9B-4 was detected by qPCR in HepG2 cells with overexpressing lnc-ATG9B-4 (a) or shRNA-lnc-ATG9B-4 (b). All data were from three independent experiments (Student’s t-test, ***P < 0.001, ****P < 0.0001). OE: overexpressing; shRNA1: shRNA1-lnc-ATG9B-4; shRNA3: shRNA3-lnc-ATG9B-4. (A color version of this figure is available in the online journal.)
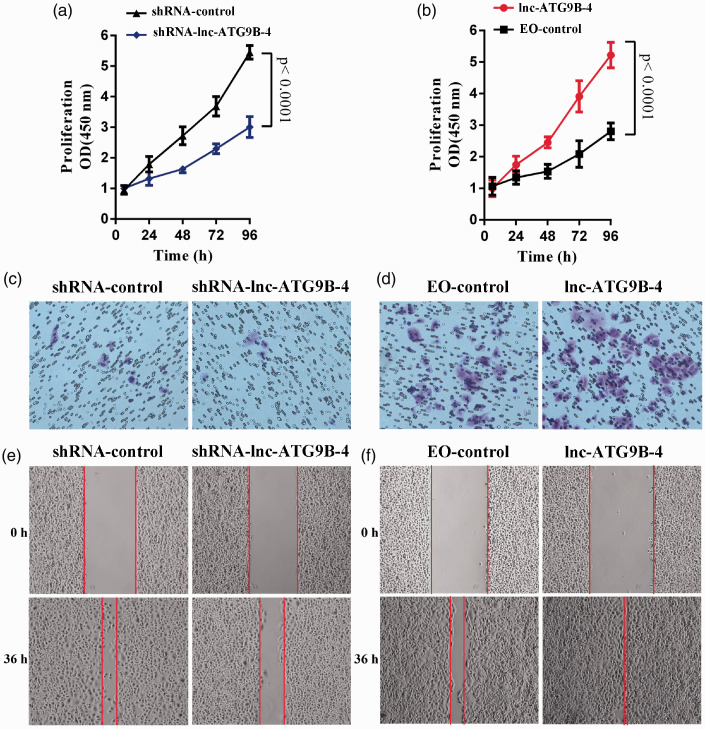
The proliferation was detected by CCK-8. The viability of HepG2-ATG9B-4 cells was apparently higher than that of control vector-transfected cells (EO-control) (P < 0.05) (Figure 4(a)). However, the viability of HepG2-shRNA-ATG9B-4 cells was lower than that of HepG2-shRNA-control cells (P < 0.05) (Figure 4(b)). The results indicated that lnc-ATG9B-4 facilitated the proliferation of HepG2 cells.
Figure 4.
Lnc-ATG9B-4 promoted the proliferation, invasion, as well as migration of HepG2 cells. The proliferation of HepG2 cells with shRNA-lnc-ATG9B-4 (a) and lnc-ATG9B-4 overexpression (b) was analyzed by CCK-8. The invasion of HepG2 cells overexpressed shRNA-lnc-ATG9B-4 (c) and lnc-ATG9B-4 (d) was analyzed by transwell assay. The migration of HepG2 cells overexpressed shRNA-lnc-ATG9B-4 (e) and lnc-ATG9B-4 (f) was analyzed by wound healing assay. All data were from three independent experiments (Student’s t-test). (A color version of this figure is available in the online journal.)
The invasion was analyzed via transwell assay. As shown in Figure 4(c) and (d), lnc-ATG9B-4 downregulation obviously inhibited the invasion of HepG2 cells, and lnc-ATG9B-4 upregulation notably accelerated the invasion of HepG2 cells, which suggested that lnc-ATG9B-4 promoted the invasion of HepG2 cells. In addition, a wound healing assay showed that lnc-ATG9B-4 facilitated migration in HepG2 cells (Figure 4(e) and (f)).
From the above, these results indicated that lnc-ATG9B-4 facilitated the proliferation, invasion, as well as migration of HepG2 cells
Lnc-ATG9B-4 upregulated CDK5 expression in cells and tissues of HCC
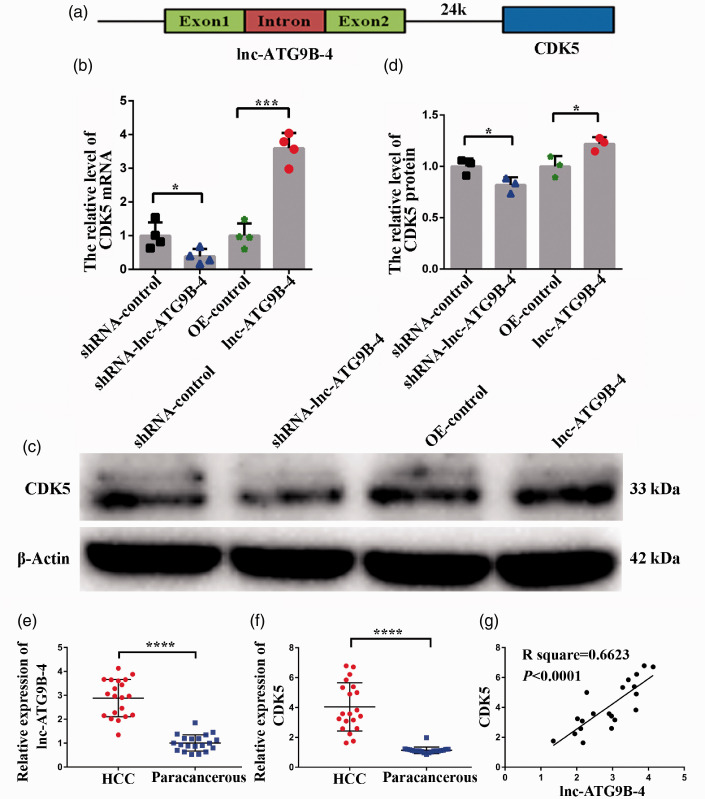
To explore the potential mechanism of lnc-ATG9B-5 promoting proliferation, invasion, and migration in HepG2 cells, the target genes were predicted to be within the vicinity of lnc-ATG9B-4. Lnc-ATG9B-4 was located on chromosome 7 (chr7: 151,028,781–151,029,754). The genes on chr7: 150,928,781–151,129,754 could be potential target genes of lnc-ATG9B-4 (Table 4). Genes expression was detected by qPCR and WB. Among them, CDK5 was located 24k downstream of the lnc-ATG9B-4 (Figure 5(a)), and the expression of CDK5 in the HepG2-shRNA-lnc-ATG9B-4 cells was lower than that in HepG2-shRNA-control cells, while the expression of CDK5 in HepG2-lnc-ATG9B-4 cells was higher than that in control HepG2 cells (Figure 5(b) to (d)), which showed that lnc-ATG9B-4 upregulated CDK5 expression in HepG2 cells.
Table 4.
Potential target genes of lnc-ATG9B-4.
| Gene names | Location |
|---|---|
| KCNH2 | chr7: 150,944,961–150,978,321 |
| NOS3 | chr7: 150,991,017–151,014,588 |
| ASIC3 | chr7:151,048,292–151,052,756 |
| ABCB8 | chr7:151,028,422–151,047,782 |
| FASTK | chr7:151,076,621–151,080,883 |
| RP11-148K1.10 | chr7:151,028,781–151,029,754 |
| TMUB1 | chr7:151,081,085–151,083,493 |
| CDK5 | chr7:151,053,815–151,057,897 |
| SLC4A2 | chr7:151,058,212–151,076,526 |
| RP11-148K1.12 | chr7:151,074,742–151,076,530 |
| AGAP3 | chr7:151,086,475–151,144,434 |
Figure 5.
Lnc-ATG9B-4 enhanced the expression of CDK5. (a) Schematic illustration of CDK5. The expression of CDK5 mRNA (b) and protein (c–d) was tested by qPCR and Western blotting, respectively, in the HepG2 cells overexpressing lnc-ATG9B-4 and shRNA-lnc-ATG9B-4. The expression of CDK 5 (e) and lnc-ATG9B-4 (f) in 20 patients with HCC. (g) CDK5 expression was positively related to the expression of lnc-ATG9B-4 by Spearman correlation analysis. All data were from three independent experiments (Student’s t-test, *P < 0.05, ***P < 0.001, ****P < 0.0001). (A color version of this figure is available in the online journal.)
In order to investigate whether CDK5 expression upregulates with increase of lnc-ATG9B-4 in HCC tissues, the expression of CDK5 and lnc-ATG9B-4 was detected by qPCR in 20 patients with HCC. Compared with paracancerous tissues, the CDK5 and lnc-ATG9B-4 expression was higher in the HCC tissues (Figure 5(e) and (f)). The relationship between the expression of CDK5 and lnc-ATG9B-4 was analyzed. The results have shown that CDK5 expression was positively correlated with lnc-ATG9B-4 expression (Figure 5(g)).
In summary, the high expression of lnc-ATG9B-4 indicates that HCC patients have a poor prognosis, while lnc-ATG9B-4 could promote the proliferation, invasion, as well as migration of HepG2 by upregulating CDK5 expression.
Discussion
HCC is one of the main malignancies, with the five-year survival rate of less than 30% on account of metastasis and recurrence.2 Metastasis is associated with a variety of molecular abnormalities. The differential protein expression of HCC has been the focus of previous research. In recent years, more and more evidences show that lncRNAs play an essential role in HCC, especially in HCC metastasis. HULC was the first lncRNA discovered to be high level expressed in HCC and to promote the proliferation of HCC cells.8 Subsequently, several lncRNAs closely connected to the progression and metastasis of HCC were identified, such as MALAT1,21 LET,22 DLEU2,23 cdh4-2,24 PDPK2P,25 DCST1-AS1,26 X91348,27 Linc01296,28 ZNF385D‑AS2,29 and PTTG3P.30 Here, we report that lnc-ATG9B-4 is correlated with the development and prognosis of HCC.
Compared to previously reported lncRNAs, lnc-ATG9B-4 better reflects the relationship between the pathological characteristics of patients with HCC and prognosis. DLEU2 is highly expressed in HCC but is associated only with tumor size, TNM stage, and PVTT.23 The differential expression of cdh4-2 was identified in various TNM stages of HCC, but it is not closely correlated with the pathological characteristics of patients with HCC.24 PDPK2P25 is highly expressed in both HCC and paracarcinoma tissue, and HCC patients with high PDPK2P expression are more prone to macrovascular invasion and poor tumor differentiation. X91348, which is expressed at low levels in HCC, is related to tumor size, HBsAg, and the Child-Pugh score.27 Moreover, HCC patients with low X91348 expression do not live as long as those with high X91348 expression. ZNF385D‑AS2, which is expressed at low levels in HCC, is related to sex, TNM stage, and the survival status.29 High expression levels of PTTG3P, which is associated with tumor size, clinical stage and metastasis, were identified in HCC.30 The above lncRNAs, which were differentially expressed in both HCC and normal liver tissues, reflect a relationship between the clinicopathological characteristics of HCC patients and lncRNA levels to some extent. Hence, they may be considered as biomarkers to predict HCC progression and prognosis.
Here, lnc-ATG9B-4 was first examined in HCC and paracancerous tissues by microarray analysis, and the difference was verified by qPCR in 176 patients with HCC. The results showed that lnc-ATG9B-4 is highly expressed in HCC. HCC patients with high lnc-ATG9B-4 expression tend to have a higher proportion of TNM stages III–IV, PVTT, an incomplete tumor capsule, poorly differentiated cancer, metastasis, and a shorter survival time than those with low lnc-ATG9B-4 expression. However, the number of tumors measuring 3–5 cm was obviously lower in patients with high lnc-ATG9B-4 expression HCC than in patients with low lnc-ATG9B-4 expression, which indicated that HCC cells with high expression were prone to metastasis. This study showed that lnc-ATG9B-4 promoted proliferation, invasion, as well as migration in HepG2 cells. Meanwhile, the results indicated that lnc-ATG9B-4 facilitated the expression of CDK5. Our results showed that CDK5 promoted proliferation, invasion, as well as migration of HepG2 cells, which was in line with the previous research. However, the specific molecular mechanism was further investigated. In summary, high expression of lnc-ATG9B-4 indicates that HCC patients have a poor prognosis. The CDK5 may be a potential target gene of lnc-ATG9B-4 in HCC and lnc-ATG9B-4 facilitated the proliferation, invasion, as well as migration of HepG2 cells by upregulating CDK5 expression. Therefore, lnc-ATG9B-4 acts as a new biomarker for predicting the prognosis of HCC patients and could be an underlying target for medical interventions of HCC, which is superior to the abovementioned lncRNAs.
ACKNOWLEDGEMENTS
Thanks to Hunan Province Key Laboratory for Antibody-based Drug and Intelligent Delivery System (Hunan University of Medicine), Guangxi Key Laboratory of Molecular Medicine in Live Injury and Repair (Guilin Medical University) and Institute of Basic Medical Sciences (Guilin Medical University).
Authors’ contributions: ML, XSH, and ZCM conceived the experiments, analyzed data, wrote, proofread, and revised the manuscript. LW and PYL conducted the qPCR, Western blotting, lentivirus packing, and cells culture. XMZ and FL conducted the proliferation, migration, and invasion analysis. FY conducted collection of clinical samples. All authors participated in data interpretation, reviewed, and approved the manuscript.
Declaration OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: The project was approved for the using of HCC patients’ biopsy by the Institution Ethics Committee of Hunan University of Medicine (HUM-2018–124) and adhered to the principles in the Declaration of Helsinki. The written informed consent was received form all participants at the time of surgery in this study
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported Guangxi Key Laboratory of Molecular Medicine in Live Injury and Repair (No. GXLIRMMKL-K202006); the Scientific Research Fund of Hunan Provincial Education Department, China (No. 18B534); the Students Innovation Pilot Scheme of Hunan University of Medicine, China (No. 24) and the PhD Research Startup Foundation of Hunan University of Medicine, China (No. 2018–01).
ORCID iD: Ming Li https://orcid.org/0000-0001-6000-8571
References
- 1.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, Ogunbiyi OJ, Azevedo ESG, Chen WQ, Eser S, Engholm G, Stiller CA, Monnereau A, Woods RR, Visser O, Lim GH, Aitken J, Weir HK, Coleman MP, Group CW. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018; 391:1023–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66:115–32 [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424 [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA A Cancer J Clin 2020; 70:7–30 [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136:629–41 [DOI] [PubMed] [Google Scholar]
- 6.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007; 316:1484–8 [DOI] [PubMed] [Google Scholar]
- 7.Wang P, Ning SW, Zhang YP, Li RH, Ye JR, Zhao ZXL, Zhi H, Wang TT, Guo Z, Li X. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res 2015; 43:3478–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xin XR, Wu MY, Meng QY, Wang C, Lu YN, Yang YX, Li XN, Zheng QD, Pu H, Gui X, Li TM, Li J, Jia S, Lu DD. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer 2018; 17:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang T, He XJ, Chen A, Tan K, Du XL. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene 2018; 670:114–22 [DOI] [PubMed] [Google Scholar]
- 10.He JH, Han ZP, Liu JM, Zhou JB, Zou MX, Lv YB, Li YG, Cao MR. Overexpression of long Non-Coding RNA MEG3 inhibits proliferation of hepatocellular carcinoma Huh7 cells via negative modulation of miRNA-664. J Cell Biochem 2017; 118:3713–21 [DOI] [PubMed] [Google Scholar]
- 11.Wang YL, Liu JY, Yang JE, Yu XM, Chen ZL, Chen YJ, Kuang M, Zhu Y, Zhuang SM. Lnc-UCID promotes G1/S transition and hepatoma growth by preventing DHX9-Mediated CDK6 down-regulation. Hepatology 2019; 70:259–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu YXX, Luo XX, He WG, Chen GC, Li Y, Li W, Wang XC, Lai Y, Ye YB. Long Non-Coding RNA PVT1/miR-150/HIG2 axis regulates the proliferation, invasion and the balance of iron metabolism of hepatocellular carcinoma. Cell Physiol Biochem 2018; 49:1403–19 [DOI] [PubMed] [Google Scholar]
- 13.Wang FQ, Zhao WX, Gao YH, Zhou JC, Li HF, Zhang GY, Guo D, Xie CR, Li J, Yin ZY, Zhang J. CDK5-mediated phosphorylation and stabilization of TPX2 promotes hepatocellular tumorigenesis. J Exp Clin Cancer Res 2019; 38:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang R, Lin P, Yang H, He Y, Dang YW, Feng ZB, Chen G. Clinical role and biological function of CDK5 in hepatocellular carcinoma: a study based on immunohistochemistry, RNA-seq and in vitro investigation. Oncotarget 2017; 8:108333–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao YY, Li YS, Hsu HW, Lin H, Wang HY, Wo RR, Cheng AL, Hsu CH. Potent activity of composite cyclin dependent kinase inhibition against hepatocellular carcinoma. Cancers 2019; 11:1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herzog J, Ehrlich SM, Pfitzer L, Liebl J, Fröhlich T, Arnold GJ, Mikulits W, Haider C, Vollmar AM, Zahler S. Cyclin-dependent kinase 5 stabilizes hypoxia-inducible factor-1alpha: a novel approach for inhibiting angiogenesis in hepatocellular carcinoma. Oncotarget 2016; 7:27108–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ardelt MA, Fröhlich T, Martini E, Müller M, Kanitz V, Atzberger C, Cantonati P, Meßner M, Posselt L, Lehr T, Wojtyniak JG, Ulrich M, Arnold GJ, König L, Parazzoli D, Zahler S, Rothenfußer S, Mayr D, Gerbes A, Scita G, Vollmar AM, Pachmayr J. Inhibition of cyclin-dependent kinase 5: a strategy to improve sorafenib response in hepatocellular carcinoma therapy. Hepatology 2019; 69:376–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–8 [DOI] [PubMed] [Google Scholar]
- 19.Li M, Wu XM, Gao J, Yang F, Zhang CL, Ke K, Wang YC, Zheng YS, Yao JF, Guan YY, Chen X, Chen J, Liu XL, Yang XY. Mutations in the P10 region of procaspase-8 lead to chemotherapy resistance in acute myeloid leukemia by impairing procaspase-8 dimerization. Cell Death Dis 2018; 9:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim AS, Lim TH. Fluorescence in situ hybridization on tissue sections. Methods Mol Biol 2017; 1541:119–25 [DOI] [PubMed] [Google Scholar]
- 21.Zhang P, Ha M, Li L, Huang X, Liu C. MicroRNA-3064-5p sponged by MALAT1 suppresses angiogenesis in human hepatocellular carcinoma by targeting the FOXA1/CD24/Src pathway. FASEB J 2020; 34:66–81 [DOI] [PubMed] [Google Scholar]
- 22.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell 2013; 49:1083–96 [DOI] [PubMed] [Google Scholar]
- 23.Guo Y, Bai M, Lin L, Huang J, An Y, Liang L, Liu Y, Huang W. LncRNA DLEU2 aggravates the progression of hepatocellular carcinoma through binding to EZH2. Biomed Pharmacother 2019; 118:109272. [DOI] [PubMed] [Google Scholar]
- 24.Gao YZ, Wang GX, Zhang CL, Lin MJ, Liu XL, Zeng YY, Liu JF. Long non-coding RNA linc-cdh4-2 inhibits the migration and invasion of HCC cells by targeting R-cadherin pathway. Biochem Biophys Res Commun 2016; 480:348–54 [DOI] [PubMed] [Google Scholar]
- 25.Pan W, Li W, Zhao J, Huang Z, Zhao J, Chen S, Wang C, Xue Y, Huang F, Fang Q, Wang J, Brand D, Zheng SG. lncRNA-PDPK2P promotes hepatocellular carcinoma progression through the PDK1/AKT/caspase 3 pathway. Mol Oncol 2019; 13:2246–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Zhai DS, Huang Q, Chen HL, Zhang Z, Tan QF. LncRNA DCST1-AS1 accelerates the proliferation, metastasis and autophagy of hepatocellular carcinoma cell by AKT/mTOR signaling pathways. Eur Rev Med Pharmacol Sci 2019; 23:6091–104 [DOI] [PubMed] [Google Scholar]
- 27.Zeng Z, Dong J, Li Y, Dong Z, Liu Z, Huang J, Wang Y, Zhen Y, Lu Y. The expression level and clinical significance of lncRNA X91348 in hepatocellular carcinoma. Artif Cells Nanomed Biotechnol 2019; 47:3067–71 [DOI] [PubMed] [Google Scholar]
- 28.Zhang LB, Hu J, Hao MH, Bu L. Long noncoding RNA Linc01296 promotes hepatocellular carcinoma development through regulation of the miR-26a/PTEN axis. Biol Chem 2020; 401:407–16 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Wang S, Liu Y, Meng Z, Chen F. Low lncRNA ZNF385DAS2 expression and its prognostic significance in liver cancer. Oncol Rep 2019; 42:1110–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Q, Zhang W, Wang Z, Liu S. Long non-coding RNA PTTG3P functions as an oncogene by sponging miR-383 and up-regulating CCND1 and PARP2 in hepatocellular carcinoma. BMC Cancer 2019; 19:731. [DOI] [PMC free article] [PubMed] [Google Scholar]