Abstract
The long non-coding RNA colon cancer-associated transcript 1 (CCAT1) has been investigated to involve in the progression of non-small cell lung cancer (NSCLC). Thus, this study aims to explore the detailed molecular mechanisms of CCAT1 in NSCLC. The expression of CCAT1, miR-216a-5p, RAP2B, Bax, Bcl-2, and cleaved caspase 3 was detected by qRT-PCR or Western blot. Cell proliferation, apoptosis, migration, and invasion were analyzed using cell counting kit-8, flow cytometry or Transwell assays, respectively. The interaction between miR-216a-5p and CCAT1 or RAP2B was analyzed by luciferase reporter, RNA immunoprecipitation, and pull-down assays. The expression of CCAT1 was elevated in NSCLC, and CCAT1 deletion could inhibit NSCLC cell proliferation, migration, and invasion but induce apoptosis in vitro as well as imped tumor growth in vivo. MiR-216a-5p was confirmed to be a target of CCAT1, and silencing miR-216a-5p could reverse CCAT1 depletion-mediated inhibitory effects on cell tumorigenesis in NSCLC. Besides that, miR-216a-5p was decreased in NSCLC, and miR-216a-5p restoration inhibited cell tumorigenesis by regulating RAP2B, which was verified to be a target of miR-216a-5p. Additionally, co-expression analysis suggested that CCAT1 indirectly regulated RAP2B level by targeting miR-216a-5p in NSCLC cells. Taken together, CCAT1 deletion could inhibit cell progression in NSCLC through miR-216a-5p/RAP2B axis, indicating a novel pathway underlying NSCLC cell progression and providing new potential targets for NSCLC treatment.
Keywords: Colon cancer-associated transcript 1, miR-216a-5p, RAP2B, proliferation, apoptosis, metastasis
Impact statement
We investigated that CCAT1 expression was elevated in NSCLC and CCAT1 deletion was identified to inhibit cell carcinogenic phenotypes in NSCLC cells via miR-216a-5p/RAP2B axis, which reveals a novel pathway underlying progression in NSCLC cells and providing potential targets for NSCLC treatment.
Introduction
Lung cancer is one of the leading causes of cancer-associated mortality in the world with about 80–85% of lung cancers being non-small cell lung cancer (NSCLC).1,2 Within the last decade, despite advances in the diagnosis and treatment strategies, the five-year overall survival rate of NSCLC remains dismal at 18%.3 Actually, more than 60% of lung cancer patients developed the locally advanced or metastatic disease at the time of diagnosis.4 Thus, it is important to better understand the mechanisms underlying NSCLC carcinogenesis and progression, and develop promising diagnostic markers and therapeutic targets.
Long non-coding RNAs (lncRNAs) are non-protein-coding RNA molecules longer than 200 nucleotides, and are now considered as a new frontier in the study of human malignant diseases.5 Increasing evidence has indicated that lncRNAs participate in various biological functions and disease processes, including cell proliferation, invasion, migration, apoptosis, and carcinogenesis, through regulating oncogenes or tumor suppressor genes.6,7 LncRNAs are thought of as prospective biomarkers and potential therapeutic targets in cancer. LncRNA colon cancer-associated transcript 1 (CCAT1), mapped at chromosome 8q24.21, was originally identified to be an oncogene in colon cancer.8 In a series of recent studies, CCAT1 was found to stimulate symmetric division of NSCLC stem cells through activating the Wnt signaling cascade, thereby facilitating the expansion of the stem cell pool, contributing to long-term relapse and therapy failure.9 Additionally, Hu et al. revealed that CCAT1 increased cell cisplatin resistance in NSCLC via regulating miR-130a-3p-mediated SOX4 repression.10 All the evidence indicates that CCAT1 may function as a significant regulator in NSCLC development.
MicroRNAs (miRNAs) belong to a class of endogenous non-coding RNA molecules with approximately 19–25 nucleotides in size, which have emerged as critical regulators in modulating target genes expression, and act as tumor suppressors or oncogenes in carcinogenesis.11 MiR-216a-5p, a member of miRNAs, has been reported to be a cancer-correlated miRNA, which involves in the development of many cancers.12,13 However, the detail effects of miR-216a-5p on NSCLC progression are still vague. RAP2B, a member of the Ras superfamily of guanosine triphosphate-binding proteins, has been suggested to implicate in the regulation of various biological processes in human cells, such as apoptosis, proliferation, signal transduction, and migration.14,15 Additionally, the recent study indicated that RAP2B participated in the modulation of multiple malignant phenotypes in lung cancer cells and might be a novel candidate oncogene in carcinogenesis in lung cancer.16
Here, this work attempted to investigate the expression patterns and effects of CCAT1, miR-216a-5p and RAP2B in NSCLC cell tumorigenesis, and explore the potential regulatory association among them in NSCLC progression.
Materials and methods
Patient specimens
Tumor tissues and adjacent normal lung tissues of 35 patients with NSCLC who underwent primary surgical resection at the Department of Respiratory Medicine, Yantai Yuhuangding Hospital were collected. All patients were diagnosed by two independent pathologists and did not receive any preoperative treatment. The specimens were immediately frozen in liquid nitrogen at −80°C. The clinical features, including age, gender, tumor size, TNM stages, and lymphatic node metastasis were collected from the recruited patients. In the first two years after surgery, follow-up was carried out regularly every three months. However, follow-up was routinely conducted once every six months from the third year. The last follow-up was performed in June 2018. The study was approved by the Ethics Committee of Department of Respiratory Medicine, Yantai Yuhuangding Hospital and written informed consents were obtained from all patients.
Cell culture and transfection
Human NSCLC cell lines (A549 and NCI-H1299) and normal cell line BEAS-2 (ATCC, Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Carlsbad, MD, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 μg/mL streptomycin, and 100 U/mL penicillin (Sigma, St. Louis, MO, USA). All cells were incubated in a humidified atmosphere at 37°C with 5% CO2.
Small interfering RNA (siRNA) targeting CCAT1 (si-CCAT1) and siRNA negative control (si-NC), the CCAT1-specific short hairpin RNA (shRNA; sh-CCAT1) and shRNA scramble control (sh-NC), pcDNA-CCAT1/RAP2B overexpression vector (pcDNA-CCAT1/pcDNA-RAP2B), and plasmid with scrambled sequence (pcDNA-NC) were synthesized by Genepharma (Shanghai, China). The miR-216a-5p mimic (miR-216a-5p), miR-216a-5p inhibitor (anti-miR-216a-5p), and their negative control (NC or anti-NC) were obtained from RIBOBIO (Guangzhou, China). All oligonucleotides or vectors were transfected into A549 and NCI-H1299 cells using Lipofectamine™2000 reagent (Invitrogen, Carlsbad, CA, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from the tissues or cell lines using TRIzol reagent (Invitrogen) and quantified by Nanodrop 2000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cDNA was generated from 5 µg of total RNA using a PrimeScript™ RT-PCR Kit (Takara, Dalian, China). Then real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s introduction on the ABI 7500 real-time PCR system (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 was as an internal control and the 2−ΔΔCt method was used to calculate the fold changes. The specific primer sequences were listed as follows: miR-216a-5p, forward 5ʹ-TGTCGCAAATCTCTGCAGG-3ʹ, reverse 5ʹ-CAGAGCAGGGTCCGAGGTA-3ʹ; U6, forward 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, reverse 5ʹ-ACGCTTCACGAATTTGCGT-3ʹ; CCAT1, forward 5ʹ-CATTGGGAAAGGTGCCGAGA-3ʹ, reverse 5ʹ-ACGCTTAGCCATACAGAGCC-3ʹ; RAP2B, forward 5ʹ-CTCTGGTGGAAATGTGGCTCT-3ʹ, reverse 5ʹ-ATGGTTCT CCCGGACTTCCTT-3ʹ; and GAPDH, forward 5ʹ-AACGGATTTGGTCGTATTGG-3ʹ, reverse 5ʹ-TTGATTTTGGAGGGATCTCG-3ʹ.
Cells proliferation assay
Transfected cells with DMEM (with 10% FBS) were seeded into 96-well plates (2 × 103 cells/well) and cultured for 24, 48, and 72 h, respectively. Subsequently, each well was incubated with 10 μL of cell counting kit-8 (CCK-8) solution (Dojindo, Tokyo, Japan) at each time point for 4 h at 37°C. The absorbance value was measured at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
Cells apoptosis analysis
After transfection, cells were re-suspended with PBS to obtain a cell density of 1 × 106 cells/mL, and then were stained with the Annexin V-FITC and PI (Solarbio, Beijing, China) in the dark. The results were detected by a flow cytometry (BD Biosciences, San Jose, CA, USA), and the data were analyzed by the ModFit LT.
Western blot assay
Western blot assay was performed as described before.10 Proteins were isolated using RNA immunoprecipitation analysis (RIPA) lysis buffer (Beyotime, Shanghai, China), and then were quantified using a bicinchoninic acid (BCA) protein assay kit (Beyotime). Equal amounts of protein were separated with SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were incubated with primary antibodies against RAP2B, Bax, Bcl-2 and cleaved caspase 3 as well as GAPDH overnight at 4°C, followed by interaction with horseradish peroxidase-labeled secondary antibodies for 2 h at room temperature after blocked with 5% non-fat milk for 1 h at 37°C. Immunoreactive signals were visualized using a commercial enhanced chemiluminescence chromogenic substrate (Beyotime).
Transwell assay
Cell migration and invasion were detected by transwell assay. For the migration assay, transfected cells were seeded in the upper chamber with serum-free DMEM and 500 μL DMEM fixed with 10% FBS was added to the lower chamber as a chemoattractant. After incubation for 24 h at 37°C, cells attached to the bottom were fixed with methanol and stained with 0.5% crystal violet for 30 min. For the invasion assay, the upper transwell chambers were pre-coated with Matrigel (BD Biosciences) for 1 h at 37°C and other philosophy of measurement was similar to the process of cell migration. Finally, migrated and invaded cells in five randomly selected fields were counted using an inverted microscope.
Luciferase reporter assay
The wild-type (WT) or mutant (MUT) CCAT1 or CDX1 3ʹ-UTR containing the predicted miR-216a-5p target sites were cloned into the pGL3 vectors basic vectors (Promega, Shanghai, China), respectively. Then, these constructed vectors were respectively co-transfected into A549 and NCI-H1299 cells with miR-216a-5p mimics, NC, anti-miR-216a-5p, or anti-NC using Lipofectamine 2000. Forty-eight hours later, a dual luciferase assay kit (Promega) was employed to analyze the luciferase activity.
RNA immunoprecipitation assay
A549 and NCI-H1299 cells were transfected with miR-216a-5p or NC for 48 h, then cells were lysed and the lysate was incubated with magnetic beads coated with Ago2 or IgG antibody (Millipore). Subsequently, the immunoprecipitated RNAs were analyzed by qRT-PCR.
Pull-down assay
Biotinylated (Bio)-miR-216a-5p (Bio-miR-216a-5p) or Bio-NC was generated by GenePharma Company (Shanghai, China), and then these biotinylated oligonucleotides were transfected into A549 and NCI-H1299 cells for 48 h. Subsequently, cells were lysed and incubated with streptavidin-coupled beads. Finally, the biotin-coupled RNA complex was isolated and was determined by qRT-PCR.
Xenograft experiments in vivo
BALB/c nude mice (5-week-old, N = 6) were bought to conduct in vivo experiments in accordance with the guidelines permitted by the Animal Research Committee of Department of Respiratory Medicine, Yantai Yuhuangding Hospital. The nude mice (3 mice per group) were subcutaneously injected in the right hip with A549 cells (106) stably infected with Lenti-sh-NC or Lenti-sh-CCAT1 to generate subcutaneous tumor model. Tumor volumes were detected and calculated every week after injection. The mice were killed on week 4 after xenograft and the tumors were weighed and collected for subsequent molecular analyses.
Statistical analysis
All statistical analyses were performed with GraphPad Prism 7. The data were expressed as the mean ± standard deviation (SD). Student’s t-test or one-way analysis of variance was used to evaluate the significant differences. P < 0.05 suggested statistically significant.
Results
CCAT1 is up-regulated in NSCLC tissues and cell lines and high CCAT1 expression shows poor overall survival rate
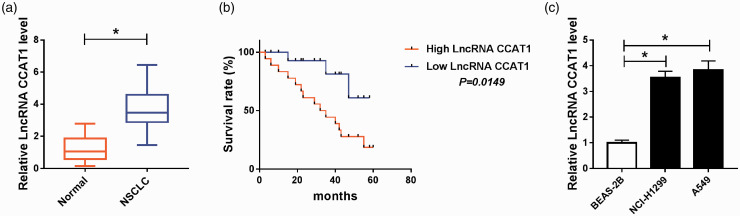
The expression of CCAT1 was firstly detected, and results showed CCAT1 expression was significantly increased in NSCLC tissues and human NSCLC cell lines (A549 and NCI-H1299) compared with the normal lung tissue and BEAS-2B cell line (Figure 1(a) and (c)). Subsequently, based on the median level of CCAT1, NSCLC patients were divided into two groups: the high CCAT1 expression group and low expression CCAT1 group. After that, we found the overall survival of patients in the low CCAT1 expression group was significantly longer than that in the high CCAT1 group (Figure 1(b), P =0.0149). Furthermore, as shown in Supplementary Table 1, higher CCAT1 expression was linked to tumor size (P = 0.0275), TNM stages (P = 0.0059), and lymph node metastasis (P = 0.0045). The data reveal that NEAT1 expression was abnormal in NSCLC and its high expression showed shorter overall survival of patients with NSCLC.
Figure 1.
CCAT1 expression is elevated in NSCLC tissues and cell lines. (a and b) The expression of CCAT1 in NSCLC tissues and matched adjacent normal lung tissue, as well as in human NSCLC cell lines (A549 and NCI-H1299) and normal human lung cell line BEAS-2B was detected using qRT-PCR. *P < 0.05. (A color version of this figure is available in the online journal.)
CCAT1 promotes cell malignant phenotypes in NSCLC
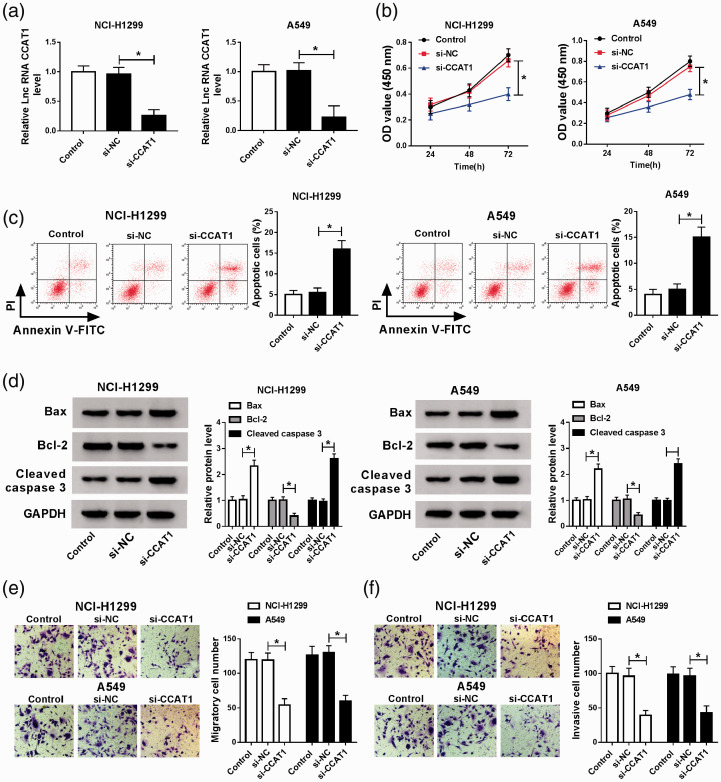
To explore the biological effects of CCAT1 on NSCLC progression, A549 and NCI-H1299 cells were transfected with si-NC or si-CCAT1, and then a visible decrease of CCAT1 in A549 and NCI-H1299 cells transfected with si-CCAT1 was observed, indicating the successful transfection (Figure 2(a)). After that, CCK-8 assay showed CCAT1 silence inhibited A549 and NCI-H1299 cell proliferation (Figure 2(b)). Then we found CCAT1 deletion induced cell apoptosis in NSCLC, reflected by the increase of apoptosis rate (Figure 2(c)) and Bax and cleaved caspase-3 levels, as well as the decrease of Bcl-2 levels (Figure 2(d)). Moreover, transwell assay suggested that knockdown of CCAT1 suppressed the migration and invasion of A549 and NCI-H1299 cells (Figure 2(e) and (f)). Besides that, A549 and NCI-H1299 cells were also transfected with CCAT1 overexpression vector (CCAT1), as expected, CCAT1 expression was significantly up-regulated in A549 and NCI-H1299 cells (Figure S1(A)). Next, functional experiments exhibited that CCAT1 overexpression showed the exact opposite effects by promoting cell proliferation, migration, invasion, and inhibiting apoptosis in NSCLC (Figure S1(C) to (F)). In sum up, we illustrated that CCAT1 facilitated NSCLC cell carcinogenesis.
Figure 2.
CCAT1 deletion inhibits cell malignant phenotypes in NSCLC. A549 and NCI-H1299 cells were transfected with si-NC or si-CCAT1. (a) CCAT1 expression was measured using qRT-PCR. Cell proliferation (b) and apoptosis (c) were detected by CCK-8 assay or flow cytometry, respectively. (d) Levels of apoptosis-related proteins were measured by Western blot. (e and f) Cell migration and invasion abilities were detected by transwell assay. *P < 0.05. (A color version of this figure is available in the online journal.)
Overexpressed miR-216a-5p inhibits cell malignant phenotypes in NSCLC
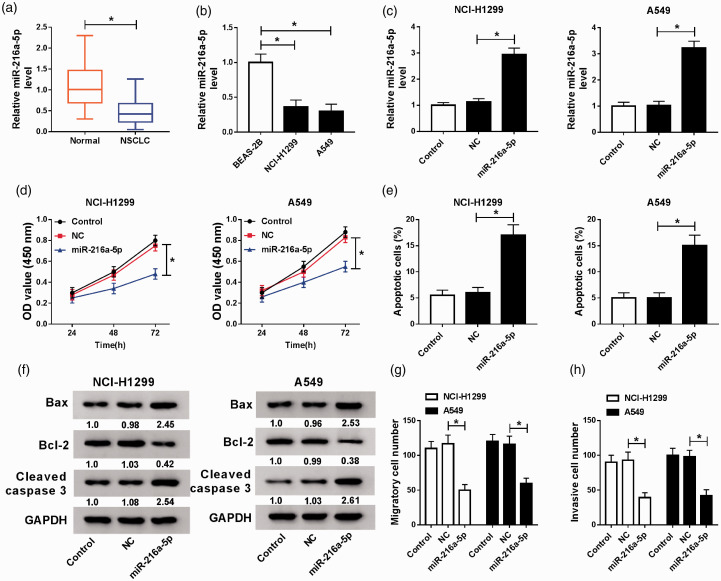
The expression of miR-216a-5p was discovered to be decreased in NSCLC tissues and human NSCLC cell lines (A549 and NCI-H1299) compared with that in controls (Figure 3(a) and (b)), indicating miR-216a-5p might be an important regulator in NSCLC development. Then to detect the potential roles of miR-216a-5p in NSCLC, A549 and NCI-H1299 cells were transfected with miR-216a-5p mimic or NC mimic. After transfection, an apparent increase of miR-216a-5p expression in A549 and NCI-H1299 was found (Figure 3(c)). Subsequently, functional experiments were performed and we discovered miR-216a-5p mimic transfection markedly suppressed the proliferation, migration, and invasion but induced apoptosis in A549 and NCI-H1299 cells (Figure 3(d) to (h)). Therefore, we demonstrated that miR-216a-5p might be an important tumor suppressor in NSCLC progression.
Figure 3.
Overexpressed miR-216a-5p inhibits cell malignant phenotypes in NSCLC. (a and b) The expression of miR-216a-5p in NSCLC tissues and matched adjacent normal lung tissues (a), as well as in human NSCLC cell lines (A549 and NCI-H1299) and BEAS-2B cells (b) was detected using qRT-PCR. A549 and NCI-H1299 cells were transfected with NC or miR-216a-5p. (c) The expression of miR-216a-5p was examined by qRT-PCR. Cell proliferation (d) and apoptosis (e) were detected by CCK-8 assay or flow cytometry, respectively. (f) Levels of apoptosis-related proteins were measured by Western blot. (g and h) Transwell assay was applied to determine cell migration and invasion abilities. *P < 0.05. (A color version of this figure is available in the online journal.)
CCAT1 directly binds to miR-216a-5p and suppresses miR-216a-5p expression
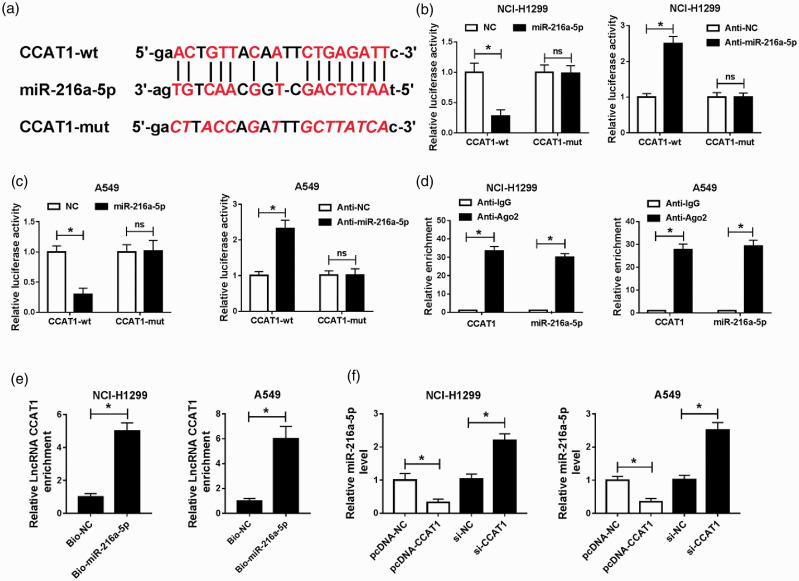
According to the prediction of starBase v.2.0 database, CCAT1 was identified to have the putative binding site in miR-216a-5p (Figure 4(a)). Then the exact copy number of CCAT1 and miR-216a-5p expression in each cell was detected, qRT-PCR analysis showed that CCAT1 was expressed in 53/45 copies and miR-216a-5p was 26/18 copies in per NCI-H1299 or A549 cell (Figure S2(A) and (B)). Thus, we suspected that CCAT1-mediated regulatory functions might operate through a competing endogenous RNAs (ceRNAs) mechanism. To validate their interaction, luciferase reporter assay was performed, and results showed miR-216a-5p overexpression reduced the luciferase activities of the CCAT1-wt reporter vector but not CCAT1-mut reporter vector, while anti-miR-216a-5p transfection showed the completely opposite effects in NCI-H1299 and A549 cells (Figure 4(b) and (c)). In the meanwhile, the RIPA and RNA pull-down assay further confirmed the direct interaction between miR-216a-5p and CCAT1, because a significant enrichment of CCAT1 was measured in both assays in NCI-H1299 and A549 cells (Figure 4(d) and (e)). Furthermore, relative quantitative PCR was performed and we found CCAT1 overexpression reduced miR-216a-5p expression, while si-CCAT1 accelerated miR-216a-5p expression in NCI-H1299 and A549 cells (Figure 4(f)). All the results suggested that CCAT1 was a sponge of miR-216a-5p and inversely modulated its expression.
Figure 4.
CCAT1 directly binds to miR-216a-5p and suppresses its expression. (a) The potential binding sites between CCAT1 and miR-216a-5p. (b and c) Luciferase reporter assay was used to demonstrate whether CCAT1 directly targeted miR-216a-5p in A549 and NCI-H1299 cells, respectively. (d) RIPA was carried out, and expression of CCAT1 and miR-216a-5p was examined in the samples bound to the Ago2 or IgG antibody. (e) CCAT1 expression level was measured by qRT-PCR in samples pulled down by Bio-miR-216a-5p or negative control. (f) qRT-PCR analysis of miR-216-5p level in A549 and NCI-H1299 cells transfected with CCAT1 or si-CCAT1 was conducted. *P < 0.05. (A color version of this figure is available in the online journal.)
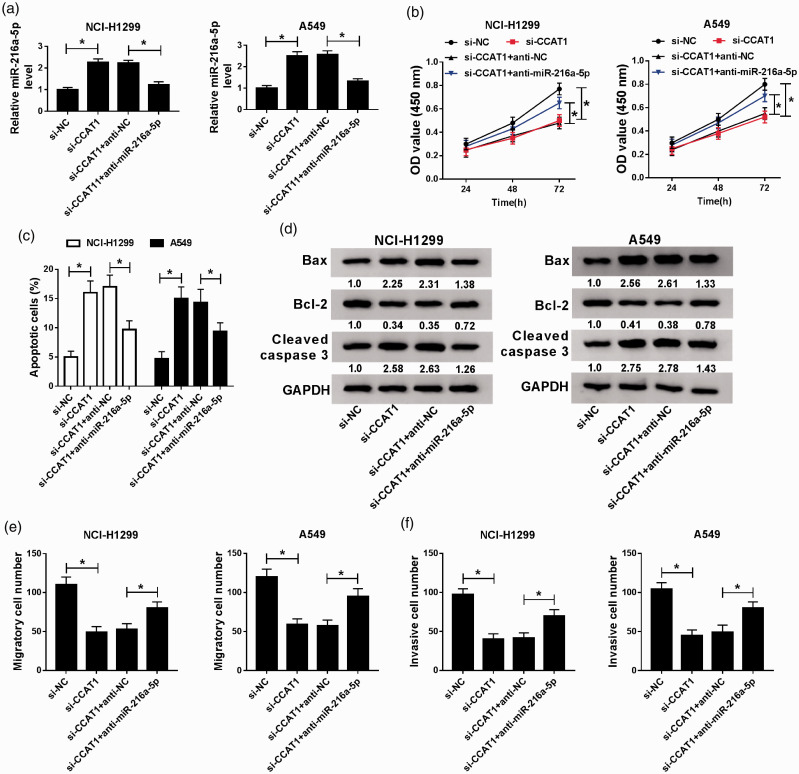
CCAT1 promotes NSCLC cell tumorigenesis by interacting with miR-216a-5p
To investigate whether CCAT1/miR-216a-5p axis was responsible for the NSCLC progression, NCI-H1299 and A549 cells were transfected with si-NC, si-CCAT1, si-CCAT1 + anti-NC, or si-CCAT1 + anti-miR-216a-5p, and then we observed the expression of miR-216a-5p was promoted by CCAT1 knockdown, but was inhibited by the following addition of miR-216a-5p inhibitor (Figure 5(a)). Subsequently, rescue assay showed that the miR-216a-5p inhibition could attenuate the inhibitory effects of si-CCAT1 on NCI-H1299 and A549 cell oncogenic phenotypes (Figure 5(b) to (f)). Additionally, it was also proved that miR-216a-5p re-expression rescued CCAT1 overexpression-induced down-regulation of miR-216a-5p level in NCI-H1299 and A549 cells (Figure S1(B)), importantly, the regulatory effects of CCAT1 on cell malignant phenotypes were markedly abrogated by miR-216a-5p up-regulation in NCI-H1299 and A549 cells (Figure S1(C) to (F)). Taken together, CCAT1 deletion exerted anti-tumor effects on NSCLC progression by sponging miR-216a-5p.
Figure 5.
CCAT1 promotes the progression of NSCLC by interacting with miR-216a-5p. NCI-H1299 and A549 cells were transfected with si-NC, si-CCAT1, si-CCAT1 + anti-NC, or si-CCAT1 + anti-miR-216a-5p. (a) qRT-PCR analysis of the level of miR-216a-5p. (b) CCK-8 assay of cell proliferation. (c) Flow cytometry of cell apoptosis. (d) Western blot analysis of the levels of apoptosis-related proteins. (e and f) Transwell assay of cell migration and invasion abilities. *P < 0.05. (A color version of this figure is available in the online journal.)
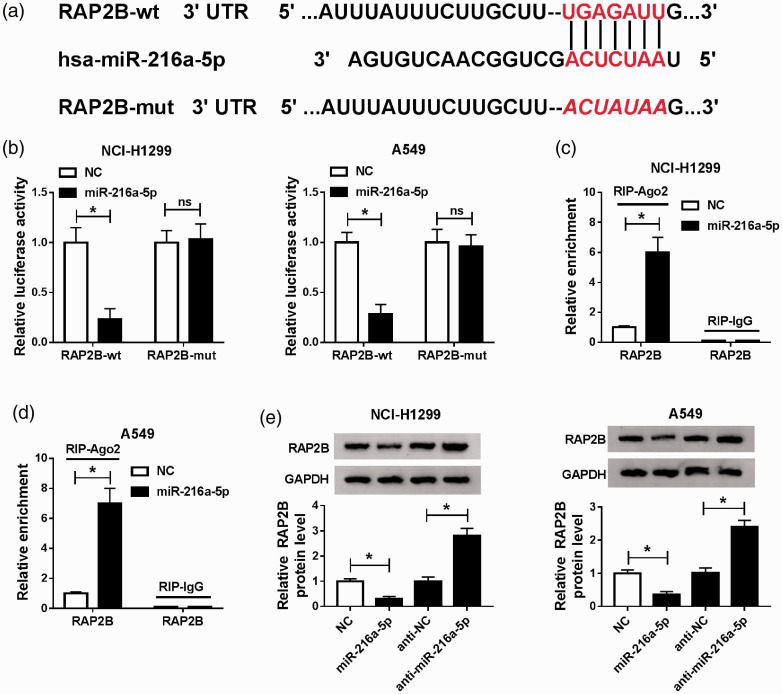
MiR-216a-5p directly targets RAP2B and negatively regulates RAP2B expression
We further explore the underlying regulatory mechanism of miR-216a-5p on NSCLC progression. The potential target genes of miR-216a-5p were predicted according to the Targetscan database, and RAP2B was identified to have the putative binding sites in miR-216a-5p (Figure 6(a)). The qRT-PCR analysis indicated RAP2B was expressed in 85/74 copies and miR-216a-5p was 26/18 copies in per NCI-H1299 or A549 cell (Figure S2(A) and (B)). Thus, the interaction of miR-216a-5p and RAP2B was explored. Results from luciferase reporter analysis exhibited the luciferase activities of CCAT1-wt reporter vector was reduced by the miR-216a-5p mimic, but no change was observed in CCAT1-mut reporter vector after miR-216a-5p overexpression in NCI-H1299 and A549 cells (Figure 6(b)). Besides, RIPA showed a great enrichment of RAP2B in NCI-H1299 and A549 cells, further confirming the direct interaction between miR-216a-5p and RAP2B (Figure 6(c) and (d)). Moreover, Western blot results indicated that RAP2B expression in NCI-H1299 and A549 cells were inhibited by overexpressed miR-216a-5p but was promoted by miR-216a-5p inhibition (Figure 6(e)). These results indicated miR-216a-5p directly targeted RAP2B and negatively regulated RAP2B expression.
Figure 6.
MiR-216a-5p directly targets RAP2B and negatively regulates RAP2B expression. (a) The potential binding sites between RAP2B and miR-216a-5p. (b) Luciferase reporter assay was used to analyze whether miR-216a-5p directly targeted RAP2B in A549 and NCI-H1299 cells, respectively. (c and d) The expression of RAP2B was detected in A549 and NCI-H1299 cells after RIPA. (e) The protein expression of RAP2B was analyzed using Western blot in A549 and NCI-H1299 cells transfected with miR-216a-5p or anti-miR-216a-5p. *P < 0.05. (A color version of this figure is available in the online journal.)
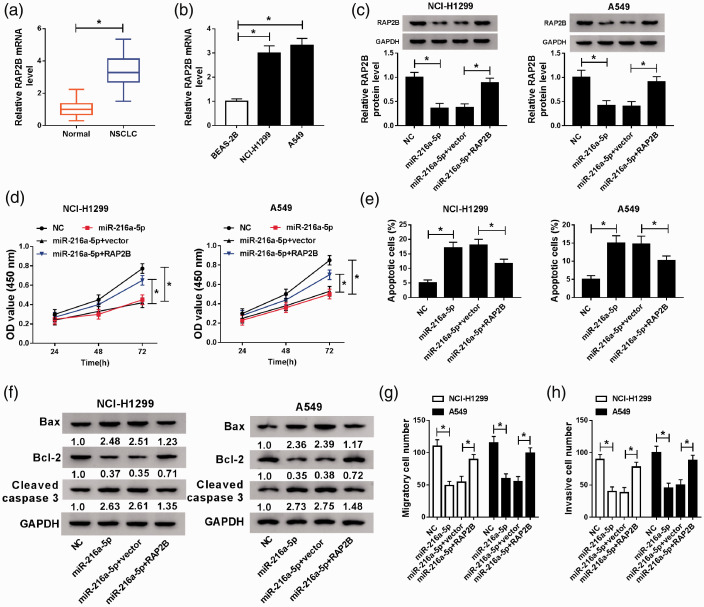
Overexpressed miR-216a-5p represses NSCLC cell tumorigenesis by targeting RAP2B
Based on the relationship between miR-216a-5p and RAP2B, whether RAP2B involved in miR-216a-5p mediated regulation on NSCLC cell tumorigenesis was then explored. First, the expression of RAP2B was detected and results showed RAP2B was elevated in NSCLC tissues and cell lines (Figure 7(a) and (b)). Then miR-216a-5p, NC, miR-216a-5p + pcDNA (vector), or miR-216a-5p + RAP2B were transfected into A549 and NCI-H1299 cells. After transfection, we discovered that the expression of RAP2B was reduced by overexpressed miR-216a-5p, while was increased by following RAP2B transfection (Figure 7(c)). After that, we found overexpressed RAP2B significantly attenuated miR-216a-5p restoration-mediated inhibition on cell proliferation, migration and invasion as well as enhancement on cell apoptosis in NSCLC (Figure 7(d) to (h)). In all, miR-216a-5p hindered NSCLC progression by targeting RAP2B.
Figure 7.
Overexpressed miR-216a-5p inhibits NSCLC progression through targeting RAP2B. RAP2B expression in NSCLC tissues and matched adjacent normal lung tissue (a), as well as in human NSCLC cell lines (A549 and NCI-H1299) and BEAS-2B cell lines (b) was detected by qRT-PCR. RAP2B expression detection by Western blot (c), cell proliferation by CCK-8 assay (d), cell apoptosis by flow cytometry (e), levels of apoptosis-related proteins analysis by Western blot (f), cell migration and invasion abilities by transwell assay (g and h) in A549 and NCI-H1299 cells transfected with NC, miR-216a-5p, miR-216a-5p + vector, or miR-216a-5p + RAP2B. *P < 0.05. (A color version of this figure is available in the online journal.)
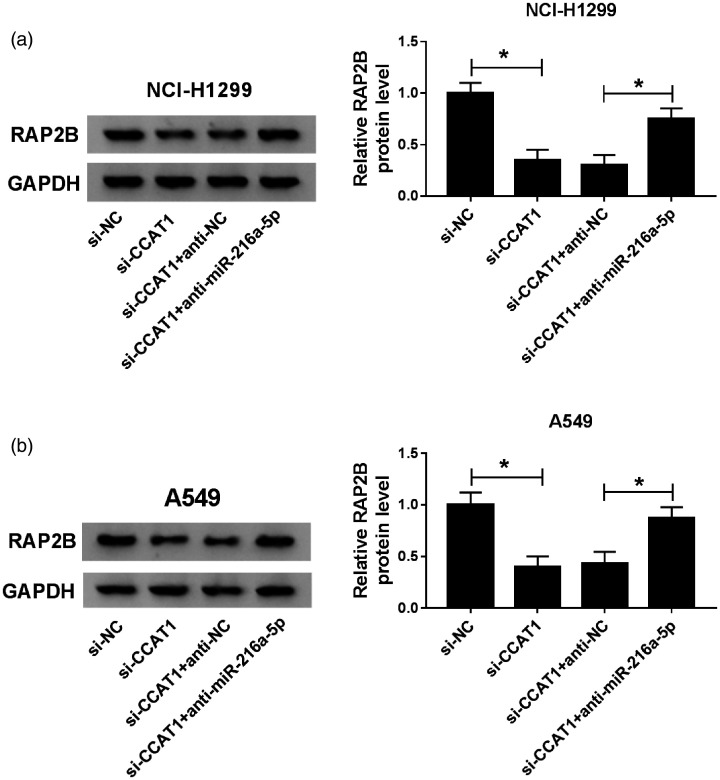
CCAT1 may promote NSCLC progression by regulating miR-216a-5p/RAP2B axis
Based on the CCAT1/miR-216a-5p axis, we investigated whether CCAT1 could regulate RAP2B via miR-216a-5p. Western blot analysis showed the expression of RAP2B was reduced by si-CCAT1 transfection, while was increased by following miR-216a-5p inhibitor transfection in A549 and NCI-H1299 cells (Figure 8(a) and (b)), indicating that CCAT1 functioned as a ceRNA to regulate RAP2B level by targeting miR-216a-5p in NSCLC cells. Therefore, a CCAT1/miR-216a-5p/RAP2B axis was identified in NSCLC cells.
Figure 8.
CCAT1 may promote NSCLC progression by regulating miR-216a-5p/RAP2B axis. (a and b) Western blot analysis of RAP2B protein expression in A549 and NCI-H1299 cells transfected with si-NC, si-CCAT1, si-CCAT1 + anti-NC or si-CCAT1 + anti-miR-216a-5p. *P < 0.05.
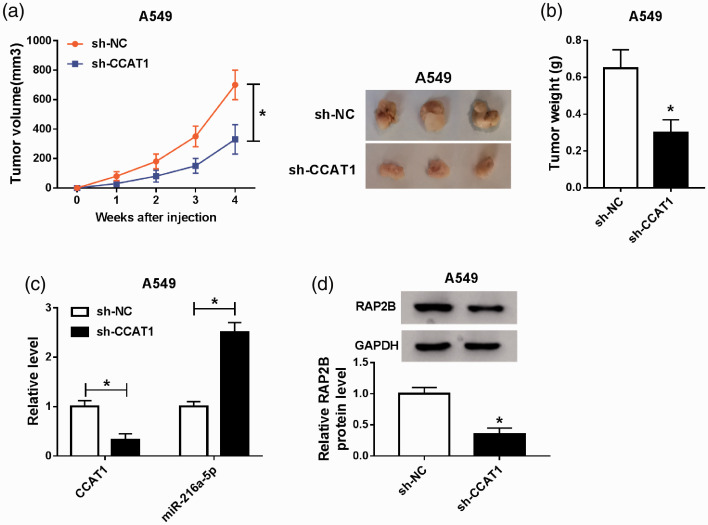
CCAT1 knockdown impedes NSCLC tumor growth in vivo by regulating miR-216a-5p and RAP2B expression
The carcinogenic roles of CCAT1 in vivo were further investigated. As illustrated in Figure 9(a) and (b), CCAT1 silence significantly repressed tumor growth, evidenced by the reduction of tumor volume and weight in sh-CCAT1 groups. Subsequent molecular analysis exhibited that CCAT1 expression was decreased in tumor masses isolated from sh-CCAT1 groups (Figure 9(c)), and decreased CCAT1 up-regulated the level of miR-216a-5p (Figure 9(c)), and down-regulated the level of RAP2B (Figure 9(d)). Collectively, these results implied that CCAT1 knockdown suppressed NSCLC tumor growth in vivo via partially modulating the expression of miR-216a-5p and RAP2B.
Figure 9.
CCAT1 knockdown impedes NSCLC tumor growth in vivo by regulating miR-216a-5p and RAP2B expression. (a) Tumor volume was calculated every week after injection. (b) Tumor masses were isolated on day 28 from each group and then were weighed. (c and d) The levels of CCAT1, miR-216a-5p, and RAP2B were detected in two groups by qRT-PCR or Western blot. *P < 0.05. (A color version of this figure is available in the online journal.)
Discussion
It has been reported that lncRNAs interacts with genes, proteins or chromatin remodeling to influence the expression levels of genes, thereby affecting malignant physiological or pathological processes.17 Recently, numerous lncRNAs, like lncRNA XIST, UCA1, PVT1, and MALAT1, have been identified to involve in the progression in NSCLC by modulating downstream target miRNAs, genes or pathways.18–21 CCAT1 was originally identified as an oncogene in colon cancer, and an increasing number of studies have showed the carcinogenic role and prognostic value of CCAT1 in various cancers.22 For example, CCAT1 induced metastasis and predicted poor outcome in epithelial ovarian cancer.23 Decrease of CCAT1 increased radiosensitivity of breast cancer cells through inversely modulating miR-148b expression.24 The c-Myc interacted with CCAT1 to promote the progression of gastric carcinoma.25 Thus, it is of great clinical significance to reveal the role of CCAT1 in NSCLC cells.
Previous studies revealed that CCAT1 functioned as an oncogene to promote docetaxel-resistant and stimulate metastasis through epithelial-to-mesenchymal transition in lung adenocarcinoma.26,27 However, the exact regulatory mechanisms of CCAT1 in NSCLC remain vague. In the current study, we recognized that CCAT1 expression was up-regulated in NSCLC, and CCAT1 functioned as an oncogene via promoting cell proliferation, migration, and invasion but inhibiting cell apoptosis to promote NSCLC progression. Furthermore, results of xenograft assay displayed that CCAT1 deletion restrained NSCLC tumor growth in vivo.
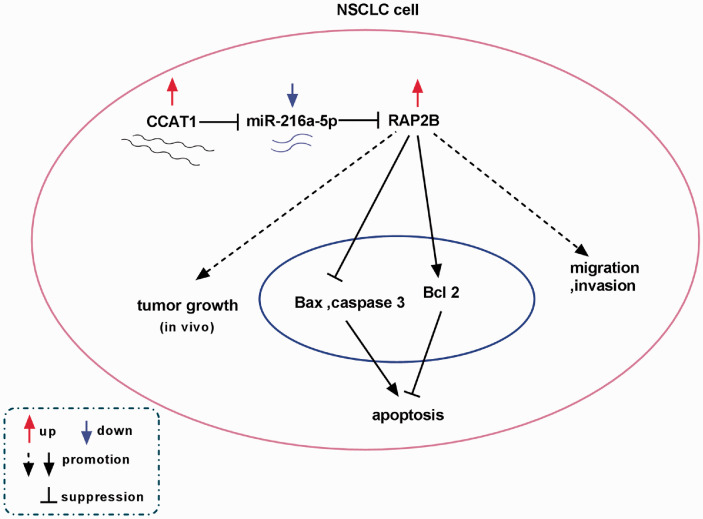
It is well documented that lncRNAs can function as ceRNAs to compete with other genes for miRNA binding, thus exerting the regulatory effects of miRNAs on targeted mRNAs.28 Thus, the mechanisms underlying CCAT1 in NSCLC progression were elucidated, and we proved that CCAT1 bound to miR-216a-5p. MiR-216a-5p has been reported to function as a tumor suppressor to involve in the progression of small cell lung cancer.29 However, there is no research on miR-216a-5p in NSCLC. In this study, miR-216a-5p expression was demonstrated to be decreased in NSCLC and was negatively regulated by CCAT1. Afterwards, rescue experiments indicated CCAT1 promoted NSCLC cell progression by interacting with miR-216a-5p. In addition, we also confirmed that miR-216a-5p targeted RAP2B in NSCLC cells. RAP2B was elevated in NSCLC, and was negatively regulated by miR-216-5p. Rescue experiments suggested that overexpressed miR-216a-5p inhibited cell tumorigenesis in NSCLC through targeting RAP2B. In the meanwhile, CCAT1 was verified to directly regulate RAP2B level by sponging miR-216a-5p in NSCLC cells, indicating a novel CCAT1/miR-216a-5p/RAP2B axis in the regulation of NSCLC cell tumorigenesis (Figure 10).
Figure 10.
Schematic model showing the role of CCAT1 in NSCLC cell tumorigenesis. CCAT1 promoted NSCLC cell oncogenic phenotypes in vitro as well as suppressed tumor growth in vivo by regulating miR-216a-5p/RAP2B axis. (A color version of this figure is available in the online journal.)
In sum up, we investigated that CCAT1 functioned as an oncogene while miR-216a-5p served as a tumor suppressor in NSCLC to regulate cell proliferation, migration, invasion, and apoptosis. Additionally, molecular analyses further identified a novel CCAT1/miR-216a-5p/RAP2B regulatory network in the progression of NSCLC in vitro and in vivo (Figure 10), indicating a new insight on NSCLC pathogenesis and novel potential targets for NSCLC treatment.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220961013 for Long non-coding RNA CCAT1 promotes non-small cell lung cancer progression by regulating the miR-216a-5p/RAP2B axis by Lingling Pang, Qianqian Zhang, Yanmin Wu, Qingru Yang, Jinghao Zhang, Yuanyuan Liu and Ruoran Li in Experimental Biology and Medicine
Supplemental material, sj-pdf-2-ebm-10.1177_1535370220961013 for Long non-coding RNA CCAT1 promotes non-small cell lung cancer progression by regulating the miR-216a-5p/RAP2B axis by Lingling Pang, Qianqian Zhang, Yanmin Wu, Qingru Yang, Jinghao Zhang, Yuanyuan Liu and Ruoran Li in Experimental Biology and Medicine
Footnotes
AUTHORS’ CONTRIBUTIONS: All authors participated in the design, interpretation of the studies and analysis of the data, and review of the manuscript.
Declaration OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Ruoran Li https://orcid.org/0000-0001-9577-2270
Supplemental material: Supplemental material for this article is available online.
REFERENCES
- 1.Powell HA, Tata LJ, Baldwin DR, Potter VA, Stanley RA, Khakwani A, Hubbard RB. Treatment decisions and survival for people with small-cell lung cancer. Br J Cancer 2014; 110:908–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosell R, Karachaliou N. Lung cancer: maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol 2013; 10:549–50 [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Lu Q, Zhu D, Han Y, Zhou X, Ren T. Lnc-SNHG1 may promote the progression of non-small cell lung cancer by acting as a sponge of miR-497. Biochem Biophys Res Commun 2018; 506:632–40 [DOI] [PubMed] [Google Scholar]
- 4.Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol 2018; 52:103–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan Y, Zang H, Feng J, Lu J, Chen L, Fan S. Long non-coding RNAs associated with non-small cell lung cancer. Oncotarget 2017; 8:69174–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017; 36:5661–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong Y, Wang T, Wang M, Zhao J, Li X, Zhang Z, Zhou Y, Liu J, Jia L, Han Y. Long non-coding RNAs function as novel predictors and targets of non-small cell lung cancer: a systematic review and Meta-analysis. Oncotarget 2018; 9:11377–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, Yang L, Chen LL. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 2014; 24:513–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu C, Xiao G, Zhang B, Wang M, Wang J, Liu D, Zhang J, Ren H, Sun X. CCAT1 stimulation of the symmetric division of NSCLC stem cells through activation of the wnt signalling Cascade. Gene Ther 2018; 25:4–12 [DOI] [PubMed] [Google Scholar]
- 10.Hu B, Zhang H, Wang Z, Zhang F, Wei H, Li L. LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance in non-small-cell lung cancer cell line by targeting SOX4. Cancer Biol Ther 2017; 18:974–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang P, Li J, Song Y, Wang X. MiR-129-5p inhibits proliferation and invasion of chondrosarcoma cells by regulating SOX4/wnt/β-catenin signaling pathway. Cell Physiol Biochem 2017; 42:242–53 [DOI] [PubMed] [Google Scholar]
- 12.Bai J, Yao B, Wang L, Sun L, Chen T, Liu R, Yin G, Xu Q, Yang W. lncRNA A1BG‐AS1 suppresses proliferation and invasion of hepatocellular carcinoma cells by targeting miR‐216a‐5p. J Cell Biochem 2019; 120:10310–22 [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Lin P, Zou J, Zou G, Wang W, Liu Y, Zhao H, Fang A. MiR-216a-5p act as a tumor suppressor, regulating the cell proliferation and metastasis by targeting PAK2 in breast cancer. Eur Rev Med Pharmacol Sci 2019; 23:2469–75 [DOI] [PubMed] [Google Scholar]
- 14.Qu D, Huang H, Di J, Gao K, Lu Z, Zheng J. Structure, functional regulation and signaling properties of Rap2B. Oncol Lett 2016; 11:2339–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canobbio I, Trionfini P, Guidetti GF, Balduini C, Torti M. Targeting of the small GTPase Rap2b, but not Rap1b, to lipid rafts is promoted by palmitoylation at Cys176 and Cys177 and is required for efficient protein activation in human platelets. Cell Signal 2008; 20:1662–70 [DOI] [PubMed] [Google Scholar]
- 16.Peng Y-G, Zhang Z-Q, Chen Y-B, Huang J-A. Rap2b promotes proliferation, migration, and invasion of lung cancer cells. J Recept Signal Transduct Res 2016; 36:459–64 [DOI] [PubMed] [Google Scholar]
- 17.Ren K, Li Y, Lu H, Li Z, Li Z, Wu K, Li Z, Han X. Long noncoding RNA HOTAIR controls cell cycle by functioning as a competing endogenous RNA in esophageal squamous cell carcinoma. Transl Oncol 2016; 9:489–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Mei Z, Hu HB, Zhang X. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol 2018; 233:6679–88 [DOI] [PubMed] [Google Scholar]
- 19.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett 2016; 371:99–106 [DOI] [PubMed] [Google Scholar]
- 20.Guo D, Wang Y, Ren K, Han X. Knockdown of LncRNA PVT1 inhibits tumorigenesis in non-small-cell lung cancer by regulating miR-497 expression. Exp Cell Res 2018; 362:172–9 [DOI] [PubMed] [Google Scholar]
- 21.Sun W, Zu Y, Fu X, Deng Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep 2017; 38:3347–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi D, Wu F, Gao F, Qing X, Shao Z. Prognostic value of long non-coding RNA CCAT1 expression in patients with cancer: a meta-analysis. PLoS One 2017; 12:e0179346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Y, Shi H, Ren F, Jia Y, Zhang R. Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in epithelial ovarian cancer. Exp Cell Res 2017; 359:185–94 [DOI] [PubMed] [Google Scholar]
- 24.Lai Y, Chen Y, Lin Y, Ye L. Down-regulation of LncRNA CCAT1 enhances radiosensitivity via regulating miR-148b in breast cancer. Cell Biol Int 2018; 42:227–36 [DOI] [PubMed] [Google Scholar]
- 25.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol 2013; 139:437–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Zhang K, Song H, Wang R, Chu X, Chen L. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget 2016; 7:62474–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin H, Cheng W, Yan H, Zhang X. Overexpression of the long noncoding RNA CCAT1 promotes metastasis via epithelial-to-mesenchymal transition in lung adenocarcinoma. Oncol Lett 2018; 16:1809–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballantyne M, McDonald R, Baker A. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther 2016; 99:494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Hu B, Wang Y, Li Z, Wu J, Yang Y, Wei Y, Peng X, Chen H, Chen R, Jiang P, Fang S, Yu Z, Guo L. miR-216a-5p inhibits malignant progression in small cell lung cancer: involvement of the bcl-2 family proteins. Cancer Manag Res 2018; 10:4735–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220961013 for Long non-coding RNA CCAT1 promotes non-small cell lung cancer progression by regulating the miR-216a-5p/RAP2B axis by Lingling Pang, Qianqian Zhang, Yanmin Wu, Qingru Yang, Jinghao Zhang, Yuanyuan Liu and Ruoran Li in Experimental Biology and Medicine
Supplemental material, sj-pdf-2-ebm-10.1177_1535370220961013 for Long non-coding RNA CCAT1 promotes non-small cell lung cancer progression by regulating the miR-216a-5p/RAP2B axis by Lingling Pang, Qianqian Zhang, Yanmin Wu, Qingru Yang, Jinghao Zhang, Yuanyuan Liu and Ruoran Li in Experimental Biology and Medicine