Abstract
The respiratory microbiome has been less explored than the gut microbiome. Despite the speculated importance of dysbiosis of the microbiome in ventilator-associated pneumonia (VAP) and acute respiratory distress syndrome (ARDS), only few studies have been performed in invasively ventilated ICU patients. And only the results of small cohorts have been published. An overlap exists between bacterial populations observed in the lower respiratory tract and the oropharyngeal tract. The bacterial microbiota is characterized by relatively abundant bacteria difficult to cultivate by standard methods. Under mechanical ventilation, a dysbiosis occurs with a drop overtime in diversity. During VAP development, lung dysbiosis is characterized by a shift towards a dominant bacterial pathogen (mostly Proteobacteria) whereas enrichment of gut-associated bacteria mainly Enterobacteriaceae is the specific feature discriminating ARDS patients. However, the role of this dysbiosis in VAP and ARDS pathogenesis is not yet fully understood. A more in-depth analysis of the interplay between bacteria, virus and fungi and a better understanding of the host-microbiome interaction could provide a more comprehensive view of the role of the microbiome in VAP and ARDS pathogenesis. Priority should be given to validate a consensual and robust methodology for respiratory microbiome research and to conduct longitudinal studies. A deeper understanding of microbial interplay should be a valuable guide for care of ARDS and VAP preventive/therapeutic strategies. We present a review on the current knowledge and expose perspectives and potential clinical applications of respiratory microbiome research in mechanically ventilated patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00134-020-06338-2.
Keywords: Lung microbiome, 16S rRNA gene, High-throughput sequencing, Dysbiosis, Metagenomics, Ventilator-associated pneumonia, Mechanical ventilation, Acute respiratory distress syndrome
Take-home message
| Interactions between fungi, bacteria and viruses (bacteriophages and eukaryotic viruses) highlight the need for a concomitant analysis of their evolution to understand the pathophysiology of ventilator-associated pneumonia and their role in ARDS evolution. | |
| A validated and accepted method of analysis (extraction, amplification, sequencing, bioinformatics analysis) should allow homogenization of microbiome studies and comparison between studies. |
Introduction
The study of microbial populations within the human body has long been hampered by the limits of conventional microbiology. Because of the poor sensitivity of these methods, the lung was considered sterile. Thanks to the development of culture-independent techniques, particularly high throughput sequencing, this hypothesis has been challenged. Many microorganisms including bacteria, viruses and fungi, coexist in the lungs of healthy individuals to constitute the lung microbiome.
The study of the lung microbiome has, until recently, been limited to the sole bacterial microbiota, using 16S rRNA genes (genes encoding 16S ribosomal RNA) sequencing. In the field of intensive care medicine, investigations are in their infancy compared to chronic respiratory diseases where studies have shown (as with the gut) that lung microbiota interactions with the host, as well as dysbiosis, likely play a key role in the pathophysiology of chronic inflammatory diseases such as asthma, chronic obstructive pulmonary disease (COPD) and cystic fibrosis. Moreover, no study has really focused on the evolution of the fungal microbiota (or mycobiota) or the virome in ventilated patients, whereas fungal lung colonization and viral reactivation are extensively described in this population [1, 2]. Addressing the dynamic evolution of the whole lung microbiome composition (including bacteria, fungi, and viruses) is thus one of the main challenges in respiratory medicine to redefine our understanding of ventilator-associated pneumonia (VAP) and acute respiratory distress syndrome (ARDS) pathophysiology.
The purposes of this review are to specify the main definitions and terminology used in microbiome research, to present current knowledge on respiratory microbiome in intensive care, and to describe current techniques used, their interests and their limits. Finally, we will show how a deeper, general understanding of the composition and the evolution of respiratory microbiome will facilitate future research projects leading to many potential clinical applications to improve prevention, diagnosis and treatment of acute lung injury in general, and ARDS and VAP in particular.
Definition
Lung bacterial microbiota
Bacterial microbiota represents the composition of all bacteria in a medium. Bacterial taxonomy classified from the domain to the species is used to describe the bacterial microbiota (example for Pseudomonas aeruginosa in Fig. 1). The presence of hypervariable sequences framed by extremely conserved sequences within the bacterial genomic DNA coding for the 16S rRNA allows identification of bacterial composition. Indeed, these conserved sequences can be amplified by polymerase chain reaction (PCR) using universal primers regardless of the bacterial species. In contrast, the hypervariable regions framed by conserved regions are specific of the genus or even the species. The sequencing of one or more hypervariable regions included between these conserved sequences permits identification of the different bacteria present within the sample (microbiota).
Fig. 1.
Bacterial taxonomy, example for Pseudomonas aeruginosa.
Adapted from Faner et al. (ref# 6 from the online supplementary material)
The lung microbiota of an individual can be studied with a lower respiratory tract sample [3]. The lung is interconnected with many reservoirs (oral cavity, digestive tract especially in intubated patients, and ambient air). Recent studies have reported that the microbiota of the oropharynx and the lower respiratory tract have a similar composition in healthy individuals, whereas upper and lower respiratory tract are clearly different in all chronic respiratory diseases (COPD, and interstitial lung diseases as idiopathic pulmonary fibrosis) [4–6]. In the field of acute pneumonia, there is no published data concerning the comparison between oropharyngeal and pulmonary microbiota.
Conflicting data exist regarding the similarity between lower respiratory tract microbiota collected from the bronchial tree or from the alveolar surface of healthy individuals [7, 8]. In the context of intensive care, bronchoalveolar lavage (BAL) has been used more often than endotracheal aspirate (ETA) [9–15]. To the best of our knowledge, there are however no studies that have compared these two sampling methods. Hence, it is not possible to make any clear recommendation on which to use preferentially.
The main singularity of the lower respiratory tract is the low bacterial density, as assessed by conventional microbiology, of about 105 colony-forming units per millilitres. This corresponds to a biomass of 105 bacteria per gram of tissue [16]. Despite this low density, a high diversity is observed, which constitutes a biomarker of respiratory health. Within this diversity can be identified a group of bacterial species present in the majority of healthy individuals, representing a pulmonary "core microbiota" [17]. This core microbiota is composed of the main genera Prevotella, Streptococcus, Veillonella, Fusobacterium and Haemophilus [7, 16, 18]. For each individual, lung microbiota composition derives from this core microbiota and results from the balance between microbial spread from the upper respiratory tract and microbial elimination by host-defense mechanisms [18, 19]. Finally, like the gut microbiota, final lung microbiota composition is probably shaped by many endogenous forces, such as mucus production and innate immune mechanism, but also exogenous forces such as environmental diversity [18, 20–23].
Mycobiota
Like the bacterial microbiota, fungal microbiota represents all the fungal species of an ecosystem or a sample of it. The fungi genomes also contain highly conserved sequences within the fungal kingdom, which framed hypervariable regions allowing their identification. These sequences encode various parts of ribosomal RNA (rRNA) or are adjacent regions such as ITS 1 (internal transcribed spacer 1) or ITS 2 [24].
Virome
The human virome corresponds to all genomes of DNA and RNA viruses in a medium. It includes two major entities: eukaryotic viruses and prokaryotic viruses, also called bacteriophages. In contrast to bacteria and fungi, no genomic sequence is conserved within the viral kingdom. Thus, there is no “virote” analysis. Only virome can be studied and this requires a deep sequencing of all nucleic acids in the sample to analyse the viral population.
Method of biota analysis
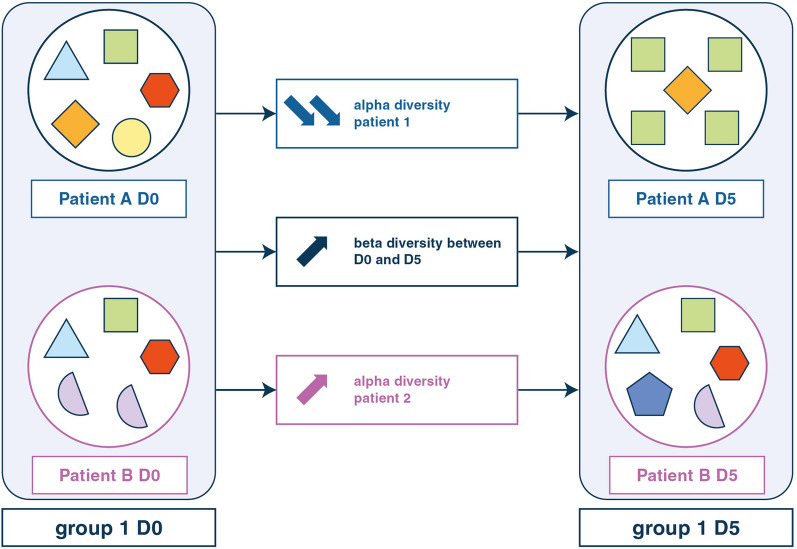
The progress made in the field of high throughput sequencing enables in-depth investigations of abundance and diversity, without resorting to conventional culture-based techniques. The Table 1 specifies the definitions necessary for a clear understanding of this field of expertise. General principles of the methods for airway microbiome analysis, including microbiota, mycobiote and virome (Fig. 2) and their limits are presented in the supplementary material (see online supplementary material). The Fig. 3 illustrates these notions of diversity with an example of evolution over time.
Table 1.
Glossary of definitions used for the evaluation of the human microbiota
| Terms | Definition |
|---|---|
| Microbiota | All microorganisms contained in a given biotope |
| Microbiome | All genomes (genetic information) and gene products of a given microbiota |
| Metagenome | All genes within a given biotope |
| Amplicon sequencing method | Method based on sequencing a DNA product of specific amplification via PCR |
| Shotgun sequencing method | Method for DNA sequencing based on random sequencing of total DNA from human and microbial origin after DNA fragmentation into short segments |
| 16S ribosomal RNA (16S rRNA) | Component of the 30S small subunit of prokaryotic ribosomes encoded by the 16SrRNA gene and used to obtain bacterial phylogenetic data. 16SrRNA gene contained constant and hypervariable sequences which allow universal amplification of hypervariable regions and microbial identification through sequencing. 16SrRNA gene is a specific taxonomic marker gene for bacteria |
| Nuclear ribosomal internal transcribed (ITS) spacer gene | Region of the nuclear ribosomal DNA gene used to obtain fungal phylogenetic data and formally proposed as the primary barcode marker |
| OTU (operational taxonomic unit) | Cluster of microorganisms identified from sequencing data and characterized by DNA sequencing similarity of 16SrRNA for bacteria or ITS for fungi |
| Taxon | Group of one or more populations of microorganisms considered to form a unit |
| Dysbiosis | Imbalance in the composition of the microbiota of a given biotope, linked to changes in local conditions, which may lead to a pathological state |
| Alpha diversity |
Appreciates the number of OTUs in a single sample. A greater alpha diversity indicates a higher number of OTUs Main alpha-diversity index used are the Shannon index and the Simpson index |
| Beta diversity | Describe the differences in OTUs diversity between samples. Widely used beta-diversity index are Bray–Curtis distance, Manhattan distance and Weighted-Unifrac distance. A greater beta diversity indicates higher differences of OTUs between samples |
| Resilience | Capacity of the microbiota to return to its initial state after various external challenges (such as antibiotics) |
Fig. 2.
Schematic view of microbiome analysis by high throughput sequencing. Specific stages for bacteria and fungi appeared in blue and those for virus in green
Fig. 3.
Alpha and beta diversity for microbiome analysis. On day 0 (D0), endotracheal aspirate of patient A (EAA0) contained five different OTUs, three of which are common with patient B whose endotracheal aspirate on D0 (EAB0) contained four different OTUs. Endotracheal aspirate of patient A on D5 (EAA5) contained only two OTUs: alpha diversity has decreased. In contrast, EAB5 had five different OTUs on D5: alpha diversity has increased. EAA and EAB contained three common OTUs on D0 and only one common OTU on D5, beta diversity has increased
State of the art
Most publications studying the pulmonary microbiome are limited to a subset of its components, mainly the bacterial microbiota.
Lung bacterial microbiota in intensive care unit (ICU)
Lung microbiota and invasive mechanical ventilation
To date, most studies on the respiratory microbiota have been conducted in patients with chronic respiratory diseases such as asthma, COPD and cystic fibrosis [25–27]. Some interesting concepts have been highlighted. Studies seem to indicate: (1) that each chronic respiratory disease is associated with a different microbiota pattern or signature, (2) an association between the severity of the dysbiosis and the clinical status [6, 28].
Only a few descriptive studies have been conducted in ICU [11, 13, 14, 29–31]. One work demonstrated the extreme diversity of microbial populations within ETA collected from ventilated and colonized patients with Pseudomonas aeruginosa [29]. Bousbia et al. also observed important bacterial diversities in BAL from ICU patients under mechanical ventilation for a community-acquired pneumonia (CAP) or a VAP. A large repertoire of 146 species belonging to 7 phyla was identified, of which 73 bacterial species had never been described in patients with pneumonia [11]. Subsequently, Smith et al. specifically focused on the microbiota of 15 uninfected ventilated patients admitted to a surgical ICU with negative BAL by conventional microbiology. Same phylum were identified and the three major were Firmicutes, Bacteroidetes and Proteobacteria [30]. Most patients had profiles with a high degree of alpha diversity (species diversity within a sample of a given individual). Inter-individual variations were mostly described at the genus level [30]. However, the question of the dynamic evolution of the airway microbiota during mechanical ventilation, which is probably the most relevant study, has just been addressed in two more recent works. In 2016, Kelly et al. reported a rapidly decreasing alpha diversity after intubation which gradually dropped with prolongation of mechanical ventilation, compared to control patients (healthy, unventilated subjects). Microbiota profiles also revealed more diversity between individuals in the ventilated patients (higher beta diversity) than in control patients. This was mainly due to the dominance of a single operational taxonomic unit (OTU) in ventilated patients [31]. The same observation was made in 35 ventilated patients, for whom 111 samples were analysed. In this population, alpha diversity decreased overtime under mechanical ventilation and, interestingly, antibiotics had no influence on the airway microbiota [13]. Recently, Emonet et al. failed to observe significant intra-individual changes in diversity between two time points (day of endotracheal intubation and a subsequent sample) in VAP patients compared to controls [14]. Reasons for this discrepancy are discussed below.
Lung microbiota and acute respiratory distress syndrome (ARDS)
Beyond the specific effect of mechanical ventilation on the lung microbiome, ARDS or severe systemic inflammatory response syndrome (SIRS) can have an impact on its composition directly or through enrichment from the gut microbiome. Only few preclinical studies have investigated these aspects in critically ill patients, but the relationship between gut and lung microbiome has been well described in asthma or in cystic fibrosis and is called the “gut-lung” axis [32, 33].
In a murine model of sepsis, Dickson et al. reported an enrichment of the lung microbiome with gut bacteria, including Bacteroidales order, Enterococccus species and Lachnospiracea species. Authors concluded that the “lung microbiome was altered following experimental sepsis, remained altered for at least 5 days, and normalizes by 2 weeks”. In BAL samples from 68 patients with ARDS, Bacteroides species were observed in 33% of cases as compared to only 3% in those from healthy controls [9]. The same authors found that gut-associated bacteria in the lung microbiome was associated with the presence of ARDS in a recent study of 91 mechanically ventilated patients [10]. They thus suggested a potential undetermined common mechanism behind the gut microbiome’s role in these two common critical illnesses. Similarly, Panzer et al. observed “significant differences in the lung microbiota composition in critically ill patients blunt trauma injuries related to both smoking status and subsequent development of ARDS, as well as a marked divergence in bacterial community composition between 0 and 48 h of ICU admission” [15].
Two other studies reported a decreased alpha diversity in ARDS patients as compared to controls [12, 34]. However, the high heterogeneity of lung microbiota in ARDS patients did not allow individualization of a specific profile [34]. The Table 2 summarizes these publications.
Table 2.
Studies concerning lung microbiota of mechanically ventilated patient in ARDS acute respiratory distress syndrome
| Authors | Study design | Number of enrolled patients | Subgroup analysis | Samples | Size effect and statistical significance |
|---|---|---|---|---|---|
| Dickson et al. 2016 | Experimental and clinical study | 68 ARDS patient |
29.4% pneumonia 20.6% aspiration |
BAL on enrollment and on D3, D7 and D14 and D21 |
33% of ARDS patient vs. 3% control had a specific gut-associated member of Bacteroides genus (OTU009) in their BAL Lung microbiome is enriched with gut associated bacteria during ARDS Positive correlation between relative abundance of Bacteroides genus (OTU009) and systemic inflammation (serum TNF-α). (R2 = 0.09, p = 0.01) Positive correlation between relative abundance of Proteobacteria phylum and alveolar TNF-α, (R2 = 0.13 p = 0.003) |
| Panzer et al. 2018 | Clinical study Prospective | 30 mechanically ventilated patients after severe blunt trauma |
8 nonsmokers, 6 passive smokers, and 16 smokers 13 ARDS patients 17 non ARDS patients |
ETA on admission and 24 h after |
Association between ARDS development and lung community composition at 48 h (r2 = 0.08, p = 0.04, Univariate PERMANOVA based on a weighted UniFrac distance matrix) ARDS patients most significantly enriched for a specific taxon belonging to Enterobacteriaceae and for specific taxa also enriched at baseline in smokers (Prevotella and Fusobacterium) |
| Kyo et al. 2019 | Prospective |
47 mechanically ventilated patients: 40 with ARDS 7 controls |
22 survivors and 13 non survivors ARDS patients |
BAL within 24 h after intubation for ARDS, Serum Ang2, IL-6 IL-8 |
Increase of 16S rRNA gene copy number in ARDS patients compared to controls (3.83 × 10^6 vs. 1.01 × 10^5 copies/mL, p = 0.06) Shannon index decreased in ARDS patient compared to controls (p = 0.031) Serum level of IL-6 increased in non-survivors (567 vs. 214 pg/mL; p = 0.026) Copy number of 16S rRNA gene of Betaproteobacteria decreased in non-surviving vs surviving patient. (106 vs 104; p < 0.05) |
| Dickson et al. 2020 | Prospective | 91 critically ill ventilated patients | ARDS and non-ARDS patient | BAL within 24 h of ICU admission |
Bacterial DNA burden was greater in patients in ARDS (p = 0.014) Patients with increased lung bacterial burden had fewer ventilator-free days; HR = 0.43 95% CI (0.21–0.88) Composition of lung microbiota was distinct in ARDS patient with increased relative abundance of Enterobacteriaceae (12.5% vs. 0.8%) in unbiased regression-specimens mvabund systeme (p = 0.002) Enterobacteriaceae family is enriched in ARDS specimens (p = 0.002) Most predominant ARDS associated Enterobacteriaceae-classified taxa are Escherichia coli, Enterobacter spp, Klebsiella pneumoniae Association between presence of gut associated bacteria in the lung microbiota and the ventilator-free 28 days after admission (p = 0.003) |
| Schmitt et al. 2020 | Prospective |
30 patients 15 patients in sepsis-induced ARDS following major abdominal surgery 15 controls undergoing esophageal resection |
BAL and blood sample at ARDS onset D0, D5, D10 |
Lower alpha diversity in BAL in ARDS patients vs controls (Shannon index 3 (2;3.6) vs 1(0.5;1.5); p = 0.007) Alpha diversity index negatively correlated to length of mechanical ventilation (p = − 0.48, p = 0.009) Decrease in anaerobic bacteria tested with the log limma2 method: Prevotella spp log2 fold change 2.28 (p = 0.0033) and Veillonella spp log2 fold change 2.73 (p = 0.0002) Different microbiotic composition in the lungs of ARDS patient comparing to control (PERMANOVA R2 = 0.07; p = 0.002) The alpha-diversity index correlated with the length of stay in the intensive care unit (p = 0.015) and the need for mechanical ventilation (p = 0.009) |
MV patients mechanically ventilated patients, ARDS acute respiratory distress syndrome, BAL bronchoalveolar lavage, ETA endotracheal aspirate, D day, IL-6 interleukin 6, IL-8 interleukin 8, Ang2 angiopoietine 2
Further studies, with comparable methodologies, are required to better characterize the role of the different actors in the vicious circle of dysbiosis, inflammation and lung injury, and to determine the actual role and impact of the enrichment with gut bacteria of the lung microbiota.
Bacterial microbiota and lung nosocomial infections
The bacterial pulmonary microbiota has received little attention in the context of acute pulmonary infections, especially in patients under mechanical ventilation. Flanagan et al. were the first in 2007 to sequence 16S rRNA gene from ETA of ventilated patients with a known airway colonization with Pseudomonas aeruginosa [29]. The most prevalent bacteria belonged to the three major phyla Firmicutes, Bacteroidetes and Proteobacteria associated with other less abundant species belonging to the flora of the oropharyngeal, nasal, and gastrointestinal tracts such as Lactobacillus, Enterococcus and Veillonella. Antibiotic therapy was associated with a decreased diversity of the microbiota and a strong predominance of Pseudomonas aeruginosa (selection of colonized patients in this study). It then appears that oropharyngeal and digestive microbiota could be the source of colonizing bacteria that induce dysbiosis during mechanical ventilation.
More recently, Zakharkina et al. compared the evolution of the respiratory microbiota of ventilated patients, irrespective of whether they developed a VAP or not [13]. The greater heterogeneity of the bacterial populations in patients who developed a VAP explained the marked increase of the beta diversity as compared to the ventilated patients without a VAP. A concept seems to emerge that the bacteria responsible for VAP which represents the dominant OTU (Staphylococcus aureus, Acinetobacter Baumannii and Pseudomonas spp in this study) would reduce other bacterial communities. From results of these studies, we could also hypothesize that some non-pathogenic species have a protective barrier effect against the development of a VAP and VAP, if developed, is reduced [9]. Moreover, Bousbia et al. and Zakharkina et al. failed to identify a specific profile that could easily distinguish CAP from VAP [11, 13].
Finally, the identification of respiratory microbiota risk-markers is probably the more pertinent question. Emonet et al. have recently tried to identify metataxonomic VAP markers early during intubation and they found no significant changes of ETA microbiota between VAP patients and control patients at any point in time. However, ETA from VAP patients contained more Gammaproteobacteria class (Pseudomonas spp, Enterobacteriaceae) 3 days before VAP and less Bacilli class (Enterococcus spp, Streptococcus spp, Lactobacillus spp and Staphylococcus spp) as soon as orotracheal intubation was performed. The authors tried to use this difference to classify patients between VAP and a control group but their results have yet to be confirmed [14]. More details concerning main studies concerning mechanically ventilated patients are presented in Table 3. Further studies are needed for a deeper insight into the pathophysiological continuum that leads from intubation to airway colonization and eventually VAP.
Table 3.
Studies concerning lung microbiota of mechanically ventilated patient and ventilator-associated pneumonia
| Authors | Study design | Number of enrolled patients | Subgroups | Samples | Size effect and statistical significance | Negative result |
|---|---|---|---|---|---|---|
| Smith et al. 2016 | Prospective study | MV patients in a surgical ICU ventilated more than 36 h |
5 MV patient with suspected VAP 10 patients without VAP |
BAL after 36 h of mechanical ventilation or in case of VAP suspicion |
55 total genera identified in the common microbiome samples 20 genera with abundance > 1% |
No comparison between groups |
| Bousbia et al. 2012 |
185 pneumonia patients 25 control patient |
32 CAP 106 VAP 22 NV-ICU pneumonia 25 aspiration pneumonia |
ETA on admission and at 24 h |
93/106 VAP patients had a positive BAL by molecular assays 48 had an association of two type of microorganisms between bacteria virus and fungi 146 different bacteria belonging to seven different phyla composed the bacterial lung microbiota of patients Fungal microbiota from pneumonia patients showed the presence of 22 different new fungal species belonging to 2 phyla not previously identified Bacilli and Gammaproteobacteria were dominant in patients, whereas anaerobic bacteria related to Bacteroidia and Clostridia were dominant in controls n bacterial microbiota (p < 0.01) |
No specific pattern depending on the type of pneumonia | |
| Kelly et al. 2016 | Prospective study |
15 MV patients from medical intensive care unit versus healthy unventilated patients 4 patients with CAP/HAP |
4 patients with CAP/HAP 4 patients with aspiration at enrollment 4 patients with VAP |
ETA and OS within 24 h of orotracheal intubation and every 72 h after |
Lower alpha diversity in intubated patients than healthy controls (p = 2.3 × 10−13) Alpha diversity decreased with time in URT of VAP patient (Shannon index = 4 on day 0 versus Shannon index = 3,1 beyond day 0: p = 0.0015) Alpha diversity decreased with time in LRT of VAP patient (Shannon index = 3 on day 0 versus Shannon index = 1,9 beyond day 0: p = 0.13) Higher beta diversity in MV patients’ group than in control group Lower alpha diversity in LRT of VAP patient compared to MV patient with prolonged courses of intubation without infection (p = 0.08) |
|
| Zakharkina et al. Thorax 2017 | Post hoc analysis of patients initially included in an international multicentre prospective observational cohort study |
11 patients with VAP 18 patients without VAP 6 HAP/CAP |
BAL for VAP suspicion ETA at ICU admission and twice a week after admission |
Association between duration of MV and decreased in Shannon diversity; fixed effect regression coefficient (β): − 0.03 CI 95% [− 0.05; − 0.005] Statistical difference in Weighted Unifrac distance between VAP patient and control patient without colonized airways 0.4 (0.25; 0.5) vs. 0.65 (0.5;0.85), p = 0.02 Increase of β diversity for VAP patients is statistically higher analyzed by PCo analysis (p = 0.03) Acinetobacter, Pseudomonas, Staphyloccocus, and Burkholderia are genera correlated with changes in β diversity |
No statistical difference in Weighted Unifrac distance between VAP patient and colonized patient | |
| Emonet et al. 2019 | Case control study nested in a prospective single center cohort study | MV adult patient intubated less than 24 h in polyvalent ICU |
16 late onset confirmed VAP patient 38 matched control |
- ETA and OPS at five time points during MV D0 (of intubation), D3 (3 days after intubation, DVAP-3 (3 days before VAP) DVAP (day of VAP diagnosis), DVAP + 3 (3 days after VAP) |
Progressive increase in Proteobacteria (25% on D0 vs. 55% on DVAP + 3) and decreased in Firmicutes (40% vs. 30%) in OS and ETA of VAP patient The absolute abundance of the class Bacilli was significantly higher in ETA from controls at D0. At D0 class Bacilli had a relative abundance > 12% in 82.8% of controls but only in 18.8% of VAP patients. (p < 0.0001) Quantity of human DNA in ETA are significantly higher for VAP patients than in controls. A cutoff of 124.7 ng/μL allowed to differentiate VAP vs controls with a sensitivity of 94.1% and a specificity of 83.3% |
General trend of changes in β-diversity during MV are not different between VAP patients and control No significant changes of ETA or OS microbiota between VAP patient and control patient at any time point No lower respiratory tract microbiota markers of VAP clearly identified |
MV patients mechanically ventilated patients, BAL bronchoalveolar lavage, ETA endotracheal aspirate, OS oropharyngeal swab, URT upper respiratory tract, LRT lower respiratory tract, VAP ventilator associated pneumonia, CAP community acquired pneumonia, HAP hospital acquired pneumonia, D day
Lung mycobiota
Limiting the study of the microbiome to that of the composition of bacterial microbiota remains restrictive because of the important interaction of bacteria with viruses, fungi, and Archaea. Recently a few studies have evaluated the pulmonary mycobiota using high-throughput sequencing [35, 36]. The study of pulmonary mycobiota in healthy individuals revealed many environmental fungi including Aspergillus sp., mold (Penicillium and Cladosporium) and yeasts belonging to the two main phyla Ascomycota (Candida) and Basidiomycota (Malassezia) [35, 37]. In contrast to the bacterial microbiota, the fungal populations identified in the URT and the LRT were very distinct in healthy subjects [38]. The study of dysbiotic mycobiota and its correlation with pulmonary diseases is in its infancy, showing a restriction of diversity [37].
In most media, an interplay between bacterial and fungal communities exists, and the evolution of one community induces a modification of the other. An airway colonization with some yeasts, notably the genus Candida, has been observed in 25 to 50% of patients after a few days of invasive mechanical ventilation [39, 40]. This colonization has been statistically associated with the occurrence of bacterial lung infections [39, 41, 42]. It is therefore plausible that bacteria-fungi interactions play a significant role in the pathogenesis of VAP even if the precise mechanisms underlying interactions between fungi, bacteria and host remain poorly understood. In a multicentre study of immunocompetent critically ill patients over a 4-year period, 214 patients (26%) with a respiratory tract colonization were matched and compared to 214 unexposed patients. Bronchial Candida colonization was found to be an independent risk factor for Pseudomonas pneumonia (9 vs 4,8%) with adjusted odds ratio (aOR) = 2.22 [1,00; 4,92] (p = 0.049) without impact on mortality [39]. One possible mechanism behind these findings is an alteration in bacterial phagocytosis induced by the presence of C. albicans. Roux et al. showed in rats, that C. albicans airway colonization elicited a Th1-Th17 immune response that favored the development of bacterial pneumonia via the inhibition of bacterial phagocytosis by alveolar macrophages [43]. They also showed in vitro that C. albicans impeded alveolar macrophage ROS production and was correlated in vivo with an increase of P. aeruginosa pneumonia prevalence in rats [44]. Tan et al. showed that the same C. albicans colonization favored A. baumannii pneumonia in rats [45]. The same team also identified Candida spp. airway colonization as an independent risk factor for A. baumannii VAP in mechanically ventilated ICU patients [42].
To date, only two studies have focused specifically on the mycobiota of ventilated patients with pneumonia in ICUs. Bousbia et al., in their pioneering study performed in 31 patients with pneumonia and in 6 controls, identified 22 fungal species belonging to 2 phyla: Ascomycota and Basidiomycota. Distinct patterns emerged with some classes found specifically in cases of VAP, CAP or controls. No specific association between the mycobiota and bacterial species was observed [11]. In the study of Krause et al. on the mycobiota of ICU patients, a decrease of fungal diversity was observed in each ventilated patient with pneumonia. Candida was frequently found in pulmonary mycobiota, representing 75% of fungi species [1].
To understand how fungi contribute to the healthy equilibrium in the respiratory tract will need an important work, notably by analysing lung mycobiota with high throughput sequencing.
Virome
Viruses are the most abundant and diverse entities and virome includes eukaryotic viruses (eukaryotic virome) and bacteriophages (prokaryotic virome). Eukaryotic viruses infect cells of the human host. They can be pathogenic, causing acute or chronic infections. They can also be reactivated in connection with a pathological state (Cytomegalovirus, Herpes simplex virus) and can persist without any pathology. Their interaction with the immune system seems to play an important role in the development of respiratory diseases [46] although no causal link can be asserted. The second group of viruses, the bacteriophages, infects bacterial cells. They represent most of the viruses identified within the human respiratory tract. It therefore appears essential to integrate all components of the virome to obtain a comprehensive view of the human microbiome [47].
Virome, invasive mechanical ventilation and VAP
The impact of mechanical ventilation on the respiratory virome remains unclear and rarely studied. Viruses of the Herpesviridae, Paramyxoviridae and Picornaviridae families have been identified in all ventilated ICU patients [11]. The human virome is strongly altered in ICU patients, due to viral reactivation especially among the herpes group. Viral reactivation in the respiratory tract of septic patients with Epstein-Barr Virus (EBV) (14.6%), HSV type 1 (33–68.3%) and CMV (17%) have been described [2, 48]. This virome dysbiosis may be associated with a longer stay in ICU and in mortality [49]. In a population of 93 mechanically ventilated patients with suspected VAP, cytomegalovirus (CMV) pulmonary infection that was defined as replicating CMV in the BAL was associated with increased duration of invasive ventilation, ICU length of stay and mortality, whereas HSV replication in the lung was not [2]. Furthermore, an increased rate of bacterial or fungal superinfections was observed in ICU patients with CMV reactivation [50]. The role of viruses in the occurrence of VAP and their impact on patient outcome depends on the virus [51]. Most of these data are obtained from targeted PCR. Thus, metagenomic studies are needed to comprehensively describe the lung virome evolution during invasive ventilation in ICUs, and to determine the role and the mechanisms of viral dysbiosis on patient prognosis and VAP development.
Perspectives
As discussed above, high-throughput sequencing and metagenomics analysis present some limitations. Notably, methodological heterogeneity between studies restricts comparability. Efforts must be made to move towards procedure standardization and a consensual definition of a robust methodology for lung microbiome research. Priorities reside at different levels. Control of all sources of contamination and the use of adequate positive and negative controls are the first step. Comparative methodological studies have to be performed to optimize sampling methods to determine the most efficient DNA extraction procedure and the hypervariable region of the 16S rRNA gene which confers the most reproductive and accurate assessment of the lung bacterial microbiota. In the specific field of intensive care and mechanical ventilation, consistent sample choices must be made for each clinical issue, between oropharyngeal swabs from the upper respiratory tract and both ETA or BAL from the lower respiratory tract. At best, the method of nucleic acid extraction should allow a study of bacterial, viral, and fungal populations. Finally, bioinformatics development of standardized pipelines and databases would improve comparability between studies.
Beyond technical aspects many pathophysiological questions need to be answered. To better understand the pathogenesis of VAP, a complex interaction between the microbiome and the host response needs to be explored. In addition, studying the density of the lung microbiota in combination to 16S rRNA gene analysis is likely more pertinent than just analysing its taxonomical composition. This can be an explanation for the lack of identification of specific patterns for VAP patients comparing to ventilated patients without VAP [14, 52]. To measure markers of host response could be an option. Previous studies have also shown good discriminative power of interleukin-8 and interleukin-1β measured in BAL or of neutrophil-derived proteases, such as human neutrophil elastases, released in excess in BAL of VAP patients, for demarcating VAP patients from suspected VAP patients [53, 54]. More recently, Emonet et al. who failed to identify a specific profile of the lung microbiota among VAP patients, found a much higher human DNA count in these patients [14]. This approach has also been used in ARDS, which is one of the most frequent complication of respiratory infections [12]. This apparent discrepancy does not rule out the link between changes in the lung microbiome composition and subsequent occurrence of VAP but highlights the importance of studying lung microbiome and host response simultaneously.
Similarly, new insights into microbial roles in VAP pathogenesis must be found from microbiome studies.
The concept of VAP due to a single bacterial pathogen could be replaced by a more hypothetic complex model in which VAP would be induced by or associated with a dysbiosis. As discuss above, changes in lung microbiota of mechanically ventilated patients are characterized by a decreased relative abundance of the two major phyla Bacteroidetes and Firmicutes, composed of non-pathogenic anaerobes, in most clinical situations: either mechanical ventilation, VAP or ARDS [14, 31, 34]. These commensal bacteria play a key role in maintaining lung immune homeostasis and contribute to immune tolerance [55]. The most convincing evidence that the lung microbiota reciprocally affects local immune response is that specific lung bacteria such as Prevotella spp or Veillonella.spp were associated with an increased number of lymphocytes in BAL, Th17 cell-mediated lung inflammation and an increased TLR4 response by alveolar macrophages [56].
Even if this lung microbiome dysregulation is well described in ARDS with an enrichment with gut associated bacteria including Enterobacteriaceae, the exact process of bacterial translocation to the lung is not clearly understood [15]. To date such dysbiosis is less well characterized in VAP patients without ARDS complication [14]. In the context of critical illness and mechanical ventilation, many factors may contribute to lung dysbiosis such as hyperoxia [57]. The Fig. 4 synthesizes the various factors that may be implicated in such lung dysbiosis (see also the online supplementary material). Lung dysbiosis could even be only a marker of a generalized dysbiosis associated to critical illness with no direct role in the occurrence of VAP [58]. On the other hand, the proven responsibility of intestinal dysbiosis in an increased susceptibility to infections gives support to the role of lung dysbiosis in VAP pathogenesis. Gut microbiota contributes to protection against pathogens through nutrients competition, production of antibacterial peptides or other antibacterial factors by epithelial cells [59]. Commensal gut microbiota also modulates systemic immunity using several mechanisms linked to its structural components otherwise known as microbe-associated molecular patterns (MAMPs) or microbial metabolites, which can translocate from the gut to systemic circulation and to the lung and induce both pro and anti-inflammatory responses [60]. For example, direct interactions between segmented filamentous bacteria can enhance mucosal immunity by up regulating T helper 17 (Th17) cells in both the gut and in the lung. Gut metabolite desaminotyrosine enhances clearance of respiratory viruses by inducing a type 1 interferon response [60]. More recently, Wilmore and colleagues have shown that enriching the microbiota with members of Proteobacteria phylum considered as commensal, led to T cell-dependent increases in serum IgA levels which specifically targeted a restricted number of pathogens that translocate from the gut [61]. Microbiota is involved in the systemic host-defense against sepsis and two hypotheses emerged: i) shifts in microbiota composition potentially predispose patients to a state of immunosuppression and ii) systemic translocation of opportunistic gut bacteria increased the risk of organ failure [62, 63]. These data thus support the idea that lung dysbiosis with gut-associated bacteria may lead to local immunosuppression and lung dysfunction allowing VAP to occur.
Fig. 4.
Influential factors on lung dysbiosis in ventilated patients. Lung microbiome can be altered by a variety of factors, either intrinsic or extrinsic, when intubation and mechanical ventilation are in place. This figure summarizes the main influential factors potentially involved
These hypotheses have to be tested in future studies. If proven correct, they will broaden our understanding of the pathophysiology of VAP and will allow for the identification of potential therapeutic or preventive targets.
Clinical applications
Although the efficacy of sampling and data processing is improving, applications of microbiome analysis in the clinical settings for medical decisions are not yet developed. Nevertheless, development of real-time sequencing technologies could influence diagnostic methods through faster and accurate species identification.
Capturing temporal dynamics and defining dysbiosis signatures and thresholds of microbial abundance are key to make a difference between airway colonization and VAP. In addition, early detection of dysbiosis profiles with a high risk of VAP would go a long way to improve clinical practices. Early targeted VAP antibiotic therapy in anticipation of conventional microbiological analysis in selected patients, instead of empiric use of broad-spectrum antibiotics, would be a step forward in the fight against antimicrobial resistance. To enable clinical applications, more longitudinal and comparative studies are now required. Much better than targeted VAP antibiotic therapy, early identification of patients with dysbiosis profiles associated with a high risk of VAP could lead to targeted preventive strategies. A proposition regarding the methodology for such studies is detailed in the online supplementary material.
Future research in metagenomics longitudinal analysis in the field of intensive care will also have to focus on microbial interactions to identify bacterial, fungal and viral markers associated with an increased risk of VAP development. Focusing in parallel on the host response will help better define the profiles of patients. Overall, a better understanding of pathophysiological insights can help us to define targeted interventions on the microbiota, the mycobiote and the virome, including prokaryotic and eukaryotic viruses. Treating the dysbiosis using probiotics or prebiotics or administration of preventive aerosolized antibiotics to patients at high risk of VAP could be an approach (see online supplementary material). Clinical benefits could also be obtained by other approaches such as antiviral therapy or phagotherapy.
Conclusion
All these data underline the necessity of a global approach to the human respiratory microbiome, including, not only the bacterial, but also the fungal microbiota and the virome. The analysis of a single type of microbial population can result in a truncated and therefore erroneous view of the impact of the microbiome on health. In the future, priority must be given to reaching an agreed methodology and to performing longitudinal studies for a better understanding of VAP pathogenesis. Developments in high-throughput sequencing and bioinformatic analysis methods, although complex and expensive, will enable us to explore the global human respiratory microbiome and to get closer to clinical applications for preventive and therapeutic strategies against nosocomial pneumonia and ARDS.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Julien Do Vale, master 2 student in our institution, for sharing his knowledge of the lung virome and Mrs. Jan Aupers for technical assistance.
Author contributions
MF reviewed and appraised the literature for methodology of samples processing and previous results concerning lung bacterial microbiota. She drafted the manuscript and approves the final version. JDR as expert in nosocomial lung infections appraised the literature and clarified clinical perspectives and potential clinical applications. He revised the article and approves the final version. DR conceived the article, reviewed the literature concerning lung virome and mycobiota in the field of intensive care, revised the article and approves the final version.
Funding
MF is the recipient of the 2017 MSD/Société de Réanimation de Langue Française Award and of the 2017 Young Investigator award from the Société de Pathologie Infectieuse de Langue Française. JDR is the recipient of the European Society of Intensive Care Medicine 2014 Established Investigator award. DR is the recipient of the European Society of Intensive Care Medicine 2016 Basic Science award.
Compliance with ethical standards
Conflict of interest
DR received personal fees from Astellas, JDR received travel support by Fisher and Paykel. MF has no conflict of interest.
Consent to participate
Images come from the medical file of a patient who gave her consent for publication respecting anonymity NA.
Consent for publication
Images come from the medical file of a patient who gave her consent for publication respecting anonymity NA.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Krause R, Halwachs B, Thallinger GG, et al. Characterisation of Candida within the mycobiome/microbiome of the lower respiratory tract of ICU Patients. PLoS ONE. 2016;11:e0155033. doi: 10.1371/journal.pone.0155033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coisel Y, Bousbia S, Forel J-M, et al. Cytomegalovirus and Herpes Simplex virus effect on the prognosis of mechanically ventilated patients suspected to have ventilator-associated pneumonia. PLoS ONE. 2012;7:e51340. doi: 10.1371/journal.pone.0051340. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassis CM, Erb-Downward JR, Dickson RP, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabrera-Rubio R, Garcia-Nunez M, Seto L, et al. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2012;50:3562–3568. doi: 10.1128/JCM.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickson RP, Erb-Downward JR, Freeman CM, et al. Bacterial topography of the healthy human lower respiratory tract. mBio. 2017;8:e02287–e2316. doi: 10.1128/mBio.02287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson RP, Erb-Downward JR, Freeman CM, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12:821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson RP, Schultz MJ, van der Poll T, et al. Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med. 2020;201:555–563. doi: 10.1164/rccm.201907-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousbia S, Papazian L, Saux P, et al. Repertoire of intensive care unit pneumonia microbiota. PLoS ONE. 2012;7:e32486. doi: 10.1371/journal.pone.0032486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kyo M, Nishioka K, Nakaya T, et al. Unique patterns of lower respiratory tract microbiota are associated with inflammation and hospital mortality in acute respiratory distress syndrome. Respir Res. 2019;20:246. doi: 10.1186/s12931-019-1203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zakharkina T, Martin-Loeches I, Matamoros S, et al. The dynamics of the pulmonary microbiome during mechanical ventilation in the intensive care unit and the association with occurrence of pneumonia. Thorax. 2017;72:803–810. doi: 10.1136/thoraxjnl-2016-209158. [DOI] [PubMed] [Google Scholar]
- 14.Emonet S, Lazarevic V, Leemann Refondini C, et al. Identification of respiratory microbiota markers in ventilator-associated pneumonia. Intensive Care Med. 2019;45:1082–1092. doi: 10.1007/s00134-019-05660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panzer AR, Lynch SV, Langelier C, et al. Lung microbiota is related to smoking status and to development of acute respiratory distress syndrome in critically ill trauma patients. Am J Respir Crit Care Med. 2018;197:621–631. doi: 10.1164/rccm.201702-0441OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathieu E, Escribano-Vazquez U, Descamps D, et al. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front Physiol. 2018;9:1168. doi: 10.3389/fphys.2018.01168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Man WH, de Steenhuijsen Piters WAA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol. 2019;20:1279–1290. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- 19.Morris A, Beck JM, Schloss PD, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kellermayer R, Dowd SE, Harris RA, et al. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 2011;25:1449–1460. doi: 10.1096/fj.10-172205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickson RP, Erb-Downward JR, Huffnagle GB. Homeostasis and its disruption in the lung microbiome. Am J Physiol Lung Cell Mol Physiol. 2015;309:L1047–1055. doi: 10.1152/ajplung.00279.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanski I, von Hertzen L, Fyhrquist N, et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci USA. 2012;109:8334–8339. doi: 10.1073/pnas.1205624109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Op De Beeck M, Lievens B, Busschaert P, et al. Comparison and validation of some its primer pairs useful for fungal metabarcoding studies. PLoS ONE. 2014;9:e97629. doi: 10.1371/journal.pone.0097629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J, Xiong L, Zhang X, et al. The role of lower airway dysbiosis in asthma: dysbiosis and asthma. Mediators Inflamm. 2017;2017:3890601. doi: 10.1155/2017/3890601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H-Y, Zhang S-Y, Yang W-Y, et al. Oropharyngeal and sputum microbiomes are similar following exacerbation of chronic obstructive pulmonary disease. Front Microbiol. 2017;8:1163. doi: 10.3389/fmicb.2017.01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sibley CD, Grinwis ME, Field TR, et al. Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS ONE. 2011;6:e22702. doi: 10.1371/journal.pone.0022702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flanagan JL, Brodie EL, Weng L, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol. 2007;45:1954–1962. doi: 10.1128/JCM.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith AD, Zhang Y, Barber RC, et al. Common lung microbiome identified among mechanically ventilated surgical patients. PLoS ONE. 2016;11:e0166313. doi: 10.1371/journal.pone.0166313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly BJ, Imai I, Bittinger K, et al. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome. 2016;4:7. doi: 10.1186/s40168-016-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. 2015;17:592–602. doi: 10.1016/j.chom.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madan JC, Koestler DC, Stanton BA, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. mBio. 2012;3:e00251–e312. doi: 10.1128/mBio.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt FCF, Lipinski A, Hofer S, et al. Pulmonary microbiome patterns correlate with the course of disease in patients with sepsis-induced ARDS following major abdominal surgery. J Hosp Infect. 2020;105:438–446. doi: 10.1016/j.jhin.2020.04.028. [DOI] [PubMed] [Google Scholar]
- 35.Charlson ES, Diamond JM, Bittinger K, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delhaes L, Monchy S, Fréalle E, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS ONE. 2012;7:e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsland BJ, Gollwitzer ES. Host–microorganism interactions in lung diseases. Nat Rev Immunol. 2014;14:827–835. doi: 10.1038/nri3769. [DOI] [PubMed] [Google Scholar]
- 38.Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Path. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azoulay E, Timsit J-F, Tafflet M, et al. Candida colonization of the respiratory tract and subsequent Pseudomonas ventilator-associated pneumonia. Chest. 2006;129:110–117. doi: 10.1378/chest.129.1.110. [DOI] [PubMed] [Google Scholar]
- 40.El-Ebiary M, Torres A, Fàbregas N, et al. Significance of the isolation of Candida species from respiratory samples in critically ill, non-neutropenic patients. an immediate postmortem histologic study. Am J Respir Crit Care Med. 1997;156:583–590. doi: 10.1164/ajrccm.156.2.9612023. [DOI] [PubMed] [Google Scholar]
- 41.Eggimann P, Garbino J, Pittet D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect Dis. 2003;3:685–702. doi: 10.1016/S1473-3099(03)00801-6. [DOI] [PubMed] [Google Scholar]
- 42.Tan X, Zhu S, Yan D, et al. Candida spp. airway colonization: a potential risk factor for Acinetobacter baumannii ventilator-associated pneumonia. Med Mycol. 2016;54:557–566. doi: 10.1093/mmy/myw009. [DOI] [PubMed] [Google Scholar]
- 43.Roux D, Gaudry S, Khoy-Ear L, et al. Airway fungal colonization compromises the immune system allowing bacterial pneumonia to prevail. Crit Care Med. 2013;41:e191–199. doi: 10.1097/CCM.0b013e31828a25d6. [DOI] [PubMed] [Google Scholar]
- 44.Roux D, Gaudry S, Dreyfuss D, et al. Candida albicans impairs macrophage function and facilitates Pseudomonas aeruginosa pneumonia in rat. Crit Care Med. 2009;37:1062–1067. doi: 10.1097/CCM.0b013e31819629d2. [DOI] [PubMed] [Google Scholar]
- 45.Tan X, Chen R, Zhu S, et al. Candida albicans airway colonization facilitates subsequent Acinetobacter baumannii pneumonia in a rat model. Antimicrob Agents Chemother. 2016;60:3348–3354. doi: 10.1128/AAC.02180-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregory AC, Sullivan MB, Segal LN, Keller BC. Smoking is associated with quantifiable differences in the human lung DNA virome and metabolome. Respir Res. 2018;19:174. doi: 10.1186/s12931-018-0878-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clokie MRJ, Millard AD, Letarov AV, Heaphy S. Phages in nature Bacteriophage. 2011;1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Libert N, Bigaillon C, Chargari C, et al. Epstein-Barr virus reactivation in critically ill immunocompetent patients. Biomed J. 2015;38:70–76. doi: 10.4103/2319-4170.132905. [DOI] [PubMed] [Google Scholar]
- 49.LImaye ABM. Cytomegalovirus reactivation in critically ill immunocompetent patients. JAMA. 2008;300:413–422. doi: 10.1001/jama.300.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalil AC, Florescu DF. Prevalence and mortality associated with cytomegalovirus infection in nonimmunosuppressed patients in the intensive care unit. Crit Care Med. 2009;37:2350–2358. doi: 10.1097/CCM.0b013e3181a3aa43. [DOI] [PubMed] [Google Scholar]
- 51.Chiche L, Forel J-M, Papazian L. The role of viruses in nosocomial pneumonia. Curr Opin Infect Dis. 2011;24:152–156. doi: 10.1097/QCO.0b013e328343b6e4. [DOI] [PubMed] [Google Scholar]
- 52.Bos LDJ, Kalil AC. Changes in lung microbiome do not explain the development of ventilator-associated pneumonia. Intensive Care Med. 2019;45:1133–1135. doi: 10.1007/s00134-019-05691-1. [DOI] [PubMed] [Google Scholar]
- 53.Conway Morris A, Kefala K, Wilkinson TS, et al. Diagnostic importance of pulmonary interleukin-1 and interleukin-8 in ventilator-associated pneumonia. Thorax. 2010;65:201–207. doi: 10.1136/thx.2009.122291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkinson TS, Conway Morris A, Kefala K, et al. Ventilator-associated pneumonia is characterized by excessive release of neutrophil proteases in the lung. Chest. 2012;142:1425–1432. doi: 10.1378/chest.11-3273. [DOI] [PubMed] [Google Scholar]
- 55.O’Dwyer DN, Dickson RP, Moore BB. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J Immunol. 2016;196:4839–4847. doi: 10.4049/jimmunol.1600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segal LN, Clemente JC, Tsay J-CJ, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashley SL, Sjoding MW, Popova AP, et al. Lung and gut microbiota are altered by hyperoxia and contribute to oxygen-induced lung injury in mice. Sci Transl Med. 2020;12:eaau9959. doi: 10.1126/scitranslmed.aau9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mukherjee S, Hanidziar D. More of the gut in the lung: how two microbiomes meet in ARDS. Yale J Biol Med. 2018;91:143–149. [PMC free article] [PubMed] [Google Scholar]
- 59.Kim S, Covington A, Pamer EG. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev. 2017;279:90–105. doi: 10.1111/imr.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haak BW, Prescott HC, Wiersinga WJ. Therapeutic potential of the gut microbiota in the prevention and treatment of sepsis. Front Immunol. 2018;9:2042. doi: 10.3389/fimmu.2018.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilmore JR, Gaudette BT, Gomez Atria D, et al. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe. 2018;23:302–311.e3. doi: 10.1016/j.chom.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krezalek MA, DeFazio J, Zaborina O, et al. The shift of an intestinal “microbiome” to a “pathobiome” governs the course and outcome of sepsis following surgical injury. Shock. 2016;45:475–482. doi: 10.1097/SHK.0000000000000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klingensmith NJ, Coopersmith CM. The gut as the motor of multiple organ dysfunction in critical illness. Crit Care Clin. 2016;32:203–212. doi: 10.1016/j.ccc.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.