Abstract
Objective
Exosome-encapsulated microRNAs (miRNAs) are being considered as either diagnostic or predictive markers in different types of diseases. Here, we discussed the effects of exosome-encapsulated miR-127-3p from bone marrow-derived mesenchymal stem cells (BM-MSCs) on osteoarthritis (OA).
Methods
BM-MSCs and primary chondrocytes were isolated from Sprague Dawley rats. IL-1β was utilized to treat chondrocytes to mimic an OA in vitro model, and exosomes extracted from BM-MSCs were utilized to treat chondrocytes so as to verify their protective effects on OA. Through online website prediction and experiments confirmation, we found the most significantly enriched miRNA in exosomes and elucidated the effect of this miRNA on the therapeutic effect of exosomes by interfering with its expression. Also, the genes targeted by the miRNA and the involved pathway were also found through bioinformatics analysis and experimental research, thereby probing into the protective mechanism of exosomes on chondrocytes.
Results
Exosomes derived from BM-MSCs restricted the IL-1β-induced chondrocytes damage. miR-127-3p was found to be enriched in exosomes, and the protective effect of exosomes was reversed by miR-127-3p inhibition. miR-127-3p targeted CDH11, and overexpressed CDH11 in chondrocytes weakened the therapeutic effect of exosomes. IL-1β treatment resulted in the activation of the Wnt/β-catenin pathway in chondrocytes. Exosomes treatment could inhibit the activation of this pathway, and overexpressed CDH11 reversed the inhibitory effect of exosomes on this pathway.
Conclusion
This study suggests that exosomal miR-127-3p derived from BM-MSCs inhibits CDH11 in chondrocytes, thereby blocking the Wnt/β-catenin pathway activation and relieving chondrocyte damage in OA.
Keywords: osteoarthritis, exosome, microRNA-127-3p, bone marrow-derived mesenchymal stem cells, CDH11, Wnt/β-catenin pathway
Introduction
Osteoarthritis (OA) is the most frequent form of arthritic disease around the world, resulting in destruction of joint tissues and debilitating pain.1 The main risk factors for OA development are manifold with the local and systemic reasons, including genetics, obesity, age, gender, previous joint trauma, as well as other underlying diseases.2 This disease is related to numerous comorbidities, which considerably lowers the quality of life for OA patients.3 In clinical, the available interventions for OA are confined to palliative care, and no pharmacologic option has been found to impact disease pathogenesis at present.4 Thus, understanding the mechanisms underlying OA can promote the development of novel therapies to meet the future clinical needs.
Mesenchymal stem cells (MSCs) are multipotent fibroblast-like cells (FLS) with the functions of self-renewal and multilineage differentiation, which are considered as a possible therapeutic regimen for bone disorders.5 MSCs have been utilized for OA treatment because of their chondrogenic and immunomodulatory potentials, along with regenerative properties.6 Exosomes are extracellular vesicles involved in intercellular communication with the capability of transferring cargo molecules (eg protein, lipids and nucleic acids) at the near and far target sites.7 Exosomes exert functions in various physiological and pathological processes through mediating cell-cell communication, and they also participate in numerous diseases, including OA.8 Bone marrow-derived mesenchymal stem cells (BM-MSCs)-released exosomes have been reported to be able to protect cartilage against damage and mitigate knee OA pain in a rat model.9 Evidence has emphasized that microRNA (miRNA) and exosomes are beneficial for early diagnosis and are the useful treatment of OA.10 miRNAs invariably serve as either onco‐miRNAs or tumor‐suppressor miRNAs for different biological processes, and their roles are primarily based on its downstream genes.11 Recent studies have revealed that miRNAs play a role in OA development and progression.12,13 More specifically, emerging articles have demonstrated that the inhibitory role of miR‐127‐5p in OA progression.14–16 Nevertheless, the functions of miR‐127‐3p have not yet been elucidated. miRNAs are reported to modulate a series of biological processes via directly interacting with their target message RNAs (mRNAs).17 In this current research, Cadherin-11 (CDH11) was determined to be a downstream target of miR-127-3p through online website prediction. CDH11 has been indicated to be highly expressed in many types of fibrotic diseases, including arthritis.18 Moreover, depletion of CDH11 prevents inflammatory cell infiltration and cartilage erosion by the pannus in collagen-induced arthritis mice.19 All these evidences lead us to expound on the value of exosomal miR‑127-3p from BM-MSCs in OA progression with the involvement of CDH11.
Materials and Methods
Ethics Statement
All animal experiments were approved by the ethics committee of The Second People’s Hospital of Hefei. All animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH Publication No.85–23, revised 1996). Great efforts were made to relieve the pain of animals.
Experimental Animals, and Isolation and Culture of Primary Cells
Three 5-week-old and three 1-week-old Sprague Dawley (SD) rats were available from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China).
As reported before,20 rat BM-MSCs were isolated from the femur of SD rats. In short, three 5-week-old rats were euthanized with an overdose of pentobarbital sodium (200 mg/kg), and the femurs were separated on a sterile operating table. The bone marrow was washed by injecting with a small amount of Dulbecco’s modified Eagle medium (DMEM, Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) into the femur. Next, the cell suspension was transferred to a 50 mL test tube containing 10 mL medium and centrifuged at 450 g for 5 minutes to remove the supernatant and resuspended in 10 mL medium. The nucleated cells were seeded into DMEM containing 10% FBS and cultured at 37°C with 5% CO2. The medium was renewed every 3 days. The cells at passage 3–5 were selected for the subsequent experiments. For phenotype characterization, BM-MSCs were dyed with antibodies to CD29, CD44, CD34, and CD45 (Abcam, Cambridge, MA, USA) or appropriate isotype control immunoglobulin G (IgG), and then analyzed by flow cytometry (FACSCanto™, BD Biosciences, San Jose, CA, USA). BM-MSCs were cultured with BM-MSC osteogenic differentiation medium and adipogenic differentiation medium (Cyagen Biosciences Inc, Guangzhou, China). Alizarin red staining kit and oil red O staining kit (LMAIBio, Shanghai, China) were utilized to confirm the differentiation abilities.
For the isolation of chondrocytes, three 1-week-old SD rats were euthanized by intraperitoneal injection of an overdose of pentobarbital sodium (200 mg/kg) to collect the proximal tibial and distal femoral articular cartilage. The cartilage samples were rinsed with sterile phosphate-buffered saline (PBS) for three times, and then isolated from adipose and other bone tissues. After that, the fresh cartilage samples were sliced into 1 mm3 pieces and detached in DMEM/F12-dissolved collagenase II (Gibco) at 37°C for 4–6 hours. Subsequently, the cells were transferred to a tube (15 mL) and centrifugated for removing the supernatant. The mixture was incubated at 37°C with 5% CO2 in a culture bottle (25 cm2) with 3 mL DMEM containing 10% FBS and 1% penicillin/streptomycin. Chondrocytes at passages 1–3 were utilized in our subsequent studies.
Extraction and Identification of Exosomes Derived from BM-MSCs
DMEM with 10% FBS was ultracentrifuged at 100,000 g overnight at 4°C to remove the exosomes in serum. BM-MSCs were cultured in this medium for 48 hours, and the supernatant was harvested. The supernatant was centrifuged at 4°C at 2000 g for 15 minutes to remove the cell debris, and then the supernatant was centrifuged at 4°C at 100,000 g for 1 hour. The precipitate was resuspended in PBS and centrifuged at 100,000 g for 1 hour at 4°C to remove the supernatant, and the precipitate was stored at −80°C for subsequent experiments.
The morphology of exosomes was viewed under a 100-kV transmission electron microscope (TEM, H-7000FA, Hitachi Co. Ltd., Tokyo, Japan). Zetasizer Nano (Malvern Instruments Ltd., Worcestershire, UK) was utilized for analyzing the particle size distribution of exosomes. Antibodies against CD63 (ProteinTech, Chicago, IL, USA) and CD9 (Abcam) were utilized to identify the expression of exosomal surface marker proteins by Western blot assay.
Uptake of Exosomes from BM-MSCs
The red fluorescent dye PKH26 was adopted to label exosomes as per the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO, USA). The labeled exosomes were suspended in 20 μg/mL PBS and co-cultured with rat chondrocytes in serum-free medium at 37°C for 12 hours or 24 hours, followed by fixing with 4% paraformaldehyde. After that, the nucleus was dyed with Hoechst 33,342 (MedChemExpress, Monmouth Junction, NJ, USA). The uptake of exosomes was viewed with a confocal laser scanning microscope (Leica, Heidelberg Germany).
Cell Treatment and Grouping
BM-MSCs were assigned into three groups: untreated BM-MSCs (the extracted exosomes were named as Exo), negative control (NC) inhibitor group (transfected with miR-127-3p NC inhibitor, the extracted exosomes were named as Exo-NC), and miR-127-3p inhibitor group (transfected with miR-127-3p inhibitor, the extracted exosomes were named as Exo-inhibitor).
Chondrocytes were divided into nine groups: control group (chondrocytes without any treatment), OA (IL-1β treatment) group, Exo group (OA chondrocytes treated with Exo), Exo-NC group (OA chondrocytes treated with Exo-NC), Exo-inhibitor group (OA chondrocytes treated with Exo-inhibitor), NC mimic group (OA chondrocytes transfected with miR-127-3p NC mimic), miR-127-3p mimic group (OA chondrocytes transfected with miR-127-3p mimic), Exo + pcDNA (OA chondrocytes treated with Exo and transfected with pcDNA empty plasmid), and Exo + pcDNA-CDH11 group (OA chondrocytes treated with Exo and transfected with pcDNA-CDH11 plasmid). The in vitro OA model was simulated by adding 10 ng/mL IL-1β (Peprotech, Rocky Hill, NJ, USA) to chondrocyte culture medium for 24 hours, and the chondrocytes were treated with 20 μg exosomes each time.
The miR-127-3p mimic/inhibitor, pcDNA-CDH11 and their respective NCs were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The transfection was performed as per the manufacturer’s protocol using Lipofectamine 3000 (Thermo Fisher Scientific, Rockford, IL, USA).
3-[4,5-Dimethylthiazol-2-yl]-2,5 Diphenyl Tetrazolium (MTT) Assay
MTT kit (Beyotime, Shanghai, China) was utilized to detect the cell viability. In brief, cells were cultured for 48 hours at a density of 5000 cells per well. After that, each well was incubated for 4 hours with 10 μL MTT solution, and then incubated with 100 μL Formazan dissolving solution until the formazan precipitate was completely dissolved through the observation under an optical microscope. A spectrophotometer (Unico Instrument Co., Ltd., Shanghai, China) was adopted to measure the optical density (OD) value at 570 nm.
5-Ethynyl-2′-Deoxyuridine (EdU) Assay
The EdU kit (Ribobio, Guangzhou, China) was utilized to detect the DNA synthesis activity of the cells. In short, cells were firstly incubated with 50 μM EdU reagent for 12 hours, fixed for 30 minutes with 4% paraformaldehyde, treated with glycine for 5 minutes, and stained with anti-EdU working solution for 30 minutes. After being permeabilized with 0.5% Triton X-100 solution, the cells were incubated with 5 μg/mL Hoechst 33,342 staining solution for 30 minutes, and captured under a confocal laser scanning microscope. The percentage of EdU positive cells was calculated by ImageJ under five random fields.
Flow Cytometry
The cell apoptosis was evaluated by flow cytometry with Annexin V-fluorescein isothiocyanate (FITC) cell apoptosis detection kit (Solarbio, Beijing, China). Briefly, the cells were washed with ice-cold PBS (Gibco), resuspended in 1× binding buffer, and then suspended with 100 μL cell suspension at room temperature. Next, the bovine serum in the test tube was appended to 5 μL Annexin V-FITC and then mixed for 10 minutes without light exposure. Then, the cells were supplemented with 5 μL propidium iodide (PI) for a 5-minute incubation under the same condition. PBS was supplemented to make the total volume into 500 μL in the test tube, and the apoptosis rate of the cells was immediately analyzed by a flow cytometer (BD Biosciences, San Diego, CA, USA). The apoptosis rate was calculated as the apoptotic cells (Annexin V+/PI− and Annexin V+/PI+)/total cells × 100%.
Western Blot Assay
The harvested cells were lysed in radioimmunoprecipitation assay lysis buffer (Millipore, Billerica, MA, USA), and the protein (35 μg) was quantified by bicinchoninic acid kit (Sigma-Aldrich) for sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After 90 minutes of electrophoresis, the Trans-Blot system (Bio-Rad, Hercules, CA, USA) was implemented to transfer the proteins from the gel on the Immun-Blot polyvinylidene difluoride membrane (Bio-Rad). To avoid the non-specific signal binding, the membrane was blocked with 5% bovine serum albumin. The membrane was probed with the primary antibody overnight at 4°C, and then re-probed with the secondary antibody for 2 hours at room temperature. Glyceraldehyde phosphate dehydrogenase (GAPDH) was selected as the internal reference protein. The color reaction was carried out using an enhanced chemiluminescence substrate kit (Abcam). Finally, the signal density was analyzed using ImageLab software version 4.1 (Bio-Rad). The antibody information was showed as follows: Collagen II (1: 1000, ab188570, Abcam), matrix metalloprotease 13 (MMP13, 1: 1000, 18,165-1-AP, Proteintech), CDH11 (1: 1000, #4442, Cell Signaling Technology, Danvers, MA, USA), β-catenin (1: 5000, ab32572, Abcam), GAPDH (1: 10,000, ab181602, Abcam), and goat anti-rabbit IgG H&L (HRP) (1: 5000, ab205718, Abcam).
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
The total RNA extraction kit (Solarbio) and Universal RT-PCR kit (Solarbio) were utilized to extract RNA and obtain the complementary DNA (cDNA) as per the manufacturer’s guidelines. RT-qPCR was performed on the ABI7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR Master Mix (Solarbio). GAPDH and U6 were selected as control genes to normalize the expression of mRNA and miRNA, respectively. Finally, the 2−∆∆Ct quantification method was used for data analysis. The primer information is listed in Table 1.
Table 1.
Sequence Used for RT-qPCR
| Targets | Forward Primer (5ʹ-3ʹ) | Reverse Primer (5ʹ-3ʹ) |
|---|---|---|
| miR-127-3p | CTGAAGCTCAGAGGGCTCTG | GAACATGTCTGCGTATCTC |
| CDH11 | ACACAGGATGGTGTGGTGAAGC | CTTGACGGTCACAGTGTCCTTG |
| U6 | CTCGCTTCGGCAGCACAT | TTTGCGTGTCATCCTTGCG |
| GAPDH | CATCACTGCCACCCAGAAGACTG | ATGCCAGTGAGCTTCCCGTTCAG |
| miR-4792 | CGGTGAGCGCTCGCTGG | GAACATGTCTGCGTATCTC |
| miR-6089 | GGAGGCCGGGGTGGGGCGGGG | GAACATGTCTGCGTATCTC |
| miR-1246 | AATGGATTTTTGGAGCAGG | GAACATGTCTGCGTATCTC |
| miR-4466 | GGGTGCGGGCCGGCGG | GAACATGTCTGCGTATCTC |
| miR-7641 | TTGATCTCGGAAGCTAAGC | GAACATGTCTGCGTATCTC |
| miR-3665 | AGCAGGTGCGGGGCGG | GAACATGTCTGCGTATCTC |
| miR-658 | AGGGAAGTAGGTCCGTT | GAACATGTCTGCGTATCTC |
| miR-1273 | GAACCCATGAGGTTGAGGCT | GAACATGTCTGCGTATCTC |
| miR-7704 | CGGGGTCGGCGGCGACG | GAACATGTCTGCGTATCTC |
| KIF3B | CGAACAGCAGAAGATCCTGGAG | TCAGTTCCAGGGTCTCCTCATC |
| ING5 | AGATCCAGAGCGCCTACAGCAA | CAGGTCAGCATCAAGTCTTCGG |
| ATP2B2 | CGGATAAGCACACGCTGGTCAA | AGCCCACATCTGCCTTCTTGAG |
| MMP13 | AACCAAGATGTGGAGTGCCTGATG | CACATCAGACCAGACCTTGAAGGC |
| IL-6 | GGTGCTAGGAGAAAACACCCA | CCACTGGCATGGGGACTAAC |
| TNF-α | TTTCTGTGGTACCCTCTGTGC | GATCGGTCCCAACAAGGAGG |
| ADAMTS-5 | TGTGGCTGAATAGCCCATCC | TTCACGTGGACATCCCTGC |
Abbreviations: miR, microRNA; GAPDH, glyceraldehyde phosphate dehydrogenase; RT-qPCR, reverse transcription quantitative polymerase chain reaction; CDH11, Cadherin-11; KIF3B, kinesin family member 3B; ING5, inhibitor of growth family 5; ATP2B2, ATPase, calcium-transporting, plasma membrane 2; MMP13, matrix metalloprotease 13; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α; ADAMTS-5, a disintegrin and metalloproteinase with thrombospondin type 1 motifs-5.
Dual-Luciferase Reporter Gene Assay
The potential binding sites of miR-127-3p and CDH11 were obtained from TargetScan. The wild-type (WT) and mutant type (MT) CDH11 luciferase reporter vectors (CDH11-WT and CDH11-MT) were constructed through the molecular cloning technology with pGL3 (Promega, Madison, WI, USA) as the basic vector. CDH11-WT had potential binding sites with miR-127-3p, while CDH11-MT contained a point mutation in the predicted binding sites of miR-127-3p. CDH11-WT and CDH11-MT were co-transfected into chondrocytes with miR-127-3p mimic or NC mimic, respectively. Forty-eight hours post-transfection, the relative luciferase activity (firefly/renilla) was measured by the dual-luciferase reporter system (Promega).
TOP/FOP Flash Luciferase Assay
The TOP/FOP flash assay was used to detect the endogenous activity of the Wnt/β-catenin pathway. The TOP flash luciferase reporter plasmid (Biovector, Beijing, China) containing TCF/LEF DNA binding sites and the control plasmid FOP flash luciferase reporter plasmid (Biovector, Beijing, China) containing mutated TCF/LEF DNA binding sites were transfected into chondrocytes. After 48 h of transfection, luciferase activity was determined by a dual-luciferase reporter assay system (Promega).
RNA Immunoprecipitation (RIP) Assay
RIP analysis was performed using Imprint RIP kit (Sigma-Aldrich). In short, the harvested cells with 80% confluence were lysed in RIP buffer, and then incubated with the magnetic beads that had been coupled with anti-argonaute-2 (Anti-Ago2) or anti-IgG (NC). After isolating the immunoprecipitated RNA from the magnetic beads, RT-qPCR was performed to confirm the levels of miR-127-3p and CDH11.
Statistical Analysis
All statistical analyses were implemented using Prism version 8.0 (GraphPad software, La Jolla, CA, USA). Data were reported as mean ± standard deviation. The unpaired t test was implemented to analyze the difference between the two experimental groups, and one-way or two-way analysis of variance (ANOVA) were performed to detect significant differences of the data among multiple groups, followed by Tukey’s multiple comparison test. All experiments were repeated three times independently. *p < 0.05 referred to statistical significance.
Results
Identification of BM-MSCs and Their Derived Exosomes
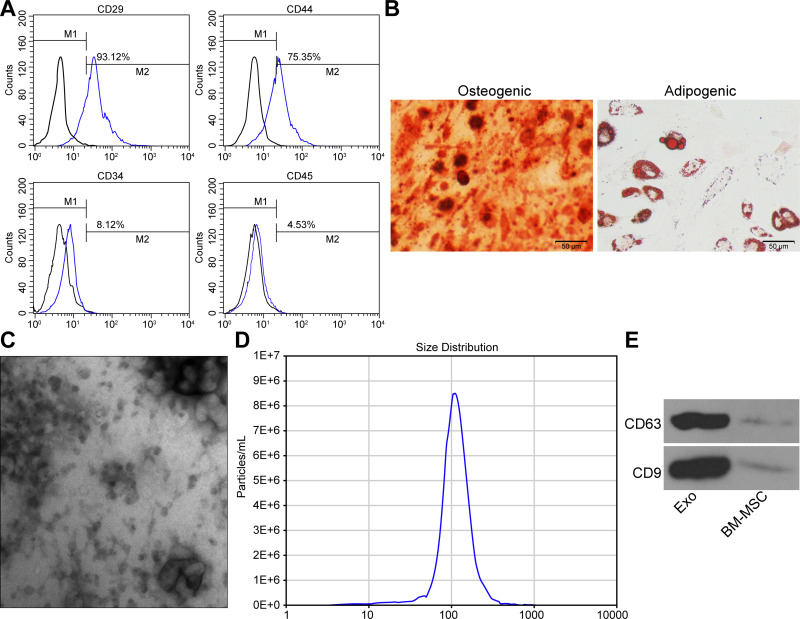
The cell surface antigens CD29, CD44, CD34 and CD45 were identified on BM-MSCs using flow cytometry (Figure 1A). The results implied that CD29 (93.12%) and CD44 (75.35%) were in positive, while CD34 (8.12%) and CD45 (4.53%) were in negative, which confirmed that BM-MSCs showed the characteristics of stem cells. Additionally, alizarin red staining and oil red O staining confirmed the differentiation abilities of BM-MSCs (Figure 1B).
Figure 1.
Extracted rat BM-MSCs and their secreted exosomes. (A) Identification of BM-MSC surface marker proteins by flow cytometry. (B) Identification of osteogenic and adipogenic differentiation capabilities of BM-MSCs. (C) Structural characteristics of exosomes-released from BM-MSCs. (D) Particle size analysis of exosomes. (E) Western blot assay utilized for the detection of the surface marker protein of exosomes. All the experiments were performed independently in triplicate and the results were the mean of three values.
Abbreviation: BM-MSCs, bone marrow-derived mesenchymal stem cells.
The shape of the extracted exosomes was observed under a TEM, which showed a typical circular or elliptical structure (Figure 1C), and the exosomes had a particle diameter of 30–120 nm (Figure 1D). The detection of CD63 and CD9 expression on the surface of exosomes by Western blot assay confirmed that the obtained precipitate was the exosomes derived from BM-MSCs (Figure 1E).
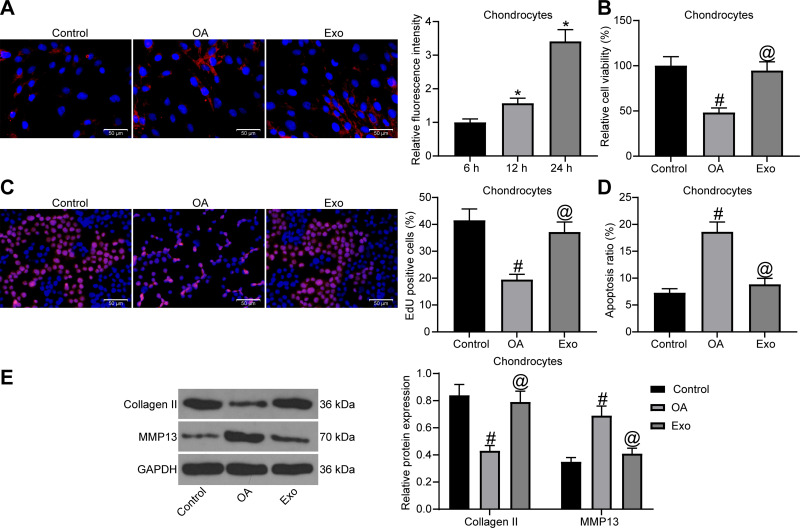
Exosomal Treatment Attenuates IL-1β-Induced Chondrocyte Damage
To test whether exosomes could act on chondrocytes, we firstly co-cultured PKH26-labeled exosomes with chondrocytes (Figure 2A). We observed that chondrocytes significantly ingested exosomes as the co-cultivation time prolonged. Next, chondrocytes with challenged with IL-1β to simulate the in vitro OA model, and then treated with exosomes. MTT assay (Figure 2B), EdU assay (Figure 2C) and flow cytometry (Figure 2D) were performed to determine cell viability, DNA synthesis activity and apoptosis rate of chondrocytes. The findings indicated that IL-1β treatment suppressed cell viability and DNA synthesis activity, and enhanced the apoptosis rate of chondrocytes. However, exosome treatment promoted cell viability and DNA synthesis activity, and suppressed the apoptosis rate. Finally, Western blot assay was conducted to detect the expression of Collagen II and MMP13 in cells (Figure 2E). We found that IL-1β treatment resulted in a reduction in Collagen II expression and an elevation in MMP13 expression in chondrocytes, and the expression of both was reversed after exosome treatment.
Figure 2.
The protective function of exosomes on chondrocytes. (A) Uptake of exosomes by chondrocytes. (B) Effects of IL-1β and exosome treatment on chondrocyte viability were confirmed by MTT assay. (C) Effects of IL-1β and exosome treatment on DNA synthesis activity of chondrocytes were confirmed by EdU assay. (D) Effects of IL-1β and exosome treatment on apoptosis rate of chondrocytes were confirmed by flow cytometry. (E) The protein expression of Collagen Ⅱ and MMP13 in chondrocytes was detected by Western blot assay. All the experiments were performed independently in triplicate and the results were the mean of three values. One-way ANOVA (A–D) or two-way ANOVA (E) was used for comparison among multiple groups. *p < 0.05 vs 6 h; #p < 0.05 vs control group; @p < 0.05 vs OA group.
Abbreviations: MTT, 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium; EdU, 5-ethynyl-2′-deoxyuridine; MMP13, matrix metalloprotease 13; ANOVA, analysis of variance; OA, osteoarthritis.
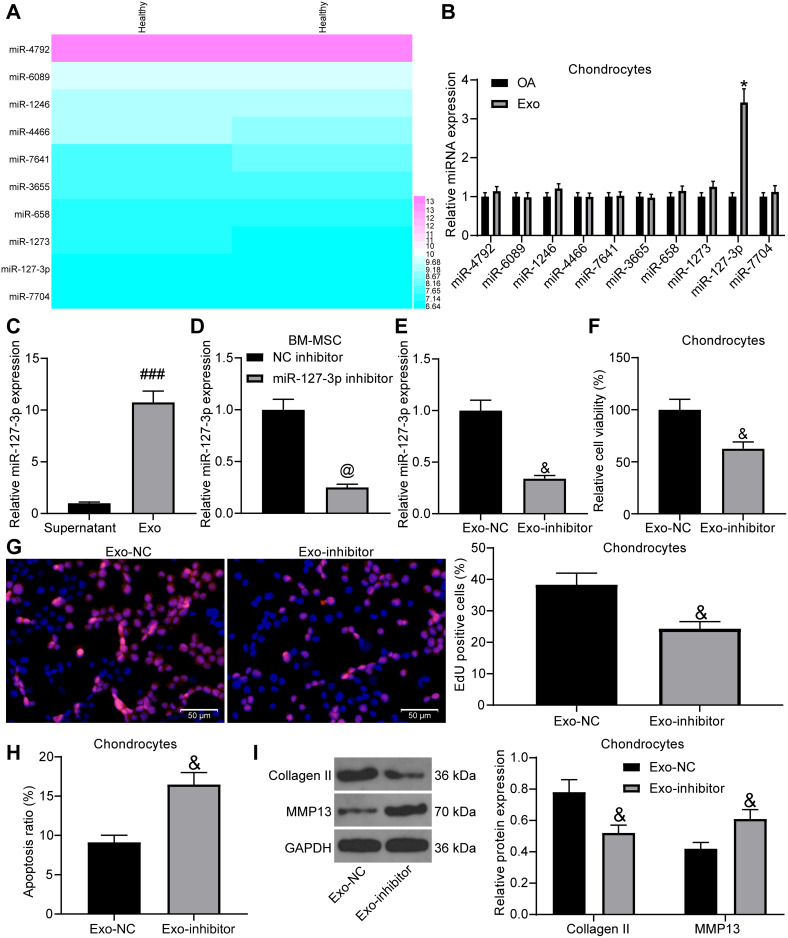
Exosomes are Protective for Chondrocytes by Increasing miR-127-3p Expression
For the purpose of probing into the mechanism of exosomes on chondrocytes, we predicted the expression of miRNAs in MSC-derived exosomes in EVmiRNA (http://bioinfo.life.hust.edu.cn/EVmiRNA/#!/), and Figure 3A presents the top-10 miRNAs (Log2 expression). We tested the expression of these miRNAs in chondrocytes before and after exosome treatment, and found that only miR-127-3p expression had a significant change (Figure 3B). Besides, we detected miR-127-3p expression in the supernatant after the exosome was extracted and in exosomes by RT-qPCR, and findings demonstrated that miR-127-3p was significantly enriched in exosomes (Figure 3C).
Figure 3.
The protective effect of exosomal miR-127-3p on chondrocytes. (A) Prediction on the miRNA (Log2 expression) that is enriched in MSC-derived exosomes. (B) The effect of extracted exosomes on the expression of related miRNAs in chondrocytes by RT-qPCR. (C) RT-qPCR utilized to detect miR-127-3p expression in the removed supernatant during the exosome extraction and in the exosomes. (D) The transfection efficiency of miR-127-3p inhibitor in BM-MSCs confirmed by RT-qPCR. (E) miR-127-3p expression in the extracted exosomes (Exo-NC and Exo-inhibitor) determined by RT-qPCR. (F) Effects of Exo-NC and Exo-inhibitor treatment on the viability of chondrocytes measured by MTT assay. (G) Effects of Exo-NC and Exo-inhibitor treatment on the DNA synthesis activity of chondrocytes measured by EdU assay. (H) Effects of Exo-NC and Exo-inhibitor treatment on the apoptosis rate of chondrocytes determined by flow cytometry. (I) Effects of Exo-NC and Exo-inhibitor treatment on the expression of Collagen II and MMP13 in chondrocytes determined by Western blot assay. All the experiments were performed independently in triplicate and the results were the mean of three values. Unpaired t test (C–H) was utilized for comparison between two groups or two-way ANOVA (B, I) was used for comparison among multiple groups. *p < 0.05 vs OA group; ###p < 0.05 vs Supernatant group; @p < 0.05 vs NC inhibitor group; &p < 0.05 vs Exo-NC group.
Abbreviations: miRNA, microRNA; MTT, 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium; EdU, 5-ethynyl-2′-deoxyuridine; RT-qPCR, reverse transcription quantitative polymerase chain reaction; Exo, exosome; NC, negative control; MMP13, matrix metalloprotease 13; ANOVA, analysis of variance.
In order to test whether the protective function of exosomes on chondrocytes was achieved by miR-127-3p, we transfected NC inhibitor and miR-127-3p inhibitor into BM-MSCs, and the effective transfection was measured by RT-qPCR (Figure 3D). Then, RT-qPCR was performed on the extracted exosomes (Exo-NC and Exo-inhibitor), and it was found that Exo-inhibitor showed reduced miR-127-3p expression (Figure 3E). The effects of Exo-NC and Exo-inhibitor on IL-1β-induced chondrocytes were verified, and Exo-inhibitor was found to decrease the viability and DNA synthesis activity, and promote the apoptosis rate of chondrocytes, accompanied by reduced expression of Collagen II and elevated expression of MMP13 in chondrocytes (Figure 3F-I).
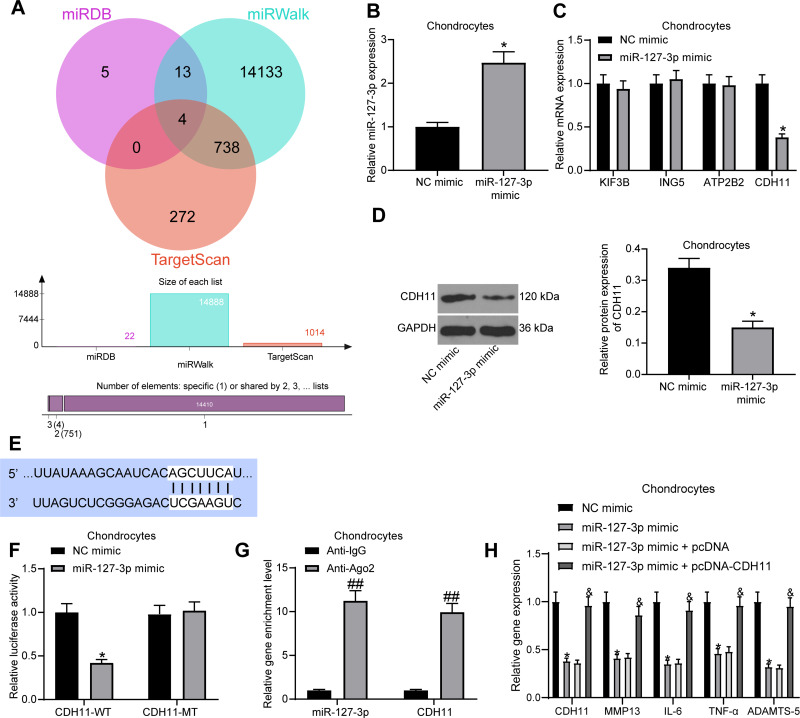
miR-127-3p Targets CDH11
For searching the downstream target genes of miR-127-3p in OA chondrocytes, we predicted the target genes in miRDB (http://www.mirdb.org/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), and TargetScan (http://www.targetscan.org/vert_71/) with rats as the species. There was a total of four intersections, namely KIF3B, ING5, ATP2B2, and CDH11 (Figure 4A). Next, we transfected miR-127-3p mimic and NC mimic into chondrocytes, and the effective transfection was measured by RT-qPCR (Figure 4B). Meanwhile, the effect of miR-127-3p mimic on the expression of four potential target genes in chondrocytes was confirmed by RT-qPCR, and we found that only CDH11 had an altered mRNA expression (Figure 4C). miR-127-3p also inhibited the protein expression of CDH11, as determined by Western blot assay (Figure 4D). The potential binding sites of miR-127-3p to CDH11 were obtained from TargetScan (Figure 4E). For the luciferase activity assay, CDH11-WT and CDH11-MT were transfected into chondrocytes with miR-127-3p mimic or NC mimic to measure the luciferase activity at 48 hours post-transfection (Figure 4F). It was showed that miR-127-3p mimic reduced the luciferase activity of CDH11-WT, but had no significant effect on the luciferase activity of CDH11-MT. Furthermore, we also performed RIP assay in chondrocytes to test whether miR-127-3p could target CDH11 through Ago2 (Figure 4G). It was indicated that compared with anti-IgG, anti-Ago2 significantly enriched miR-127-3p and CDH11.
Figure 4.
CDH11 is a downstream target gene of miR-127-3p. (A) The potential target genes of miR-127-3p in rats predicted in miRDB (http://www.mirdb.org/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), and TargetScan (http://www.targetscan.org/vert_71/). (B) RT-qPCR detection of the transfection efficiency of miR-127-3p mimic in chondrocytes. (C) Effect of miR-127-3p mimic on mRNA expression of potential target genes by RT-qPCR. (D) Effect of miR-127-3p mimic on CDH11 protein expression determined by Western blot assay. (E) Potential binding sites of miR-127-3p and CDH11. (F) Effect of miR-127-3p on CDH11-WT/CDH11-MT luciferase activity confirmed by luciferase activity assay. (G) RIP assay utilized to detect the binding relationship between miR-127-3p and CDH11. (H) Effects of miR-127-3p mimic and pcDNA-CDH11 on the expression of CDH11 and OA-related genes MMP13, IL-6, TNF-α, and ADAMTS-5 in chondrocytes examined by RT-qPCR. All the experiments were performed independently in triplicate and the results were the mean of three values. Unpaired t test (B, D) was utilized for comparison between two groups or two-way ANOVA (C–H) was used for comparison among multiple groups. *p < 0.05 vs NC mimic group; ##p < 0.05 vs Anti-IgG group; &p < 0.05 vs miR-127-3p mimic + pcDNA group.
Abbreviations: miRNA, microRNA; RT-qPCR, reverse transcription quantitative polymerase chain reaction; CDH11, Cadherin-11; WT, wild type; MT, mutant; MMP13, matrix metalloprotease 13; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α; ADAMTS-5, a disintegrin and metalloproteinase with thrombospondin type 1 motifs-5.
To investigate whether miR-127-3p can regulate the expression of OA-related genes MMP-13, IL-6, TNF-α, and ADAMTS-5 by targeting CDH11 in chondrocytes, we transfected miR-127-3p mimic with pcDNA-CDH11 into chondrocytes and detected the expression of CDH11 and OA-related genes by RT-qPCR (Figure 4H). We found that overexpression of miR-127-3p significantly suppressed the expression of CDH11 and OA-related genes, while their expression was significantly restored after overexpression of CDH11. This demonstrates the possibility that miR-127-3p affects phenotypic modulation of chondrocytes by targeting CDH11.
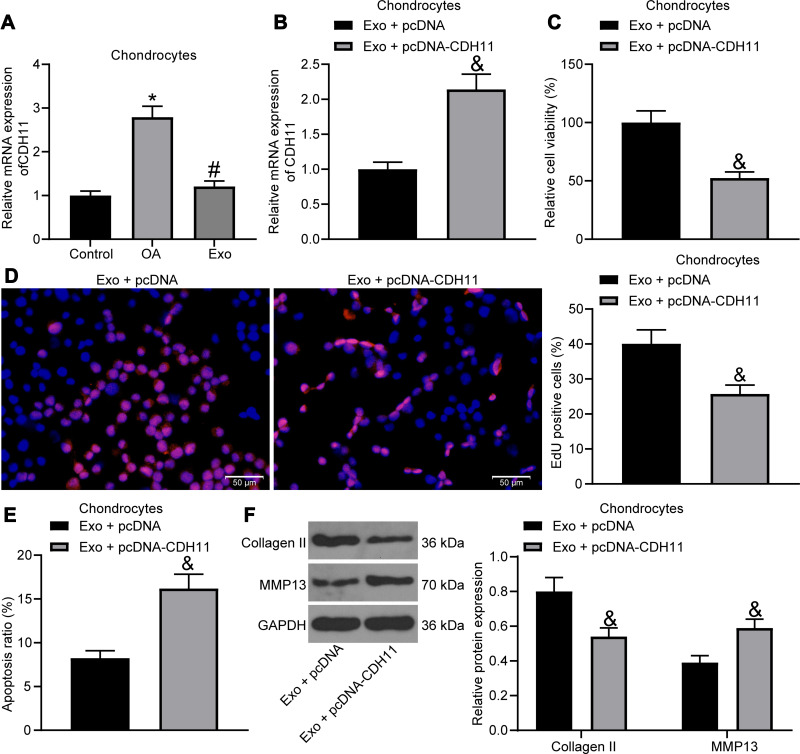
Upregulated CDH11 in Chondrocytes Weakens the Therapeutic Effect of Exosomes
In a previous report, CDH11 was found to be highly expressed in OA cartilage versus its expression in normal cartilage,21 but its effect on OA has seldomly been studied. We detected CDH11 expression by RT-qPCR in chondrocytes (Figure 5A), and it was found that IL-1β treatment led to an increase in CDH11 expression in chondrocytes, and CDH11 was reduced after exosome treatment. Then, we transfected pcDNA-CDH11 into chondrocytes after exosome treatment, and the effective transfection was confirmed by RT-qPCR (Figure 5B). The effects of upregulated CDH11 on IL-1β-induced chondrocytes were verified by MTT assay, EdU assay, flow cytometry and Western blot assay, and upregulated CDH11 was observed to suppress the viability and DNA synthesis activity, and increase the apoptosis rate of chondrocytes, accompanied by reduced expression of Collagen II and elevated expression of MMP13 in chondrocytes (Figure 5C-F), suggesting that the alleviating effects of exosome on IL-1β-treated chondrocytes were reversed by overexpression of CDH11.
Figure 5.
Exosomes carrying miR-127-3p inhibit CDH11 expression in chondrocytes. (A) Effects of IL-1β and exosome treatment on the expression of CDH11 in chondrocytes detected by RT-qPCR. (B) RT-qPCR for detecting the transfection efficiency of pcDNA-CDH11. (C) Effects of upregulated CDH11 on the viability of chondrocytes measured by MTT assay. (D) Effects of upregulated CDH11 on the DNA synthesis activity of chondrocytes measured by EdU assay. (E) Effects of upregulated CDH11 on the apoptosis rate of chondrocytes determined by flow cytometry. (F) Effects of upregulated CDH11 on the expression of Collagen II and MMP13 in chondrocytes determined by Western blot assay. All the experiments were performed independently in triplicate and the results were the mean of three values. Unpaired t test (B–E) was utilized for comparison between two groups or one-way ANOVA (A)/two-way ANOVA (F) was used for comparison among multiple groups. *p < 0.05 vs control group; #p < 0.05 vs OA group; &p < 0.05 vs Exo + pcDNA group.
Abbreviations: miRNA, microRNA; RT-qPCR, reverse transcription quantitative polymerase chain reaction; CDH11, Cadherin-11; MTT, 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium; EdU, 5-ethynyl-2′-deoxyuridine; Exo, exosome; NC, negative control; MMP13, matrix metalloprotease 13; ANOVA, analysis of variance; OA, osteoarthritis.
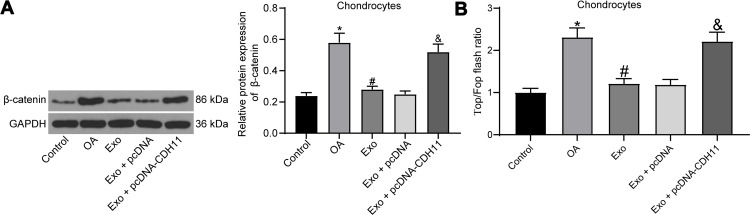
Exosome Treatment Improves IL-1β-Induced Chondrocytes Damage by Inhibiting the Wnt/β-Catenin Pathway Activation
The activation of the Wnt/β-catenin pathway can contribute to chondrocyte damage and promote the progression of OA.22,23 The effects of IL-1β, exosome treatment and overexpression of CDH11 on the Wnt/β-catenin pathway activation in chondrocytes were then verified by Western blot assay (Figure 6A). We found that IL-1β treatment resulted in an increase in β-catenin expression, and β-catenin expression was inhibited after exosome treatment. Meanwhile, overexpressed CDH11 enhanced the expression of β-catenin. To detect changes in the endogenous activity of the Wnt/β-catenin pathway, we also performed TOP/FOP flash experiments in chondrocytes (Figure 6B). We found that IL-1β treatment significantly increased the TOP/FOP ratio, which means IL-1β treatment significantly elevated the endogenous activity of the Wnt/β-catenin pathway in chondrocytes. Exosome treatment remarkably repressed the endogenous activity of Wnt/β-catenin pathway induced by IL-1β. The inhibitory effect of exosomes on the endogenous activity of the Wnt/β-catenin pathway was significantly attenuated after overexpression of CDH11. Based on all the aforementioned experiments, we confirm that exosomes carrying miR-127-3p can inhibit CDH11 expression in chondrocytes, thereby blocking the activation of Wnt/β-catenin pathway and ameliorating the IL-1β-induced chondrocytes damage.
Figure 6.
Exosome treatment alleviates IL-1β-induced chondrocytes damage by blocking the Wnt/β-catenin pathway activation. (A) Effects of IL-1β, exosome treatment and overexpression of CDH11 on the expression of β-catenin (a downstream protein of the Wnt/β-catenin pathway) determined by Western blot assay. (B) TOP/FOP Flash assay for detecting endogenous activity of the Wnt/β-catenin pathway. All the experiments were performed independently in triplicate and the results were the mean of three values. One-way ANOVA was used for comparison among multiple groups. *p < 0.05 vs control group; #p < 0.05 vs OA group; &p < 0.05 vs Exo + pcDNA group.
Abbreviation: ANOVA, analysis of variance.
Discussion
OA is an active, progressive and multifactorial disease featured by pain and physical disability.24 Indeed, all the currently accepted treatments are focused on symptom control, instead of disease prevention.25 MSC-derived exosomes are revealed to be efficacious against various diseases, which are implied to modulate cartilage repair and regeneration.26 In this study, an in-vitro model of OA was mimicked by IL-1β induction in chondrocytes to make clear the underlying mechanisms of exosomal miR‑127-3p from BM-MSCs in OA progression. To conclude, this study provides evidence that exosomal miR-127-3p from BM-MSCs alleviates chondrocyte damage in OA through downregulating CDH11 and blocking the Wnt/β-catenin pathway activation.
Since evidence has shown that MSC-derived exosomes could mitigate OA through repairing and regenerating the injured articular cartilage, the focus of the OA pathogenesis,27 we initially tested whether exosomes from BM-MSCs acted on IL-1β-treated chondrocytes. The findings indicated that BM-MSCs-derived exosomes could promote cell viability and DNA synthesis activity, and suppress the apoptosis rate of chondrocytes. Similarly, it has been exhibited that exosomes secreted by MSCs could attenuate OA through the promotion of chondrocyte migration and proliferation.28 Cosenza et al have declared that MSCs-derived exosomes are able to protect cartilage from degradation in OA via enhancing expression of chondrocyte markers, decreasing catabolic markers and inflammatory markers, blocking of macrophage activation, as well as protecting chondrocytes from apoptosis.29 These findings demonstrate that BM-MSCs-derived exosomes are beneficial against the OA development.
With the aim to probe into the mechanism of exosomes on chondrocytes, we predicted the expression of miRNAs in BM-MSC-derived exosomes. miR-127-3p was found to be enriched in exosomes through online website prediction and experiments, and the therapeutic effect of exosomes was diminished by inhibition of miR-127-3p in BM-MSC. As a crucial miRNA, miR-127-5p, which is decreased by IL-1β in human chondrocytes, functions as a vital regulator of the catabolic signaling pathways.30 There are articles highlighting that miR‐127‐5p is downregulated in OA cartilage tissues and chondrocytes, and rescuing miR-127-5p expression might be a useful strategy for OA therapy.14,15 Meanwhile, through bioinformatics analysis and experimental research, we found that CDH11 was a downstream gene of miR-127-3p, and upregulated CDH11 in chondrocytes weakened the therapeutic effect of exosomes. In a previous study, it is suggested that umbilical cord-derived MSC transplantation could decrease CDH11 expression in the synovium, implying that umbilical cord-derived MSCs inhibit CDH11 expression in FLSs of rheumatoid arthritis (RA) by secreting IL-10.31 In FLSs from RA patients, CDH11 has been demonstrated to enhance the secretion of IL-6 and other proinflammatory factors through the involvement of the key signaling pathways.32,33 In addition, activated chondrocytes also release MMP-13 and ADAMTS-5, while TNF-α and IL-6 may cause OA indirectly by mediating the production of adiponectin and leptin.34 While our cytokine-level measurement revealed that miR-127-3p notably repressed the levels of these factors, which were elevated by CDH11 overexpression. These data suggest that CDH11 suppression is an essential therapeutic target in arthritis. Although the investigation on the binding relationship of miR-127-3p and CDH11 is still in its infancy, other researchers have concentrated on the binding relationship of miR-127-3p with other genes during OA progression. Zhou et al have confirmed that miR-127-5p could target MMP-13 and modulate the expression of Collagen II in IL-1β-induced mouse chondrocytes.16 Many of the functions of miR-127-3p and CDH11 have not yet been clarified, and further exploration is required in this regard.
Currently available researches have demonstrated the involvement of many pivotal molecules and signaling pathways in the therapeutic capabilities of MSCs-derived exosomes on bone disorders, including the Wnt/β-catenin pathway.5 The Wnt/β-catenin pathway is unique in modulating arthritis development, and the molecules in this pathway act as significant targets for the treatment of arthritis.22,35 Based on these evidences, we then aimed to elucidate the effects of IL-1β, exosome treatment and overexpression of CDH11 on the Wnt/β-catenin pathway activation in chondrocytes. The obtained results suggested that exosome treatment could suppress the Wnt/β-catenin pathway activation, and upregulated CDH11 reversed the suppressive effect of exosomes on this pathway. Furthermore, Liu et al have stated that exosomes from platelet-rich plasma could alleviate knee OA by promoting chondrocyte proliferation and suppressing chondrocyte apoptosis via regulating the Wnt/β-catenin pathway.36
Conclusion
In summary, this study indicates that exosomal miR-127-3p derived from BM-MSCs inhibits CDH11 in chondrocytes, thereby blocking the Wnt/β-catenin pathway activation and protecting chondrocytes against damage in OA. Further research into the network of exosomal miR-127-3p modulation could accelerate the development of a new therapeutic intervention or preventive approach in OA.
Disclosure
The authors declare no potential conflicts of interest.
References
- 1.Thudium CS, Lofvall H, Karsdal MA, Bay-Jensen AC, Bihlet AR. Protein biomarkers associated with pain mechanisms in osteoarthritis. J Proteomics. 2019;190:55–66. doi: 10.1016/j.jprot.2018.04.030 [DOI] [PubMed] [Google Scholar]
- 2.Anderson KL, Zulch H, O’Neill DG, Meeson RL, Collins LM. Risk factors for canine osteoarthritis and its predisposing arthropathies: a systematic review. Front Vet Sci. 2020;7:220. doi: 10.3389/fvets.2020.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bijlsma JW, Berenbaum F, Lafeber FP. Osteoarthritis: an update with relevance for clinical practice. Lancet. 2011;377(9783):2115–2126. doi: 10.1016/S0140-6736(11)60243-2 [DOI] [PubMed] [Google Scholar]
- 4.Brown S, Kumar S, Sharma B. Intra-articular targeting of nanomaterials for the treatment of osteoarthritis. Acta Biomater. 2019;93:239–257. doi: 10.1016/j.actbio.2019.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu S, Xu X, Liang S, et al. The application of MSCs-derived extracellular vesicles in bone disorders: novel cell-free therapeutic strategy. Front Cell Dev Biol. 2020;8:619. doi: 10.3389/fcell.2020.00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousnaki M, Bakopoulou A, Kritis A, Koidis P. The efficacy of stem cells secretome application in osteoarthritis: a systematic review of in vivo studies. Stem Cell Rev Rep. 2020. [DOI] [PubMed] [Google Scholar]
- 7.D’Agnelli S, Gerra MC, Bignami E, Arendt-Nielsen L. Exosomes as a new pain biomarker opportunity. Mol Pain. 2020;16:1744806920957800. doi: 10.1177/1744806920957800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni Z, Zhou S, Li S, et al. Exosomes: roles and therapeutic potential in osteoarthritis. Bone Res. 2020;8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L, He T, Xing J, et al. Bone marrow mesenchymal stem cell-derived exosomes protect cartilage damage and relieve knee osteoarthritis pain in a rat model of osteoarthritis. Stem Cell Res Ther. 2020;11(1):276. doi: 10.1186/s13287-020-01781-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie F, Liu YL, Chen XY, et al. Role of MicroRNA, LncRNA, and exosomes in the progression of osteoarthritis: a review of recent literature. Orthop Surg. 2020;12(3):708–716. doi: 10.1111/os.12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92. [PMC free article] [PubMed] [Google Scholar]
- 12.Swingler TE, Niu L, Smith P, et al. The function of microRNAs in cartilage and osteoarthritis. Clin Exp Rheumatol. 2019;37 Suppl 120(5):40–47. [PubMed] [Google Scholar]
- 13.Yu Y, Zhao J. Modulated autophagy by MicroRNAs in osteoarthritis chondrocytes. Biomed Res Int. 2019;2019:1484152. doi: 10.1155/2019/1484152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Yuan B, Pei Z, et al. Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. J Cell Mol Med. 2019;23(10):6554–6564. doi: 10.1111/jcmm.14400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang J, Xu L, Zhou F, et al. MALAT1/miR-127-5p Regulates Osteopontin (OPN)-mediated proliferation of human chondrocytes through PI3K/Akt pathway. J Cell Biochem. 2018;119(1):431–439. doi: 10.1002/jcb.26200 [DOI] [PubMed] [Google Scholar]
- 16.Zhou ZB, Huang GX, Fu Q, et al. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol Ther. 2019;27(3):531–541. doi: 10.1016/j.ymthe.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pu M, Chen J, Tao Z, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76(3):441–451. doi: 10.1007/s00018-018-2940-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowler MA, Bersi MR, Ryzhova LM, Jerrell RJ, Parekh A, Merryman WD. Cadherin-11 as a regulator of valve myofibroblast mechanobiology. Am J Physiol Heart Circ Physiol. 2018;315(6):H1614–H1626. doi: 10.1152/ajpheart.00277.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DM, Kiener HP, Agarwal SK, et al. Cadherin-11 in synovial lining formation and pathology in arthritis. Science. 2007;315(5814):1006–1010. doi: 10.1126/science.1137306 [DOI] [PubMed] [Google Scholar]
- 20.Lennon DP, Caplan AI. Isolation of rat marrow-derived mesenchymal stem cells. Exp Hematol. 2006;34(11):1606–1607. doi: 10.1016/j.exphem.2006.07.015 [DOI] [PubMed] [Google Scholar]
- 21.Karlsson C, Dehne T, Lindahl A, et al. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage. 2010;18(4):581–592. doi: 10.1016/j.joca.2009.12.002 [DOI] [PubMed] [Google Scholar]
- 22.Gao JB, Lin L, Men XQ, et al. Fibulin-5 protects the extracellular matrix of chondrocytes by inhibiting the Wnt/beta-catenin signaling pathway and relieves osteoarthritis. Eur Rev Med Pharmacol Sci. 2020;24(10):5249–5258. doi: 10.26355/eurrev_202005_21307 [DOI] [PubMed] [Google Scholar]
- 23.Gu Y, Ren K, Wang L, Yao Q. Loss of Klotho contributes to cartilage damage by derepression of canonical Wnt/beta-catenin signaling in osteoarthritis mice. Aging (Albany NY). 2019;11(24):12793–12809. doi: 10.18632/aging.102603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie F, Tanvejsilp P, Campbell K, Gaebel K. Cost-effectiveness of pharmaceutical management for osteoarthritis pain: a systematic review of the literature and recommendations for future economic evaluation. Drugs Aging. 2013;30(5):277–284. doi: 10.1007/s40266-013-0062-3 [DOI] [PubMed] [Google Scholar]
- 25.Freitag J, Bates D, Boyd R, et al. Mesenchymal stem cell therapy in the treatment of osteoarthritis: reparative pathways, safety and efficacy - a review. BMC Musculoskelet Disord. 2016;17:230. doi: 10.1186/s12891-016-1085-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Chu WC, Lai RC, Lim SK, Hui JH, Toh WS. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthritis Cartilage. 2016;24(12):2135–2140. doi: 10.1016/j.joca.2016.06.022 [DOI] [PubMed] [Google Scholar]
- 27.Toh WS, Lai RC, Hui JHP, Lim SK. MSC exosome as a cell-free MSC therapy for cartilage regeneration: implications for osteoarthritis treatment. Semin Cell Dev Biol. 2017;67:56–64. doi: 10.1016/j.semcdb.2016.11.008 [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Wang Y, Zhao B, et al. Comparison of exosomes secreted by induced pluripotent stem cell-derived mesenchymal stem cells and synovial membrane-derived mesenchymal stem cells for the treatment of osteoarthritis. Stem Cell Res Ther. 2017;8(1):64. doi: 10.1186/s13287-017-0510-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosenza S, Ruiz M, Toupet K, Jorgensen C, Noel D. Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Sci Rep. 2017;7(1):16214. doi: 10.1038/s41598-017-15376-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SJ, Cheon EJ, Lee MH, Kim HA. MicroRNA-127-5p regulates matrix metalloproteinase 13 expression and interleukin-1beta-induced catabolic effects in human chondrocytes. Arthritis Rheum. 2013;65(12):3141–3152. doi: 10.1002/art.38188 [DOI] [PubMed] [Google Scholar]
- 31.Zhao C, Zhang L, Kong W, et al. Umbilical cord-derived mesenchymal stem cells inhibit cadherin-11 expression by fibroblast-like synoviocytes in rheumatoid arthritis. J Immunol Res. 2015;2015:137695. doi: 10.1155/2015/137695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang SK, Noss EH, Chen M, et al. Cadherin-11 regulates fibroblast inflammation. Proc Natl Acad Sci U S A. 2011;108(20):8402–8407. doi: 10.1073/pnas.1019437108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumura T, Saito Y, Suzuki T, et al. Phosphorylated platelet-derived growth factor receptor-positive cells with anti-apoptotic properties accumulate in the synovium of patients with rheumatoid arthritis. Front Immunol. 2019;10:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T, He C. Pro-inflammatory cytokines: the link between obesity and osteoarthritis. Cytokine Growth Factor Rev. 2018;44:38–50. doi: 10.1016/j.cytogfr.2018.10.002 [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y, Wang T, Hamilton JL, Chen D. Wnt/beta-catenin signaling in osteoarthritis and in other forms of arthritis. Curr Rheumatol Rep. 2017;19(9):53. doi: 10.1007/s11926-017-0679-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Wang L, Ma C, Wang G, Zhang Y, Sun S. Exosomes derived from platelet-rich plasma present a novel potential in alleviating knee osteoarthritis by promoting proliferation and inhibiting apoptosis of chondrocyte via Wnt/beta-catenin signaling pathway. J Orthop Surg Res. 2019;14(1):470. doi: 10.1186/s13018-019-1529-7 [DOI] [PMC free article] [PubMed] [Google Scholar]