Abstract
Cerebellum plays an important role in several key functions including control of movement, balance, cognition, reward, and affect. Imaging studies indicate that distinct cerebellar regions contribute to these different functions. Molecular studies examining regional cerebellar differences are lagging as they are mostly done on whole cerebellar extracts thereby masking any distinctions across specific cerebellar regions. Here we describe a technique to reproducibly and quickly dissect four different cerebellar regions: the deep cerebellar nuclei (DCN), anterior and posterior vermal cerebellar cortex, and the cerebellar cortex of the hemispheres. Dissecting out these distinct regions allows for the exploration of molecular mechanisms that may underlie their unique contributions to balance, movement, affect and cognition. This technique may also be used to explore differences in pathological susceptibility of these specific regions across various mouse disease models.
Keywords: Cerebellum, Regionalization, Deep Cerebellar Nuclei, Cerebellar Cortex, RNA, RTqPCR
SUMMARY:
Different cerebellar regions have been implicated to play a role in distinct behavioral outputs, yet the underlying molecular mechanisms remain unknown. This work describes a method to reproducibly and quickly dissect cerebellar cortex of the hemispheres, anterior and posterior regions of the vermis, and the deep cerebellar nuclei in order to probe for molecular differences by isolating RNA and testing for differences in gene expression.
INTRODUCTION:
The cerebellum contains over half of the neurons in the brain and has historically been referred to as a motor control and balance center in the brain1. More recently, studies have demonstrated that the cerebellum plays a key role in various other functions including cognition, reward processing, and affect2–5.
The cerebellum has well-described anatomy: the cortex region is composed of granule, Purkinje, and molecular layers. Granule cells form the granule cell layer and send input via parallel fibers to the Purkinje cell dendrites of the molecular layer which also receive input from climbing fibers that originated in the inferior olive. Purkinje cells send inhibitory projections to cells in the deep cerebellar nuclei (DCN), which serves as the main output from the cerebellum. The output of this cerebellar circuit is further modulated by the activity of the inhibitory interneurons in the cerebellar cortex, including Golgi, stellate, and basket cells4. This cerebellar functional unit is distributed throughout all the lobules of the cerebellar cortex. Despite this relatively uniform circuitry across the cerebellum, evidence from human neuroimaging literature and patient studies indicates functional heterogeneity of the cerebellum6, 7.
The cerebellar cortex can be divided into two main regions: the midline-defined vermis, and the lateral hemispheres. The vermis can be further divided into anterior and posterior lobules. These distinct regions of the cerebellum have been implicated in contributing to different behaviors. Task-evoked or task-free activity patterns implicated that anterior regions of the vermis contribute more to motor function while posterior vermis contributes more to cognition 7,6. The vermis is also linked with affect and emotions, while cerebellar hemispheres contribute to executive, visual-spatial, language, and other mnemonic functions8. In addition, anatomical studies provided evidence that functionally distinct cerebellar regions are connected with different cortical regions9. Lesion-symptom mapping revealed that patients with strokes affecting the anterior lobules (extending into lobule VI) had poorer performance on fine motor tasks, while patients with damage to posterior lobe regions and hemispheres exhibited cognitive deficits in the absence of cerebellar motor syndrome10. Finally, regional cerebellar pathology in disease indicates that functionally distinct cerebellar regions are also differently susceptible to disease 11,12
While much less explored, preliminary evidence demonstrates distinct gene expression signatures across cerebellar cortical regions. Purkinje cell expression of Zebrin II shows region specific patterning in the vermis such that there are more Zebrin II positive cells in the posterior lobules and fewer in the anterior lobules13. This also correlates with regionally distinct physiological function as Zebrin II negative Purkinje cells display higher frequency of tonic firing than Purkinje cells that are Zebrin II positive14.
In addition to the cerebellar cortex, the cerebellum includes the deep cerebellar nuclei (DCN) which serve as the primary output for the cerebellum. The nuclei are made up of the medial (MN), interposed (IN), and lateral nuclei (LN). Functional imaging and patient studies have demonstrated that the DCN also participate in various behaviors15, but very few studies examine gene expression change in DCN.
Advances in molecular techniques have made it possible to assess regional gene expression in the brain and have uncovered heterogeneity across and within different brain regions in both physiological and disease states16. Such studies implicate that the cerebellum is different from other brain regions. For example, the ratio of neurons to glial cells is inverted in the cerebellum compared to other brain regions 1. Even in normal physiological conditions, the expression of proinflammatory genes is upregulated in the cerebellum compared to the other brain regions17. Molecular techniques have also been very useful in identifying the pathways that contribute to the pathogenesis of cerebellar diseases. For instance, RNA sequencing of the whole cerebellar extracts identified genes altered in a Purkinje cell specific transgenic mouse model of spinocerebellar ataxia type 1 (SCA1) as compared to their wild type controls. Such evidence has revealed key molecular pathways underlying pathogenesis in cerebellar Purkinje cells and has helped identify potential therapeutic targets18. However, recent studies suggest that there are differences in the vulnerability to diseases across the cerebellar regions 11,12,19. This could indicate that there are key changes occurring in distinct cerebellar regions, which may be masked or undetected with whole cerebellar extracts. Thus, there is a need to develop techniques which allow researchers to examine molecular profiles in different cerebellar regions.
The technique proposed here describes a reproducible method to dissect four distinct regions of the mouse cerebellum in order to isolate RNA from those regions and explore regional differences in gene expression. The schematic of the mouse cerebellum in Figure 1A highlights the vermis in blue, and hemispheres in yellow. Specifically, this technique makes it possible to isolate four regions: deep cerebellar nuclei (DCN) (red-dotted boxes in Figure 1A), the cerebellar cortex of anterior vermis (CCaV) (dark blue in Figure 1A), the cerebellar cortex of the posterior vermis (CCpV) (light blue in Figure 1A), and the cerebellar cortex of the hemispheres (CCH) (yellow in Figure 1A). By assessing gene expression of these regions separately, it will be possible to investigate molecular mechanisms underlying discrete functions of these different regions as well as potential differences in their vulnerability in disease.
Figure 1: Representative images of cerebellar dissection.
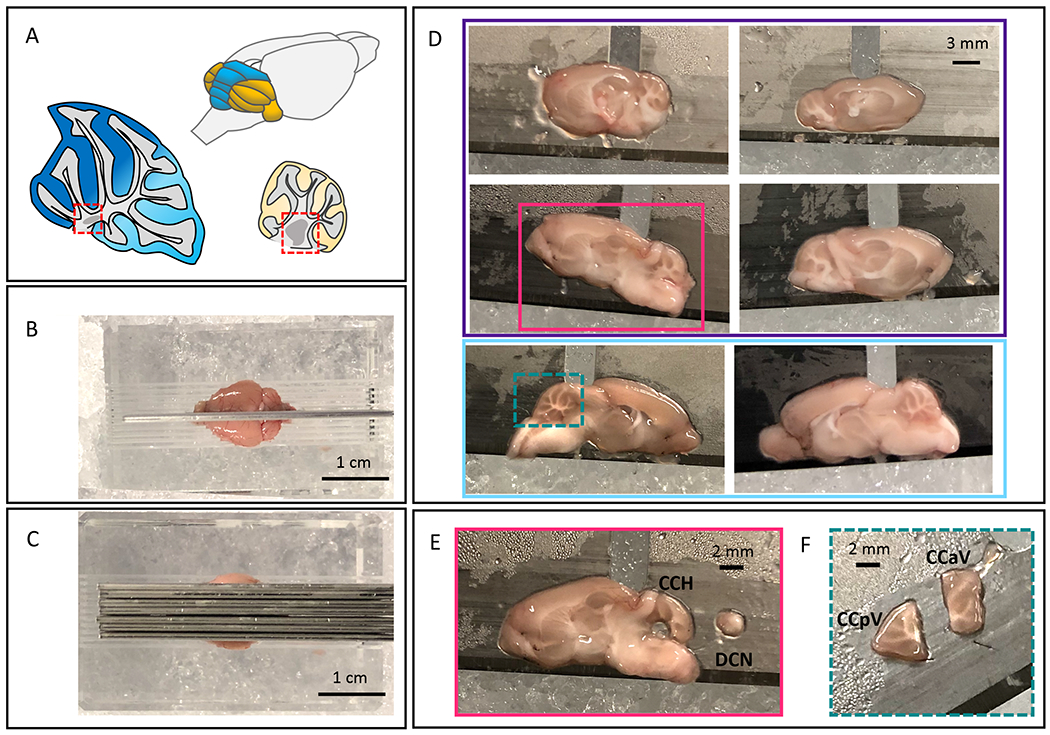
A. Schematic of mouse cerebellum with vermis in blue and hemispheres in yellow. Sagittal cerebellar schematics of both vermis and hemisphere. Vermis is in blue, with dark blue marking the anterior vermis, and light blue marking posterior vermis. Hemisphere in yellow. The DCN are marked in each with red-dotted boxes. B. Full brain in sagittal mouse brain matrix, with razor blade down midline. C. Placement of three razor blades 1 mm apart on either side of the midline. D. The resulting six sagittal brain sections. Four contain lateral/hemisphere cerebellar sections (the top four images outlined in purple). Two contain medial/vermal cerebellar sections (bottom two images, outlined in light blue). E. Representative lateral/hemisphere cerebellar section with DCN punch dissected to the right of the section (section outlined in pink in Figure 1D). F. Cerebellar section in Figure 1D with square dotted turquois around it, dissected into anterior and posterior vermal lobules.
PROTOCOL:
1. Set up
1.1. Gather necessary equipment including decapitation scissors, blunt forceps, dissection scissors, vascular scissors, microspatula, sagittal mouse brain matrix, razor blades, 200 μl pipet tips, glass petri dish, glass slide, and ice bucket. Lay out all equipment on an absorbent pad.
1.2. Place petri dish, glass plate, and brain matrix on ice.
1.3. Using a razor blade at a perpendicular angle, trim about 5 mm off the tip of one 200 μl pipet tip. This makes the size of the opening at the end of the tip about 1 mm wide. This should be sufficient to punch out the DCN. But this can also be adjusted as needed depending on the dissection. Have many tips ready for easy replacement and adjustment.
1.4. Label 1.5 ml microfuge tubes with animal identification and cerebellar region (four tubes per animal).
1.5. Fill cryosafe container with liquid nitrogen to flash freeze tissue after extraction.
2. Brain extraction and dissection
All experiments were conducted in accordance with the guidelines of the Animal Care Committees of the University of Minnesota.
2.1. Euthanize the mouse using 5% CO2 exposure. Once breathing has ceased, perform cervical dislocation. Decapitate the mouse with the decapitation scissors and discard the carcass in the appropriate receptacle.
2.2. Make an incision with a razor blade along the medial sagittal line of the head starting at the nose and continuing all the way back. Separate the skin, parting to either side of the midline. Use the razor blade to cut away the muscle on each side, cutting down past the ear canals.
2.3. Using dissecting scissors, trim any spinal cord regions, up to where the brain stem meets the cerebellum, careful to not damage the cerebellum.
2.4. Insert one of the vascular scissor blades into the space between the brainstem and vertebral column and cut toward the ear canal, lifting up on the scissors to cleanly cut the bone but limit damage to the tissue.
2.5. Continue to cut along the edge of the skull up toward the olfactory bulbs, continuing to lift up while cutting to limit damage to the brain tissue.
2.6. Using the blunt forceps, gently peel off the back of the skull uncovering the posterior region of the brain and cerebellum.
2.7. Using the blunt forceps along the edge of the skull that was just cut, peel the rest of the skull up and over the brain. This step should remove the majority of the skull cap, revealing the brain.
2.8. Trim the rest of the skull with the vascular scissors and blunt forceps, clearing most of the skull from the top of the brain.
2.9. Using the microspatula, lift the brain slightly and scoop under and slide up to remove the olfactory bulbs from the remaining skull and disconnect optic tract fibers. Brain should come free easily at this point.
2.10. Place the brain into the petri dish sitting on ice and remove any remaining skull or other debris.
2.11. Using the microspatula, gently place the brain into the brain matrix with dorsal side up. Take time to make sure it is set level in the matrix, especially that the midline falls center in the matrix. This matrix is designed for adult mouse brain tissue, tissue from younger or diseased animals may rest lower in the matrix but it should still be possible to achieve reproducible results across samples.
2.12. Place one razor blade along the sagittal midline, making sure that the blade pushes all the way to the bottom of the matrix (Figure 1B).
2.13. Place another razor blade 1 mm to the side of the first blade (Figure 1B). Place two more blades 1 mm apart from each other. The end result should be three blades placed on one side of the brain, all 1mm apart. Do the same to the other side. In total, 7 blades will be placed 1mm apart (Figure 1C).
2.14. Carefully, grab the front and back ends of the razor blades and lift straight up out of the matrix. The tissue on the outside of the razor blades can be discarded.
2.15. Slowly, separate one razor blade at a time from the others, being careful not to damage the tissue sections.
2.16. Carefully slide the tissue section off of the razor blade and onto the glass slide with the microspatula. In total there will be six sagittal brain sections (Figure 1D).
2.17. The 4 most lateral sections will have DCN visible (Purple box, Figure 1D). To isolate the DCN, hold the trimmed 200 μl pipet tip perpendicularly over the DCN and push down through the tissue firmly, rocking in all directions to fully dissect the DCN from surrounding tissue. Lift straight back up to cleanly remove the DCN, and visually confirm the presence of the tissue in the tip.
2.18. Place one finger at the top of the tip and push down, causing the tissue to bulge out. Place the tip into correctly labeled microfuge tube and ensure that tissue punch is placed in the bottom of the tube. Repeat 2.17 for the remaining three sections, placing the DCN punches in the same tube. Place the tube into liquid nitrogen to flash freeze. Representation of size of each punch in Figure 1E.
2.19. Sections that had DCN extracted qualify as cerebellar hemisphere. Push away the rest of the brain tissue around the cerebellum in these sections. Use blunt forceps and gently pick up these hemisphere cerebellar cortex sections and place into respective microfuge tube. Flash freeze.
2.20. For the last two vermal sections (light blue box, Figure 1D), push away surrounding brain tissue leaving only the cerebellum. Using a razor blade, make a cut separating the anterior lobules from the posterior lobules. Cut should be just after the formation of lobule 6 and should not include lobule 10 (Figure 1F).
2.21. Using blunt forceps, carefully place the anterior cerebellar cortex sections and posterior cerebellar cortex sections into their respective microfuge tubes and flash freeze by leaving the tubes in liquid nitrogen for 5 minutes. From here move forward to RNA extraction, or may store the tubes at −80 °C.
3. RNA extraction
3.1. This protocol is modified from the Cold Spring Harbor Protocol for RNA Extraction with TRIzol20. TRIzol solubilizes biological material, making it possible to extract RNA.
3.2. Place the microfuge tubes in ice to keep the tissue from thawing too quickly, apply 150μl of cold TRIzol in the microfuge tube. Homogenize with a sterilized pestle. Once tissue is homogenized, pipet the solution up and down to ensure there is no remaining tissue intact. Further break up any small tissue pieces by pulling it up into an insulin syringe a few times.
3.3. Add another 350 μl of TRIzol and pipet up and down to mix thoroughly. Let sit at room temp for 5 minutes.
3.4. Add 150 μl of chloroform to the tube and shake vigorously and then let rest for 2-3 minutes. The chloroform separates the homogenized tissue solution into phases (RNA, DNA, and protein).
3.5. Centrifuge at 12,000 x g, at 15°C, for 10 minutes. Ensure that all tubes are at the same orientation.
3.6. Carefully remove the tubes and set the temperature of the centrifuge to 4°C. Remove only the clear aqueous phase into a new tube (this is the RNA), careful not to disrupt the opaque interphase (the DNA). The lowest phase will be red and contains protein. Remaining solution in the tubes can be saved or discarded.
3.7. Add 100% isopropyl alcohol at a 1:2 ratio (if removed 200 ul of aqueous phase, add 100 ul of isopropyl alcohol). Mix thoroughly by pipetting up and down. Let rest at room temperature for 10 minutes. The isopropyl alcohol precipitates the RNA out of solution.
3.8. Centrifuge at 12,000 × g, at 4°C, for 10 minutes. Be sure to place all the tubes at the same orientation to make it easier to visualize the pellet. The resulting pellet will be the extracted RNA.
3.9. Carefully remove the tubes, remove the supernatant with a pipet being careful not to disrupt the pellet. The pellet is somewhat gel-like and will be difficult to see, but should be able to estimate where it is based on the orientation of the tubes in the centrifuge.
3.10. After removing all of the supernatant, add 500 μl of 75% ethanol, vortex briefly, and centrifuge at 7500 × g, at 4°C, for 5 minutes. The ethanol further washes the pellet.
3.11. Remove the supernatant carefully, without disrupting the pellet. Leave caps open to dry the sample. This usually takes 5-10 minutes but can vary depending on how much ethanol was left on the pellet. Do not over dry.
3.12. Once dry, resuspend the pellet in DNase free water. Add 20 μl to samples for DCN, and 30 μl for all others.
3.13. Once resuspended, samples can be stored at −80 °C or proceed to further testing.
4. Real Time quantitative Polymerase Chain Reaction (RTqPCR)
4.1. Before this step it will be necessary to DNase treat the RNA to remove any genomic DNA, and generate cDNA utilizing iScript from BioRad. Be sure to normalize the concentration of RNA before making the cDNA. In addition, it is important to optimize primers for qPCR. Follow the methods described here to complete this step21. Brief description of the qPCR below.
4.2. Primers are all purchased from IDT. Forward and reverse sequences are both in the same sample tube, and stored at 20x concentration.
4.3. Rps18 (control gene): Forward – 5’-CCTGAGAAGTTCCAGCACAT-3’
Reverse – 5’-ACACCACATGAGCATATCTCC-3’
4.4. Parvalbumin (Calcium binding protein, inhibitory cells):
Forward – ‘5-ATGAGGTGAAGAAGGTGTTCC-3’
Reverse – ‘5-AGCGTCTTTGTTTCTTTAGCAG-3’
4.5. Kcng4 (Potassium channel subunit): Forward – 5’-CTGTCTTTTCCTGGTCAGTGA-3’
Reverse – 5’ -GCATTGCCTCAGACTGTCAG-3’
4.6. Aldolase C (Zebrin II, differentially expressed across the cerebellar cortex):
Forward – 5’-AGAGGACAAAGGGATAATGCTG-3’
Reverse – 5’-TCAGTAGGCATGGTTGGC-3’
4.7. qPCR conditions were as follows. Pre-incubation period, 5 minutes, at 95°C. Amplification period, 50 cycles: 10 minutes at 95°C, 10 minutes at 62°C, and 10 minutes at 72°C. Melting curve period, 5 seconds at 95°C, one minute at 65°C, and set ramp rate to .07°C/second until target temp of 97°C. Then cooling period for 10 minutes to 40°C. All qPCR reactions were performed in triplicate and gene expression was analyzed using 2− ΔΔCt with Rps18 as a loading control to normalize gene expression. Bulk cerebellar extract was used as a reference.
REPRESENTATIVE RESULTS:
For these experiments, four eleven-week-old female wild type C57/Black6 mice were used. One mouse was used to conduct a full cerebellar dissection which is referred to as ‘bulk cerebellum’ and allowed for the comparison of RNA levels in dissected regions to a full dissection. The other three mice were used to conduct the cerebellar dissection described in this protocol. Using three mice makes it possible to ensure that the trends detected in the levels of RNA are reproducible across mice.
Figure 1A represents the mouse cerebellum – vermis in blue and hemispheres in yellow. Sagittal schematics of the vermis (anterior regions in dark blue, posterior regions in light blue) and hemisphere (in yellow). The DCN are highlighted in red dotted boxes. A successful dissection will start with the initial razor blade placement down the midline of the brain (Figure 1B); this guides the successful placement of the following six blades (Figure 1C). This leaves six sagittal sections (Figure 1D), four lateral/hemisphere sections (outlined in purple) and two midline/vermal sections (outlined in light blue). The 4 lateral sections contain the DCN; figure 1E depicts a successful DCN punch dissection. The midline vermal sections will most likely have half of the most medial DCN present in the 1 mm thick section, however it will not be present all the way through and isn’t possible to reproducibly dissect out. The midline vermal sections are separated into anterior and posterior lobules depicted in figure 1F. A successful dissection sets up the rest of the experiments for success.
Real Time qualitative Polymerase Chain Reaction (RTqPCR) results demonstrate the feasibility of assessing the gene expression levels of each individual region as well as serve to validate dissections. We used primers detecting genes that show gradient expression from anterior to posterior cerebellar cortex and enrichment in the DCN and compared their expression to the bulk cerebellar lysates.
We assessed the expression levels of three genes: aldolase C, parvalbumin, and Kcng4. Aldolase C, also known as zebrin II, is an enzyme that is expressed in a consistent banding pattern through the cerebellum. It is expressed more highly in the posterior vermis than the anterior vermis. There are also bands in the hemispheres13,14. Parvalbumin which is a calcium binding protein that is expressed in inhibitory cells. Based on the Allen Brain Atlas, parvalbumin seems relatively uniformly expressed throughout the cerebellar cortex and in the DCN (http://mouse.brain-map.org/gene/show/19056). Kcng4, a potassium voltage gated channel subunit, appears to be enriched in the DCN and in the anterior compared to the posterior lobes (https://mouse.brain-map.org/gene/show/42576). Quantitative expression analysis showed that, as expected, aldolase C is more highly expressed in the posterior cerebellar vermis (CCpV) but lower in the DCN and the anterior region of the vermis (CCaV) when compared to the bulk cerebellar dissection (Figure 2A). Parvalbumin is similarly present in the DCN, anterior vermis, posterior vermis, and hemisphere cerebellar cortices as in the bulk cerebellar extracts (Figure 2B). Kcng4 is significantly enriched in the DCN and the anterior vermis (CCaV) and it is not significantly enriched in the posterior vermis (CCpV) or hemispheres (CCH) when compared to the bulk extraction (Figure 2C). This result follows what was expected based on the pattern seen in the Allen Brain Atlas. Thus, gene expression analysis validates the dissection protocol and confirms that good quality RNA can be obtained and tested.
Figure 2: Relative gene expression in isolated specific regions of the cerebellum.
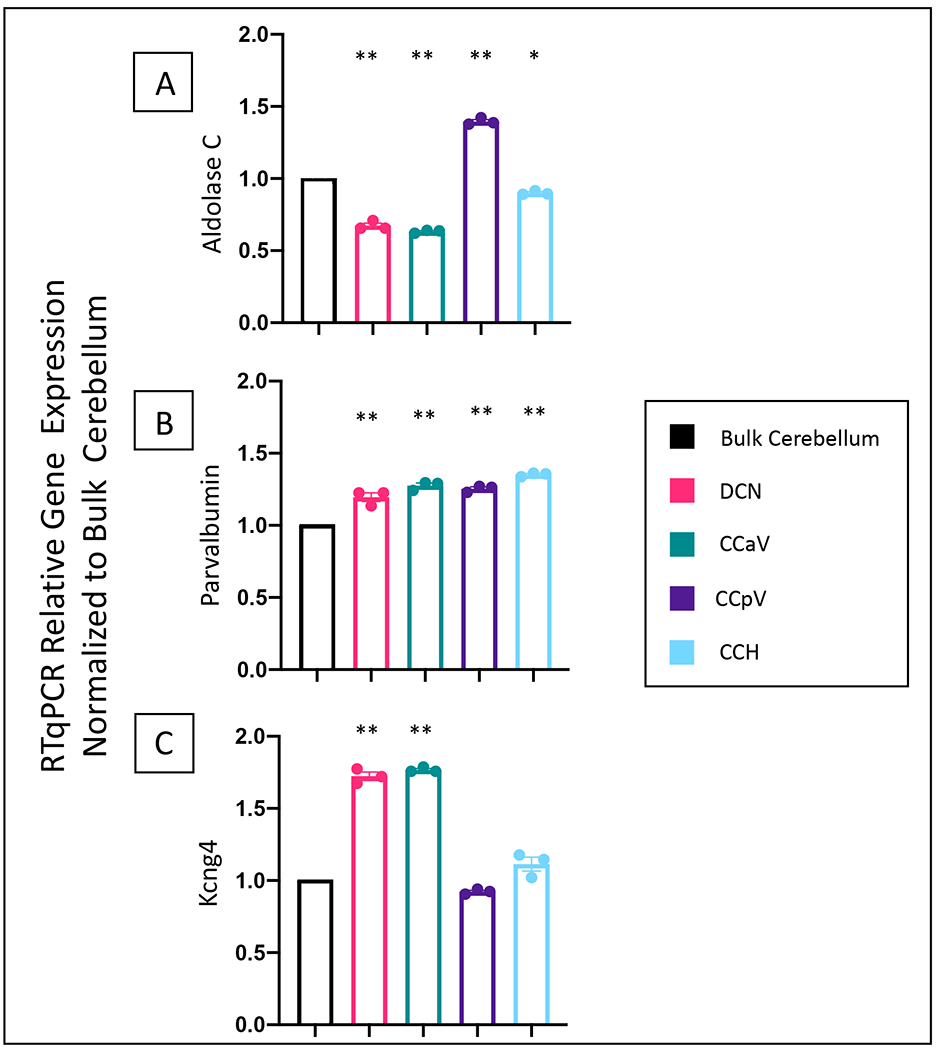
Relative expression of aldolase C (2A), parvalbumin (2B), and Kcng4 (2C) normalized to Rps18 (protein associated with ribosomal RNA, expressed in all cells), and using bulk cerebellar extract as a reference. As expected, aldolase C expression was higher in the posterior vermis and lower in the DCN and anterior vermis (2A). As expected, based on Allen Brain Atlas, parvalbumin expression is uniformly expressed across each extracted region, while Kcng4 expression is significantly enriched in the DCN and CCaV. One-way ANOVA, with a Tukey’s post hoc test. *p<.005, ** p<.0001 relative to the bulk cerebellar extract. Histograms represent average values for N=3 with values for each individual mouse shown as dots. Error bars represent standard error of the mean. DCN (Deep cerebellar nuclei), CCaV (the cerebellar cortex of anterior vermis), CCpV (the cerebellar cortex of the posterior vermis), and CCH (the cerebellar cortex of the hemispheres). N=3 mice for cerebellar dissection regions. N=1 bulk extract. Experiment done in triplicate.
To directly compare the expression of aldolase C across the cerebellar cortex, the expression levels were compared to where it is supposed to be lowest, the anterior vermis (CCaV) (Figure 3). The expression level of aldolase C was significantly higher in the posterior vermis (CCpV), and trending higher in the cerebellar hemispheres (CCH), though not quite significantly so. This trend in the cerebellar hemispheres is likely because there are bands of aldolase C in the hemispheres, and the dissection captures aldolase negative and positive bands.
Figure 3: RTqPCR Relative gene expression of aldolase C across cerebellar cortex.
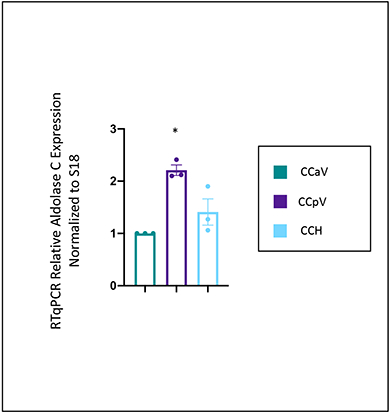
Relative expression of aldolase C in specific regions of the cerebellar cortex – anterior vermis (CCaV), posterior vermis (CCpV), and hemispheres (CCH). Gene expression level of aldolase C was normalized to Rps18 and compared to the expression level in the anterior vermis. As expected aldolase C expression was enriched in the posterior vermis. One-way ANOVA, with a Tukey’s post hoc test. *p<.005 relative to CCaV. Error bars represent standard error of the mean. CCaV (the cerebellar cortex of anterior vermis), CCpV (the cerebellar cortex of the posterior vermis), and CCH (the cerebellar cortex of the hemispheres). N=3 mice, experiment done in triplicate.
DISCUSSION:
The method described here makes it possible to assess the underlying gene expression and molecular mechanisms within four distinct cerebellar regions – the deep cerebellar nuclei (DCN), the anterior cerebellar cortex of the vermis (CCaV), the posterior cerebellar cortex of the vermis (CCpV), and the cerebellar cortex of the hemispheres (CCH). The ability to assess these regions separately will expand our knowledge of the heterogeneity of specific cerebellar regions and possibly shed light on their contribution to various behaviors.
By sectioning the full brain tissue sagittally it is possible to easily visualize and identify these four regions of the cerebellum, allowing for a quick dissection. To the best of the author’s knowledge this is the first description of full cerebellar regional dissection for molecular analysis. In a paper recently published, researchers used bulk dissection of the anterior lobules of the cerebellum and the nodular lobule of the cerebellum for molecular analysis22. However, with the described method they would not be able to also dissect the DCN. Attempts to isolate DCN punches using a vibratome to cut 300um thick slices usually run into three critical problems. First, due to slight differences in mounting of the cerebella and the angle at which the tissue is cut, it is not easy to reproducibly isolate the same regions across animals. Second, mounting and slicing takes time, increasing the chances of RNA degradation and decreasing quality of RNA sample for downstream applications. Lastly, punches from 300um thick slices produce low yield of RNA. Data shown here suggests that it is possible to use this technique to assess relative gene expression across cerebellar regions. Aldolase C is highly upregulated in the CCpV. Kcng4 is highly upregulated in the DCN and CCaV, and parvalbumin is expressed relatively equally across all regions in the cerebellum. While these are only three genes, these results demonstrate that this dissection method can be used to identify the underlying molecular signatures of these distinct regions.
There are a few critical things to pay attention to throughout this protocol. Positioning the brain in the matrix is an important step. In some cases, the brain may have not been set perfectly level in the matrix or exactly on the midline. This would become clear when looking at each of the sagittal sections. For example, if cut exactly at the midline, the most medial cerebellar sections will look almost identical and have no DCN visible. Because these landmarks are identifiable by eye it is possible to troubleshoot how many sections qualify as vermal or hemisphere sections, and they can be combined together in the same microfuge tube. Another critical step is the extraction of the DCN. In some instances, depressing a finger to the top of the 200 μl pipet tip is not enough pressure to extract the tissue punch. If this is the case, it will be necessary to scoop out the punch with a needle nose forceps and place the punch in the correct tube. The next critical step of this protocol is the RNA extraction. While the volumes listed have been optimized for maximal RNA extraction, it may also be necessary to adjust the amounts of solution in the RNA extraction protocol to fit the lab’s individual needs. Because there is little tissue to work with, there is a higher chance of phenol contamination in the final RNA product. This can be managed by adjusting the amount of TRIzol used to homogenize the tissue and ensuring that the homogenized tissue is thoroughly mixed.
Limitations of this protocol are that while the DCN are divided into three separate nuclei (lateral, interposed, and medial) it is difficult to visualize and reproducibly punch each of these out of 1 mm sections, let alone have enough tissue from which to extract RNA. Along with this limitation, half of the most medial DCN appears in the vermis; however, with this dissection it is difficult to visualize and reproducibly dissect out. As the protocol is currently, the other half of the medial DCN and the remaining DCN are dissected out. There are also ten individual vermal lobules and eight lobules in the hemispheres, but to dissect each of these individually and in a reproducible manner would be difficult and lead to very low RNA concentration yields. While this method does make it possible to delve more specifically into regions of the cerebellum, it does still combine distinct anatomical regions into groups which could be masking unique changes on the level of individual lobules or sub-lobule units.
In conclusion, this method makes it possible to simultaneously explore regional molecular differences four specific regions of the cerebellum. By assessing the cerebellum in this region-specific manner, it is possible to tease apart underlying molecular mechanisms and gene expression that characterize those regions. This will further our understanding of the role that the distinct cerebellar regions play in different behaviors and allow for future work to focus specifically at one cerebellar region and explore its role in disease or target treatment.
Table of Materials
| Name of Material/ Equipment | Company | Catalog Number |
|---|---|---|
| 1.5 Microcentrifuge tubes | ThermoScietific | 3456 |
| 100% Isopropyl Alcohol | VWR Life sciences | 1106C361 |
| 200 ul Pipet tips | GeneMate | P-1237-200 |
| Adult Mouse Brain Matrix Sagittal | Kent Scientific Corporation | RBMA-200S |
| Blunt forceps | ||
| Chloroform | Macron | 220905 |
| Decapitation Scissors | ||
| Dissecting Scissors | ||
| Ethyl Alcohol | Pharmco | 111000200 |
| Glass Slide (for electrophoresis) | BIORAD | |
| Homogenizer | Kimble | 6HAZ6 |
| Ice Bucket | ||
| Insulin Syringe (.5ml) | BD | 329461 |
| iScript Adv cDNA kit for RT-qPCR | BIORAD | 1725037 |
| Micro Spatula | ||
| Needle Nose forceps | ||
| Petri Dish | Pyrex | |
| Primetime Primer for Aldolase C | IDT | Mm.PT.58>43415246 |
| Primetime Primer for Kcng4 | IDT | Mm.PT.56a.9448518 |
| Primetime Primer for Parvalbumin | IDT | Mm.PT.58.7596729 |
| Primetime Primer Rps18 | IDT | Mm.PT.58.12109666 |
| Single Edge Rzor Blades | Personna GEM | |
| Sterile, sigle-use pestles | FisherScientific | 12141364 |
| TRIzol Reagent | Ambion by Life technologies | 15596018 |
| Vascular Scissors | ||
| Comments/Description |
ACKNOWLEDGMENTS:
We are grateful to Austin Ferro and Juao-Guilherme Rosa in the Cvetanovic lab for their help in troubleshooting dissections and in RNA extraction and RTqPCR. This research is funded by M. Cvetanovic, R01 NS197387; HHS | National Institutes of Health (NIH).
Footnotes
DISCLOSURES:
The authors have nothing to disclose.
REFERENCES:
- 1.Herculano-Houzel S The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia. 62 (9), 1377–1391, doi: 10.1002/glia.22683 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Schmahmann JD, Caplan D Cognition, emotion and the cerebellum. Brain. 129 (2), 290–292, doi: 10.1093/brain/awh729 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Badura A et al. Normal cognitive and social development require posterior cerebellar activity. eLife. 7, e36401, doi: 10.7554/eLife.36401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diedrichsen J, King M, Hernandez-Castillo C, Sereno M, Ivry RB Universal Transform or Multiple Functionality? Understanding the Contribution of the Human Cerebellum across Task Domains. Neuron. 102 (5), 918–928, doi: 10.1016/j.neuron.2019.04.021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strick PL, Dum RP, Fiez JA Cerebellum and Nonmotor Function. Annual Review of Neuroscience. 32 (1), 413–434, doi: 10.1146/annurev.neuro.31.060407.125606 (2009). [DOI] [PubMed] [Google Scholar]
- 6.King M, Hernandez-castillo CR, Poldrack RA, Ivry RB, Diedrichsen J Functional boundaries in the human cerebellum revealed by a multi-domain task battery. Nature Neuroscience. 22 (August), 1371–1378, doi: 10.1038/s41593-019-0436-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BTT The organization of the human cerebellum estimated by intrinsic functional connectivity. Journal of neurophysiology. 106 (5), 2322–45, doi: 10.1152/jn.00339.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmahmann JD From movement to thought: Anatomic substrates of the cerebellar contribution to cognitive processing. Human Brain Mapping. 4 (3), 174–198, doi: 1996. [DOI] [PubMed] [Google Scholar]
- 9.Kelly RM, Strick PL Cerebellar Loops with Motor Cortex and Prefrontal Cortex of a Nonhuman Primate. The Journal of Neuroscience. 23 (23), 8432–8444 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoodley CJ, Macmore JP, Makris N, Sherman JC, Schmahmann JD Clinical Location of lesion determines motor vs . cognitive consequences in patients with cerebellar stroke. NeuroImage: Clinical. 12, 765–775, doi: 10.1016/j.nicl.2016.10.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo CC, Tan R, Hodges JR, Hu X, Sami S, Hornberger M Network-selective vulnerability of the human cerebellum to Alzheimer’s disease and frontotemporal dementia. Brain. 139 (5) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bocchetta M, Cardoso MJ, Cash DM, Ourselin S, Warren JD, Rohrer JD Patterns of regional cerebellar atrophy in genetic frontotemporal dementia. NeuroImage: Clinical. 11, 287–290, doi: 10.1016/j.nicl.2016.02.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sillitoe RV, Fu Y, Watson C Cerebellum. The Mouse Nervous System. 360–397, doi: 10.1016/B978-0-12-369497-3.10011-1 (2012). [DOI] [Google Scholar]
- 14.Nguyen-Minh VT, Tran-Anh K, Luo Y, Sugihara I Electrophysiological Excitability and Parallel Fiber Synaptic Properties of Zebrin-Positive and -Negative Purkinje Cells in Lobule VIII of the Mouse Cerebellar Slice. Frontiers in Cellular Neuroscience. 12, 513, doi: 10.3389/fncel.2018.00513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manto M, Oulad Ben Taib N Cerebellar Nuclei: Key Roles for Strategically Located Structures. The Cerebellum. 9 (1), 17–21, doi: 10.1007/s12311-010-0159-8 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Driessen TM, Lee PJ, Lim J Molecular pathway analysis towards understanding tissue vulnerability in spinocerebellar ataxia type 1. eLife. 7, doi: 10.7554/eLife.39981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabert K et al. Microglial brain region - dependent diversity and selective regional sensitivities to aging. Nat Neurosci. 19 (3), 504, doi: 10.1038/nn.4222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram M et al. Cerebellar Transcriptome Profiles of ATXN1 Transgenic Mice Reveal SCA1 Disease Progression and Protection Pathways. Neuron. 89 (6), 1194–1207, doi: 10.1016/j.neuron.2016.02.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cendelin J From mice to men : lessons from mutant ataxic mice. 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rio DC, Ares M, Hannon GJ, Nilsen TW Purification of RNA Using TRIzol (TRI Reagent). Cold Spring Harbor Protocols. 2010 (6), pdb.prot5439-pdb.prot5439, doi: 10.1101/pdb.prot5439 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Kim JH, Lukowicz A, Qu W, Johnson A, Cvetanovic M Astroglia contribute to the pathogenesis of spinocerebellar ataxia Type 1 (SCA1) in a biphasic, stage-of-disease specific manner. Glia. 66 (9), 1972–1987, doi: 10.1002/glia.23451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chopra R et al. Altered Capicua expression drives regional Purkinje neuron vulnerability through ion channel gene dysregulation in spinocerebellar ataxia type 1. Human Molecular Genetics. 10.1093/hmg/ddaa212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


