Preface
Inflammatory bowel disease (IBD) is a complex genetic disease that is instigated and amplified by the confluence of multiple genetic and environmental variables that perturb the immune- microbiome axis. The challenge of dissecting pathological mechanisms underlying IBD has spawned the development of transformative approaches in human genetics and functional genomics. Here, we describe IBD as a model disease in the context of leveraging human genetics to dissect cellular and molecular pathway interactions that control homeostasis of the mucosal immune system. Finally, we synthesize emerging insights from multiple experimental approaches into pathway paradigms and discuss future prospects for disease subtype classification and therapeutic intervention.
Introduction
The contemporary view of inflammatory bowel disease (IBD) has evolved rapidly as a result of recent advancements in human genetics, mucosal immunology, and microbiome research. IBD is a chronic relapsing and remitting disease associated with dysregulation of the mucosal immune system and the commensal ecosystem. As such, IBD is regarded as a model disease that exemplifies the complexities of interactions between genetic, immune and environmental variables that coordinately impact disease. IBD genetics has benefited significantly from studying common variants in large cohorts of well-phenotyped patients and rare variants in cases associated with Mendelian inheritance. Functional studies inspired by IBD genetics helped to uncover fundamental mechanisms of immunity, host-microbe interactions, and offer actionable insights into therapeutic innovation. Accordingly, the last decade of IBD research exposed genetic vulnerabilities in an expansive interconnected network of host pathways that intimately interface with the microbiome. This work has helped to shape the emerging view of IBD as a system-level perturbation of the mucosal immune system and commensal ecosystem. This network dyshomeostasis model defies simple cause and effect explanations of IBD triggers; rather, it invokes many environmental variables acting cumulatively over time on the backdrop of many genetic variants to explain the evolution of the pathological relationship between host and microbiome. Below, we highlight significant advancements in our understanding of IBD pathogenesis that provide novel insights into human biology with relevance across the spectrum of health and disease.
Clinical Features of IBD.
IBD is viewed as two subtypes, ulcerative colitis (UC) and Crohn’s disease (CD), with UC affecting the colon and CD affecting any region of the gastrointestinal tract, but primarily the terminal ileum of the small intestine1. IBD onset typically occurs in early adulthood, although it can occur at any age and is increasing in prevalence at all ages, including in early onset and geriatric populations. IBD is driven by genetics and environment, such that dysregulated mucosal immune function is associated with a dysbiotic commensal microbiome that coordinately drives a waxing and waning pathological inflammatory cycle. Although IBD has historically been considered a disease of the West, incidences in the Eastern hemisphere are increasing dramatically and follow geographical patterns of industrialization/westernization2.
Heterogeneity of Disease.
Accumulating evidence indicates that IBD is more heterogeneous than the traditional UC/CD dichotomy. Thus, IBD likely comprises many disease subtypes that are distinguishable based on the natural history of disease, response to treatment, and distinct genetic risk factors. Clear distinctions based on molecular phenotypes remain elusive. With bourgeoning clinical datasets comprising genotype data, molecular diagnostics, and metagenomics, the field is poised to define a clinically informative framework for classifying and treating IBD subtypes3. New biomarkers have emerged from similar efforts and prognostic markers in blood have been identified to classify IBD patients based on risk of frequent relapse and complicated clinical outcomes. In particular, a CD8 T cell exhaustion gene expression signature predicts positive outcomes in IBD and autoimmunity4.
Clinical studies provide novel insights into heterogeneity of disease at the level of pathogenesis and treatment response rates. Accordingly, inception cohorts offer a unique view of IBD at the time of diagnosis, prior to therapeutic intervention, and enable longitudinal monitoring of disease progression5. It is important to note the inherent challenges and potential power of patient-driven research and reverse translation of clinical observations into hypotheses for validation in the laboratory. Indeed, the field is in early stages of translating gene expression signatures and multi’omic data into a tangible understanding of disease. Nevertheless, these data function as a snapshot of disease that can be mined and integrated with other data, including genetics and functional biology experiments. For example, expression of genes within the IL-7R pathway were elevated prior to treatment in patients that subsequently failed anti-TNF6. In experimental models, IL-7R signaling induced expression of ɑ4β7 integrin and imprinted gut homing ability upon T cells. In humanized mice, IL-7R blockade reduced T cell homing to the gut and subsequent intestinal inflammation6. These studies shed light on why subsets of patients may respond to treatment while others lose responsiveness or fail to respond initially. Given that a significant percentage of patients do not respond to anti-TNF and other biologics, there is a pressing need to identify new therapeutic mechanisms of action.
Intestinal Complications and Learning Opportunities.
IBD pathology escalates and often leads to the development of heterogeneous comorbidities and other complications that shed light on mechanisms underlying intestinal homeostasis. In some cases, colectomy in UC patients can evolve into a CD-like disease associated with ileal pouch inflammation (pouchitis), demonstrating a level of plasticity in disease progression that is poorly understood. Additionally, UC can be associated with complications affecting the gut-liver axis such as primary sclerosing cholangitis (PSC). PSC is a chronic idiopathic cholangiopathy that can progress to cirrhosis and liver disease and is often associated with liver, gallbladder, or colon carcinogenesis. Thus, the burden of IBD over time is particularly evident in the elevated risk of cancer associated with both CD and UC. Conversely, intestinal pathologies can arise as comorbidities associated with other clinical contexts. For example, colitis associated with checkpoint inhibition tumor immunotherapy highlights the importance of T cell coinhibitory pathways in maintaining intestinal tolerance7. Furthermore, IL-17 has emerged as a surprising arbiter of intestinal homeostasis based on clinical observations of exacerbated IBD symptoms in psoriasis patients treated with biologics that inhibit the IL-17 pathway8. Taken together, studying intestinal complications arising from IBD and other diseases can reveal unexpected mechanisms of immunoregulation.
Genetics of IBD
Common Variant Association Studies and Fine Mapping Disease Traits.
The post-human genome era rapidly ushered in efforts to map the genetic heterogeneity of human populations with the objective of functionally assigning genotype-phenotype relationships, particularly in the context of complex genetic diseases. Although GWAS have been remarkably successful at implicating IBD risk loci, identifying the causal genes and variants within these loci requires additional approaches (Figure 1). GWAS have been empowered by expanding arrays to allow dense genotyping of key immune-related loci9. As the number of IBD cases in GWAS increased, so has the power to identify additional risk-associated loci. Ongoing efforts to incorporate diverse ancestry in GWAS shows promise in capturing global genetic diversity that contributes to disease risk, although this may require genotyping strategies that capture genetic variation beyond what is typically captured in European-derived ancestry10. Trans-ancestry association studies of IBD have identified differential risk associations for NOD2 and TNFSF15 wherein the former dominates in European and the latter in East Asian populations10. The most recent integrative analysis of IBD GWAS analyzed nearly 60,000 subjects (including over 25,000 IBD cases) and identified approximately 240 loci statistically associated with risk of developing IBD11. Fine-mapping GWAS studies have helped to refine risk loci to implicate specific causal SNPs associated with IBD12,13. Although this approach is powerful, it has limitations for risk haplotypes that comprise many noncoding SNPs that exist in linkage disequilibrium and cooperatively function to impact gene expression through epigenetic mechanisms14,15. With a growing number of loci associated with IBD risk and progression, the field is poised to convert these associations into causal disease mechanisms16.
Figure 1. Strategies in human genetics and functional genomics to dissect mechanisms of disease.
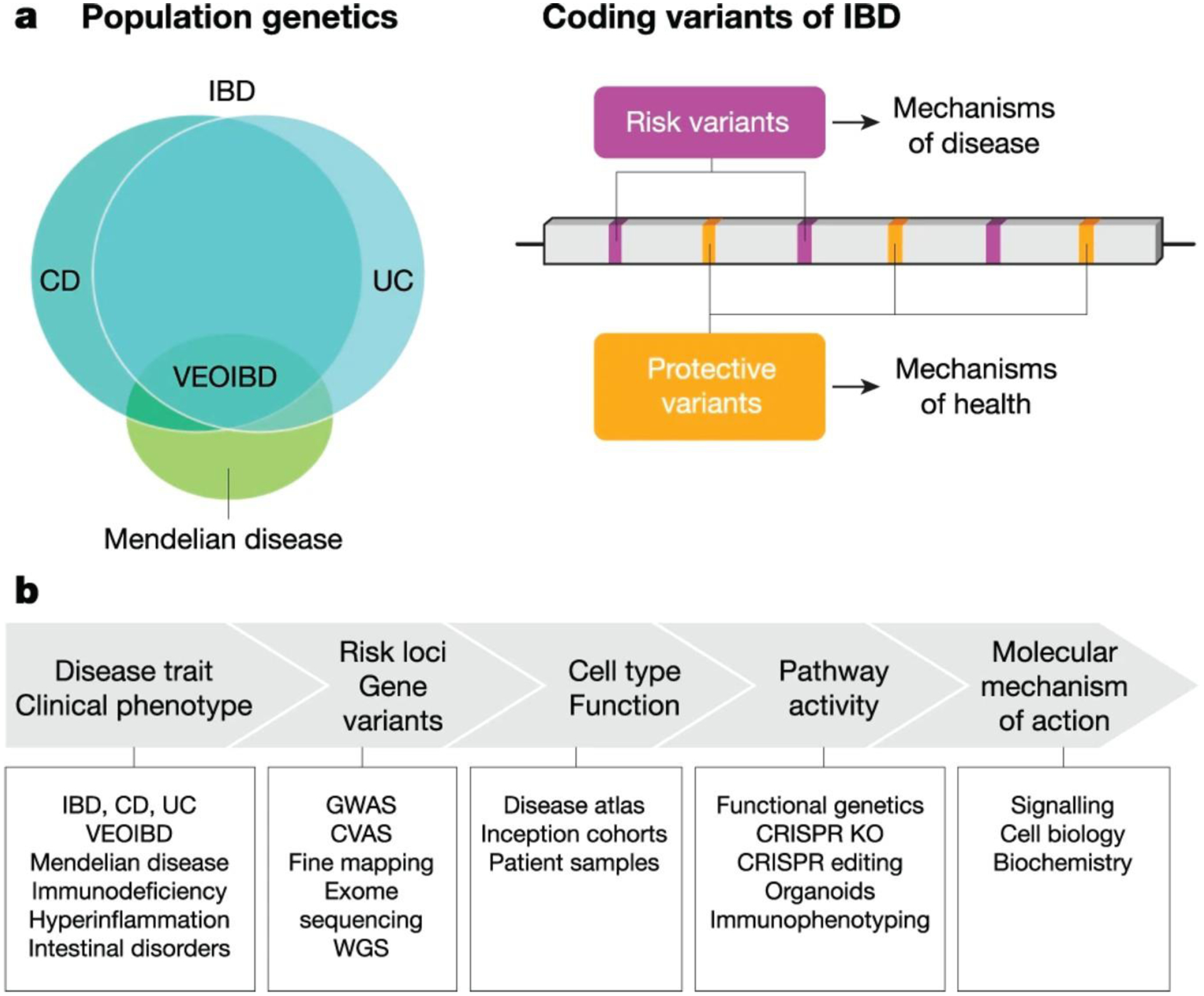
a, Left, IBD is a complex genetic disease that is affected by many genetic risk factors defined in common variant association studies (CVAS). These genetic risk factors partially distinguish disease phenotypes associated with CD versus UC. Rare genetic variants associated with severe IBD and very early-onset (VEO)IBD exhibit Mendelian inheritance patterns and have helped to identify genes that control intestinal homeostasis. Similar insights have been gained from rare genetic variants associated with primary immunodeficiencies and hyperinflammatory disorders that manifest with intestinal pathologies or those that confer protection from these conditions. Right, mechanistic study of individual coding variants comprising an allelic series for a gene can reveal molecular functions at the protein level. Understanding the molecular function of a risk variant can reveal mechanisms of disease and conversely, study of protective variants can reveal mechanisms of health. b, Functional genetics has helped to mechanistically link risk genes and genetic variants with cellular and molecular functions underlying the disease process. KO, knockout; MOA, mechanism of action; WGS, whole genome sequencing.
Exome Sequencing Identifies Risk and Protective Variants.
Genotyping SNPs in GWAS assigns a binary risk or non-risk phenotype to haplotypes spanning genomic loci comprised of SNPs across multiple genes and intergenic regions. In contrast, exome sequencing is not constrained by genotyping known variants, and is powered for discovery of rare coding variants that act independently of common haplotypes and often influence disease risk with larger effect sizes. Searching for rare causal variants by exome sequencing is constrained by the requirement for large cohorts. One way around this potential limitation is to study populations that have been subject to a founder effect, or genetic bottleneck, in which rare alleles and their associated phenotypes may be enriched17–19. One example is the Ashkenazi Jewish (AJ) population, in which the incidence of CD is 2–4 times higher than non-Jewish (NJ) European populations17. Exome sequencing studies comparing AJ vs NJ European populations revealed novel coding variants in GWAS loci and in novel genes that may explain the increased incidence of CD in the AJ population17. CD risk in the AJ population is enriched in genes related to autophagy, whereas in the NJ population, it is enriched for genes involved in the IL-23/17 pathway17. Genetic studies in cohorts of different ethnicities hold great potential for identifying the heterogeneity underlying genetic risk factors and suggest the existence of discrete disease subtypes potentially driven by distinct mechanisms.
Exome sequencing has identified multiple independent coding variants in genes that collectively represent an allelic series associated with a spectrum of phenotypes, ranging from risk to protection from disease. For example, exome sequencing identified risk and protective variants comprising allelic series in several genes including CARD920,21, IL23R20, and RNF18619,21. These findings highlight the importance of innate microbial sensing pathways, cytokine networks, and barrier function in IBD risk. Additional functional studies are required to reveal the mechanistic basis of risk versus protection, and as discussed below, these efforts are important for guiding the development of therapeutic strategies to inhibit risk-related mechanisms or mimic protective mechanisms (Figure 1).
Mendelian Genetics.
Genome sequencing has revolutionized mapping of genetic variants to monogenic traits in the context of Mendelian disorders. This is particularly true in cases of primary immunodeficiencies associated with intestinal manifestations and in very early onset IBD (VEOIBD), which typically occurs in kindred prior to the age of six. The discovery of causal genes and variants associated with severe intestinal pathologies uncovered unique mechanisms controlling intestinal homeostasis. One of the first genes associated with VEOIBD was IL10RA, in which severe hypomorphic alleles reduced IL-10 signaling, resulting in impaired tolerance22. IL-10R signaling in macrophages is essential for limiting intestinal inflammation23–25. Similarly, GWAS studies identified noncoding SNPs near IL10 and a weaker signal in the IL10RA locus associated with adult onset IBD. Importantly, Mendelian disorders uncovered novel genes and pathways associated with intestinal pathology not detected in GWAS (Figure 1). To date, more than 350 primary immunopathologies have been identified and linked with distinct molecular etiologies, roughly one third of which are associated with gastrointestinal symptoms26. These immunopathologies span the phenotypic spectrum from immunodeficiencies to hyperinflammatory disorders. For example, lymphoproliferative disorders associated with deficiencies in cell death regulating genes, such as Caspase 8 and XIAP, or genes involved in T cell regulatory function such as FOXP3 or CTLA426 typically elicit intestinal inflammation. On the opposite end of the spectrum, mutations linked to immunodeficiencies revealed insights into gut mucosal immunity. Defective TCR repertoire selection was observed in kindred bearing a novel mutation in MALT1 that impaired NFkB signaling27. Additionally, intestinal manifestations occur in innate immunodeficiencies such as chronic granulomatous disease (CGD), which is associated with mutations in CYBB28. Similarly, the lysosomal storage disease Niemann-Pick disease type C1 manifests with CD like symptoms due to impaired antibacterial activity29. Taken together, emerging insights suggest that IBD is associated with hyperinflammatory conditions related to cytokine imbalance and also immunodeficiencies related to defective antimicrobial responses. Immunodeficiencies may lead to an unhealthy relationship with the microbiome, thus offering a potential explanation for why these pathologies target the intestine.
A number of genetic variants have been linked with Mendelian diseases associated with intestinal pathologies that directly impact epithelial function30. In particular, regulators of epithelial polarity such as TTC7A are associated with VEOIBD. TTC7A is thought to function as a critical component of the PI4KIIIα complex to maintain epithelial apico-basal polarity31. Other examples highlight genes controlling epithelial barrier functions, such as the epithelial adhesion molecule EPCAM and SPINT232, which are associated with congenital tufting enteropathy and congenital sodium diarrhea, respectively. In fact, mechanisms controlling intestinal electrolyte balance are important genetic vulnerabilities for intestinal inflammation. Familial GUCY2C diarrhea syndrome, which is linked to a high prevalence of IBD, is associated with gain of function variants that induce intestinal secretion of NaCl and water through CFTR and inhibition of sodium hydrogen exchanger 3 (NHE3)33. GUCY2C encodes an epithelial receptor that recognizes endogenous ligands (uroguanylin and guanylin) and induces production of cyclic guanosine monophosphate (cGMP), which regulates CFTR through protein kinase GII activation. Taken together, these findings implicate novel genes that regulate pathways, such as electrolyte balance, epithelial polarity, and barrier function.
Functional Genomics.
With a rapidly expanding number of genetic associations statistically linked with IBD and intestinal pathologies, the field is moving to convert these insights into a mechanistic understanding of disease risk or pathological mechanisms. Complicating matters, genetic associations occur in a background of thousands of genetic variants per individual that collectively interact with environmental variables to impact disease. Traditionally, implicating a gene or variant as causal with respect to a phenotype involves gene knock-out studies to demonstrate the requirement for a gene, or alternatively, knock-in to determine sufficiency for a variant to induce a phenotype (Figure 1). Knock-out or knock-down screening strategies have been employed to demonstrate the requirement for candidate IBD risk genes in pathways such as autophagy34, antimicrobial function35, Th17 development36, and inflammatory cytokine production37,38. CRISPR- based approaches have also been developed to functionally map noncoding regions of the genome, such as enhancers, that are associated with diseases such as IBD39. CRISPR-mediated genome editing technologies enable functional characterization of coding variants implicated in IBD in isogenic cells and mouse models that differ from their corresponding controls only in the introduced variant. These approaches interrogate the phenotype of a single coding variant, and emerging CRISPR methodologies using base editing technology suggest the possibility of phenotyping many IBD risk variants in parallel, in screening mode.
Mouse models have been a mainstay for modeling intestinal inflammation, while mechanistic in vitro studies have benefitted from advancements in organoid technology. Organoid models recapitulate key intestinal epithelial features and can be engineered using CRISPR technology to functionally characterize disease-associated genes40. These organoid systems have been exploited to define gene function in epithelial-intrinsic cellular processes and also to dissect crosstalk between immune and epithelial lineages41. Increasingly sophisticated coculture systems are being developed to mimic intestinal biology in vitro in gut-on-a-chip platforms that incorporate multiple cell types, microbial communities, and physiological conditions such as oxygen gradients and forces associated with fluid flow42. These model systems hold great potential for identifying functional roles for IBD risk genes and variants.
Single Cell Technology and the Disease Atlas.
As in vitro cellular models become more sophisticated and capable, new technologies in single cell genomics offer an unprecedented view of the disease process in patients. Single cell transcriptomics allows for an unbiased census of cell lineages and their functional states in health and disease. Perhaps more importantly, single cell studies are powered to deconvolute cellular interactions and pathway crosstalk underlying the pathophysiology of disease. Several recent studies performed single cell transcriptomics on intestinal biopsies derived from UC patients43–45, CD patients46, and mouse models of enteric infection47. Accordingly, a cellular atlas was constructed from paired biopsies derived from UC patients and healthy controls resulting in identification of 51 cell types/states based on transcriptomic profiles45. These profiles capture important cellular lineages and biological functions impacting IBD and intestinal homeostasis (Figure 2). During inflammation, the cellular composition of the mucosa changes, with a notable influx of inflammatory monocytes and differentiation of inflammatory fibroblasts45. Modeling intercellular communication by mapping expression of receptor-ligand pairs revealed a potential functional link between inflammatory monocytes, which produce the IL-6 family cytokine OSM and inflammation-associated fibroblasts, which express the receptor heterodimer OSMR-IL6ST45. In fact, this myeloid cell-derived OSM circuit with fibroblasts induces expression of inflammatory chemokines and is implicated in severe IBD associated with resistance to anti-TNF therapy48. Integrating IBD GWAS and disease atlases can be leveraged to identify precise cell type(s) in which candidate genes are expressed, whether disease status correlates with their transcriptional regulation, or if they are transcriptionally coregulated within functionally related gene modules that hint at mechanisms of action (Figure 2).
Figure 2. IBD genes and pathways controlling mucosal immunity.

IBD risk genes regulate a complex network of interconnected functional pathways. IBD genes (red text) have been implicated in key biological functions (grey circles) that are controlled by interconnected molecular pathways (coloured rectangles). Lines connecting nodes reflect overlapping molecular regulation by common genes. Several IBD risk genes regulate several distinct biological functions depending on their cell type-specific activities.
Regulation of gene expression is cell type-specific and context-dependent. Thus, quantification at single cell resolution is a powerful approach for determining how common genetic variants associated with IBD impact gene expression and cellular function. Future studies leveraging single cell RNAseq for eQTL studies in healthy versus inflamed intestinal biopsies may provide a deeper understanding of the impact of genetic variation at the population level on gene expression in IBD risk and progression. Thus, gene expression datasets derived from intestinal tissues can be integrated with IBD genetics to establish gene-trait associations. In this context, single cell transcriptomic datasets comprising the disease atlas offer a high-resolution view of individual cellular transcriptomes. Building the disease atlas as a reference dataset yields a powerful tool for iteratively modeling, testing, and revising mechanistic models of disease in experimental settings, animal models, and clinical studies.
Pathways
IBD as a Model Disease.
Studies of IBD genetics have implicated genes and pathways underlying inflammatory pathology. Many core cytokine pathways implicated in IBD GWAS overlap amongst inflammatory diseases and autoimmunity, while IBD genetics provides a unique view of pathways that control mucosal immunity49. As such, this core set of pathways represents vulnerabilities to disease development that suggest potential opportunities for intervention (Figure 3).
Figure 3. Pathway paradigms highlighted by IBD genetics.

IBD risk genes listed at the bottom of each panel represent genetic vulnerabilities that perturb key pathways underlying intestinal homeostasis and drive inflammatory pathology in the gut mucosa (centre). a, Phagocytes have evolved numerous mechanisms to detect microorganisms and elicit effector responses that promote inflammation through cytokines and antimicrobicidal responses such as oxidative burst and xenophagy. b, Intestinal epithelial cells maintain barrier integrity through dynamic remodelling of junctional complexes. c, Coordinated interactions between dendritic cells, T cells and B cells facilitate the induction of antigen-specific immune responses and immunological memory directed against commensal microorganisms. d, Stromal cells—such as fibroblasts—are key mediators of tissue remodelling and healing that dynamically respond to inflammatory conditions. e, Cell-extrinsic and -intrinsic stressors associated with inflammation sensitize cells to death. The integrated stress response facilitates adaptation to these stressors by coordinating cellular responses to ER stress, oxidative stress, hypoxia, autophagy and cell death pathways. f, Intercellular communication among immune cells, stroma and epithelial cells is tightly controlled by elaborate cytokine networks. g, Microbe-sensing pathways that operate in the cytosol of host cells function to detect and respond to intracellular infection or exposure to bacterial toxins.
Distinct inflammasome complexes mediate activation and secretion of IL-1β or IL-18 and elicit pyroptosis, a proinflammatory form of programmed cell death.
The Epithelial Barrier and Dynamic Remodeling of Junctional Complexes.
The gut mucosa comprises specialized cell types that functionally interconnect host physiological systems with extrinsic commensal communities, pathogens, metabolites, and dietary factors. While epithelial cells maintain a physical barrier separating the host from its environment, they simultaneously interact with their environment to relay information throughout the body and coordinate an appropriate host response. Specialized intestinal epithelial cell lineages derive from ISCs positioned at the base of invaginated crypt structures47. The self-renewing stem cell compartment is located within a unique niche that is supported by signals from the microbiota and growth factors derived from stroma and accessory epithelial cells. Chronic inflammation associated with IBD damages crypts and impairs stem cell reprogramming during epithelial restitution. Insights from IBD genetics implicate the aryl hydrocarbon receptor (AHR) as a critical sensor of the luminal microenvironment capable of protecting the stem cell niche from inflammatory damage50. This niche is also regulated by Paneth cells in the small intestine, which secrete growth factors that promote ISC regeneration during restitution or inhibit it during chronic inflammation51,52. In addition, Paneth cells participate in innate immunity by producing antimicrobial peptides that protect the crypt from microbial infestation. Similarly, goblet cells also link innate immunity with barrier function by producing and secreting a protective mucus layer on the luminal surface of the intestinal epithelium. Goblet cells and Paneth cells uniquely express ITLN1, an IBD risk gene encoding a lectin thought to bind carbohydrate moieties on the surface of microbiota and promote host innate immunity45.
The most abundant intestinal epithelial cell type is the absorptive enterocyte, which helps establish the physical barrier of the epithelium, mediate nutrient and water uptake, and possesses innate pathogen-sensing capabilities. In active CD, enterocytes exhibit morphological defects in brush border and microvillus structure consistent with impaired nutrient absorption and barrier function53. Additionally, several IBD risk genes are enriched in expression in enterocytes and contribute to lineage specification (HNF4A), junctional integrity (C1orf106), and innate immunity (GSDMB)45. Specialized epithelial cells also contribute to adaptive immunity. Microfold cells (M cells), are directly apposed to secondary lymphoid structures in the mucosa, and transport luminal antigens to lymphoid follicles. M cells expand in number in inflamed regions of the colon of UC patients45 and perform critical functions in immune-microbiome homeostasis54. Similarly, Tuft cells interface with the microbiome by acting as chemosensory sentinels that detect pathogens and toxins and subsequently elicit type 2 immune responses through the production of IL-2555. Another important class of sentinels in the epithelium are Enteroendocrine cells, which function as sensors and effectors that produce hormones to coordinate with the enteric nervous system (ENS) and other digestive organs. Subsets of regionally-specific Enteroendocrine cells are characterized by the unique repertoires of hormones they produce56,57 and how their functions change during inflammation47. For example, serotonin-producing Enteroendocrine cells sense changes in the microbiome milieu through G protein-coupled receptors (GPCRs) that initiate serotonin secretion and subsequent engagement of serotonergic neurons, which control gut motility58.
Genetic studies of IBD have identified novel regulators of diverse epithelial functions. In particular, genes controlling barrier integrity, such as C1orf106, RNF186, and HNF4A represent key risk factors for UC11 (Figure 3). C1orf106 was first implicated in UC GWAS, and exome sequencing subsequently identified a coding variant associated with UC risk20. In functional studies, C1orf106 functions as an adaptor that regulates ubiquitination and degradation of cytohesin proteins, which are guanine nucleotide exchange factors for the small G protein ARF659. ARF6 plays a critical role in endocytic recycling of cell surface receptors, including cadherin proteins, which form adherens junctions that physically link epithelial cells together in monolayers59. The IBD risk variant of C1orf106 Y33F is associated with elevated levels of cytohesins, defective remodeling of adherens junctions, and subsequent impairment of barrier function, which is thought to sensitize to chronic microbiota-driven intestinal inflammation59,60.
IBD GWAS identified another novel regulator of epithelial barrier integrity, the ubiquitin ligase RNF1869. Exome sequencing subsequently identified an allelic series of coding variants for RNF186, including the A64T missense variant associated with UC risk21 and the R179X nonsense variant associated with protection from UC19. Rnf186-deficient mice and A64T knock-in mice exhibit increased intestinal permeability indicative of epithelial barrier dysfunction61. Rnf186 may regulate turnover of tight junctions by ubiquitinating occludin61, although it is likely that it regulates additional epithelial cell functions such as ER stress responses62. While Rnf186 A64T is associated with epithelial barrier dysfunction, it remains unclear how the protective variant R179X impacts intestinal epithelial biology. In humans, RNF186 R179X is associated with elevated serum creatinine63, derived from the decomposition of creatine, which functions as an important cellular energy shuttle. Within epithelial cells, creatine transports high energy phosphates from the mitochondria to the apical surface of polarized epithelial cells, where ATPase myosin motor proteins regulate assembly of microvilli and contraction of the actomyosin belt to maintain dynamic barrier integrity. Thus, dissecting pathways that control intestinal barrier function may reveal novel targets for therapeutic development.
Innate Microbial Sensor Pathways.
The epithelial barrier sequesters commensal microorganisms from the host innate immune system, which is equipped with an arsenal of microbe-sensing mechanisms. NOD2 was one of the first genes conclusively linked to IBD12,20,64,65. NOD2 functions as an intracellular cytosolic sensor of muramyl dipeptide (MDP), a component of peptidoglycan derived from bacterial cell walls. Engagement of NOD2 by MDP activates NFkB via RIPK2, and cooperates with inflammasomes to promote IL-1b secretion66,67. A mouse model for Nod2 fs1007insC demonstrated impaired cytokine production in macrophages in response to MDP68. Separate studies suggest that CD-associated NOD2 variants impair NFkB activation, indicating that attenuated NOD2 signaling may result in unproductive antibacterial responses leading to hyperinflammatory phenotypes. In fact, Nod2-deficient mice exhibited increased pathology in epithelial injury models, suggesting that impaired Nod2 signaling may allow inflammatory signaling to persist through alternative pathways69, and/or that microbiome pathobionts can tip the balance towards chronic inflammation70. Specifically, Nod2-deficiency resulted in excess Nlrp3 microbiome-driven activation, and a small molecule inhibitor of Nlrp3 abrogated intestinal inflammation in Nod2-deficient mice69. Thus, NOD2 functions in the intestine as a critical microbial sensor and inflammatory effector impinged upon by additional IBD risk genes, including RIPK211, XIAP71,72, TRIM2273, and TNFAIP374. While variants in NOD2 and NOD2-regulating pathways represent genetic vulnerabilities to intestinal pathology, they are also potential drug targets75,76.
Like NOD2, CARD9 functions in microbial sensing pathways highlighted by IBD genetics11 (Figure 3). As an adaptor protein, CARD9 is required for NFkB activation and cytokine production downstream of myeloid receptors that signal through immunoreceptor tyrosine-based activation motifs (ITAMs)77. In particular, CARD9 is required for inducing cytokine production in phagocytes after engagement of Dectin-1 by fungal ligands, which secondarily promotes Th17 immunity. Several loss of function CARD9 alleles are associated with susceptibility to lethal fungal infections in humans, while in IBD, an allelic series was identified for CARD9 that comprises risk and protective alleles. The missense variant CARD9 S12N is associated with IBD risk20,21. Conversely, a protective variant CARD9 IV11+1G>C (c.1434+1G>C)(rs141992399) disrupts mRNA splicing and is thought to result in exon skipping and a frameshift that truncates CARD9 prior to its C- terminal domain (CTD)20,21 (Figure 3). A mouse model for Card9 S12N revealed an unexpected function for the risk variant in alveolar macrophages associated with increased cytokine production in response to fungal ligands78. In CD patients, commensal fungi such as Malassezia restricta colonize the intestines of S12N carriers at higher rates compared to S12 patients, and this unusual colonization is proposed to amplify chronic inflammation associated with CD79. While the CARD9 risk allele S12N is associated with enhanced inflammatory cytokine production in myeloid cells, the protective truncated CARD9 (deltaCTD) functions as a dominant negative and is associated with reduced cytokine production in dendritic cells after stimulation with fungal ligands80. Mechanistically, the truncated CARD9 deltaCTD protein is unable to physically interact with the ubiquitin ligase TRIM6280. In overexpression systems, TRIM62 is sufficient to ubiquitinate CARD9 at K125, and this post-translational modification is required for CARD9-mediated activation of NFkB downstream of fungal receptors80. Taken together, these findings suggest that mimicking the function of the protective IV11+1G>C allele by disrupting ubiquitination of CARD9 may represent a safe and effective therapeutic intervention in IBD81.
Humoral Immunity and Antibodies in IBD.
Innate antimicrobial immune responses are complemented by functions of the adaptive immune system. B lymphocytes comprise a major arm of the mucosal immune system responsible for immunity to commensal microbes and pathogens. The mucosa is enriched in IgA antibodies that both promote and limit colonization by commensals to maintain intestinal homeostasis82. While IgA antibodies are considered to be neutralizing, other isotypes found in intestinal tissue can engage host cellular effector mechanisms such as the complement system and Fc receptors. For example, UC patients exhibit unusually high levels of commensal-reactive IgG in the colonic mucosa83. In turn, IgG-commensal immune complexes can engage Fc gamma receptors on macrophages and induce IL-1b and neutrophil-specific chemokines83. A missense coding variant in FCGR2A resulting in H131R substitution9 is protective with respect to UC risk and reduces the affinity of Fc gamma receptor for immune complexes, thus attenuating macrophage inflammatory responses and subsequent type 17 immunity83. With a greater understanding of the molecular composition of the human intestinal microbiome3, the field is poised to identify novel bioactives and antigens that impact host pathways in health and IBD84.
Cellular Immunity and Lymphocytes.
Antigen presentation operates at the epicenter of immunity, linking phagocytosis and microbial killing with T cell activation and differentiation. Indeed, many of the phagocyte effector pathways highlighted by IBD genetics, and discussed herein, directly impact processing and presentation of antigenic peptides on MHCII for activation of conventional alpha/beta T cells (Figure 3). Fine mapping the human leukocyte antigen (HLA) locus directly implicated MHCII alleles in risk of developing CD or UC85. Moreover, MHCII alleles were identified in a GWAS for CD prognosis that strongly associate with severe clinical outcomes86. In addition, IBD risk is associated with CIITA and RFX5, two transcription factors that control expression of MHCII9,17. In the context of IBD, it remains to be determined how MHCII alleles or perturbations in their expression impact T cell function and pathological inflammation. MHCII variants may qualitatively alter the spectrum of peptides displayed to T cells, thus impacting central tolerance, peripheral tolerance, T cell maintenance, attrition, and/or functional diversification. Interactions between the intestinal microbiome and host adaptive immune system require early life exposure for establishing tolerance87. Defining how commensal epitopes impact T cell development and function, is an emerging new field. The development of new technologies for TCR sequencing and profiling antigen-specific T cells, opens opportunities for specifically studying the unique functional attributes of pathogenic versus protective T cells and tracking the evolution of the TCR repertoire during the course of disease88,89.
While IBD genetic studies clearly implicate pathways regulating development and function of conventional alpha/beta T cells, emerging evidence indicates that mucosal immunity relies on several unique lymphocyte lineages that recognize conserved microbe-associated ligands presented by nonclassical MHC molecules and/or that respond to cytokines independently of antigen recognition. The cytokine profiles associated with gamma/delta T cells, NKT, and MAIT cells are diverse and overlap with key effectors implicated by IBD genetics such as IFN-γ, IL-17, and IL-13. These cytokines are also produced by specialized innate lymphocyte subsets that lack antigen receptors, such as ILCs, which function as cytokine sensors that rapidly respond to cytokine stimulation by secreting cytokines of their own90. For example, local production of IL-23 in the lamina propria by mononuclear phagocytes exposed to commensal bacteria can stimulate ILC3 cells to produce IL-17 and IL-22, which in turn act on epithelial cells to coordinately amplify inflammation and promote healing. In addition to this IL-23 mediated cytokine circuit, IL-1b produced by mononuclear phagocytes can stimulate ILC3 cells to produce IL-2, which functions as a survival and expansion signal for intestinal Tregs91. Many additional examples of lymphocyte cytokine networks have been described in the context of intestinal homeostasis and inflammation92.
Redefining Cytokine Networks.
Efforts to define the cellular heterogeneity of the immune system have revealed an astounding number of cell types that coordinate their unique functions through cytokine-mediated intercellular communication networks92. Genes associated with IBD are well represented within these cytokine networks and provide a unique perspective on how genetic vulnerabilities to inflammatory pathology impact cellular crosstalk. Some of the first clues came from genetic associations of IL10RA and IL23R with IBD93. Mechanistic and functional studies have subsequently demonstrated that macrophage-specific deletion of Il10ra results in severe colitis in mice25. Thus, in the absence of IL10R signaling, macrophages produced elevated levels of IL-23 that induced IL-22 production by conventional T cells. In turn, IL-22 induced intestinal epithelial cells to express chemokines that subsequently recruited neutrophils to the intestine and amplified inflammatory pathology25 (Figure 3). Counteracting this cytokine loop, Tregs were shown to suppress macrophage IL-23 and IL-1b production, thus limiting inflammation94. The IL- 23 cytokine network is particularly well represented in IBD genetics. Exome sequencing identified coding variants in IL23R that are associated with protection from IBD onset20, and GWAS identified the IL12B locus as associated with IBD risk12. IL-23 is a heterodimer comprised of the protein products of the IL12B and IL23A genes. IL-23 is primarily produced by mononuclear phagocytes and acts on several target cells including Th17, ILC3, and innate-like lymphocytes. In T cells, IL-23 and IL-6 cooperate to induce the Th17 transcription factor RORgt95, a nuclear receptor encoded by RORC, a recently identified IBD risk gene. In addition to RORC, IL23R, and IL12B, additional IBD risk genes may function to stabilize or promote Th17 differentiation, such as IL6ST, TYK2, GPR65, and STAT3 which are all located in IBD risk loci9. In fact, patients bearing TYK2 variants that are associated with protection from IBD exhibit impaired T cell responses to IL-23 stimulation and reduced STAT3 phosphorylation13. Collectively, several IBD risk genes complement IL-23 stimulation in lymphocytes with a primary effect on promoting production of IL-17 family cytokines and IL-22. These examples illustrate the value of functionally mapping genetic vulnerabilities to cellular communication networks.
Studies of IBD genetics inspired therapeutic approaches aimed at blocking the IL-23/17 axis. Paradoxically, antibodies against IL-12 and/or IL-23 show efficacy in IBD and psoriasis, while inhibition of IL-17 or IL17RA shows efficacy in psoriasis and may exacerbate IBD8,96. These observations suggest that IL-17 has important ancillary effects in mucosal tissues related to host defense and barrier function96. In mouse models, IL-17F-deficiency was associated with impaired antimicrobial peptide production in the epithelium, leading to a bloom of commensal bacteria (Clostridium cluster XIVa) that promoted Treg development97. Thus, IL-17F knock-out mice with elevated Treg frequencies exhibited reduced pathology in models of colitis97. Additional studies have provided a different view of IL-17 cytokines demonstrating redundancy in IL-17A and IL-17F for induction of colitis in mouse models98. It remains unclear how IL-17R signaling promotes inflammation versus healing in the mucosa, and which cells mediate these differential effects. Functional dissection of genetic risk factors in the IL17RA signaling pathway may reveal the cell types and biological contexts in which the pathway is dysregulated in disease.
In addition to the IL-23/17 axis, IBD genetics highlights specialized cytokine networks that are adapted for immunity at barrier tissues, namely the IL-1 family of cytokines. IL-1b is primarily produced by phagocytes in response to microbe-associated molecular patterns (MAMPs) that induce transcription of IL-1b mRNA, while inflammasomes promote proteolytic activation of IL-1b and assembly of gasdermin pores, facilitating noncanonical secretion of the active cytokine and induction of pyroptosis99. In IBD patients with IL10RA loss of function variants, excess IL-1b produced by macrophages led to excess inflammation, and treatment with the IL1R antagonist anakinra ameliorated symptoms in subjects that were previously refractory to treatment100. IL1R antagonism also reduced IBD symptoms in CGD patients with perianal disease, although it is important to note that these were case studies rather than controlled clinical trials101. Additional IBD genetic risk factors impact IL-1b signaling. The CD risk allele ATG16L1 T300A is associated with elevated IL-1b production in phagocytes stimulated with MAMPs, thus implicating autophagy in lysosomal degradation of inflammasome macromolecular signaling complexes102–104. Similarly, ATG16L1 has been suggested to control autophagy-dependent turnover of TRIF oligomers induced by TLR signaling, and the ATG16L1 T300A allele was shown to augment IFNb and IL- 1b production in response to TLR engagement105.
Multiple distinct inflammasome complexes regulate IL-1 family cytokines in the context of mucosal immunity and IBD. In addition to IL-1b, inflammasomes control the activation of IL-18, and exome sequencing identified variants in the IL-18 receptor signaling chain (IL18RAP) that are associated with IBD20. Moreover, mutations in NLRC4 are associated with elevated IL-18 levels and severe infantile enterocolitis30,106. Additional examples of inflammasome hyperactivation have been described for the Pyrin inflammasome, which is encoded by the MEFV gene107,17 (Figure 3) and NLRP7108.
Intestinal pathologies associated with IL-1 family cytokines extend beyond the inflammasome- dependent cytokines to include family members that function as alarmins in mucosal tissues. Accordingly, IL-33 is an IL-1 family member that is expressed intracellularly in epithelial cells and stroma. Upon physical tissue damage or cellular necrosis, IL-33 is released into the tissue microenvironment where it acts on a diversity of immune cell types expressing its receptor ST2, which is encoded by IL1RL1, a gene implicated by IBD fine mapping GWAS12. Thus, IL-33 is thought to contribute protective and pathological functions in the mucosa by promoting Treg- mediated tolerance109, inducing type 2 cytokine production in ILC2 cells110, and promoting mast cell activation111. Similar to IL-33, the IL-36 subfamily of cytokines function as tissue alarmins. IL- 36 family cytokines perform important immune functions at barrier tissues such as skin and gut, thus implicating these cytokines in psoriasis and IBD92. Specifically, CD patients with fibrostenotic disease exhibited elevated levels of IL-36 in intestinal biopsies112. Inhibition of IL36R by administration of blocking antibody or genetic deletion of the receptor (Il1rl2) reduced inflammation in mouse models of epithelial injury112. Further functional mapping of these alarmin networks will help identify mechanisms of intestinal homeostasis and provide insights into mechanisms of disease.
Intrinsic Cell Stress Pathways.
Chronic inflammation driven by cytokines and inflammatory mediators in the intestine imposes environmental and metabolic stress on tissues and cells. Early evidence linking IBD to cellular stress pathways was derived from GWAS identifying polymorphisms in autophagy-related genes IRGM and ATG16L1113–115. Autophagy is a cellular disposal system that directs cytoplasmic cargo into lysosomes for proteolytic degradation. Given the central role of autophagy in recycling biomass such as organelles, autophagy integrates cellular metabolism and catabolism to meet the energetic demands of the cell. In addition, autophagy machinery plays diverse roles in several cellular processes including intracellular host defense, lysosome homeostasis, and cellular secretion. Thus, autophagy and lysosome homeostasis intersect with multiple cellular systems including regulators of vesicular trafficking such as the kinase LRRK2. The LRRK2 G2019S coding variant is associated with CD and Parkinson’s disease, diseases with different clinical manifestations but surprising genetic overlap17,116. LRRK2 variants may impart IBD risk by impairing antibacterial phagolysosomal disposal in intestinal phagocytes117 and impart risk of Parkinson’s disease by impairing phagolysosomal disposal in microglia or mitophagy in neurons116. Indeed, several additional IBD risk genes, including GPR65, are associated with lysosome dysfunction and autophagy34.
Defining the complex roles of autophagy in IBD has been facilitated by the generation of knock- in mice that model the CD risk variant ATG16L1 T300A. These mice exhibit several cell type- specific phenotypes that are thought to cumulatively contribute to IBD risk in humans. For example, Atg16l1 T300A knock-in mice exhibit defective antibacterial autophagy against enteric pathogens104. Macrophages derived from T300A mice produce elevated levels of IL-1b in response to inflammasome stimulation103. Finally, Paneth and goblet cells derived from Atg16l1 T300A knock-in mice exhibit granule anomalies associated with impaired secretion of antimicrobial peptides103 and mucus. Similarly, CD patients bearing ATG16L1 T300A alleles exhibit abnormal numbers and morphology of Paneth cell granules that segregate CD into distinct pathological subsets118,119. Taken together, the ATG16L1 T300A allele conspires with environmental triggers to induce several cell type-specific phenotypes resulting in tissue dysregulation and predisposition to inflammatory pathology.
The autophagy pathway is intertwined with the integrated stress response. The ATG16L1 T300A substitution creates a caspase 3 cleavage site, rendering ATL16L1 susceptible to caspase cleavage, thus functionally connecting autophagy with apoptosis103,104. Cell-extrinsic and -intrinsic stressors prime genetic susceptibility to intestinal pathology in the context of ATG16L1 insufficiency. CD patients bearing ATG16L1 T300A alleles exhibited pathological ER stress in Paneth cells, which was specific to the T300A genotype and was even evident in quiescent disease120. ER stress responses control epithelial barrier function, thus coordinating interactions of the adaptive immune system with luminal antigens. Healthy subjects bearing ATG16L1 T300A alleles exhibit signs of elevated epithelial ER stress and ensuing polyreactive IgA responses121. Thus, ATG16L1 T300A functions to integrate autophagy and ER stress pathways in the intestinal mucosa.
The ER stress pathway plays a central role in epithelial barrier function. Conditional deletion of key ER stress sensors and effectors in the intestinal epithelium results in pathological, unresolved ER stress and spontaneous enterocolitis122. Thus, secretory epithelial cells such as Paneth cells and goblet cells require highly efficient ER secretory machinery to maintain barrier integrity and innate immunity123,124. Genetic vulnerability in cell stress pathways can predispose secretory epithelial cells to developing pathological ER stress culminating in apoptosis, barrier breach, and chronic inflammation. Recent studies have identified genetic risk factors associated with IBD that act through regulation of ER stress, such as TMEM258, which is located in a locus associated with CD9. Mechanistically, TMEM258 functions as an essential subunit of the oligosaccharyltransferase complex, which controls N-linked protein glycosylation in the ER and is essential for directing export of nascent proteins through the secretory pathway125. Several additional IBD risk genes have been associated with defective protein glycosylation defects and congenital disorders of glycosylation such as SLC39A8126 and RFT1127 (Figure 3). These findings demonstrate the important role for protein glycosylation in facilitating protein trafficking through the secretory pathway, thus linking ER stress to epithelial function in the intestine.
Metabolism.
The inflamed intestinal mucosa in IBD is paradoxically associated with elevated oxidative stress and signs of hypoxia37,128. Inflammation can rapidly disrupt oxygen gradients in the mucosa and lead to fluctuations that perturb homeostasis and exacerbate tissue damage. Innate immune responses are highly oxygen-consumptive and generate toxic free radicals. Thus, host cells and tissues adapt to oxidative stress and hypoxia in a coordinated manner by engaging two interrelated transcriptional responses controlled by NRF2 and HIF-1a respectively.
While superoxide and reactive oxygen intermediates (ROI) are potent antimicrobicidal agents, they induce oxidative stress, and their production consumes local oxygen leading to hypoxia and HIF-1a activation129. In turn, HIF-1a induces a metabolic shift towards glycolysis and initiates angiogenesis resulting in a rebound in tissue oxygenation that can help reestablish normoxia and healing or exacerbate oxidative stress. HIF-1a activation by means of an inhibitor of CUL2 neddylation, which stabilizes HIF-1a, enhances epithelial barrier integrity and reduces pathology in models of epithelial injury130. IBD GWAS identified CUL2 as a risk gene, and exome sequencing identified a rare variant that disrupts mRNA splicing that is associated with protection from IBD20. In contrast to HIF-1a, transgenic overexpression of HIF-2a exacerbated pathology associated with epithelial injury131,132. HIF-2a uniquely controls expression of genes in the creatine metabolism pathway, including creatine kinases and the creatine transporter SLC6A8, which collectively regulate the phosphocreatine shuttle to provide energy required for remodeling of epithelial junctional complexes133. Together, HIF-1a and HIF-2a control cellular metabolism and adaptation to environmental demands imposed by inflammation, suggesting that rebalancing HIF- 1a and HIF-2a locally may be a viable therapeutic strategy.
Leukocytes must rapidly switch from quiescent to activated states to elicit effector function in a controlled manner. This requirement necessitates metabolic versatility to fuel effector responses and conserve energy during quiescence. In macrophages, TLR engagement leads to a shift from oxidative phosphorylation to glycolysis, resulting in accumulation of the tricarboxylic acid cycle (TCA) intermediate succinate, which promotes stabilization of HIF-1a to enhance transcription of IL-1b134 and also regulates type 2 immunity through its effects on tuft cells135. Another TCA intermediate, aconitate, is converted to itaconate by Irg1, which is an enzyme that is induced as part of the interferon response136. Itaconate is an electrophilic compound that alkylates key cysteine residues in Keap1, thus relieving inhibition of Nrf2, which elicits the antioxidant response and downregulates inflammatory cytokine production137. Thus, immunometabolism is intimately linked with inflammatory and cell stress pathways associated with IBD. In this context, the enigmatic enzyme LACC1 (C13orf31) was identified in genetic studies as associated with CD, leprosy, and other inflammatory diseases9. Lacc1 knock-out mice were shown to be highly susceptible to severe histopathology in response to the enteric pathogen Citrobacter rodentium or collagen-induced arthritis (CIA)138. In these models, elevated levels of IL-17A were observed in Lacc1-deficient mice, although it remains unclear if this phenotype is a primary or secondary effect of Lacc1-deficiency. Given expression patterns of Lacc1, it is not likely to be a T cell intrinsic phenotype138. Instead, LACC1 is highly expressed in myeloid cells and is thought to regulate immunometabolism to promote antibacterial responses and cytokine production139,140. The CD risk variant LACC1 I254V was shown to exhibit impaired function associated with defective antibacterial responses139,140. Although the specific metabolic function of LACC1 is incompletely understood, it has been proposed to regulate mitochondrial respiration and/or fatty acid oxidation140. Many more metabolic processes and intermediates are likely to be discovered and attributed with immunomodulatory properties that impact IBD.
Tissue Stroma, Inflammation, and Fibrosis.
Inflammation and metabolic stress regulate tissue homeostasis and immunity at barrier sites112,141. In this context, chronic inflammation associated with autoimmune diseases frequently elicits a pathological inflammation-healing cycle leading to tissue damage from fibrosis and scarring. This core pathological pathway is conserved across disparate organ-specific autoimmune diseases but has distinctly tissue-specific effects in IBD. Although significant progress has been made in targeting inflammatory pathways in IBD, a significant proportion of patients are refractory to these treatments and develop severe complications associated with fibrosis, which ultimately require surgical intervention. Thus, there is an unmet need for therapeutic strategies aimed at preventing the onset of fibrosis, suggesting that we are missing key target pathways underlying fibrosis.
Several clinical studies have examined treatment-refractory patients and derived gene expression signatures from intestinal biopsies to define the nonresponsive state5,48. While these studies examining gene expression signatures in bulk tissue identified a clear signature associated with treatment resistance, they do not directly reveal the cellular source(s) of this pathogenic response. By overlaying the treatment-resistance signature onto a single cell transcriptomic map of UC, fibroblasts were shown to be the dominant cell type of origin for this signature45. Specifically, the anti-TNF-resistance signature was enriched in inflammation associated fibroblasts (IAFs) and included genes such as IL13RA2, TNFRSF11B, and IL1145. These findings corroborate previous studies delineating a cellular signaling circuit that is actively engaged in the TNF-resistant state, wherein IAFs are activated by the IL-6 family cytokine OSM to produce chemokines that recruit neutrophils to the inflamed mucosa48 (Figure 3). Thus, inflammatory myeloid cells communicate with fibroblasts through OSM, which signals through a heterodimeric receptor encoded by OSMR and IL6ST48, two genes implicated in IBD GWAS.
Based on the link between IL6ST and IBD, and the fact that IL6ST (gp130) functions as a common signaling receptor for multiple IL-6 family members, the pathological effects of IL6ST variants may span multiple cytokine networks and cellular functions. For example, IL6ST and IL11RA together encode the receptor for IL-11, which is highly expressed in IAFs and associated with the anti- TNF-resistance signature45. IL-11 is induced in fibroblasts in response to stimulation with TGF-b, and subsequently engages an autocrine loop in fibroblasts resulting in deposition of extracellular matrix142. Inhibition of IL-11 signaling ameliorates cardiovascular fibrosis in mouse models142, suggesting that IL-11 may control the arm of the TGF-b response that drives fibrosis. Indeed, TGF-b exhibits pleiotropic effects on tissue development and extracellular matrix remodeling. This is exemplified by the complex phenotypes associated with TGFB1-deficiency in humans, which include severe IBD and encephalopathy143. Thus, targeting fibrosis by blocking TGF-b may result in adverse effects, but identifying alternative targets controlling defined pathways in fibrosis may offer viable alternatives. For example, IL-11 and IGFBP3 are highly enriched in intestinal fibroblasts derived from UC patients44,45 and have been previously implicated as profibrogenic factors142,144. Moreover, IGFBP1–3 were identified as genetic risk factors for severe complications associated with CD prognosis in GWAS86. Taken together, evidence from genetics and clinical studies continue to provide clues into the contribution of fibrosis pathways in IBD pathogenesis.
Therapeutics
Influence of Genetics on IBD Therapies.
IBD genetic studies have been instrumental in identifying pathways associated with disease risk and that function as mechanistic drivers of disease pathology. Thus, insights from genetics have direct implications for classifying IBD into clinical subsets based on underlying molecular pathomechanisms and devising more focused therapeutic interventions. The current standard of care for CD and UC is similar, despite recognized differences in clinical and molecular features. Corticosteroids, aminosalicylates, occasionally thiopurines and antibiotics remain the first line treatment for mild to moderate CD and UC. Treatment of moderate to severe cases of IBD has benefited from the recent development of biologics targeting cytokines (TNF or IL-12/23) or leukocyte trafficking receptors (alpha4 beta7 integrin). Historically, therapeutic strategies deployed for IBD have focussed on treating inflammation. Decades of clinical practice have led to the optimization of treatment regimens, although it is not entirely clear how and when these approaches will succeed or fail to achieve efficacy in individual patients. New insights are emerging from IBD genetics that show great potential for illuminating the mechanisms of disease, defining mechanisms of action for treatment interventions, and nominating new candidate targets for therapeutic development.
The majority of therapeutics that are currently utilized to treat IBD were developed prior to the maturation of IBD genetics. We anticipate that genetics and functional genomics will continue to play an important role in advancing the development of novel therapeutic strategies. With that said, emerging evidence from IBD genetics provides rationale that helps explain the efficacy of therapeutics that were developed many years ago. For example, anti-TNF has been a mainstay for the treatment of IBD and several autoimmune diseases. While it is still not entirely clear how TNF inhibition leads to remission in IBD, there are clues from genetics that TNF shedding mediated by ADAM17 proteases17,145 and TNFR signaling through RIPK1146 are important contributors to pathogenesis. These observations provide insights into how anti-TNF may function in patients, what pharmacodynamic markers might help in evaluating efficacy, and how alternative targets in these pathways may be exploited.
Some of the newer biologics used to treat IBD, can be rationalized by genetics. The most recent IBD GWAS implicated a number of novel risk genes encoding integrin subunits11. This finding supports clinical observations that inhibition of leukocyte trafficking through integrin blockade (anti-alpha4beta7 antibodies) or antagonists of S1PR exhibit efficacy in IBD147,148. Additionally, protective variants in IL23R predict the observed efficacy of anti-IL-23 in IBD patients20,21,93. Although IL-23 is a major driver of IL-17 production by T cells and ILCs, biologics targeting IL-17 or IL17RA have not demonstrated convincing efficacy in IBD96. Thus, IL-17 appears to have pleiotropic effects on mucosal homeostasis, some of which are beneficial, and clues from IBD genetics may help dissect the differential mechanisms of IL-17 signaling in target cells. Perhaps emerging insights from genetics will help pinpoint optimal points of entry within complex cytokine pathways. For example, fine mapping IBD GWAS loci implicated JAK2 as a causal risk gene12, thus suggesting that targeting multiple inflammatory cytokine receptor pathways through JAK inhibition may be a viable therapeutic strategy. Indeed, pan-JAK inhibitors have shown efficacy in clinical trials for IBD147,148.
Taken together, IBD genetics has identified genetic vulnerabilities within integrated pathway networks that regulate homeostasis between the immune system, mucosal barrier tissues, and the microbiome. These genetic vulnerabilities represent key regulatory nodes in the network that may be “tunable” in response to targeted therapeutic interventions that aim to restore mucosal homeostasis. Here, the challenging therapeutic objective is to restore intestinal homeostasis in individuals, regardless of the genetic and environmental perturbations that may have initially triggered network dyshomeostasis. Recent insights from IBD genetics and functional genomics suggest novel strategies for targeting genetic vulnerabilities and mimicking protective variants.
Outlook
The Future.
The field of IBD genetics has advanced quickly in the genomics era. Ongoing efforts have made great strides in synthesizing human genetics with functional genomics, patient-based research, animal models, and mechanistic studies. Accordingly, the genetic architecture of IBD has been elaborated to provide functional context by mapping genes-to-pathways. Deeper insights into disease pathogenesis are emerging from exome sequencing and subsequent experimental conversion of variants-to-mechanisms. Collectively, these efforts offer new directions for solving outstanding problems in clinical management of IBD, and more broadly, for shedding light on mechanisms of immunoregulation. Indeed, IBD can be viewed as a model disease to dissect mechanisms of mucosal immunity; however, IBD is a complex phenotype that is impacted by multiple pathways. Parsing and condensing common variants into functionally related polygenic pathway scores may help reclassify patients based on molecular phenotypes. Clinical studies examining cohorts defined by pathway risk scores, and individuals at the extreme ends of the risk spectrum, may facilitate biomarker discovery and identify disease mechanisms that inform targeted therapeutic interventions. Moreover, developing high fidelity biomarkers and clinical assays to quantify key pathways in IBD will be instrumental in redefining genotype- phenotype relationships. These efforts are catalyzed by public databases such as the UK Biobank and FIMM, which include large cohorts of individuals that have undergone exome sequencing and incorporate extensive clinical metrics with genotype data. These biobanks can be powerful engines for discovery, especially if they grow to incorporate informative clinical metrics of IBD- relevant pathways, such as intestinal permeability, immune function, and serum titers for common vaccines, to name just a few.
Much of the work to date in human genetics has focused on heritable genetic variants, while studying somatic variants may offer another opportunity to identify mutations in pathways that impact pathology. For example, biopsies from inflamed regions of the colon may contain more and qualitatively different somatic mutations compared to non-inflamed regions. In this regard, inflammation may positively select for somatic mutations in signaling pathways that render epithelial and immune cells resistant to cytokine toxicity. While it remains to be determined if somatic mutations associated with inflammation contribute to the pathogenesis of IBD, there is a clear precedent for inflammation inducing somatic mutations that drive carcinogenesis. Future studies aimed at finding genes that are under positive selection pressure in the context of intestinal inflammation may help identify key pathways associated with IBD pathology.
Genetics has provided unique insights into functional pathways that mediate homeostasis between the immune system and mucosal tissues. Recent research has shed light on how the microbiome interacts with host genetics to modulate intestinal homeostasis84. There are clear associations between host genetic variables and features of microbial dysbiosis in the microbiome that are downstream consequences of perturbed immune function. The challenge moving forward will be to identify compelling statistical associations between host genetics and metagenomic pathways that reveal direct mechanistic interactions between microbes and the host. Integrating host and microbiome genetics remains an area of active investigation3, which has benefited from years of research functionally annotating the human genome. In this context, functional gene annotations are subject to revision over time as experimental evidence accumulates. Thus, there is a need to accelerate the generation of high-quality empirical data such as protein-protein interaction networks derived from relevant cell types and functional genetic screens in the context of key IBD pathways. Defining more accurate gene-pathway annotations, will significantly accelerate the process of assigning function to disease-associated variants. It is important to note that the genetic variants associated with human disease may exhibit similar or different phenotypes compared to null alleles. While genetic knockout studies are invaluable for assigning gene function, it is essential to study the mechanisms of action for variants associated with human disease.
Figure 4.

Exome sequencing has identified many IBD risk genes (middle) with allelic series associated with a spectrum of phenotypes (left to right). Top, for example, CARD9S12N is associated with risk of IBD, whereas CARD9ΔCTD is associated with protection and CARD9Q295X is associated with an immunodeficiency linked with chronic life-threatening fungal infections. Other IBD genes contain coding variants with distinct phenotypes and disease associations that may offer clues to understanding mechanisms of IBD pathology. FMF, familial Mediterranean fever; CGD, chronic granulomatous disease; CVD, cardiovascular disease; CDG, congenital disorder of glycosylation.
Acknowledgements
This work is supported by grants from the Helmsley Charitable Trust and NIH (to R.J.X.). We thank Heather Kang for valuable scientific input, editorial assistance, and illustrative design.
Footnotes
Competing interests
R.J.X. is a consultant to Novartis and Nestle.
References
- 1.Xavier RJ, and Podolsky DK (2007). Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan GG, and Ng SC (2017). Understanding and Preventing the Global Increase of Inflammatory Bowel Disease. Gastroenterology 152, 313–321.e2. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKinney EF, Lee JC, Jayne DRW, Lyons PA, and Smith KGC (2015). T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyams JS, Davis Thomas S, Gotman N, Haberman Y, Karns R, Schirmer M, Mo A, Mack DR, Boyle B, Griffiths AM, et al. (2019). Clinical and biological predictors of response to standardised paediatric colitis therapy (PROTECT): a multicentre inception cohort study. Lancet 393, 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belarif L, Danger R, Kermarrec L, Nerrière-Daguin V, Pengam S, Durand T, Mary C, Kerdreux E, Gauttier V, Kucik A, et al. (2019). IL-7 receptor influences anti-TNF responsiveness and T cell gut homing in inflammatory bowel disease. J. Clin. Invest 130, 1910–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martins F, Sofiya L, Sykiotis GP, Lamine F, Maillard M, Fraga M, Shabafrouz K, Ribi C, Cairoli A, Guex-Crosier Y, et al. (2019). Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat. Rev. Clin. Oncol Available at: 10.1038/s41571-019-0218-0. [DOI] [PubMed] [Google Scholar]
- 8.Emond B, Ellis LA, Chakravarty SD, Ladouceur M, and Lefebvre P (2019). Real- world incidence of inflammatory bowel disease among patients with other chronic inflammatory diseases treated with interleukin-17a or phosphodiesterase 4 inhibitors. Curr. Med. Res. Opin, 1–9. [DOI] [PubMed] [Google Scholar]
- 9.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]; The Immunochip GWAS identified several new loci associated with IBD risk and offered new insights into underlying pathways driving disease risk.
- 10.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. (2015). Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet 47, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]; This GWAS analyzed individuals across diverse ancestries, with larger cohorts, collectively implicated new loci associated with IBD.
- 11.de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J, Ji S-G, et al. (2017). Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet 49, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]; This GWAS and meta-analyis captured approximately 240 loci associated with IBD.
- 12.Huang H, Fang M, Jostins L, Umićević Mirkov M, Boucher G, Anderson CA, Andersen V, Cleynen I, Cortes A, Crins F, et al. (2017). Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]; This fine-mapping GWAS implicates putative causal SNPs associated with a number of IBD risk loci.
- 13.Dendrou CA, Cortes A, Shipman L, Evans HG, Attfield KE, Jostins L, Barber T, Kaur G, Kuttikkatte SB, Leach OA, et al. (2016). Resolving TYK2 locus genotype-to- phenotype differences in autoimmunity. Sci. Transl. Med 8, 363ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Momozawa Y, Dmitrieva J, Théâtre E, Deffontaine V, Rahmouni S, Charloteaux B, Crins F, Docampo E, Elansary M, Gori A-S, et al. (2018). IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat. Commun 9, 2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calderon D, Nguyen MLT, Mezger A, Kathiria A, Müller F, Nguyen V, Lescano N, Wu B, Trombetta J, Ribado JV, et al. (2019). Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet Available at: 10.1038/s41588-019-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, Gazal S, Loh P-R, Lareau C, Shoresh N, et al. (2018). Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet 50, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivas MA, Avila BE, Koskela J, Huang H, Stevens C, Pirinen M, Haritunians T, Neale BM, Kurki M, Ganna A, et al. (2018). Insights into the genetic epidemiology of Crohn’s and rare diseases in the Ashkenazi Jewish population. PLoS Genet. 14, e1007329. [DOI] [PMC free article] [PubMed] [Google Scholar]; This exome sequencing study identified coding variants associated with IBD risk and leveraged ancestry to identify rare strong-acting genetic variants.
- 18.Arnadottir GA, Norddahl GL, Gudmundsdottir S, Agustsdottir AB, Sigurdsson S, Jensson BO, Bjarnadottir K, Theodors F, Benonisdottir S, Ivarsdottir EV, et al. (2018). A homozygous loss-of-function mutation leading to CYBC1 deficiency causes chronic granulomatous disease. Nat. Commun 9, 4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivas MA, Graham D, Sulem P, Stevens C, Desch AN, Goyette P, Gudbjartsson D, Jonsdottir I, Thorsteinsdottir U, Degenhardt F, et al. (2016). A protein-truncating R179X variant in RNF186 confers protection against ulcerative colitis. Nat. Commun 7, 12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, et al. (2011). Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet 43, 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beaudoin M, Goyette P, Boucher G, Lo KS, Rivas MA, Stevens C, Alikashani A, Ladouceur M, Ellinghaus D, Törkvist L, et al. (2013). Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 9, e1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glocker E-O, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, et al. (2009). Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med 361, 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]; This exome sequencing study identified risk and protective coding variants associated with IBD.
- 23.Shouval DS, Biswas A, Goettel JA, McCann K, Conaway E, Redhu NS, Mascanfroni ID, Al Adham Z, Lavoie S, Ibourk M, et al. (2014). Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti- inflammatory macrophage function. Immunity 40, 706–719. [DOI] [PMC free article] [PubMed] [Google Scholar]; This exome sequencing study identified risk and protective coding variants associated with IBD.
- 24.Zigmond E, Bernshtein B, Friedlander G, Walker CR, Yona S, Kim K-W, Brenner O, Krauthgamer R, Varol C, Müller W, et al. (2014). Macrophage-restricted interleukin- 10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733. [DOI] [PubMed] [Google Scholar]
- 25.Bernshtein B, Curato C, Ioannou M, Thaiss CA, Gross-Vered M, Kolesnikov M, Wang Q, David E, Chappell-Maor L, Harmelin A, et al. (2019). IL-23-producing IL- 10Rα-deficient gut macrophages elicit an IL-22-driven proinflammatory epithelial cell response. Sci Immunol 4 Available at: 10.1126/sciimmunol.aau6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartono S, Ippoliti MR, Mastroianni M, Torres R, and Rider NL (2018). Gastrointestinal Disorders Associated with Primary Immunodeficiency Diseases. Clin. Rev. Allergy Immunol Available at: 10.1007/s12016-018-8689-9. [DOI] [PubMed] [Google Scholar]
- 27.Frizinsky S, Rechavi E, Barel O, Najeeb RH, Greenberger S, Lee YN, Simon AJ, Lev A, Ma CA, Sun G, et al. (2019). Novel MALT1 Mutation Linked to Immunodeficiency, Immune Dysregulation, and an Abnormal T Cell Receptor Repertoire. J. Clin. Immunol 39, 401–413. [DOI] [PubMed] [Google Scholar]
- 28.Denson LA, Jurickova I, Karns R, Shaw KA, Cutler DJ, Okou DT, Dodd A, Quinn K, Mondal K, Aronow BJ, et al. (2018). Clinical and Genomic Correlates of Neutrophil Reactive Oxygen Species Production in Pediatric Patients With Crohn’s Disease. Gastroenterology 154, 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwerd T, Pandey S, Yang H-T, Bagola K, Jameson E, Jung J, Lachmann RH, Shah N, Patel SY, Booth C, et al. (2017). Impaired antibacterial autophagy links granulomatous intestinal inflammation in Niemann-Pick disease type C1 and XIAP deficiency with NOD2 variants in Crohn’s disease. Gut 66, 1060–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charbit-Henrion F, Parlato M, Hanein S, Duclaux-Loras R, Nowak J, Begue B, Rakotobe S, Bruneau J, Fourrage C, Alibeu O, et al. (2018). Diagnostic Yield of Next- Generation Sequencing in Very Early-Onset Inflammatory Bowel Diseases: A Multicenter Study. J. Crohns. Colitis Available at: 10.1093/ecco-jcc/jjy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jardine S, Dhingani N, and Muise AM (2019). TTC7A: Steward of Intestinal Health. Cell Mol Gastroenterol Hepatol 7, 555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holt-Danborg L, Vodopiutz J, Nonboe AW, De Laffolie J, Skovbjerg S, Wolters VM, Müller T, Hetzer B, Querfurt A, Zimmer K-P, et al. (2019). SPINT2 (HAI-2) missense variants identified in congenital sodium diarrhea/tufting enteropathy affect the ability of HAI- 2 to inhibit prostasin but not matriptase. Hum. Mol. Genet 28, 828–841. [DOI] [PubMed] [Google Scholar]
- 33.Tronstad RR, Polushina T, Brattbakk H-R, Stansberg C, von Volkmann HL, Hanevik K, Ellinghaus E, Jørgensen SF, Ersland KM, Pham KD-C, et al. (2018). Genetic and transcriptional analysis of inflammatory bowel disease-associated pathways in patients with GUCY2C-linked familial diarrhea. Scand. J. Gastroenterol 53, 1264–1273. [DOI] [PubMed] [Google Scholar]
- 34.Lassen KG, McKenzie CI, Mari M, Murano T, Begun J, Baxt LA, Goel G, Villablanca EJ, Kuo S-Y, Huang H, et al. (2016). Genetic Coding Variant in GPR65 Alters Lysosomal pH and Links Lysosomal Dysfunction with Colitis Risk. Immunity 44, 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graham DB, Becker CE, Doan A, Goel G, Villablanca EJ, Knights D, Mok A, Ng ACY, Doench JG, Root DE, et al. (2015). Functional genomics identifies negative regulatory nodes controlling phagocyte oxidative burst. Nat. Commun 6, 7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, et al. (2015). Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell 163, 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham DB, Jasso GJ, Mok A, Goel G, Ng ACY, Kolde R, Varma M, Doench JG, Root DE, Clish CB, et al. (2018). Nitric Oxide Engages an Anti-inflammatory Feedback Loop Mediated by Peroxiredoxin 5 in Phagocytes. Cell Rep. 24, 838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. (2015). A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell 162, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simeonov DR, Gowen BG, Boontanrart M, Roth TL, Gagnon JD, Mumbach MR, Satpathy AT, Lee Y, Bray NL, Chan AY, et al. (2017). Discovery of stimulation- responsive immune enhancers with CRISPR activation. Nature 549, 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeda H, Kataoka S, Nakayama M, Ali MAE, Oshima H, Yamamoto D, Park JW, Takegami Y, An T, Jenkins NA, et al. (2019). CRISPR-Cas9-mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proc. Natl. Acad. Sci. U. S. A Available at: 10.1073/pnas.1904714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su CW, Smillie C, Shekhar K, et al. (2018). T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 175, 1307–1320.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, Chalkiadaki A, Kim HJ, and Ingber DE (2018). Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell Mol Gastroenterol Hepatol 5, 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, et al. (2019). Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55. [DOI] [PubMed] [Google Scholar]; This study provides a single cell transcriptional map of epithelial cells in UC.
- 44.Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, et al. (2018). Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 175, 372–386.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, et al. (2019). Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides a holistic single cell transcriptional map of UC.
- 46.Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA, Gettler K, Chuang L-S, Nayar S, Greenstein AJ, et al. (2019). Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 178, 1493–1508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides a single cell transcriptional map of CD.
- 47.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.West NR, Hegazy AN, Owens BMJ, Bullers SJ, Linggi B, Buonocore S, Coccia M, Görtz D, This S, Stockenhuber K, et al. (2017). Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat. Med 23, 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study utilizes transcriptomic signatures of treatment resistance to identify and functionally validate cytokine circuits associated with severe IBD.
- 49.Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, Park YR, Raychaudhuri S, Pouget JG, Hübenthal M, et al. (2016). Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet 48, 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study compares GWAS risk loci across distinct inflammatory diseases to identify common and disease-specific genes.
- 50.Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, Li V, Maradana MR, Schiering C, and Stockinger B (2018). The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity 49, 353–362.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pentinmikko N, Iqbal S, Mana M, Andersson S, Cognetta AB 3rd, Suciu RM, Roper J, Luopajärvi K, Markelin E, Gopalakrishnan S, et al. (2019). Notum produced by Paneth cells attenuates regeneration of aged intestinal epithelium. Nature 571, 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serra D, Mayr U, Boni A, Lukonin I, Rempfler M, Challet Meylan L, Stadler MB, Strnad P, Papasaikas P, Vischi D, et al. (2019). Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.VanDussen KL, Stojmirović A, Li K, Liu T-C, Kimes PK, Muegge BD, Simpson KF, Ciorba MA, Perrigoue JG, Friedman JR, et al. (2018). Abnormal Small Intestinal Epithelial Microvilli in Patients With Crohn’s Disease. Gastroenterology 155, 815–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramakrishnan SK, Zhang H, Ma X, Jung I, Schwartz AJ, Triner D, Devenport SN, Das NK, Xue X, Zeng MY, et al. (2019). Intestinal non-canonical NFκB signaling shapes the local and systemic immune response. Nat. Commun 10, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider C, O’Leary CE, and Locksley RM (2019). Regulation of immune responses by tuft cells. Nat. Rev. Immunol Available at: 10.1038/s41577-019-0176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, and van Oudenaarden A (2015). Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255. [DOI] [PubMed] [Google Scholar]
- 57.Beumer J, Artegiani B, Post Y, Reimann F, Gribble F, Nguyen TN, Zeng H, Van den Born M, Van Es JH, and Clevers H (2018). Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell Biol 20, 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gehart H, van Es JH, Hamer K, Beumer J, Kretzschmar K, Dekkers JF, Rios A, and Clevers H (2019). Identification of Enteroendocrine Regulators by Real-Time Single- Cell Differentiation Mapping. Cell 176, 1158–1173.e16. [DOI] [PubMed] [Google Scholar]
- 59.Mohanan V, Nakata T, Desch AN, Lévesque C, Boroughs A, Guzman G, Cao Z, Creasey E, Yao J, Boucher G, et al. (2018). C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science 359, 1161–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manzanillo P, Mouchess M, Ota N, Dai B, Ichikawa R, Wuster A, Haley B, Alvarado G, Kwon Y, Caothien R, et al. (2018). Inflammatory Bowel Disease Susceptibility Gene C1ORF106 Regulates Intestinal Epithelial Permeability. Immunohorizons 2, 164–171. [DOI] [PubMed] [Google Scholar]
- 61.Fujimoto K, Kinoshita M, Tanaka H, Okuzaki D, Shimada Y, Kayama H, Okumura R, Furuta Y, Narazaki M, Tamura A, et al. (2017). Regulation of intestinal homeostasis by the ulcerative colitis-associated gene RNF186. Mucosal Immunol. 10, 446–459. [DOI] [PubMed] [Google Scholar]
- 62.Tong X, Zhang Q, Wang L, Ji Y, Zhang L, Xie L, Chen W, and Zhang H (2018). RNF186 impairs insulin sensitivity by inducing ER stress in mouse primary hepatocytes. Cell. Signal 52, 155–162. [DOI] [PubMed] [Google Scholar]
- 63.Sveinbjornsson G, Mikaelsdottir E, Palsson R, Indridason OS, Holm H, Jonasdottir A, Helgason A, Sigurdsson S, Jonasdottir A, Sigurdsson A, et al. (2014). Rare mutations associating with serum creatinine and chronic kidney disease. Hum. Mol. Genet 23, 6935–6943. [DOI] [PubMed] [Google Scholar]
- 64.Mukherjee T, Hovingh ES, Foerster EG, Abdel-Nour M, Philpott DJ, and Girardin SE (2018). NOD1 and NOD2 in inflammation, immunity and disease. Arch. Biochem. Biophys Available at: 10.1016/j.abb.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 65.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. (2001). A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606. [DOI] [PubMed] [Google Scholar]
- 66.Hsu L-C, Ali SR, McGillivray S, Tseng P-H, Mariathasan S, Humke EW, Eckmann L, Powell JJ, Nizet V, Dixit VM, et al. (2008). A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc. Natl. Acad. Sci. U. S. A 105, 7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinon F, Agostini L, Meylan E, and Tschopp J (2004). Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol 14, 1929–1934. [DOI] [PubMed] [Google Scholar]
- 68.Kim Y-G, Shaw MH, Warner N, Park J-H, Chen F, Ogura Y, and Núñez G (2011). Cutting edge: Crohn’s disease-associated Nod2 mutation limits production of proinflammatory cytokines to protect the host from Enterococcus faecalis-induced lethality. J. Immunol 187, 2849–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Umiker B, Lee H-H, Cope J, Ajami NJ, Laine J-P, Fregeau C, Ferguson H, Alves SE, Sciammetta N, Kleinschek M, et al. (2019). The NLRP3 inflammasome mediates DSS-induced intestinal inflammation in Nod2 knockout mice. Innate Immun. 25, 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caruso R, Mathes T, Martens EC, Kamada N, Nusrat A, Inohara N, and Núñez G (2019). A specific gene-microbe interaction drives the development of Crohn’s disease-like colitis in mice. Sci Immunol 4 Available at: 10.1126/sciimmunol.aaw4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chirieleison SM, Marsh RA, Kumar P, Rathkey JK, Dubyak GR, and Abbott DW (2017). Nucleotide-binding oligomerization domain (NOD) signaling defects and cell death susceptibility cannot be uncoupled in X-linked inhibitor of apoptosis (XIAP)-driven inflammatory disease. J. Biol. Chem 292, 9666–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Latour S, and Aguilar C (2015). XIAP deficiency syndrome in humans. Semin. Cell Dev. Biol 39, 115–123. [DOI] [PubMed] [Google Scholar]
- 73.Li Q, Lee CH, Peters LA, Mastropaolo LA, Thoeni C, Elkadri A, Schwerd T, Zhu J, Zhang B, Zhao Y, et al. (2016). Variants in TRIM22 That Affect NOD2 Signaling Are Associated With Very-Early-Onset Inflammatory Bowel Disease. Gastroenterology 150, 1196–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zammit NW, Siggs OM, Gray PE, Horikawa K, Langley DB, Walters SN, Daley SR, Loetsch C, Warren J, Yap JY, et al. (2019). Denisovan, modern human and mouse TNFAIP3 alleles tune A20 phosphorylation and immunity. Nat. Immunol 20, 1299–1310. [DOI] [PubMed] [Google Scholar]
- 75.Hrdinka M, Schlicher L, Dai B, Pinkas DM, Bufton JC, Picaud S, Ward JA, Rogers C, Suebsuwong C, Nikhar S, et al. (2018). Small molecule inhibitors reveal an indispensable scaffolding role of RIPK2 in NOD2 signaling. EMBO J. 37 Available at: 10.15252/embj.201899372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haile PA, Casillas LN, Votta BJ, Wang GZ, Charnley AK, Dong X, Bury MJ, Romano JJ, Mehlmann JF, King BW, et al. (2019). Discovery of a First-in-Class Receptor Interacting Protein 2 (RIP2) Kinase Specific Clinical Candidate, 2-((4-(Benzo[ d]thiazol-5-ylamino)-6-( tert-butylsulfonyl)quinazolin-7-yl)oxy)ethyl Dihydrogen Phosphate, for the Treatment of Inflammatory Diseases. J. Med. Chem Available at: 10.1021/acs.jmedchem.9b00575. [DOI] [PubMed] [Google Scholar]
- 77.Hara H, Ishihara C, Takeuchi A, Imanishi T, Xue L, Morris SW, Inui M, Takai T, Shibuya A, Saijo S, et al. (2007). The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat. Immunol 8, 619–629. [DOI] [PubMed] [Google Scholar]
- 78.Xu X, Xu J-F, Zheng G, Lu H-W, Duan J-L, Rui W, Guan J-H, Cheng L-Q, Yang D-D, Wang M-C, et al. (2018). CARD9S12N facilitates the production of IL-5 by alveolar macrophages for the induction of type 2 immune responses. Nat. Immunol 19, 547–560. [DOI] [PubMed] [Google Scholar]
- 79.Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, et al. (2019). Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 25, 377–388.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao Z, Conway KL, Heath RJ, Rush JS, Leshchiner ES, Ramirez-Ortiz ZG, Nedelsky NB, Huang H, Ng A, Gardet A, et al. (2015). Ubiquitin Ligase TRIM62 Regulates CARD9-Mediated Anti-fungal Immunity and Intestinal Inflammation. Immunity 43, 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leshchiner ES, Rush JS, Durney MA, Cao Z, Dančík V, Chittick B, Wu H, Petrone A, Bittker JA, Phillips A, et al. (2017). Small-molecule inhibitors directly target CARD9 and mimic its protective variant in inflammatory bowel disease. Proc. Natl. Acad. Sci. U. S. A 114, 11392–11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bunker JJ, and Bendelac A (2018). IgA Responses to Microbiota. Immunity 49, 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Castro-Dopico T, Dennison TW, Ferdinand JR, Mathews RJ, Fleming A, Clift D, Stewart BJ, Jing C, Strongili K, Labzin LI, et al. (2019). Anti-commensal IgG Drives Intestinal Inflammation and Type 17 Immunity in Ulcerative Colitis. Immunity 50, 1099–1114.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Plichta DR, Graham DB, Subramanian S, and Xavier RJ (2019). Therapeutic Opportunities in Inflammatory Bowel Disease: Mechanistic Dissection of Host-Microbiome Relationships. Cell 178, 1041–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review covers the role of the microbiome and environmental factors that impact IBD.
- 85.Goyette P, Boucher G, Mallon D, Ellinghaus E, Jostins L, Huang H, Ripke S, Gusareva ES, Annese V, Hauser SL, et al. (2015). High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet 47, 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]; This fine-mapping GWAS implicated specific HLA alleles in IBD risk.
- 86.Lee JC, Biasci D, Roberts R, Gearry RB, Mansfield JC, Ahmad T, Prescott NJ, Satsangi J, Wilson DC, Jostins L, et al. (2017). Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat. Genet 49, 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S, Kim D, Hsieh C-S, Hogan SP, Elson CO, et al. (2017). Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol 2 Available at: 10.1126/sciimmunol.aao1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu J, Pendegraft AH, Byrne-Steele M, Yang Q, Wang C, Pan W, Lucious T, Seay T, Cui X, Elson CO, et al. (2018). Expanded TCRβ CDR3 clonotypes distinguish Crohn’s disease and ulcerative colitis patients. Mucosal Immunol. 11, 1487–1495. [DOI] [PubMed] [Google Scholar]
- 89.Christophersen A, Lund EG, Snir O, Solà E, Kanduri C, Dahal-Koirala S, Zühlke S, Molberg Ø, Utz PJ, Rohani-Pichavant M, et al. (2019). Distinct phenotype of CD4+ T cells driving celiac disease identified in multiple autoimmune conditions. Nat. Med 25, 734–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. (2018). Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066. [DOI] [PubMed] [Google Scholar]
- 91.Zhou L, Chu C, Teng F, Bessman NJ, Goc J, Santosa EK, Putzel GG, Kabata H, Kelsen JR, Baldassano RN, et al. (2019). Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 568, 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Friedrich M, Pohin M, and Powrie F (2019). Cytokine Networks in the Pathophysiology of Inflammatory Bowel Disease. Immunity 50, 992–1006. [DOI] [PubMed] [Google Scholar]
- 93.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. (2006). A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bauché D, Joyce-Shaikh B, Jain R, Grein J, Ku KS, Blumenschein WM, Ganal- Vonarburg SC, Wilson DC, McClanahan TK, Malefyt R de W., et al. (2018). LAG3+ Regulatory T Cells Restrain Interleukin-23-Producing CX3CR1+ Gut-Resident Macrophages during Group 3 Innate Lymphoid Cell-Driven Colitis. Immunity 49, 342–352.e5. [DOI] [PubMed] [Google Scholar]
- 95.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, and Littman DR (2007). IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol 8, 967–974. [DOI] [PubMed] [Google Scholar]
- 96.Maxwell JR, Zhang Y, Brown WA, Smith CL, Byrne FR, Fiorino M, Stevens E, Bigler J, Davis JA, Rottman JB, et al. (2015). Differential Roles for Interleukin-23 and Interleukin-17 in Intestinal Immunoregulation. Immunity 43, 739–750. [DOI] [PubMed] [Google Scholar]
- 97.Tang C, Kakuta S, Shimizu K, Kadoki M, Kamiya T, Shimazu T, Kubo S, Saijo S, Ishigame H, Nakae S, et al. (2018). Suppression of IL-17F, but not of IL-17A, provides protection against colitis by inducing Treg cells through modification of the intestinal microbiota. Nat. Immunol 19, 755–765. [DOI] [PubMed] [Google Scholar]
- 98.Leppkes M, Becker C, Ivanov II, Hirth S, Wirtz S, Neufert C, Pouly S, Murphy AJ, Valenzuela DM, Yancopoulos GD, et al. (2009). RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL- 17F. Gastroenterology 136, 257–267. [DOI] [PubMed] [Google Scholar]
- 99.Malik A, and Kanneganti T-D (2017). Inflammasome activation and assembly at a glance. J. Cell Sci 130, 3955–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shouval DS, Biswas A, Kang YH, Griffith AE, Konnikova L, Mascanfroni ID, Redhu NS, Frei SM, Field M, Doty AL, et al. (2016). Interleukin 1β Mediates Intestinal Inflammation in Mice and Patients With Interleukin 10 Receptor Deficiency. Gastroenterology 151, 1100–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de Luca A, Smeekens SP, Casagrande A, Iannitti R, Conway KL, Gresnigt MS, Begun J, Plantinga TS, Joosten LAB, van der Meer JWM, et al. (2014). IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc. Natl. Acad. Sci. U. S. A 111, 3526–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang B-G, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. (2008). Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456, 264–268. [DOI] [PubMed] [Google Scholar]
- 103.Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu T-C, Heath RJ, et al. (2014). Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc. Natl. Acad. Sci. U. S. A 111, 7741–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, et al. (2014). A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456–462. [DOI] [PubMed] [Google Scholar]
- 105.Samie M, Lim J, Verschueren E, Baughman JM, Peng I, Wong A, Kwon Y, Senbabaoglu Y, Hackney JA, Keir M, et al. (2018). Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling. Nat. Immunol 19, 246–254. [DOI] [PubMed] [Google Scholar]
- 106.Weiss ES, Girard-Guyonvarc’h C, Holzinger D, de Jesus AA, Tariq Z, Picarsic J, Schiffrin EJ, Foell D, Grom AA, Ammann S, et al. (2018). Interleukin-18 diagnostically distinguishes and pathogenically promotes human and murine macrophage activation syndrome. Blood 131, 1442–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong Y-N, Peng X, Xi JJ, Chen S, et al. (2014). Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513, 237–241. [DOI] [PubMed] [Google Scholar]
- 108.Khare S, Dorfleutner A, Bryan NB, Yun C, Radian AD, de Almeida L, Rojanasakul Y, and Stehlik C (2012). An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schiering C, Krausgruber T, Chomka A, Fröhlich A, Adelmann K, Wohlfert EA, Pott J, Griseri T, Bollrath J, Hegazy AN, et al. (2014). The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA, Bucks C, Kane CM, Fallon PG, Pannell R, et al. (2010). Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.He Z, Song J, Hua J, Yang M, Ma Y, Yu T, Feng J, Liu B, Wang X, Li Y, et al. (2018). Mast cells are essential intermediaries in regulating IL-33/ST2 signaling for an immune network favorable to mucosal healing in experimentally inflamed colons. Cell Death Dis. 9, 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Scheibe K, Kersten C, Schmied A, Vieth M, Primbs T, Carlé B, Knieling F, Claussen J, Klimowicz AC, Zheng J, et al. (2019). Inhibiting Interleukin 36 Receptor Signaling Reduces Fibrosis in Mice With Chronic Intestinal Inflammation. Gastroenterology 156, 1082–1097.e11. [DOI] [PubMed] [Google Scholar]
- 113.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. (2007). Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet 39, 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al. (2008). Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet 40, 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. (2007). A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet 39, 207–211. [DOI] [PubMed] [Google Scholar]
- 116.Hui KY, Fernandez-Hernandez H, Hu J, Schaffner A, Pankratz N, Hsu N-Y, Chuang L-S, Carmi S, Villaverde N, Li X, et al. (2018). Functional variants in the LRRK2 gene confer shared effects on risk for Crohn’s disease and Parkinson’s disease. Sci. Transl. Med 10 Available at: 10.1126/scitranslmed.aai7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Takagawa T, Kitani A, Fuss I, Levine B, Brant SR, Peter I, Tajima M, Nakamura S, and Strober W (2018). An increase in LRRK2 suppresses autophagy and enhances Dectin-1-induced immunity in a mouse model of colitis. Sci. Transl. Med 10 Available at: 10.1126/scitranslmed.aan8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. (2008). A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.VanDussen KL, Liu T-C, Li D, Towfic F, Modiano N, Winter R, Haritunians T, Taylor KD, Dhall D, Targan SR, et al. (2014). Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology 146, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Deuring JJ, Fuhler GM, Konstantinov SR, Peppelenbosch MP, Kuipers EJ, de Haar C, and van der Woude CJ (2014). Genomic ATG16L1 risk allele-restricted Paneth cell ER stress in quiescent Crohn’s disease. Gut 63, 1081–1091. [DOI] [PubMed] [Google Scholar]
- 121.Grootjans J, Krupka N, Hosomi S, Matute JD, Hanley T, Saveljeva S, Gensollen T, Heijmans J, Li H, Limenitakis JP, et al. (2019). Epithelial endoplasmic reticulum stress orchestrates a protective IgA response. Science 363, 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kaser A, Lee A-H, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EES, Higgins DE, Schreiber S, Glimcher LH, et al. (2008). XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Adolph TE, Tomczak MF, Niederreiter L, Ko H-J, Böck J, Martinez-Naves E, Glickman JN, Tschurtschenthaler M, Hartwig J, Hosomi S, et al. (2013). Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hosomi S, Grootjans J, Tschurtschenthaler M, Krupka N, Matute JD, Flak MB, Martinez-Naves E, Gomez Del Moral M, Glickman JN, Ohira M, et al. (2017). Intestinal epithelial cell endoplasmic reticulum stress promotes MULT1 up-regulation and NKG2D-mediated inflammation. J. Exp. Med 214, 2985–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Graham DB, Lefkovith A, Deelen P, de Klein N, Varma M, Boroughs A, Desch AN, Ng ACY, Guzman G, Schenone M, et al. (2016). TMEM258 Is a Component of the Oligosaccharyltransferase Complex Controlling ER Stress and Intestinal Inflammation. Cell Rep. 17, 2955–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Park JH, Hogrebe M, Grüneberg M, DuChesne I, von der Heiden AL, Reunert J, Schlingmann KP, Boycott KM, Beaulieu CL, Mhanni AA, et al. (2015). SLC39A8 Deficiency: A Disorder of Manganese Transport and Glycosylation. Am. J. Hum. Genet 97, 894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bastaki F, Bizzari S, Hamici S, Nair P, Mohamed M, Saif F, Malik EM, Al-Ali MT, and Hamzeh AR (2018). Single-center experience of N-linked Congenital Disorders of Glycosylation with a Summary of Molecularly Characterized Cases in Arabs. Ann. Hum. Genet 82, 35–47. [DOI] [PubMed] [Google Scholar]
- 128.Taylor CT, and Colgan SP (2017). Regulation of immunity and inflammation by hypoxia in immunological niches. Nat. Rev. Immunol 17, 774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Campbell EL, Bruyninckx WJ, Kelly CJ, Glover LE, McNamee EN, Bowers BE, Bayless AJ, Scully M, Saeedi BJ, Golden-Mason L, et al. (2014). Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 40, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Curtis VF, Ehrentraut SF, Campbell EL, Glover LE, Bayless A, Kelly CJ, Kominsky DJ, and Colgan SP (2015). Stabilization of HIF through inhibition of Cullin-2 neddylation is protective in mucosal inflammatory responses. FASEB J. 29, 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xue X, Ramakrishnan S, Anderson E, Taylor M, Zimmermann EM, Spence JR, Huang S, Greenson JK, and Shah YM (2013). Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology 145, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Solanki S, Devenport SN, Ramakrishnan SK, and Shah YM (2019). Temporal induction of intestinal epithelial hypoxia-inducible factor-2α is sufficient to drive colitis. Am. J. Physiol. Gastrointest. Liver Physiol 317, G98–G107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Glover LE, Bowers BE, Saeedi B, Ehrentraut SF, Campbell EL, Bayless AJ, Dobrinskikh E, Kendrick AA, Kelly CJ, Burgess A, et al. (2013). Control of creatine metabolism by HIF is an endogenous mechanism of barrier regulation in colitis. Proc. Natl. Acad. Sci. U. S. A 110, 19820–19825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. (2013). Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Fontana MF, et al. (2018). Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 49, 33–41.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nair S, Huynh JP, Lampropoulou V, Loginicheva E, Esaulova E, Gounder AP, Boon ACM, Schwarzkopf EA, Bradstreet TR, Edelson BT, et al. (2018). Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection. J. Exp. Med 215, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E, et al. (2018). Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Skon-Hegg C, Zhang J, Wu X, Sagolla M, Ota N, Wuster A, Tom J, Doran E, Ramamoorthi N, Caplazi P, et al. (2019). LACC1 Regulates TNF and IL-17 in Mouse Models of Arthritis and Inflammation. J. Immunol 202, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lahiri A, Hedl M, Yan J, and Abraham C (2017). Human LACC1 increases innate receptor-induced responses and a LACC1 disease-risk variant modulates these outcomes. Nat. Commun 8, 15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cader MZ, Boroviak K, Zhang Q, Assadi G, Kempster SL, Sewell GW, Saveljeva S, Ashcroft JW, Clare S, Mukhopadhyay S, et al. (2016). C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat. Immunol 17, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Molofsky AB, Savage AK, and Locksley RM (2015). Interleukin-33 in Tissue Homeostasis, Injury, and Inflammation. Immunity 42, 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schafer S, Viswanathan S, Widjaja AA, Lim W-W, Moreno-Moral A, DeLaughter DM, Ng B, Patone G, Chow K, Khin E, et al. (2017). IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 552, 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kotlarz D, Marquardt B, Barøy T, Lee WS, Konnikova L, Hollizeck S, Magg T, Lehle AS, Walz C, Borggraefe I, et al. (2018). Human TGF-β1 deficiency causes severe inflammatory bowel disease and encephalopathy. Nat. Genet 50, 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Flynn RS, Mahavadi S, Murthy KS, Grider JR, Kellum JM, Akbari H, and Kuemmerle JF (2011). Endogenous IGFBP-3 regulates excess collagen expression in intestinal smooth muscle cells of Crohn’s disease strictures. Inflamm. Bowel Dis 17, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Geesala R, Schanz W, Biggs M, Dixit G, Skurski J, Gurung P, Meyerholz DK, Elliott D, Issuree PD, and Maretzky T (2019). Loss of RHBDF2 results in an early-onset spontaneous murine colitis. J. Leukoc. Biol 105, 767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li Y, Führer M, Bahrami E, Socha P, Klaudel-Dreszler M, Bouzidi A, Liu Y, Lehle AS, Magg T, Hollizeck S, et al. (2019). Human RIPK1 deficiency causes combined immunodeficiency and inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A 116, 970–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pagnini C, Pizarro TT, and Cominelli F (2019). Novel Pharmacological Therapy in Inflammatory Bowel Diseases: Beyond Anti-Tumor Necrosis Factor. Front. Pharmacol 10, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ma C, Battat R, Dulai PS, Parker CE, Sandborn WJ, Feagan BG, and Jairath V (2019). Innovations in Oral Therapies for Inflammatory Bowel Disease. Drugs. Available at: 10.1007/s40265-019-01169-y. [DOI] [PubMed] [Google Scholar]


