Abstract
Background
This study was designed to explore the incompletely investigated role of the complement component 3a receptor 1 (C3AR1) in the prognosis of stomach adenocarcinomas (STAD).
Material/Methods
Using bioinformatic methods, we systematically determined the expression and prognosis value of C3AR1 in various cancers by using the TIMER (Tumor Immune Estimation Resource) database, UALCAN platform, GEPIA (Gene Expression Profiling Interactive Analysis) server, and the OncoLnc tool. The biological processes influenced by C3AR1 were determined using the GSEA (Gene Set Enrichment Analysis) software (Copyright 2004–2020 Broad Institute, Inc., Massachusetts Institute of Technology, and Regents of the University of California). The correlation between C3AR1 expression and the immune-infiltrating cells as well as the correlation analysis between C3AR1 expression and the corresponding immune-marker sets were conducted using the TIMER and GEPIA databases.
Results
The expression of C3AR1 was significantly (P<0.001) differentially expressed on several tumor types, while its prognosis value could only be determined on STAD, with a high expression of C3AR1 closely correlated with a poor prognosis. The GSEA analysis revealed that the differential expression of C3AR1 profoundly affected the immune-related biological processes. The expression of C3AR1 was strongly and positively correlated with the infiltration of monocytes, tumor-associated macrophages, M2 macrophages, dendritic cells, and exhausted T cells.
Conclusions
Our results have revealed that a high expression of C3AR1 is positively correlated with a poor prognosis and increased tumor-immune infiltration. C3AR1 can promote the polarization of M2 macrophages and T cell exhaustion, leading to the immune escape of STAD. These findings suggest that C3AR1 could be used as a prognostic and immune-infiltration marker in the pathogenesis of STAD.
Keywords: Biological Markers; Lymphocytes, Tumor-Infiltrating; Prognosis; Receptors, Complement; Stomach Neoplasms
Background
Stomach cancer is a global health problem and the 3rd leading cause of cancer-related death [1]. According to global cancer statistics, more than 1 million people were newly diagnosed with stomach cancer in 2018, making it the 5th most common neoplasm [2]. Although there have been great advances in endoscopic and surgical therapies, chemotherapy, and systemic treatments, the 5-year survival rate for stomach cancer (~25%) remains unsatisfactory [3]. Therefore, further therapeutic exploration of stomach cancer treatments are of great importance, including research on the tumor-immune microenvironment for cell-based therapy, identifying the potential diagnostic and prognostic markers for molecular therapy, and novel agents for direct tumor elimination.
Stomach adenocarcinomas (STAD) are the most common stomach cancers (~95%) [3]. Advanced STAD patients, especially those unsuitable for surgical treatment, have a median survival of <10 months, further suggesting the importance and urgency of novel therapy development. Reports have proved that an obtuse immune system profoundly affects the progression of STAD. In 2011, the FDA approved ipilimumab, which targets cytotoxic T lymphocyte-associated protein 4 (CTLA-4) for the treatment of advanced melanomas. Ipilimumab is the first antibody drug to target the immune checkpoints. Ipilimumab had promising results [4], which further promoted research on the developments in tumor immunotherapy. Currently, 3 kinds of immunotherapy are being developed for STAD treatment: immune checkpoint inhibitors (ICIs), including ipilimumab, chimeric antigen receptor (CAR) T cell therapy, and tumor-antigen vaccines [5,6]. No beneficial effects were observed when ipilimumab was compared with chemotherapy in a phase II clinical trial for advanced STAD treatment (NCT01585987) [7]. Negative results were reported for another ICI (pembrolizumab, PD-l inhibitor) for STAD treatment in a clinical trial (NCT01848834) [8]. Although approved for other tumor treatments, the CAR T cell therapy and tumor-antigen vaccines for STAD treatment are still being developed. The clinical trials have shown that immune cell infiltration and immune checkpoint expression levels vary in the tumors between patients, and many patients develop drug resistance and adverse reactions. Therefore, it is urgent to explore novel therapeutic targets or precise prognostic markers of immune infiltration to improve the efficiency of the STAD treatment.
A sustained inflammatory response can increase the risk of gastric cancer [9], while complement activation is an important method to induce inflammation [10]. In addition, there is increasing evidence that complement activation in the tumor microenvironment plays a role in tumor promotion by locally suppressing the immune effect of T cells and maintaining chronic inflammation, ultimately promoting tumor immune escape, growth, and distant metastasis [11–13]. Acute C3a-C3AR1 activation decreased inflammation while chronic activation was shown to promote the progression of autoimmune-related disease. C3AR1, a protein-coding gene, is a G-protein-coupled transmembrane spanning the receptor of C3a in the complement system [14,15]. C3a is a 78-amino acid peptide derived from the protein cleavage of the complement protein C3, a well-recognized danger signal-associated molecular pattern. C3AR1 is predominantly expressed on the leukocytes of myeloid lineage, including neutrophils and monocyte/macrophages; however, it can also be detected on endothelial cells and neurons. Depending on the cell type and environmental cues, C3AR1 mediates pro-inflammatory or immunomodulatory functions. An activated-C3a signal could contribute to tumorigenesis, while the role of C3AR1 in the development of STAD remains unexplored [15,16].
In the present study, the correlation between C3AR1 and the prognosis of STAD was analyzed via the public databases GEPIA (Gene Expression Profiling Interactive Analysis) and UALCAN (http://ualcan.path.uab.edu/index.html), and the web tool, OncoLnc (http://www.oncolnc.org/). The relationships between C3AR1 and the tumor-infiltrating immune cells in different tumor microenvironments were investigated via TIMER (Tumor Immune Estimation Resource). Our results could improve the understanding of the possible role of C3AR1 in STAD. Moreover, our findings could provide a potential correlation between C3AR1 and tumor immune infiltration to lay a theoretical foundation for considering C3AR1 as a prognosis marker for STAD.
Material and Methods
Data Acquisition
In this study, all the data applied to each online analysis platform were from The Cancer Genome Atlas (TCGA), including the transcriptome analysis, biological sample information, original sequencing data, copy number variations, deoxyribonucleic acid methylation, and clinical information. Although the samples used for each platform were not identical, the data set used for the single-gene analysis included raw sequencing data for all 443 cases of gastric adenocarcinoma in the TCGA database [17]. The TCGA project, a landmark cancer genomics project administered by the National Cancer Institute and National Human Genome Research Institute of the United States government, is the largest cancer genomics online database and is valued for its inclusiveness of cancer cases, cancer types, and sequenced-data bigness. In the present study, various online analysis platforms (TIMER, UALCAN, GEPIA, and OncoLnc) were used to mine and analyze the TCGA data, including differential expression analysis, survival analysis, and immune cell infiltration analysis.
Expression and survival analysis
The expression of C3AR1 in various tumors was first evaluated by TIMER [18], and further validated by UALCAN [19] and GEPIA [20]. TIMER is a web server for tumor-immune interactions, investigation, and visualization. It provides modules for differential expression analysis (DiffExp modules) to compare the expression of the gene of interest (C3AR1) between the tumor and the adjacent tissues across the TCGA tumors. The tumor types with differentially expressed C3AR1 were further verified by UALCAN and GEPIA. P values <0.05 were considered statistically significant.
The prognostic values of the C3AR1 mRNA expression in the selected tumor types were evaluated by GEPIA, TIMER, and OncoLnc. The patients were divided into 2 groups according to their relative expression level of C3AR1: the high-expression group and the low-expression group. In the GEPIA analysis platform, the high-expression group included 20% of cases with the highest C3AR1 expression ranking from high to low; and the low-expression group included the remaining 80% of cases. In the TIMER database, the high-expression group included 30% of cases of C3AR1 expression ranked from high to low, and the low-expression group included 30% of the lowest C3AR1 expression cases. In the OncoLnc analysis platform, the high-expression group included the top 25% cases with the highest C3AR1 expression ranked from high to low, and the low-expression group included the remaining 75% of cases. The effect of C3AR1 expression on the prognosis was evaluated by comparing the survival curves of the 2 groups (high- and low-expression) using the log-rank test on the indicated online web tools. P<0.05 was considered statistically significant.
Gene Correlation by TIMER and GEPIA Databases
The STAD tumor tissue markers and immune-infiltrating cell markers identified from the references were input into the “correlation” module of TIMER to analyze their correlation with the C3AR1 expression level. Then the correlation between these markers and C3AR1 was verified using the GEPIA database.
Analysis of Tumor-infiltrating Immune Cells
The statistics between the tumor-infiltrating immune cells (TIICs) and C3AR1 were analyzed via the “Gene” module in the TIMER database. The TIIC includes 6 subsets of B cells, CD4 T cells, CD8 T cells, macrophages, neutrophils, and dendritic cells (DCs). After a purity correction, Spearman’s correlation (via scatterplots) and statistical significance was calculated. The left-most panel always shows the gene expression level of tumor purity, then, the correlation between the B cells, CD8+T cells, CD4+T cells, macrophages, neutrophils, and DC, and the gene expression level is listed. If the absolute value of the correlation coefficient was >0.3, it was considered to be correlated. P<0.05 was considered statistically significant.
Gene set Enrichment Analysis (GSEA)
GSEA (Copyright 2004–2020 Broad Institute, Inc., Massachusetts Institute of Technology, and Regents of the University of California) is a powerful instrument for evaluating the microarray data and identifying the potential functions of genes [21]. The distribution of the single genes from the TCGA STAD datasets was verified using GSEA. The 443 samples from the TCGA database were divided into 2 groups according to their expression of C3AR1. Then, the GSEA V3.0 software was used to verify whether the gene set in the V6.2 data set of the Molecular Signature Database (MSigDB) (Copyright 2004–2020 Broad Institute, Inc., Massachusetts Institute of Technology, and Regents of the University of California) was related to the expression of C3AR1 [22]. We selected the packet C5 BP from the biological process part of gene ontology from the MSigDB for functional annotation. To identify the biological processes that warranted further investigation, we selected the statistical significance P<0.05, a false discovery rate <0.25, and a normalized enrichment score (NES) >1.0 [23].
Statistical Analysis
The log-rank test was used to evaluate the relationship between the C3AR1 expression level and STAD prognosis. A P value <0.05 was considered statistically significant. The calculation of Spearman’s rho was used to assess the correlation between the 2 genes or between C3AR1 and the level of immune cell infiltration. The absolute values of the correlation coefficient >0.3, 0.6, and 0.9 represent low, medium, and high correlations, respectively.
Results
mRNA Expression of C3AR1 in Diverse Cancers
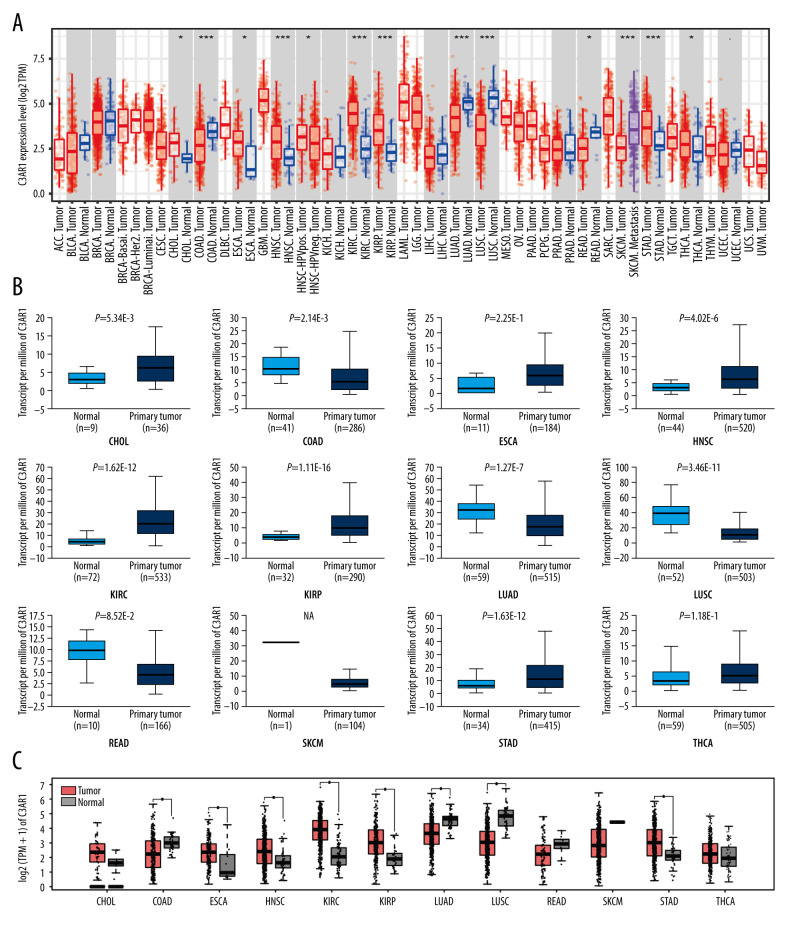
Since the use of the ICIs for STAD treatment by targeting CTLA4 and PD1/PDL1 were not as promising as previously thought, and a panel of immune checkpoints (PDL2, TIM-3, and VSIG4) [17,24,25] were reported to participate in the pathogenesis of STAD, we anticipated that other prognostic markers can also exist. The Pearson correlation analysis was performed using an online analyzing tool (LinkedOmics http://www.linkedomics.org/login.php) [26] to determine the genes highly associated with those immune checkpoints, including PD-L2, TIM-3, and VSIG4. Three lists of genes highly related to PDL2, TIM-3, and VSIG4 in STAD patients were selected, with each list containing 20 genes. Four genes (FPR3, LILRB4, C3AR1, LAIR1) co-exist in the 3 lists (Figure 1). However, only the expression of C3AR1 was associated with the prognosis of STAD. To assess the C3AR1 expression in human cancers, the mRNA expression of C3AR1 was determined in all the TCGA cancers and the corresponding normal tissues. As presented in Figure 2A, the mRNA expression of C3AR1 was significantly upregulated in cholangiocarcinomas (CHOL), esophageal carcinomas (ESCA), head and neck squamous cell carcinomas (HNSC), kidney renal clear cell carcinomas (KIRC), kidney renal papillary cell carcinomas (KIRP), STAD, and thyroid carcinomas (THCA). It was downregulated in colon adenocarcinomas (COAD), lung adenocarcinomas (LUAD), lung squamous cell carcinomas (LUSC), and rectum adenocarcinomas (READ), when compared with the corresponding adjacent normal tissues using the TIMER database. The differential expression of C3AR1 was further verified in the UALCAN (Figure 2B) and GEPIA databases (Figure 2C). The analysis revealed that the expression of C3AR1 was differentially expressed only in COAD, HNSC, KIRC, KIPP, LUAD, LUSC, and STAD (Figure 2B, 2C).
Figure 1.
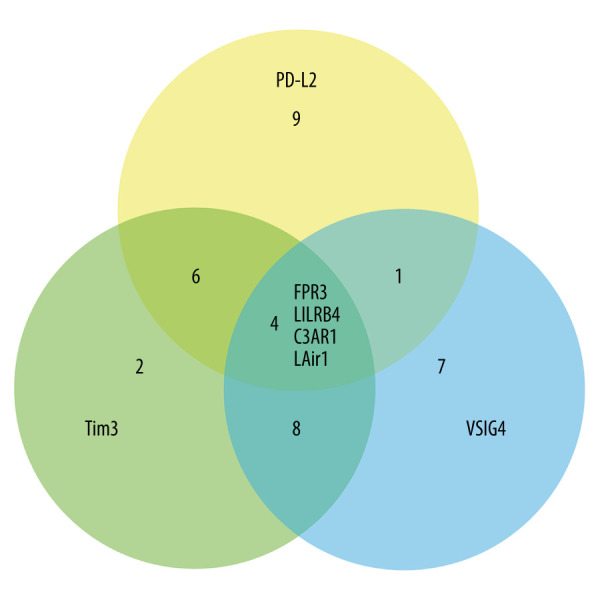
Venn diagram showing the common genes highly associated with the expression of PDL2, TIM3, and VSIG4.
Figure 2.
mRNA expression of C3AR1 in various cancers. (A) Comparison of the C3AR1 expression levels between the tumor tissue and paracarcinomic tissue across the TCGA database by the TIMER platform. (B) Expression of C3AR1 determined by the UALCAN platform. (C) Expression of C3AR1 determined by the GEPIA database. P<0.1, * P<0.05, ** P<0.01, *** P<0.001. GEPIA – Gene Expression Profiling Interactive Analysis; TCGA – The Cancer Genome Atlas; TIMER – Tumor Immune Estimation Resource.
Correlation Between C3AR1 Expression and Survival Rates in Diverse Cancers
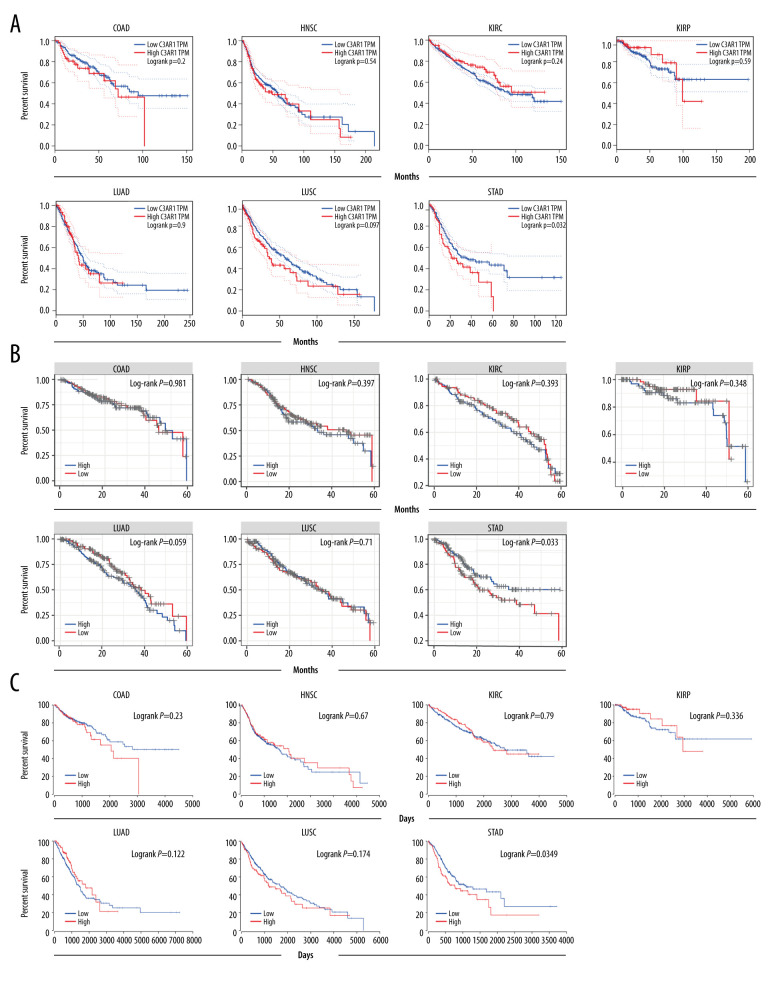
To determine the prognostic value of C3AR1 in previously identified tumor types, the overall survival rates of the higher expression levels of C3AR1 were compared with the lower-expression cancers using a Cox regression model with the online TIMER, GEPIA, and OncoLnc tools. The diagnostic value of C3AR1 expression can only be identified on STAD, with a higher C3AR1 expression closely associated with poor diagnosis as first determined by GEPIA analysis (Figure 3A). This result was revalidated by the other 2 TCGA-based online analysis databases, TIMER (Figure 3B) and OncoLnc (Figure 3C); they showed the same prognostic potential of C3AR1 in STAD. These results confirmed that C3AR1 expression is specifically associated with the prognosis of STAD among the selected cancer types.
Figure 3.
Survival curve of C3AR1 in various cancers. (A) Overall survival obtained by analysis through the GEPIA database. (B) Survival curve obtained from analysis using the TIMER database. (C) Survival curve obtained from analysis using the OncoLnc platform. GEPIA – Gene Expression Profiling Interactive Analysis; TIMER – Tumor Immune Estimation Resource.
Analysis of Biological Processes Influenced by C3AR1 in GSEA Using Gene Ontology (GO)
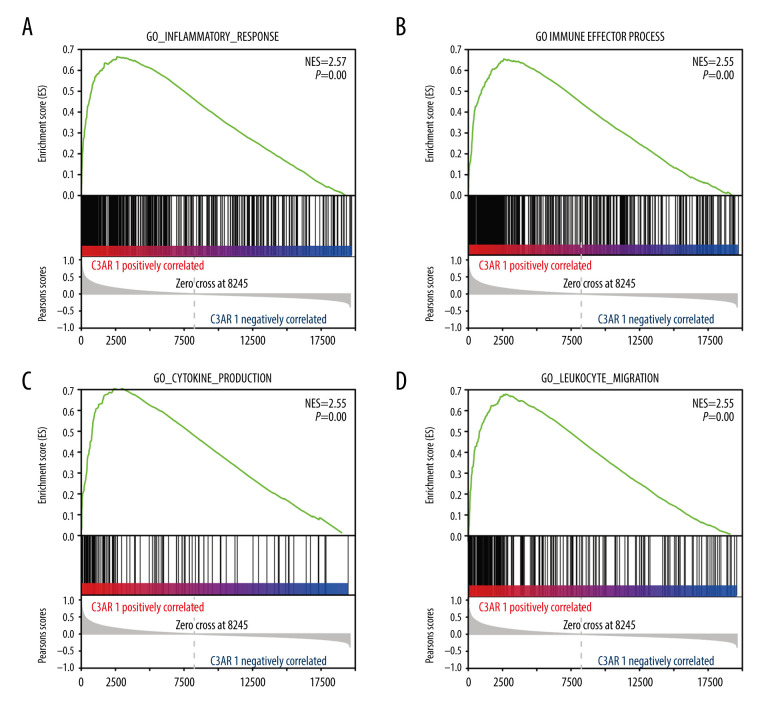
To explore the underlying mechanisms by which C3AR1 affects the prognosis of STAD, GSEA was used to study the biological processes affected by C3AR1. The data downloaded from TCGA was grouped according to the level of C3AR1 expression, processed into the format required by the GSEA software, and imported into the GSEA software. C3AR1 was selected for single-gene analysis and the system automatically grouped the data according to the level of C3AR1 expression. As presented in Figure 4A–4D, the biological processes with a significant positive correlation with C3AR1 expression were enriched, typically including the inflammatory response (NES=2.57, NOM P [normalized P value]=0.00), immune-effector process (NES=2.55, NOM P=0.00), production of cytokines (NES=2.55, NOM P=0.00), and the process of leukocyte migration (NES=2.55, NOM P=0.00). More than half of the biological functions of C3AR1, which ranked in the top 30 on the data list, were related to the immune response process (Table 1). As C3AR1 is an important component of the complement system, it is reasonable to assume that C3AR1 can promote STAD progression via regulating the tumor-immune microenvironment.
Figure 4.
GSEA-analyzed biological processes data positively correlated with C3AR1 expression. (A) Inflammatory response. (B) Immune-effector process. (C) Production of cytokines. (D) Leukocyte migration. GSEA – Gene Set Enrichment Analysis.
Table 1.
Biological processes of C3AR1 were analyzed by GSEA software.
| Name | SIZE | ES | NES | NOM p-val | FDR q-val |
|---|---|---|---|---|---|
| GO_INFLAMMATORY_RESPONSE | 442 | 0.665513 | 2.568941 | 0 | 0 |
| GO_IMMUNE_EFFECTOR_PROCESS | 449 | 0.654583 | 2.5537 | 0 | 0 |
| GO_CYTOKINE_PRODUCTION | 118 | 0.719826 | 2.551827 | 0 | 0 |
| GO_LEUKOCYTE_MIGRATION | 256 | 0.678578 | 2.550392 | 0 | 0 |
| GO_POSITIVE_REGULATION_OF_CYTOKINE_PRODUCTION | 364 | 0.649705 | 2.537406 | 0 | 0 |
| GO_ACTIVATION_OF_IMMUNE_RESPONSE | 388 | 0.682517 | 2.526013 | 0 | 0 |
| GO_REGULATION_OF_IMMUNE_EFFECTOR_PROCESS | 418 | 0.606066 | 2.525585 | 0 | 0 |
| GO_CYTOKINE_MEDIATED_SIGNALING_PATHWAY | 439 | 0.647561 | 2.517916 | 0 | 0 |
| GO_POSITIVE_REGULATION_OF_DEFENSE_RESPONSE | 360 | 0.645124 | 2.514322 | 0 | 0 |
| GO_REGULATION_OF_CYTOKINE_SECRETION | 142 | 0.701544 | 2.498589 | 0 | 0 |
| GO_POSITIVE_REGULATION_OF_RESPONSE_TO_EXTERNAL_STIMULUS | 293 | 0.633304 | 2.492779 | 0 | 0 |
| GO_LEUKOCYTE_MEDIATED_IMMUNITY | 157 | 0.670396 | 2.48643 | 0 | 0 |
| GO_RESPONSE_TO_INTERFERON_GAMMA | 139 | 0.77279 | 2.483442 | 0 | 0 |
| GO_LEUKOCYTE_ACTIVATION | 409 | 0.667228 | 2.480178 | 0 | 0 |
| GO_REGULATION_OF_CELL_ACTIVATION | 451 | 0.654573 | 2.479348 | 0 | 0 |
| GO_CELLULAR_RESPONSE_TO_BIOTIC_STIMULUS | 159 | 0.680143 | 2.477039 | 0 | 0 |
| GO_NEGATIVE_REGULATION_OF_IMMUNE_SYSTEM_PROCESS | 361 | 0.609162 | 2.474501 | 0 | 0 |
| GO_MYELOID_LEUKOCYTE_MIGRATION | 96 | 0.707282 | 2.470155 | 0 | 0 |
| GO_REGULATION_OF_INFLAMMATORY_RESPONSE | 285 | 0.627035 | 2.469305 | 0 | 0 |
| GO_LEUKOCYTE_CHEMOTAXIS | 114 | 0.724023 | 2.469249 | 0 | 0 |
| GO_GRANULOCYTE_MIGRATION | 72 | 0.743939 | 2.464845 | 0 | 0 |
| GO_REGULATION_OF_INNATE_IMMUNE_RESPONSE | 353 | 0.649503 | 2.462347 | 0 | 0 |
| GO_CELLULAR_RESPONSE_TO_INTERFERON_GAMMA | 117 | 0.786396 | 2.456372 | 0 | 0 |
| GO_REGULATION_OF_LEUKOCYTE_PROLIFERATION | 201 | 0.689642 | 2.45446 | 0 | 0 |
| GO_ADAPTIVE_IMMUNE_RESPONSE | 251 | 0.730522 | 2.450656 | 0 | 0 |
| GO_MYELOID_LEUKOCYTE_ACTIVATION | 95 | 0.741534 | 2.45024 | 0 | 0 |
| GO_REGULATION_OF_LEUKOCYTE_MEDIATED_IMMUNITY | 156 | 0.705238 | 2.444989 | 0 | 0 |
| GO_POSITIVE_REGULATION_OF_IMMUNE_EFFECTOR_PROCESS | 156 | 0.686433 | 2.441367 | 0 | 0 |
| GO_REGULATION_OF_T_CELL_PROLIFERATION | 143 | 0.697275 | 2.440364 | 0 | 0 |
| GO_RESPONSE_TO_BACTERIUM | 499 | 0.571084 | 2.435284 | 0 | 0 |
SIZE – number of genes; ES – enrichment score; NES – normalized enrichment scores; NOM p-val – normalized P-value; FDR q-val – q-value of false discovery rate. We generally assume that the absolute value of NES >1.0, NOM P-val <0.05, FDR q-val <0.25 was statistically significant.
Correlation Between C3AR1 Expression and Immune Infiltration Cells in Stomach Adenocarcinoma
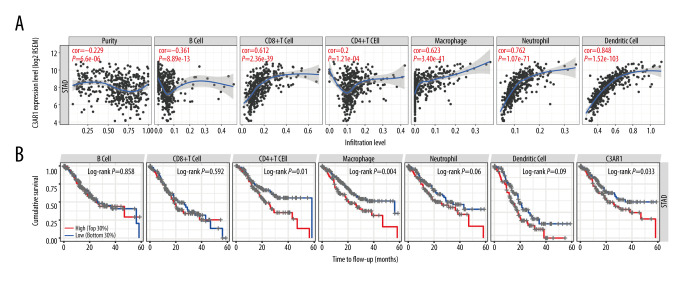
The biological processes affected by C3AR1 expression led us to further explore whether the C3AR1 expression has effects on the tumor infiltration of lymphocytes since tumor immune infiltration is an independent predictor of sentinel-lymph node status and survival in cancers [27,28]. The correlation analysis was performed using the TIMER database (Figure 5A). Among all the selected cell types, the expression of C3AR1 was positively and significantly correlated with the infiltration of CD8+ T cells (r=0.612, P=2.36e-39), macrophages (r=0.623, P=3.40e-41), neutrophils (r=0.762, P=1.07e-71), and DCs (r=0.848, P=1.52e-103). We also analyzed the effect of various immune cell infiltrates on the prognosis of STAD (Figure 5B). The results showed that the infiltration of the CD4+T cells and the macrophages could significantly affect the prognosis of STAD. According to the reported and analyzed biological functions, a high C3AR1 expression can promote STAD in a myeloid cell infiltration-dominated manner.
Figure 5.
Correlation between the C3AR1 expression in stomach adenocarcinomas and the infiltration degree of immune cells. (A) Expression of C3AR1 was negatively correlated with tumor purity, and positively correlated with CD8+ T cells, monocytes, neutrophils, and dendritic cells. (B) Influence of the infiltration degree of B cells, CD8+T cells, CD4+T cells, macrophages, neutrophils, and dendritic cells on stomach adenocarcinoma prognosis.
Correlation Between C3AR1 Expression and Immune Marker Sets
To study the relationship between C3AR1 and the various immune-infiltrating cells, the correlation of C3AR1 expression with the various immune marker sets in STAD were analyzed in TIMER and GEPIA. The immune marker sets were classified as CD8+ T cells, B cells, monocytes, tumor-associated macrophages (TAMs), M1 and M2 macrophages, DCs, natural killer cells, T cells, and neutrophils (Table 2). The T-cell marker sets with different functions were analyzed, including helper T-cell subtypes (Th1 cells, Th2 cells, Tfh cells, Th17 cells), Treg cells, and exhausted T cells. It was discovered that the majority of immune marker sets have a significant correlation with the C3AR1 expression levels.
Table 2.
Correlation between C3AR1 and immune markers in STAD by TIMER and GEPIA database.
| Description | Gene markers | TIMER | GEPIA | ||
|---|---|---|---|---|---|
| r | P-value | r | P-value | ||
| CD8+T Cell | CD8A | 0.552 | *** | 0.57 | *** |
| CD8B | 0.342 | *** | 0.37 | *** | |
| B Cell | CD19 | 0.285 | *** | 0.24 | *** |
| CD79A | 0.373 | *** | 0.34 | *** | |
| Monocyte | CD86 | 0.909 | *** | 0.92 | *** |
| CD115(CSF1R) | 0.902 | *** | 0.9 | *** | |
| TAMs | CCL2 | 0.522 | *** | 0.55 | *** |
| CD68 | 0.661 | *** | 0.73 | *** | |
| IL10 | 0.704 | *** | 0.75 | *** | |
| M1 macrophage | INOS(NOS2) | 0.113 | * | 0.15 | ** |
| IRF5 | 0.342 | *** | 0.4 | *** | |
| COX2(PTGS2) | 0.074 | . | 0.17 | *** | |
| M2 macrophage | CD163 | 0.903 | *** | 0.83 | *** |
| VSIG4 | 0.865 | *** | 0.87 | *** | |
| MS4A4A | 0.923 | *** | 0.93 | *** | |
| Dendritic Cells | HLA-DPB1 | 0.661 | *** | 0.69 | *** |
| HLA-DQB1 | 0.515 | *** | 0.4 | *** | |
| HLA-DRA | 0.698 | *** | 0.71 | *** | |
| HLA-DPA1 | 0.696 | *** | 0.7 | *** | |
| BDCA-1(CD1C) | 0.46 | *** | 0.45 | *** | |
| BDCA-4(NRP1) | 0.587 | *** | 0.64 | *** | |
| CD11c (ITGAX) | 0.814 | *** | 0.8 | *** | |
| NK Cells | KIR2DL1 | 0.366 | *** | 0.37 | *** |
| KIR2DL3 | 0.326 | *** | 0.46 | *** | |
| KIR2DL4 | 0.299 | *** | 0.3 | *** | |
| KIR3DL1 | 0.286 | *** | 0.35 | *** | |
| KIR3DL2 | 0.31 | *** | 0.37 | *** | |
| KIR3DL3 | 0.077 | . | 0.048 | . | |
| KID2DS4 | 0.253 | *** | – | – | |
| T Cell (general) | CD3D | 0.566 | *** | 0.55 | *** |
| CD3E | 0.53 | *** | 0.54 | *** | |
| CD2 | 0.652 | *** | 0.67 | *** | |
| Th1 | T-bet (TBX21) | 0.563 | *** | 0.58 | *** |
| STAT4 | 0.624 | *** | 0.62 | *** | |
| STAT1 | 0.343 | *** | 0.45 | *** | |
| IFN-γ (IFNG) | 0.423 | *** | 0.45 | *** | |
| TNF-α (TNF) | 0.279 | *** | 0.33 | *** | |
| Th2 | GATA3 | 0.417 | *** | 0.44 | *** |
| STAT6 | 0.019 | . | 0.016 | ** | |
| STAT5A | 0.465 | *** | 0.55 | *** | |
| IL13 | 0.133 | ** | 0.19 | *** | |
| Tfh | BCL6 | 0.3 | *** | 0.4 | *** |
| IL21 | 0.435 | *** | 0.49 | *** | |
| Th17 | STAT3 | 0.365 | *** | 0.45 | *** |
| IL17A | 0.007 | . | 0.031 | . | |
| Treg cells | FOXP3 | 0.562 | *** | 0.59 | *** |
| CCR8 | 0.665 | *** | 0.71 | *** | |
| STAT5B | 0.346 | *** | 0.48 | *** | |
| TGF-β | 0.453 | *** | 0.5 | *** | |
| Neutrophils | CD66B | 0.064 | . | 0.069 | . |
| CD11b | 0.784 | *** | 0.77 | *** | |
| CCR7 | 0.492 | *** | 0.51 | *** | |
| Exhaustion T cells | TIM-3 (HAVCR2) | 0.907 | *** | 0.92 | *** |
| TIGIT | 0.57 | *** | 0.6 | *** | |
| GZMA | 0.553 | *** | 0.56 | *** | |
| LAG-3 | 0.501 | *** | 0.49 | *** | |
| CTLA4 | 0.478 | *** | 0.54 | *** | |
| PD-1 (PDCD1) | 0.467 | *** | 0.49 | *** | |
TAMs – tumor-associated macrophages; NK cells – natural killer cells; Th1, Th2, Tfh, Th17 – helper T cell subtypes. P≥0.05,
P<0.05,
P<0.01,
P<0.001.
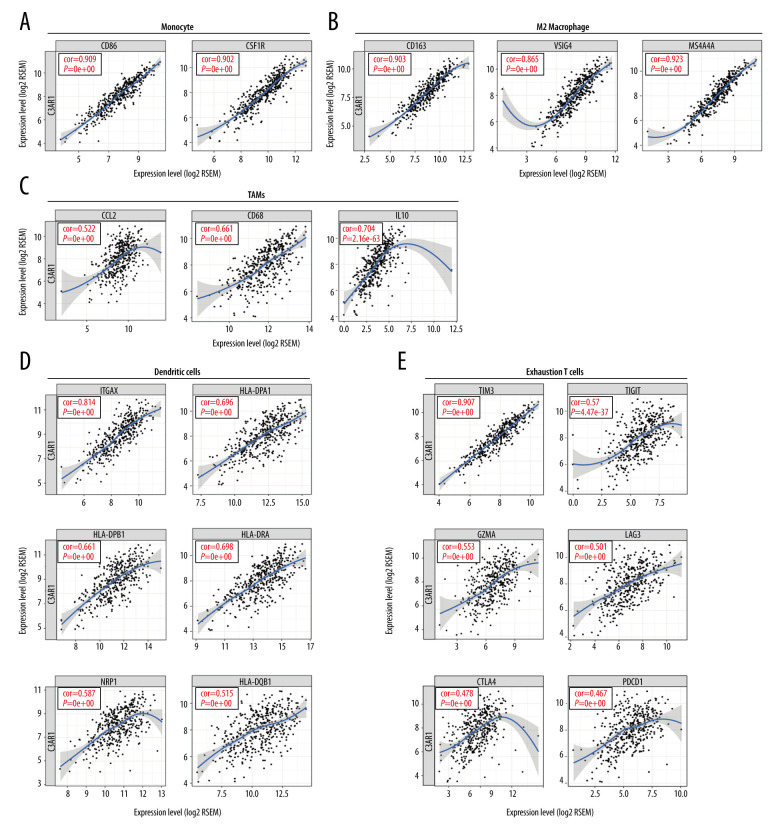
A majority of the immune markers for monocytes, TAMs, and M2 macrophages showed strong correlations with the C3AR1 expression levels in STAD. The scatterplots illustrate the correlation between CD86, CSF1R of monocytes, CCL2, CD68, IL10 of TAM, CD163, VSIG4, MS4A4A of M2 macrophages, and the C3AR1 expression levels in STAD (Figure 6A–6C). It can be speculated that C3AR1 regulates the polarization of the macrophages in STAD.
Figure 6.
Correlation between the C3AR1 expression and the markers of immune cells in STAD. (A) Immune markers of monocytes correlated with the expression of C3AR1. (B) Immune markers of M2 macrophages significantly correlated with the expression of C3AR1. (C) Immune markers of tumor-associated macrophages correlated with the expression of C3AR1. (D) Immune markers of dendritic cells correlated with the expression of C3AR1. (E) Immune markers of exhaustion T cells correlated with the expression of C3AR1.
The C3AR1 expression levels related to the infiltration level of DCs in STAD (Figure 5A). HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DPA1, BDCA-4 (NRP1), and CD11c (ITGAX), which are markers of DCs showed significant correlations with C3AR1 expression (Figure 6D). This is of great value in proving that the high expression of C3AR1 is related to the high infiltration of DCs.
It is important to note that C3AR1 expression has a significant correlation with immune markers of exhausted T cells, including TIM-3 (HAVCR2), TIGIT, GZMA, LAG-3, CTLA4, PD-1 (PDCD1) (Figure 6E). The expression of TIM-3 is strongly correlated with the expression level of C3AR1. As an immune checkpoint, TIM-3 plays an important role in the regulation of T-cell activity. It suggested that C3AR1 plays a vital role in TIM-3-mediating T-cell exhaustion. These results show that C3AR1 has a good correlation with immune-infiltrating cells, and it can be inferred that C3AR1 plays a key role in the immune-escape process of STAD.
Discussion
The current immunotherapy for tumor treatment was designed for reactivating the T cells in the tumor microenvironment, which only benefits some patients owing to the limitations of single-gene targeting in this therapy. It is of great importance to explore the other possible targets for cancer immune therapy. In the present study, we validated the expression of C3AR1 in the various tumor types; a high expression of C3AR1 correlates with a poor prognosis of STAD. The C3AR1 signaling mostly affected the immune-related pathways. Further analysis showed that the C3AR1 expression was related to a set of immune markers and immune infiltration in STAD. Therefore, our research revealed the potential role of C3AR1 in STAD immunotherapy and its possibility as a prognosis marker.
To identify the possible role of C3AR1 in STAD, firstly we analyzed the C3AR1 expression in different cancer tissues compared with adjacent normal tissues in the TIMER online analysis database based on the TCGA data, and further verified this role in the UALCAN and GEPIA servers. The results showed that C3AR1 expression was significantly different in the 7 cancers and statistically significant in all 3 databases. The prognosis values of C3AR1 in these 7 cancers were evaluated in the 3 databases. The results in all 3 databases revealed that the C3AR1 expression was related with tumor prognosis only in STAD. To further identify the potential relations between C3AR1 expression and STAD progression, a set of well-recognized STAD markers, vascular endothelial growth factor receptor 2 (VEGFR-2) [29], transforming growth factor beta 1 (TGFB1) [30], matrix metalloproteinase 9 (MMP9) [31], indoleamine-pyrrole 2,3-dioxygenase (IDO1) [32], nuclear factor kappa B subunit 1 (NFKB1) [33], snail family transcriptional repressor 2 (SNAI2) [34], zinc finger E-box binding homeobox 1 (ZEB1), and zinc finger E-box binding homeobox 2 (ZEB2) [35] were chosen to conduct the correlation analysis with C3AR1 expression through the TIMER database. The high expression of these markers promoted the development of STAD to some extent. The significant positive correlation between C3AR1 and these markers indicates that the high expression of C3AR1 can promote the progression of STAD and can be used as a prognostic marker for STAD.
The complement system, which consists of extensive molecules, is an important regulator in innate and adaptive immunity. Through the 3 canonical pathways (classical, alternative, and lectin pathways) of activation, the complement pathways were activated to promote the pro-inflammatory or immunomodulatory processes depending on the triggers and the local microenvironments. The C3 cleavage is a common consequence of all the pathway signal transductions. C3AR1 as a receptor of the complement effector C3a is important in mediating the downstream signal transduction of the complement activation. In order to reveal how the expression of C3AR1 affects the prognosis of STAD, we used GSEA software to analyze the biological processes related to C3AR1. The results show that C3AR1 is involved in many immune-response processes. In addition to being an important factor regulating the inflammatory response, C3AR1 also affects the biological processes of cytokine production and secretion, immune cell activation, proliferation, differentiation, and migration. These immune cells, including lymphocytes, monocytes/macrophages, granulocytes, and DC, play different roles in the occurrence and development of cancer. Thus, the high expression of C3AR1 can affect the prognosis of STAD by regulating the immune signal-related pathways.
Previous research on antibody-to-tumor-antigen-based immunotherapy has found that complement activation can promote tumor elimination via complement-dependent cytotoxicity and complement-dependent phagocytosis [36]. However, recent clinical observations and animal studies revealed that complement activation within the tumor microenvironment can play tumor-promoting roles. For example, a high concentration of C3a in the serum of esophageal cancer patients was shown to be associated with a poor prognosis [37]. A C3AR1 deficiency in mice resulted in retarded melanoma tumorigenesis when compared with wild mice [38]. A further mechanistical exploration found that C3a-C3AR1 signaling impairs neutrophil responses, which are the main drivers of melanoma tumorigenesis. As C3AR1 affects certain biological processes, we analyzed the relationship between C3AR1 and the tumor-infiltrating lymphocytes through the TIMER online analysis database. The results showed that a high expression of C3AR1 was strongly correlated with the high infiltration of CD8+ T cells, macrophages, neutrophils, and DCs. Therefore, it can be speculated that C3AR1 has a regulatory effect on these cells. Specifically, we analyzed the correlation between C3AR1 and the various immune gene marker sets. It showed that C3AR1 has a strong correlation with monocytes, TAM, and M2 macrophages. The TAM exhibit 2 major phenotypes, M1 and M2, which have opposing effects on tumor progression [39]. The M1 macrophages are typically activated macrophages and can be polarized by a lipopolysaccharide (LPS) and interferon-H (IFN-H). The M1 macrophages produce interleukin-1β (IL-1β), IL-12, and cytotoxins, including inducible nitric oxide synthase (iNOS) [40]. The M2 macrophages are alternately activated macrophages that can be polarized through IL-4, IL-10, or IL-13, and produce IL-10, IL-6, and angiogenic factors, including the vascular endothelial growth factor [41]. The M2 macrophages are related to tumor growth, tissue remodeling, angiogenesis promotion, and adaptive immunity suppression, and their high level of infiltration is related to a poor prognosis [42]. The TAM differentiate into M1 or M2 phenotypes through regulation by the tumor microenvironment [43–46]. The high correlation between the expression of C3AR1 and the immune marker set of M2 macrophages suggests that C3AR1 is likely to regulate the differentiation of TAM into M2 macrophages, and is an important factor regulating the polarization of M2 macrophages. C3AR1 has a highly significant correlation with TIM-3, which is another checkpoint independent of PD-1. C3AR1 has similar biological functions to PD-1. TIM-3/Gal-9 inhibits tumor immunity by negatively regulating T cell immunity [47]. Therefore, we speculated that C3AR1 affects tumor immunity by regulating the TIM-3 expression, leading to tumor immune escape and promoting the development of STAD.
Conclusions
In summary, these results indicate that C3AR1 plays an important role in tumor immunity. A high expression of C3AR1 is associated with a poor prognosis of STAD and a high infiltration of the immune cells. C3AR1 promotes the polarization of M2 macrophages and participates in the regulation of TIM-3 expression. It affects the infiltration of immune cells, leads to the immune escape of the tumor cells, and promotes the development of STAD. Therefore, the research on C3AR1 as a target has high potential value. Further experiments should be designed to study the specific mechanism of C3AR1’s regulation of macrophage polarization and TIM-3 expression, in order to find potential novel targets for tumor immunotherapy.
Footnotes
Conflicts of Interest
None.
Source of support: Departmental sources
References
- 1.Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol. 2020;3:534–42. doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Ajani JA, Lee J, Sano T, et al. Gastric adenocarcinoma. Nat Rev Dis Primers. 2017;3:17036. doi: 10.1038/nrdp.2017.36. [DOI] [PubMed] [Google Scholar]
- 4.Eggermont AM, Robert C. New drugs in melanoma: It’s a whole new world. Eur J Cancer. 2011;47(14):2150–57. doi: 10.1016/j.ejca.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Q, Cao L, Guan L, et al. Dilemmas and prospect. Brief Funct Genomics. 2019;18(2):107–12. doi: 10.1093/bfgp/ely019. [DOI] [PubMed] [Google Scholar]
- 6.Coutzac C, Pernot S, Chaput N, Zaanan A. Immunotherapy in advanced gastric cancer, is it the future? Crit Rev Oncol Hematol. 2019;133:25–32. doi: 10.1016/j.critrevonc.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Bang YJ, Cho JY, Kim YH, et al. Efficacy of sequential ipilimumab monotherapy vs best supportive care for unresectable locally advanced/metastatic gastric or gastroesophageal junction cancer. Clin Cancer Res. 2017;23(19):5671–78. doi: 10.1158/1078-0432.CCR-17-0025. [DOI] [PubMed] [Google Scholar]
- 8.Nanda R, Chow L, Dees E, et al. Phase Ib KEYNOTE-012 Study. J Clin Oncol. 2016;34(21):2460–67. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engstrand L, Graham DY. Microbiome and gastric cancer. Dig Dis Sci. 2020;65(3):865–73. doi: 10.1007/s10620-020-06101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ricklin D, Reis ES, Lambris JS. Complement in disease: A defence system turning offensive. Nat Rev Nephrol. 2016;12(7):383–401. doi: 10.1038/nrneph.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markiewski MM, DeAngelis RA, Benencia F, et al. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9(11):1225–35. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pio R, Ajona D, Lambris J. Complement inhibition in cancer therapy. Semin Immunol. 2013;25(1):54–64. doi: 10.1016/j.smim.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonavita E, Gentile S, Rubino M, et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. 2015;160(4):700–14. doi: 10.1016/j.cell.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Ajona D, Ortiz-Espinosa S, Pio R. Complement anaphylatoxins C3a and C5a: Emerging roles in cancer progression and treatment. Semin Cell Dev Biol. 2019;85:153–63. doi: 10.1016/j.semcdb.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 15.Pio R, Ajona D, Ortiz-Espinosa S, et al. Complementing the cancer-immunity cycle. Front Immunol. 2019;12:774. doi: 10.3389/fimmu.2019.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reis ES, Mastellos DC, Ricklin D, et al. Untangling an intricate relationship. Nat Rev Immunol. 2018;18(1):5–18. doi: 10.1038/nri.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Fan J, Wang B, et al. A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–10. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Li C, Kang B, et al. A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Hu J, Xu H, et al. Long non-coding RNA expression profiling in biopsy to identify renal allograft at risk of chronic damage and future graft loss. Appl Biochem Biotechnol. 2020;190(2):660–73. doi: 10.1007/s12010-019-03082-2. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Liu L, Liu X, et al. The role of upregulated DDX11 as a potential prognostic and diagnostic biomarker in lung adenocarcinoma. J Cancer. 2019;10(18):4208–16. doi: 10.7150/jca.33457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li N, Zhao L, Guo C, et al. Identification of a novel DNA repair-related prognostic signature predicting survival of patients with hepatocellular carcinoma. Cancer Manag Res. 2019;11:7473–84. doi: 10.2147/CMAR.S204864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Zhao E, Zhang Z, et al. Association between Tim-3 and Gal-9 expression and gastric cancer prognosis. Oncol Rep. 2018;40(4):2115–26. doi: 10.3892/or.2018.6627. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Roh J, Lee H, et al. Expression of the immune checkpoint molecule V-set immunoglobulin domain-containing 4 is associated with poor prognosis in patients with advanced gastric cancer. Gastric Cancer. 2020;14:1–4. doi: 10.1007/s10120-020-01120-1. [DOI] [PubMed] [Google Scholar]
- 26.Vasaikar S, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–63. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn GP, Dunn IF, Curry WT. Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human glioma. Cancer Immun. 2007;13:7–12. [PMC free article] [PubMed] [Google Scholar]
- 28.Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30(21):2678–83. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 29.Abbas M, Faggian A, Sintali DN, et al. Current and future biomarkers in gastric cancer. Biomed Pharmacother. 2018;103:1688–700. doi: 10.1016/j.biopha.2018.04.178. [DOI] [PubMed] [Google Scholar]
- 30.Batlle E, Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50(4):924–40. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alaseem A, Alhazzani K, Dondapati P, et al. A challenging paradigm of cancer management. Semin Cancer Biol. 2019;56:100–15. doi: 10.1016/j.semcancer.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Kim JW, Nam KH, Ahn SH, et al. Prognostic implications of immunosuppressive protein expression in tumors as well as immune cell infiltration within the tumor microenvironment in gastric cancer. Gastric Cancer. 2016;19(1):42–52. doi: 10.1007/s10120-014-0440-5. [DOI] [PubMed] [Google Scholar]
- 33.Yin Y, Si X, Gao Y, et al. The nuclear factor-kappaB correlates with increased expression of interleukin-6 and promotes progression of gastric carcinoma. Oncol Rep. 2013;29(1):34–38. doi: 10.3892/or.2012.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savagner P, Yamada KM, Thiery JP. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137(6):1403–19. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comijn J, Berx G, Vermassen P, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7(6):1267–78. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 36.Reis ES, Mastellos DC, Ricklin D, et al. Untangling an intricate relationship. Nat Rev Immunol. 2018;18(1):5–18. doi: 10.1038/nri.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maher SG, McDowell DT, Collins BC, et al. Serum proteomic profiling reveals that pretreatment complement protein levels are predictive of esophageal cancer patient response to neoadjuvant chemoradiation. Ann Surg. 2011;254(5):809–17. doi: 10.1097/SLA.0b013e31823699f2. discussion 816–17. [DOI] [PubMed] [Google Scholar]
- 38.Nabizadeh JA, Manthey HD, Steyn FJ, et al. The complement C3a receptor contributes to melanoma tumorigenesis by inhibiting neutrophil and CD4+ T cell responses. J Immunol. 2016;196(11):4783–92. doi: 10.4049/jimmunol.1600210. [DOI] [PubMed] [Google Scholar]
- 39.Biswas SK, Allavena P, Mantovani A. Tumor-associated macrophages: Functional diversity, clinical significance, and open questions. Semin Immunopathol. 2013;35(5):585–600. doi: 10.1007/s00281-013-0367-7. [DOI] [PubMed] [Google Scholar]
- 40.Ye Y, Huang X, Zhang Y, et al. Calcium influx blocked by SK&F 96365 modulates the LPS plus IFN-gamma-induced inflammatory response in murine peritoneal macrophages. Int Immunopharmacol. 2012;12(2):384–93. doi: 10.1016/j.intimp.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Xiao X, Gaffar I, Guo P, et al. M2 macrophages promote beta-cell proliferation by up-regulation of SMAD7. Proc Natl Acad Sci USA. 2014;111(13):E1211–20. doi: 10.1073/pnas.1321347111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66(2):605–12. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 43.Jia W, Kidoya H, Yamakawa D, et al. Galectin-3 accelerates M2 macrophage infiltration and angiogenesis in tumors. Am J Pathol. 2013;182(5):1821–31. doi: 10.1016/j.ajpath.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 44.Na YR, Yoon YN, Son DI, Seok SH. Cyclooxygenase-2 inhibition blocks M2 macrophage differentiation and suppresses metastasis in murine breast cancer model. PLoS One. 2013;8(5):e63451. doi: 10.1371/journal.pone.0063451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Y, Zhu Y, Xu J, et al. PKN2 in colon cancer cells inhibits M2 phenotype polarization of tumor-associated macrophages via regulating DUSP6-Erk1/2 pathway. Mol Cancer. 2018;17(1):13. doi: 10.1186/s12943-017-0747-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: Implications for new anticancer therapies. J Pathol. 2002;196(3):254–65. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 47.Cao E, Zang X, Ramagopal UA, et al. T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface. Immunity. 2007;26(3):311–21. doi: 10.1016/j.immuni.2007.01.016. [DOI] [PubMed] [Google Scholar]