SUMMARY
Uveal melanoma (UM), the most common ocular malignancy, is characterized by GNAQ/11 mutations. Hippo/YAP and Ras/mitogen-activated protein kinase (MAPK) emerge as two important signaling pathways downstream of G protein alpha subunits of the Q class (GαQ/11)-mediated transformation, although whether and how they contribute to UM genesis in vivo remain unclear. Here, we adapt an adeno-associated virus (AAV)-based ocular injection method to directly deliver Cre recombinase into the mouse uveal tract and demonstrate that Lats1/2 kinases suppress UM formation specifically in uveal melanocytes. We find that genetic activation of YAP, but not Kras, is sufficient to initiate UM. We show that YAP/TAZ activation induced by Lats1/2 deletion cooperates with Kras to promote UM progression via downstream transcriptional reinforcement. Furthermore, dual inhibition of YAP/TAZ and Ras/MAPK synergizes to suppress oncogenic growth of human UM cells. Our data highlight the functional significance of Lats-YAP/TAZ in UM initiation and progression in vivo and suggest combination inhibition of YAP/TAZ and Ras/MAPK as a new therapeutic strategy for UM.
Graphical Abstract
INTRODUCTION
Uveal melanoma (UM), the most common primary intraocular malignancy in adults, arises from the melanocytes of the uveal tract, which consists of the choroid, ciliary body, and iris (Singh et al., 2005). Unlike cutaneous melanoma, UM is genetically characterized by distinct mutations in GANQ and GNA11. More than 80% of human UMs harbor activating mutations in GNAQ or GNA11, which encode the heterotrimeric G protein alpha subunits of the Q class (Gαq/11) (Van Raamsdonk et al., 2009, 2010). The highly conserved glutamine 209 (Q209) or arginine 183 (R183) mutations of GNAQ and GNA11 render the guanosine triphosphatase (GTPase) of these proteins defective and lead to constitutive activation of downstream pathways in UM cells (Van Raamsdonk et al., 2009, 2010).
Ras/mitogen-activated protein kinase (MAPK) and Hippo/YAP are two signaling pathways implicated downstream of GNAQ/11 during UM pathogenesis (Chen et al., 2014, 2017; Feng et al., 2014; Moore et al., 2018; Van Raamsdonk et al., 2009; Yu et al., 2014). In UM cells, activated Gαq/11 proteins are thought to interact with their direct downstream effector, PLCβ, resulting in activation of protein kinase C (PKC) and further downstream, in activation of Ras/MAPK signaling (Hubbard and Hepler, 2006). A recent report identifies PKCδ/ε as the relevant PKC isoforms in UM (Chen et al., 2017), which provides a molecular link on how PKC signaling relays to MAPK activation. In human UM cells, PKC kinases phosphorylate and activate the Ras guanine nucleotide exchange factors leading to activation of the Rasmitogen-activated protein kinase kinase-extracellular signal-regulated kinase (MEK-ERK) signal cascade (Chen et al., 2017; Moore et al., 2018). Despite the evidence of its functional importance, the ineffectiveness of targeting the MAPK pathway in UM clinical trials (Carvajal et al., 2014; Komatsubara et al., 2016) suggests the involvement of additional oncogenic pathways in GNAQ/11-mutated UM cells.
Hippo/YAP signaling, originally identified in Drosophila as an organ size control pathway, recently emerged as another key pathway in UM tumorigenesis (Feng et al., 2014; Yu et al., 2014). In the mammalian Hippo pathway, activation of the core kinase cascade, which comprises the Mst1/2 and Lats1/2 kinases, leads to phosphorylation, cytosolic retention, and degradation of the transcriptional coactivators, YAP and TAZ. Upon Hippo pathway inactivation, YAP and TAZ translocate into the nucleus and interact with the Tead family of transcription factors, thereby inducing downstream gene transcription (Halder and Camargo, 2013; Pan, 2010; Yu et al., 2015; Zanconato et al., 2016). Mis-regulation of the Hippo pathway has been implicated in a variety of human cancers, including UM (Feng et al., 2014; Halder and Camargo, 2013; Pan, 2010; Yu et al., 2014, 2015; Zanconato et al., 2016). A prior study showed strong regulation of the Hippo pathway by G protein-coupled receptors (GPCRs) through interaction with different G proteins, including Gαq/11, the oncogenic drivers of UM (Yu et al., 2012). Two subsequent studies reported the functional role of YAP in UM cells carrying GNAQ/11 mutations (Feng et al., 2014; Yu et al., 2014).
Although YAP and Ras/MAPK are potentially critical for UM genesis, it is still not clear whether their activation is able to drive UM formation in vivo and how they functionally interact during UM initiation and progression. Two recent studies generated mouse models of UM by crossing the conditional mice carrying the mutated GNAQ or GNA11 alleles in the Rosa26 locus to two different mouse melanocyte Cre lines (Huang et al., 2015; Moore et al., 2018). However, the broad Cre expression in the melanocytes or melanocyte-like cells outside the eye, including organs such as skin, CNS, lung, and inner ear, leads to early lethality and complicates UM phenotypic analysis (Huang et al., 2015; Moore et al., 2018). Here, we adapted an adeno-associated virus (AAV)-based ocular injection method to allow specific induction of Cre recombination in uveal melanocytes and used this platform to manipulate Hippo/YAP and Ras/MAPK signaling directly in mouse uveal tract. Our data revealed a distinct role of Lats1/2-YAP/TAZ in UM initiation and progression, and suggest a transcriptional reinforcement mechanism underlying the functional interplay between Hippo/YAP and Ras/MAPK during UM development.
RESULTS
A Robust UM Mouse Model via AAV-Cre Uveal Tract Delivery
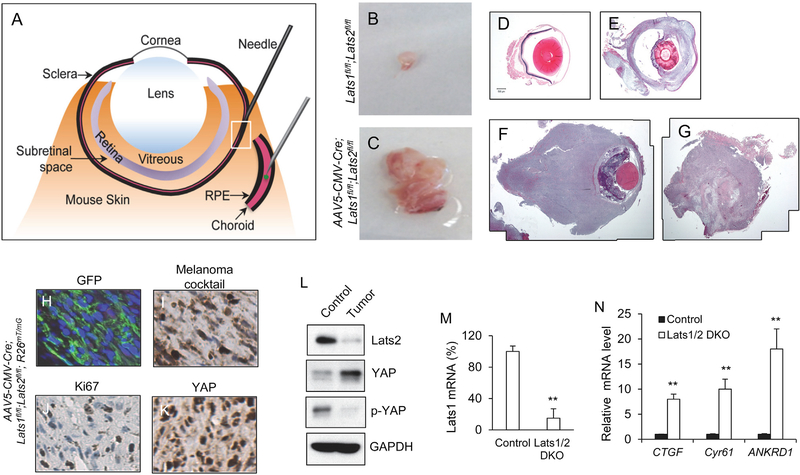
In order to examine the role of Hippo signaling in UM genesis in vivo, we adapted an AAV-based eye injection method (Venkatesh et al., 2013) for local delivery of AAV5-CMV-Cre viral vector directly into the choroid region of the mouse uveal tract (Figure 1A). We then used this uveal tract injection method to deliver Cre recombinases into adult mice carrying the floxed alleles for both Lats1 and Lats2 genes, Lats1fl/fl;Lats2fl/fl (Yi et al., 2016), or together with the reporter R26mT/mG allele (Muzumdar et al., 2007) that drives the expression of membrane-bound GFP proteins upon Cre recombination. We found that deletion of the core Hippo kinases Lats1/2 through AAV-CMV-Cre injection efficiently induced tumor formation. Within 2 months following ocular Cre delivery, the majority of the mice exhibited bulging eyes, and at 6 months after Cre injection, most of the mice developed eye tumors (Figures 1B–1G and 3A). Tumors developed in the mice carrying the R26mT/mG reporter allele were GFP-positive (Figure 1H), suggesting that they originated from the cells that underwent Cre recombination. Tumor cells also stained positively for the melanoma antibody cocktail Melan A/Mart1-HMB45 (Figure 1I), indicating that the tumors are UM in nature and arise from the melanocytes. In addition, these highly proliferative Lats1/2-deleted tumor cells exhibited nuclear YAP staining, downregulation of YAP phosphorylation, and upregulation of YAP target genes, including CTGF, Cyr61, and ANKRD1 (Figures 1J–1N). These data suggest that Hippo pathway inactivation by Lats1/2 deletion drives UM formation, and that the AAV-based eye local injection is an effective method to induce Cre recombination in the mouse uveal tract that can be used for UM tumor modeling.
Figure 1. A Mouse Model of UM via AAV-Based Cre Delivery Directly into the Uveal Tract.
(A) Illustration of AAV-based delivery of Cre recombinases into the choroid region of a mouse uveal tract.
(B–G) UM genesis in Lats1fl/fl;Lats2fl/fl mice following AAV5-CMV-Cre injection. Eye dissected from control mice (B), and UM isolated from Lats1fl/fl;Lats2fl/fl mice 6 months after Cre injection (C).
(D–G) Representative histological images of wild-type eye (D) and UMs detected 3 months (E) and 6 months (F and G) after Cre injection.
(H) GFP expression in UM derived from AAV-Cre-injected Lats1fl/fl;Lats2fl/fl;R26mT/mG mice.
(I–K) Representative IHC images of (I) Melanoma cocktail, (J) Ki67, and (K) YAP in UM derived from AAV-Cre-injected Lats1fl/fl;Lats2fl/fl;R26mT/mG mice.
(L) Immunoblot analysis of protein expression of Lats2, YAP, phospho-YAP (p-YAP), and GAPDH in control and UM tissues using the antibodies against Lats2, YAP, phospho-YAP (Ser127), and GAPDH.
(M and N) qPCR analysis of mRNA expression of Lats1, CTGF, Cyr61, and ANKRD1 in control and UM tissues.
Data are mean ± SD. **p ≤ 0.01.
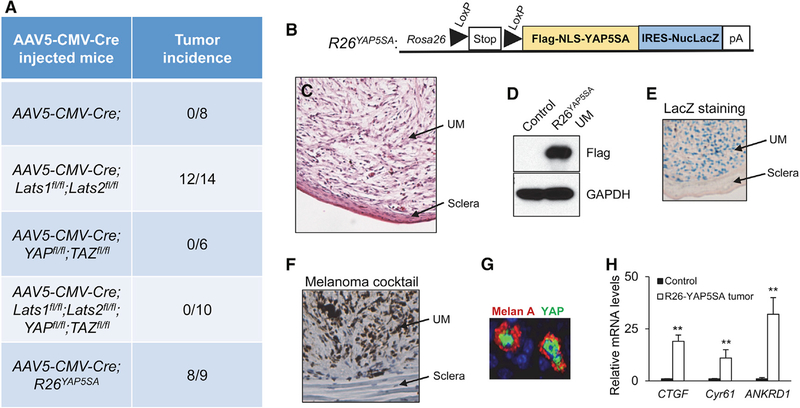
Figure 3. YAP Activation Alone Is Sufficient to Drive UM Formation.
(A) Tumor incidence in AAV5-CMV-Cre-injected mice with various genotypes.
(B) Schematic diagram of the R26YAP5SA allele.
(C) Histological images of UM developed in AAV5-CMV-GFPCre-injected R26YAP5SA mice.
(D) Immunoblot analysis of the expression of FLAG-tagged YAP5SA protein in tumors using an anti-FLAG antibody.
(E) LacZ staining shows YAP5SA transgene expression within tumor (UM), but not adjacent sclera tissue.
(F) IHC staining of Melan A in UM from AAV5-CMV-GFPCre-injected R26YAP5SA mice, using a Melanoma antibody cocktail.
(G) Immunofluorescence staining of Melan A and YAP showing nuclear YAP expression in Melan A-expressing tumor cells.
(H) qPCR analysis of mRNA levels of the YAP target genes, CTGF, Cyr61, and ANKRD1, in control tissue and YAP5SA-expressing UM.
Data are mean ± SD. **p ≤ 0.01. See also Figure S2.
Lats1/2 Kinases Specifically Suppress UM Formation in Mouse Uveal Melanocytes
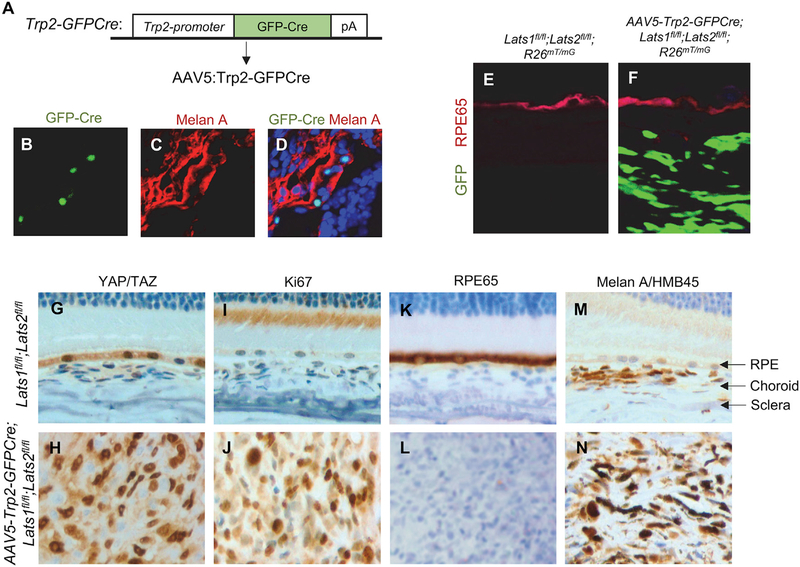
To achieve specific deletion of Lats1/2 kinases in uveal melanocytes, we generated an AAV expression vector that drives the expression of the nuclear GFP-Cre fusion protein under the control of a 1.7-kb mouse tyrosinase-related protein 2 (Trp2) promoter (Figure 2A). Trp2, an enzyme involved in an intermediate step of melanin synthesis, is expressed in the adult uveal melanocytes (Li et al., 2006), and the 1.7-kb mouse Trp2 promoter has been shown to be able to direct transgene expression (Zhao and Overbeek, 1999). Following AAV5-Trp2-GFPCre injection, we detected sporadic expression of GFP-Cre within the uveal tract, and the nuclear GFP signals were colocalized with the melanocytes expressing Melan A (Figures 2B–2D), confirming the specificity of Trp2-GFPCre expression in uveal melanocytes. Like what we observed in AAV5-CMV-Cre-injected mice (Figure 1), we found that most of the Lats1/2 conditional mice following AAV5-TRP2-GFPCre uveal tract injection developed UMs (Figures 2 and 5A). We showed that the GFP-positive tumor cells carrying the R26mT/mG reporter allele were stained negatively for RPE65 (Figures 2E and 2F), a specific marker for the retinal pigmented epithelium (RPE). In addition, our immunohistochemistry (IHC) analysis showed that the Melan A-positive, RPE65-negative tumor cells exhibited strong YAP/TAZ nuclear staining (Figures 2G–2N). Interestingly, our data also revealed that the cells in the RPE layer showed much higher nuclear YAP/TAZ levels than the cells in the adjacent choroid region of control animals (Figure 2G), suggesting that Hippo signaling and Lats1/2 kinases normally function to suppress YAP/TAZ activity in wild-type uveal melanocytes, and Lats1/2 removal in the melanocytes leads to YAP/TAZ activation, thereby inducing oncogenic transformation and subsequent UM formation.
Figure 2. Lats1/2 Kinases Suppress UM Formation Specifically in Uveal Melanocytes.
(A) Diagram of the new Trp2-GFPCre allele. In Trp2-GFPCre, expression of the nuclear GFP-Cre fusion protein is under the control of a 1.7-kb mouse tyrosinase-related protein 2 (Trp2) promoter.
(B–D) Sporadic nuclear GFP-Cre expression in uveal tract co-localizes with Melan A-expressing melanocytes in AAV5-Trp2-GFP-Cre-injected control mice. Expression of Melan A is detected by immunofluorescence using an anti-Melan A/HMB45 antibody.
(E and F) UM developed in AAV5-Trp2-GFPCre-injected Lats1fl/fl;Lats2fl/fl;R26mT/mG mice does not express RPE65. GFP expression in tumors is mutually exclusive from the RPE layer expressing the RPE65 marker. Note that GFP signals detected in tumor cells were generated from both GFP-Cre and membrane-tethered GFP expression from the R26mT/mG allele after Cre recombination. Immunofluorescence against RPE65 is performed using an anti-RPE65 antibody in both control (E) and AAV5-Trp2-GFPCre-injected Lats1fl/fl;Lats2fl/fl;R26mT/mG (F) mice.
(G–N) Representative IHC images of YAP/TAZ, Ki67, RPE65, and Melan A/HMB45 in control uveal tracts (G, I, K, and M) or UMs generated from AAV5-Trp2-GFPCre-injected mice (H, J, L, and N).
See also Figure S1.
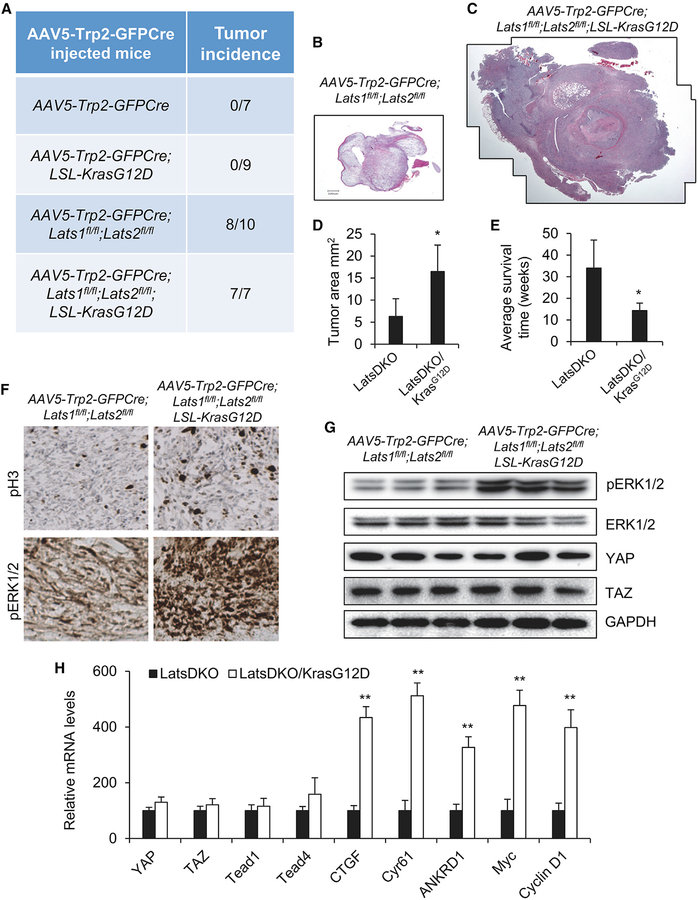
Figure 5. Lats1/2 Deletion Synergizes with Kras Activation to Promote UM Progression.
(A) Tumor incidence in AAV5-Trp2-GFPCre-injected mice with various genotypes.
(B–E) UM formation in AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl and AAV-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl;LSL-KrasG12D mice. H&E-stained sections of representative tumors from each group are shown (B and C).
(D) The tumor areas from five injected mice 4 months after Cre recombination were quantified.
(E) Average survival time of the mice with different genotypes after Cre injection.
(F) Representative IHC images of phospho-Histone H3 (pH3) and phospho-ERK1/2 (pERK1/2) in UMs derived from AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl and AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl;LSL-KrasG12D mice.
(G) Immunoblot analysis of pERK1/2, total ERK1/2, YAP, TAZ, and GAPDH in UMs derived from AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl and AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl;LSL-KrasG12D mice.
(H) qPCR analysis of transcription levels of YAP, TAZ, Tead1, Tead4, CTGF, Cyr61, ANKRD1, Myc, and Cyclin D1 in UMs derived from AAV5-Trp2-GFPCre; Lats1fl/fl;Lats2fl/fl and AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl;LSL-KrasG12D mice.
Data are mean ± SD. *p ≤ 0.05, **p ≤ 0.01. See also Figure S4.
YAP/TAZ Are Required and Sufficient for UM Initiation
Next, we set out to test whether YAP/TAZ is genetically required for UM formation induced by Lats1/2 deletion. We crossed the YAP and TAZ conditional alleles, YAPfl and TAZfl (Xin et al., 2011, 2013), into the mice carrying Lats1/2 conditional alleles in order to delete all four proteins simultaneously. We found that the mice carrying the YAP/TAZ conditional alleles failed to develop UM up to 8 months after Cre delivery (Figure 3A). Consistent with the reported role of YAP/TAZ in mediating Lats1/2 function, YAP/TAZ deletion abolished upregulation of downstream target gene transcription induced by Lats1/2 deletion (Figure S1), and the uveal tract appeared phenotypically normal in the mice carrying Lats1/2 and YAP/TAZ conditional alleles after Cre injection (Figure S1). These data suggest that YAP/TAZ function is critical for UM development following Lats1/2 inactivation in uveal melanocytes.
To examine whether YAP activation alone is able to drive UM formation, we utilized a Rosa26 conditional allele we recently generated, R26YAP5SA, which allows Cre-mediated expression of a constitutively active form of YAP, YAP5SA (Cotton et al., 2017). YAP5SA has five canonical LATS phosphorylation sites mutated from serine to alanine to prevent Hippo/Lats-mediated inhibition and degradation (Zhao et al., 2007). The R26YAP5SA allele also has a nuclear localization signal (NLS) and FLAG tag at its N terminus and an IRES-nuclear LacZ tag at its C terminus (Figure 3B). Consistent with the results in Cre-injected Lats1/2 conditional mice, most of the R26YAP5SA mice developed Ums following Cre injection (Figures 3A and 3C). The YAP5SA transgene expression in tumor cells was confirmed by western blot using an anti-FLAG antibody (Figure 3D), as well as by the positive LacZ staining in tumor cells (Figure 3E). We found that the tumor, but not the adjacent sclera tissue, expressed Melan A (Figure 3F), and immunofluorescence staining showed YAP nuclear expression in Melan A-expressing tumor cells (Figure 3G). Furthermore, we showed the elevated expression of the YAP target genes, CTGF, Cyr61, and ANKRD1, in the tumors developed in the R26YAP5SA mice following AAV5-CMV-Cre injection (Figure 3H). Taken together, these genetic data suggest that activation of YAP/TAZ is likely necessary and sufficient for UM induction in the uveal tract.
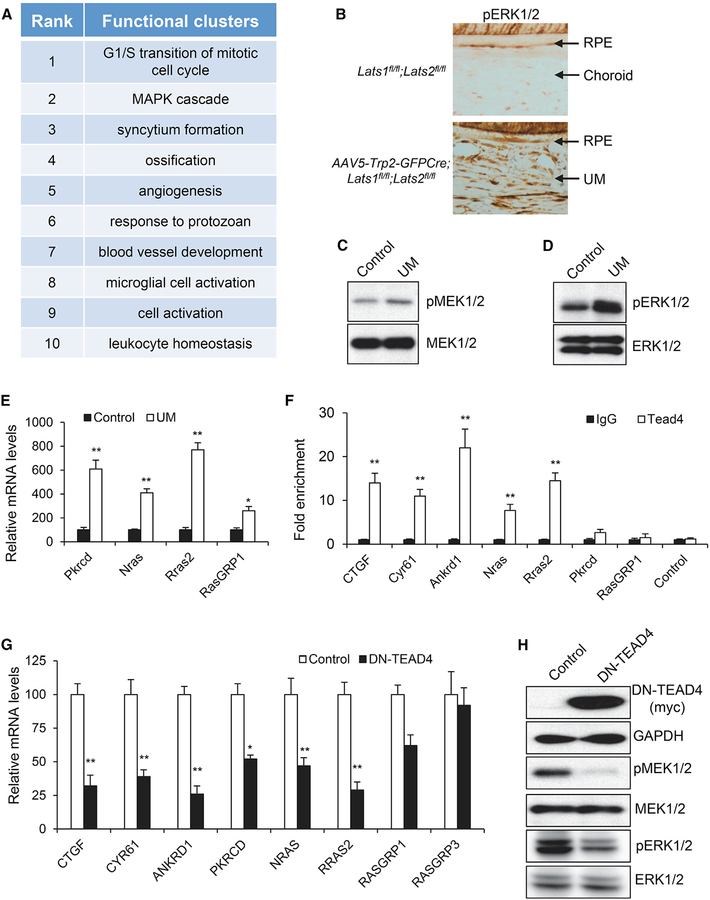
YAP/Tead Promote Ras/MAPK Activation via Downstream Transcription
To explore the downstream mechanism underlying YAP/TAZ during UM genesis, RNA-sequencing (RNA-seq) analysis was performed to profile the transcriptome of UMs from our mouse models (Table S1). Functional clustering analysis of the significantly upregulated genes identified the MAPK cascade as the second-highest ranked functional cluster, right behind the top-ranked “G1/S transition of mitotic cell cycle” (Figure 4A), suggesting a possible activation of the MAPK pathway in UM induced by YAP activation. Consistent with this notion, the IHC analysis identified the upregulation of phosphorylated ERK levels in UM, in comparison with the control uveal tract (Figure 4B). Western blot analysis also showed the increased phosphorylation of both MEK1/2 and ERK1/2 kinases (Figures 4C and 4D), further confirming the activation of the Ras/MAPK cascade.
Figure 4. YAP/Tead Promote Ras/MAPK Activation via Downstream Transcription.
(A) Functional clustering of significantly upregulated genes in UM cells from Lats1/2 knockout (KO) mice.
(B) Representative IHC images of phosphorylated ERK1/2 (pERK1/2) in normal mouse uveal tract and UM from AAV5-Trp2-GFPCre-injected Lats1fl/fl;Lats2fl/fl mice.
(C and D) Immunoblot analysis of protein levels of phospho-MEK1/2 (pMEK1/2), MEK1/2, phospho-ERK1/2 (pERK1/2), and ERK1/2 in control or UM tissues.
(E) qPCR analysis of mRNA levels of Pkrdc, Nras, Rras2, and RasGRP1 in control and UM tissues.
(F) Tead4 occupancy in the promoter regions of the CTGF, Cyr61, ANKRD1, Nras, Rras2, Prkcd, RasGRP1, and GAPDH (Control) genes in UM. Tead4 ChIP-qPCR analysis was performed in isolated mouse UM cells, and enrichment of Tead4 was calculated based upon qPCR relative to immunoglobulin G (IgG) control.
(G) qPCR analysis of mRNA levels of CTGF, CYR61, ANKRD1, PKRCD, NRAS, RRAS2, RASGRP1, and RASGRP3 in 92–1 UM cells with and without DN-TEAD4 expression.
(H) Immunoblot analysis of DN-TEAD4 (myc tag), GAPDH, pMEK1/2, MEK1/2, pERK1/2, and ERK1/2 in 92–1 UM cells with and without DN-TEAD4 expression.
Data are mean ± SD. *p ≤ 0.05, **p ≤ 0.01. See also Figure S3 and Table S1.
Among the genes associated with the Ras/MAPK signaling cascade, we found that transcription of Prkcd, Rras2, NRas, and RasGRP1 was significantly upregulated in UM cells (Figure 4E; Table S1). PKC isoform δ (PKCδ), which is encoded by the Prkcd gene, was recently identified as a key signaling component linking Gαq activation to Ras/MAPK activation in UM (Chen et al., 2017). Nras, Rras2 (TC21), and RasGRP1 (a Ras-specific guanine nucleotide exchange factor) have been shown to activate MAPK in different cellular settings (Cox and Der, 2010; Golec et al., 2016; Graham et al., 1994; Larive et al., 2012). One of the Ras guanine nucleotide exchange factors, RasGRP3, was recently reported to play functional roles in Ras/MAPK activation in UM cells (Chen et al., 2017; Moore et al., 2018). To further examine whether these genes are possible downstream targets of the YAP/Tead transcriptional complex, we intersected our data with the TEAD4 chromatin immunoprecipitation-sequencing (ChIP-seq) datasets available at the ENCODE project (https://www.encodeproject.org/) and found Tead4 occupancy in the promoter or enhancer regions of the human RRAS2, NRAS, and PRKCD genes (Encode project), suggesting that they are potential direct targets of Tead. Thus, we performed Tead4 ChIP-qPCR in mouse UM cells with Lats1/2 deletion and found that, in addition to the known YAP/Tead direct targets CTGF, Cyr61, and Ankrd1, Tead4 also occupied the promoter regions of Rras2 and Nras in mouse UM cells (Figure 4E). Although we did not detect significant enrichment of Tead4 occupancy in the promoter regions of Prkcd and RasGRP1 (Figure 4F), it remains possible that they are directly regulated by YAP/Tead through distal enhancers, as shown in the PRKCD locus in human embryonic stem cells (ESCs) (Encode project).
To further examine YAP/TAZ regulation of Ras/MAPK signaling in UM cells, we generated a lentiviral-based Tead4 repressor construct, DN-TEAD4. DN-TEAD4 lacks the N-terminal DNA binding domain but retains the entire YAP/TAZ binding domain that allows its interaction with endogenous YAP/TAZ to block downstream transcriptional activation (Figure S3A). Similar truncated repressor constructs of Tead2 and Sd (Drosophila Tead) have been reported before (Chow et al., 2004; Liu-Chittenden et al., 2012). We demonstrated that DN-TEAD4 could effectively inhibit both YAP- and TAZ-induced downstream gene transcription, measured by the activity of a Tead binding site-driven luciferase reporter (8XGIITC-Luc) (Figure S3B). More importantly, DN-TEAD4 expression by lentiviral infection was able to block endogenous transcription of the YAP/TAZ target genes, CTGF, CYR61, and ANKRD1, in human 92–1 UM cells (Figure 4G). 92–1 cells are a human UM cell line that carries the characteristic GαqQL mutation and was previously shown to be sensitive to YAP inhibition (Yu et al., 2014). In agreement with our data from mouse UMs with Lats1/2 deletion or YAP activation, we found that Tead inhibition by DN-TEAD4 in human 92–1 cells also decreased RRAS2, NRAS, and PRKCD gene transcription (Figure 4G). Western blot analysis showed downregulation of Ras/MAPK activity in 92–1 cells by DN-TEAD4, as evidenced by decreased phosphorylation of both MEK1/2 and ERK1/2 kinases (Figure 4H). Together, our data in mouse and human UM cells suggest that YAP/TAZ activation regulates MAPK signaling by promoting transcription of a subset of downstream targets associated with the Ras/MAPK cascade.
Kras Activation Promotes UM Progression In Vivo
To test whether Ras activation in the mouse uveal tract can induce UM in vivo, we utilized the well-characterized KrasG12D knockin allele, LSL-KrasG12D, which enables Cre-dependent expression of activated Kras at the endogenous Kras locus (Jackson et al., 2001). We found that, unlike Lats1/2 deletion or YAP activation, Kras activation alone in uveal melanocytes was not sufficient to drive UM formation after Cre injection (Figure 5A). However, the combination of Lats1/2 deletion and Kras activation significantly accelerated tumor progression, measured by tumor sizes and overall survival time of Cre-injected mice (Figures 5B–5E). After Cre injection, most of the mice carrying both LSL-KrasG12D and Lats1/2 conditional alleles died within 4 months because of exuberant UM growth and deteriorated health (Figure 5E). Not surprisingly, the tumors with Kras activation and Lats1/2 removal were more proliferative, as seen by phospho-Histone H3 (pH3) IHC (Figure 5F), and exhibited higher phospho-ERK1/2 levels than the tumors with only Lats1/2 removal, measured by both IHC and western blot analysis (Figures 5F and 5G). These data suggest that genetic Kras activation alone is not sufficient to induce UM initiation; rather, it promotes UM progression in conjunction with YAP/TAZ activation.
AP1 Transcriptional Upregulation in UMs with Kras Activation
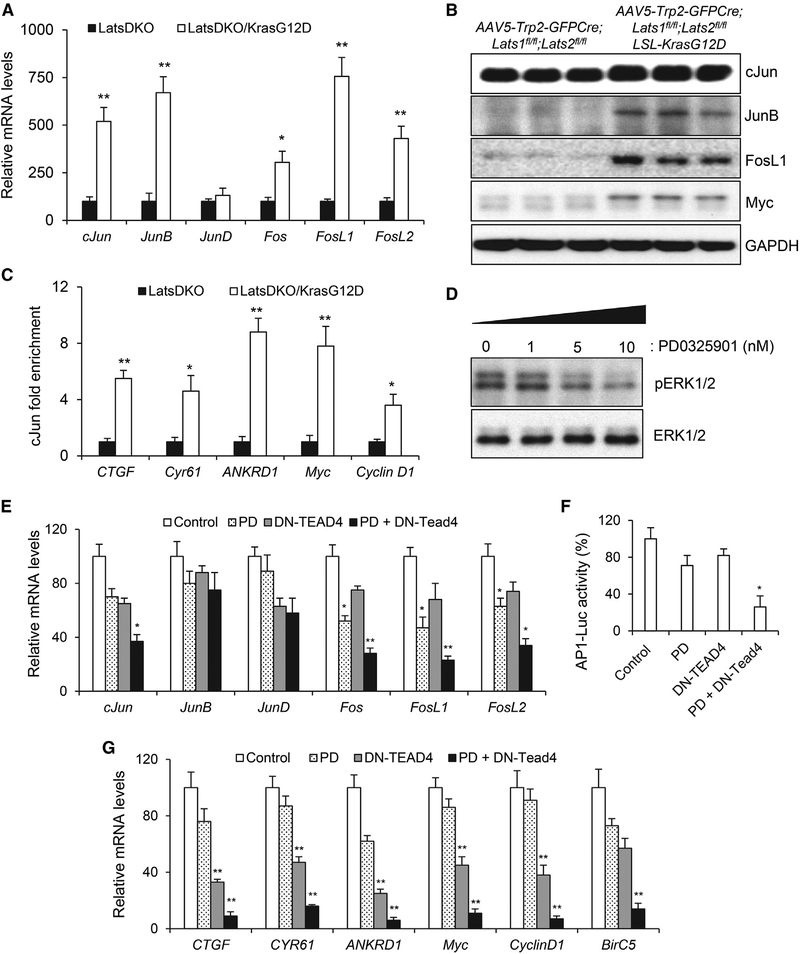
After comparing the tumors with Lats1/2 deletion with those with Lats1/2 deletion and Kras activation, we noticed that the tumors with both YAP/TAZ and Kras activation exhibited significantly higher mRNA levels of the YAP/Tead target genes, CTGF, Cyr61, and ANKRD1 (Figure 5H). It raised an intriguing possibility that Ras/MAPK activation might in turn promote YAP/Tead-mediated transcriptional output in UM.
To test this hypothesis, we first looked at YAP, TAZ, and Tead expression in mouse UM tumors with Lats1/2 deletion or Kras activation and did not detect significant change in their mRNA or protein levels, measured by qPCR or western blot analysis (Figures 5G, 5H, and S4B). Additionally, there was no change of YAP intracellular localization, because it remained in the nuclei (Figure S4A). To further understand the underlying mechanism of the increased transcriptional output of YAP/Tead, we focused on the activator protein 1 (AP1) factors. We and others recently reported the widespread AP1-Tead co-occupancy at the promoter or enhancer regions in the majority of the YAP/Tead target genes (Liu et al., 2016; Zanconato et al., 2015). We demonstrated that AP1-Tead cooperation acts as a major regulatory mechanism to coordinate downstream gene expression in various cancer cells (Liu et al., 2016; Zanconato et al., 2015). Thus, we examined AP1 expression and found that both mRNA and protein expression of several AP1 factors, including c-Jun, JunB, Fos, FosL1, and FosL2, were significantly upregulated in UMs with both Kras activation and Lats1/2 removal (Figures 6A and 6B). Furthermore, we performed c-Jun ChIP-qPCR on the promoter regions of the YAP/Tead downstream genes, including CTGF, Cyr61, ANKRD1, Myc, and CyclinD1, and found that there was significant enrichment of c-Jun occupancy in their promoter regions (Figure 6C), consistent with the elevated levels of AP1 proteins in UM cells (Figure 6B). These results suggest a possible mechanism involving AP1 upregulation to enhance YAP/Tead downstream transcriptional output.
Figure 6. AP1 Transcriptional Upregulation in UM Cells with YAP/TAZ and Kras Activation.
(A) qPCR analysis of mRNA levels of c-Jun, JunB, JunD, Fos, FosL1, and FosL2 in UMs derived from AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl (LatsDKO) and AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl;LSL-KrasG12D (LatsDKO/KrasG12D) mice.
(B) Immunoblot analysis of c-Jun, JunB, FosL1, Myc, and GAPDH in UMs developed in AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl and AAV5-Trp2-GFPCre;Lats1fl/fl; Lats2fl/fl;LSL-KrasG12D mice.
(C) ChIP-qPCR analysis shows significantly more enrichment of c-Jun at the promoter regions of the CTGF, Cyr61, ANKRD1, Myc, and CyclinD1 genes in UMs from AAV5-Trp2-GFPCre;Lats1fl/fl;Lats2fl/fl;LSL-KrasG12D mice.
(D) Immunoblot analysis of pERK1/2 and total ERK1/2 levels in 92–1 cells treated with PD0325901 at different concentrations.
(E) qPCR analysis of mRNA levels of c-Jun, JunB, JunD, Fos, FosL1, and FosL2 in 92–1 UM cells with or without DN-TEAD4 expression or PD0325901 (PD) treatment at 10 nM.
(F) Relative AP1-luciferase reporter (AP1-Luc) activity in 92–1 UM cells with or without DN-TEAD4 expression or PD0325901 (PD) treatment.
(G) qPCR analysis of mRNA levels of CTGF, CYR61, ANKRD1, Myc, Cyclin D1, and Birc5 in 92–1 UM cells with or without DN-TEAD4 expression or PD0325901 (PD) treatment.
Data are mean ± SD. *p ≤ 0.05, **p ≤ 0.01.
Dual Inhibition of TEAD and MEK in Human UM Cells
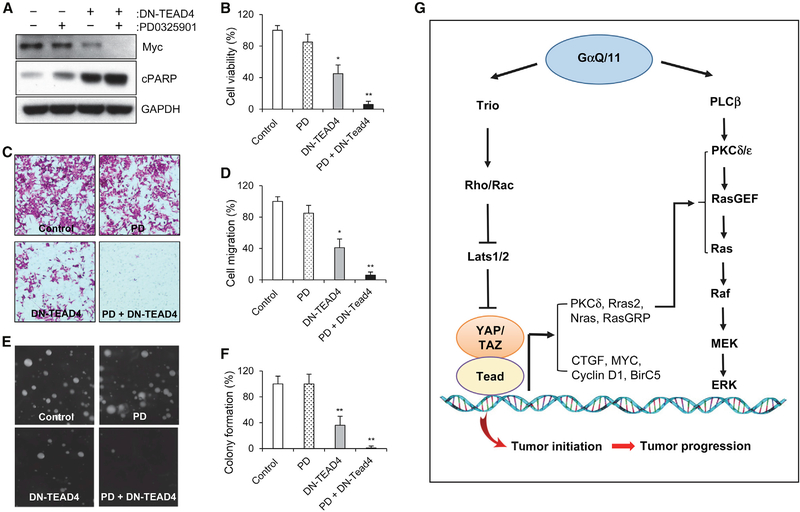
To further explore the interplay between YAP/Tead and Ras/MAPK in human UM cells, we treated 92–1 cells with a MEK inhibitor, PD0325901. PD0325901 inhibited ERK phosphorylation in a dose-dependent manner in 92–1 cells (Figure 6D). The expression of several AP1 genes, including c-Jun, Fos, FosL1, and FosL2, was downregulated after PD0325901 treatment, and their transcription was synergistically suppressed when both MEK and TEAD activation was inhibited (Figure 6E). Overall AP1 transcriptional activity in 92–1 cells was also significantly decreased by dual inhibition of TEAD and MEK, measured by the AP1-dependent luciferase reporter (AP1-Luc) assay (Figure 6F). Consistent with the notion of transcriptional cooperation between the two pathways, we found that ectopic expression of DN-TEAD4 and treatment of PD0325901 synergized to inhibit the transcription of CTGF, CYR61, and ANKRD1 (Figure 6G), as well as the YAP/TAZ target genes involved in regulation of cell proliferation and apoptosis, Myc, Cyclin D1, and BirC5 (Figures 6G and 7A).
Figure 7. Combination Inhibition of YAP/TEAD and MAPK in Human UM Cells.
(A) Immunoblot analysis of Myc and cleaved PARP (cPARP) in 92–1 UM cells with or without DN-TEAD4 expression or PD0325901 treatment.
(B–F) Cell viability (B), migration (C), and soft agar colony formation (E) in 92–1 UM cells with or without DN-TEAD4 expression or PD0325901 treatment. Cell viability was measured by MTT assay, and migration was measured by transwell migration assay. Quantitation of cell migration and soft agar colony formation assays was shown in (D) and (F). Data are mean ± SD. *p ≤ 0.05, **p ≤ 0.01.
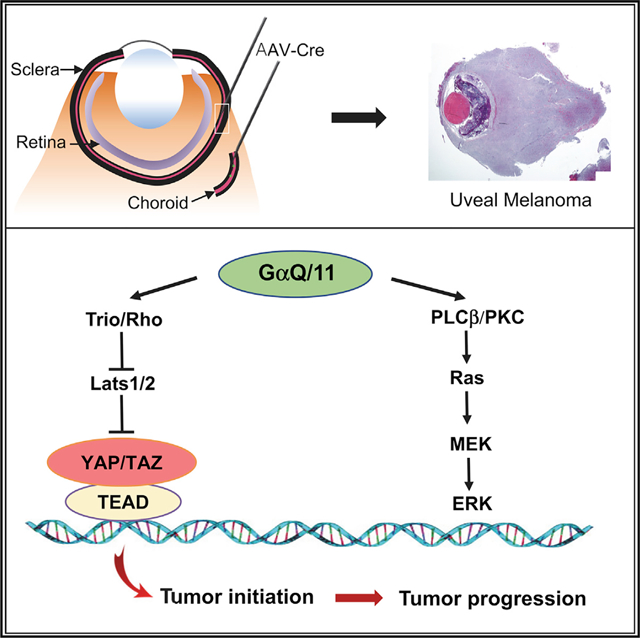
(G) A schematic model showing distinct roles of YAP/TAZ and Ras/MAPK in Gq/11-driven UM initiation and progression. YAP/TAZ activation induces UM initiation, and the cooperation of YAP/TAZ and Ras/MAPK via downstream transcriptional programs further promotes UM progression.
Further analysis showed PD0325901 treatment at 10 nM, a concentration that effectively blocked ERK phosphorylation in 92–1 UM cells (Figure 6D), had largely no effect on cell viability, measured by the MTT assay (Figure 7B). MEK inhibition by PD0325901 also did not significantly induce apoptosis in 92–1 UM cells, detected by western blot analysis of cleaved poly(ADP-ribose) polymerase (PARP) (Figure 7A). These data suggest that MEK inhibition alone has little or no effect on UM cells, consistent with previous reports on the failure of MAPK inhibition in clinical trials (Carvajal et al., 2014; Komatsubara et al., 2016). However, we found that blocking MEK activity could sensitize the inhibitory activity of DN-TEAD4 in UM cells. YAP/TEAD inhibition by DN-TEAD4 in 92–1 cells was sufficient to decrease cell viability, block cell migration, and inhibit their ability to undergo anchorage-independent growth (Figures 7B–7F), measured by transwell migration and soft agar colony formation assays. Importantly, we also observed the synergy of dual inhibition of TEAD and MEK in these assays (Figures 7A–7F), suggesting the potential therapeutic benefit of targeting both YAP/TEAD and Ras/MAPK pathways in human UM cells.
DISCUSSION
In this study, we developed an AAV-based ocular injection method to deliver melanocyte-specific Cre directly into mouse uveal tract to facilitate genetic mouse modeling of UM. Our ocular-specific Cre delivery method provides clear advantages over the general melanocyte Cre lines, such as Mitf-Cre or Tyr-CreER, that have been used to model UM (Huang et al., 2015; Moore et al., 2018). These Cre lines also drive Cre expression in melanocytes or melanocyte-like cells in tissues outside the uveal tract, including skin, CNS, lung, and inner ear, which often leads to early lethality and additional phenotypes in other organs that complicate phenotypic analysis of UM (Huang et al., 2015; Moore et al., 2018). In contrast, our AAV-based uveal tract Cre delivery allows manipulation of oncogene or tumor suppressor activity specifically in uveal melanocytes. Using this robust platform, we demonstrated the ability of Lats1/2 deletion or YAP activation to initiate UM and uncovered its cooperation with Kras to promote UM progression. Our data also revealed that Kras activation alone is not sufficient to induce UM, consistent with a recent report showing the inability of Braf activation to initiate UM (Moore et al., 2018). These data highlight the intrinsic difference of oncogenic transformation between the melanocytes in uveal tract and skin, given the fact that Braf/MAPK activation is the dominant oncogenic driver in cutaneous melanomas. Because of the inability of Kras activation alone to induce UM, it also suggests that Ras/MAPK may not play a significant role in UM initiation. The mechanism underlying GNAQ/11-driven oncogenic transformation of uveal melanocytes is likely complex. Our data focus on the role of dysregulated Hippo signaling via Lats1/2 inactivation or YAP activation in UM genesis; however, additional studies are needed to explore the possible interactions between Hippo/Yap and other oncogenic pathways downstream of GNAQ/11, such as PLCβ and PKC (Chen et al., 2014, 2017).
Our studies here reveal the cooperation between YAP/TAZ and Ras/MAPK during UM progression and suggest a possible mechanism of transcriptional reinforcement between the two pathways in UM cells. First, we showed that YAP directs a subset of its downstream transcriptional program to promote the expression of Prkcd, Rras2, Nras, and RasGRP1, directly or indirectly, in UM cells (Figure 7G). It likely forms a part of the molecular basis of YAP/TAZ promotion of Ras/MAPK signaling in cancer cells, and our data agree with a recent report of YAP regulation of Ras gene transcription in NF2-mutated thyroid cancers (Garcia-Rendueles et al., 2015). In addition, our results suggest that YAP/Tead-related transcriptional output is further augmented in UM cells with both YAP/TAZ and Ras/MAPK activation, and likely involves AP1 upregulation. We recently showed AP1-Tead co-operation as a critical mechanistic node to coordinate downstream transcription in cancer cells (Liu et al., 2016). Our results here linked AP1 to Tead-mediated transcription in UM. Ras activation is associated with YAP regulation in different tumor contexts (Kapoor et al., 2014; Shao et al., 2014; Garcia-Rendueles et al., 2015). Two prior studies on acquired resistance to Kras suppression in colon, lung, and pancreatic cancers show that YAP can rescue cell viability and sustain tumor growth in the setting of Kras suppression (Kapoor et al., 2014; Shao et al., 2014). However, different mechanisms were proposed, and one of the studies suggests that Kras and YAP converge on one of the AP1 factors, Fos, in a Tead-independent manner (Shao et al., 2014). Our data provide an alternative model of Ras-YAP interaction in UM cells, in which upregulation of AP1 factors leads to their further engagement on the Tead-occupied enhancer or promoter regions, therefore amplifying YAP/Tead-mediated downstream oncogenic output. It is intriguing to test whether such Ras-AP1-YAP/Tead regulatory axis functions in other tumors associated with Hippo dysregulation and Ras activation.
Our data from genetically modified mouse models and human UM cells highlight the potential of YAP/Tead as the valid therapeutic targets for UM. The cooperation of YAP/TAZ and Ras/MAPK on UM progression (Figure 7G) likely has important therapeutic implication as well. Clinical trials with MEK inhibitors targeting Ras signaling in UM failed in phase 3 trials (Carvajal et al., 2014; Komatsubara et al., 2016), and combination targeting of additional pathways including PKC has already been proposed (Chen et al., 2014, 2017). Our results argue that dual inhibition of both YAP/Tead and Ras/MAPK may have greater therapeutic benefit in UM. In light of our data on the potential involvement of AP1 factors in YAP-Ras interaction, targeting AP1 will also be an intriguing strategy for UM treatment, which is consistent with a previous report suggesting AP1’s role in UM cells (Vaqué et al., 2013). Clearly further functional and pre-clinical studies on the interplay among these pathways in UM are warranted.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Junhao Mao (junhao.mao@umassmed.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animals use protocols were reviewed and approved by The University of Massachusetts Medical School Institutional Animal Care and Use Committee. R26-YAP5SA (Cotton et al., 2017), Lats1flox and Lats2flox (Yi et al., 2016) mice were described previously. LSL-KrasG12D (Jackson et al., 2001) and R26mT/mG (Muzumdar et al., 2007) mice were obtained from the Jackson laboratory. Yapflox (Xin et al., 2011) and Tazflox (Xin et al., 2013) mice were kindly provided by Dr. EN Olson. The mice used in this study were maintained in a genetic background with Swiss Webster (CFW) as the major component, and both male and female mice between 2–4 months of age were used for AAV injection.
METHOD DETAILS
AAV Injection in Mouse Uveal Tract
AAV5-CMV-Cre vector was obtained from UMass Vector Core. For AAV5-TRP2-GFPCre, the 1.7Kb TRP2 promoter region (Zhao and Overbeek, 1999) was PCR amplified from mouse genomic DNA, and cloned into the pssAAV packaging plasmid together with a GFP-Cre fragment (a gift from Fred Gage, Addgene # 49056) to generate the pssAAV-TRP2-GFPCre construct. AAV5-TRP2-GFPCre vector was then produced by UMass Vector Core. For AAV uveal tract injection, recombinant AAV injections into the uveal track were performed as previously described (Venkatesh et al., 2013) with the following modification. In brief, mice were anesthetized with a mixture of ketamine/xylazine (100mg/kg and 10mg/kg). Eyes were cleaned with betadine followed by water and 70% ethanol. Thereafter 0.5 ml of virus was injected directly into the choroid by inserting a beveled glass needle through the sclera. In order to not push the needle across the sclera into the sub retinal space the needle was inserted at a flat angle. Successful targeting was visualized by bulging of the scleral/chorodial tissue, which does not occur if the needle crosses into the sub retinal space. Injections were performed using an air pressured injection pump (FemtoJet: Eppendorf). After removal of the needle corneal lubricant was applied and animals were kept at 37°C until fully recovered. All mouse experiments were conducted according to the University of Massachusetts Medical School IACUC guidelines.
Tissue Collection and Histology
Following euthanasia, tumors or eyes were dissected and fixed in 10% Neutral Buffered Formalin (NBF) at 4°C overnight. For paraffin sections, tissue was dehydrated, embedded in paraffin, and sectioned at 6 mm. For frozen sections, tissue was dehydrated in 30% sucrose overnight at 4°C, embedded in OCT, and sectioned at 12 mm. Paraffin sections were stained using standard hematoxylin & eosin reagents.
Immunohistochemistry, Immunofluorescence and b-Galactosidase Staining
For immunohistochemistry (IHC), sections were deparaffinized and rehydrated before undergoing heat-induced antigen retrieval in 10mM sodium citrate buffer (pH 6.0) for 30 minutes. Slides were blocked for endogenous peroxidase for 20 minutes, then blocked for 1 hour in 5% BSA, 1% goat serum, 0.1% Tween-20 buffer in PBS, and incubated overnight at 4°C in primary antibody diluted in blocking buffer or SignalStain® Antibody Diluent (Cell Signaling). Slides were incubated in biotinylated secondary antibodies for 1 hour at room temperature and signal was detected using the Vectastain Elite ABC kit (Vector Laboratories). Hematoxylin was used for counterstaining in IHC. For β-galactosidase staining, frozen sections were cut at 12μM intervals and subjected to standard β-galactosidase staining. For immunofluorescence (IF) cells or tissue sections were fixed by 4% paraformaldehyde for 5 minutes, blocked for 1 hour and incubated overnight at 4°C in primary antibody diluted in blocking buffer. Slides were then incubated for 1 hour at room temperature in Alexa Fluor-conjugated secondary antibodies (Invitrogen) in blocking buffer and mounted using mounting media with DAPI (EMS). All primary and secondary antibodies used for IHC/IF were described in the KEY RESOURCES TABLE.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-YAP | Cell Signaling | Cat# 14074; RRID:AB_2650491 |
| Rabbit anti-YAP/TAZ | Cell Signaling | Cat# 8418; RRID:AB_10950494 |
| Rabbit anti-Phospho-YAP (Ser127) | Cell Signaling | Cat# 13008; RRID:AB_2650553 |
| Mouse anti-TAZ | BD PharMingen | Cat# 560235; RRID:AB_1645338 |
| Rabbit anti-Lats2 | Cell Signaling | Cat# 5888; RRID:AB_10835233 |
| Rabbit anti-Ki67 | Abcam | Cat# ab15580; RRID:AB_443209 |
| Rabbit anti-phospho Histone H3 | Cell Signaling | Cat# 9701; RRID:AB_331535 |
| Rabbit anti-ERK1/2 | Cell Signaling | Cat# 4695; RRID:AB_390779 |
| Rabbit anti-phospho-ERK1/2 | Cell Signaling | Cat# 4370; RRID:AB_2315112 |
| Rabbit anti-MEK1/2 | Cell Signaling | Cat# 9126; RRID:AB_331778 |
| Rabbit anti-phospho-MEK1/2 | Cell Signaling | Cat# 9154; RRID:AB_2138017 |
| Rabbit anti-cJun | Cell Signaling | Cat# 9165; RRID:AB_2130165 |
| Rabbit anti-JunB | Bethyl | Cat# A302–704A; RRID:AB_10749029 |
| Rabbit anti-FosL1 | SCBT | Cat# sc183; RRID:AB_2106928 |
| Rabbit anti-Myc | Cell Signaling | Cat# 5605; RRID:AB_1903938 |
| Rabbit anti-Myc tag | Cell Signaling | Cat# 2278; RRID:AB_490778 |
| Rabbit anti-cleaved PARP (Asp214) | Cell Signaling | Cat# 9541; RRID:AB_331426 |
| Mouse anti-Melanoma cocktail (Melan A/Mart-1) | Abcam | Cat# ab732; RRID:AB_305844 |
| Mouse anti-TEAD4 | SCBT | Cat# sc-101184; RRID:AB_2203086 |
| Rabbit anti-GAPDH | Bethyl | Cat# A300–641A;RRID:AB_513619 |
| Normal Rabbit IgG | Cell Signaling | Cat# 272AB_3907799; RRID:AB_1031062 |
| Normal Mouse IgG | SCBT | Cat# sc-2025; RRID:AB_737182 |
| HRP conjugated goat anti-rabbit | Promega | Cat# W4011; RRID:AB_430833 |
| HRP conjugated goat anti-mouse | Promega | Cat#W4021; RRID:AB_430834 |
| Anti-Rabbit HRP | Cell Signaling | Cat# 7074S; RRID:AB_2099233 |
| Anti-Mouse HRP | Cell Signaling | Cat# 7076S; RRID:AB_330924 |
| Mouse TrueBlot® ULTRA: Anti-Mouse Ig HRP | Rockland | Cat# 18–8817–33; RRID:AB_2610851 |
| Rabbit TrueBlot® ULTRA: Anti-Mouse Ig HRP | Rockland | Cat# 18–8816–33; RRID:AB_2610848 |
| Alexa Fluor 568, goat anti-rabbit | Invitrogen | Cat#A-11011; RRID:AB_143157 |
| Alexa Fluor 633, goat anti-rabbit | Invitrogen | Cat#A-21070; RRID:AB_2535731 |
| Alexa Fluor 568, goat anti-mouse | Invitrogen | Cat#A-11004; RRID:AB_2534072 |
| Alexa Fluor 633, goat anti-mouse | Invitrogen | Cat#A-21050; RRID:AB_141431 |
| Bacterial and Virus Strains | ||
| AAV5-CMV-Cre | UMass Vector Core | N/A |
| AAV5-TRP2-GFPCre | UMass Vector Core | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease inhibitor | Promega | Cat# G653A |
| Phosphatase inhibitor cocktail 2 | Sigma | Cat# P5726 |
| PD0325901 | Selleckchem | Cat#S1036 |
| Critical Commercial Assays | ||
| Vectastain Elite ABC kit | Vector Lab | Cat# 1725124 |
| Dual-Luciferase Reporter Assay System | Promega | Cat# PK-6011 |
| ChIP-IT Express ChIP Kit | Active Motif | Cat#E1910 |
| Trizol reagent | Invitrogen | Cat#BP-111R |
| Deposited Data | ||
| RNaseq data of Lats1/2 KO mouse UM | This paper | GEO: GSE115181 |
| Experimental Models: Cell Lines | ||
| Human HEK293 cell line | ATCC | Cat#CRL-1573 |
| Human 92–1 Uveal melanoma cell line | Sigma | Cat# 13012458–1VL |
| Experimental Models: Organisms/Strains | ||
| Mouse: Lats1flox | Yi et al., 2016 | JAX# 024941 |
| Mouse: Lats2flox | Yi et al., 2016 | JAX# 025428 |
| Mouse: YAPflox | Xin et al., 2011 | N/A |
| Mouse: TAZflox | Xin et al., 2013 | N/A |
| Mouse: R26-YAP5SA | Cotton et al., 2017 | N/A |
| Mouse: R26mT/mG | Muzumdar et al., 2007 | JAX# 007676 |
| Mouse: LSL-KrasG12D | Jackson et al., 2001 | JAX# 008179 |
| Oligonucleotides | ||
| Primers for qPCR | This paper | Table S2 |
| Primers for ChIP-qPCR | This paper | Table S2 |
| Recombinant DNA | ||
| AAV5-TRP2-GFPCre | This paper | N/A |
| PGIPZ-DN-TEAD4 | This paper | N/A |
| pGIPZ-FLAG-nls-YAP5SA | Cotton et al., 2017 | N/A |
| pBABE-TAZ4SA | Gift of Dr. Kun-liang Guan, UCSD | N/A |
| AAV-GFP-Cre | Addgene | Addgene #49056 |
| 8XGTIIC-LUC (TBS-Luc) | Addgene | Addgene #34615 |
| AP1-Luc | Addgene | Addgene #40432 |
| Software and Algorithms | ||
| Fiji (ImageJ) | Schindelin et al., 2012 | Fiji (ImageJ) |
Immunoblotting Analysis
Mouse tumor and control eye tissues were dissected and lysed, and the lysate was then incubated with indicated antibody overnight. The immunoprecipitates were washed five times with RIPA buffer, before subjecting to immunoblot analysis. All primary and secondary antibodies used for IHC/IF were described in the KEY RESOURCES TABLE.
RNaseq Analysis
For mouse UM, tissue was homogenized in the Trizol reagent (Thermo Fisher) and RNA was extracted according to manufacturer’s instructions. For control tissue, eyes were enuclated and cornea, lens, retina, optic nerve and muscle attachments were removed to obtain tissue largely containing the uveal tract, RPE, and sclera. The integrity of isolated RNA was analyzed using Bioanalyzer (Agilent Technologies). RNA-seq libraries were made with Illumina Truseq RNA Sample Prep protocol by UMass Deep Sequencing Core and sequenced on Illumina HiSeq2000. All libraries have around 20–30 million reads sequenced. Quality assessment of the raw reads (single end 75bp) was performed using fastqc (FastQC: a quality control tool for high throughput sequence data. Available online at: (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), followed by alignment to the reference mouse genome (mm10) using tophat (Kim et al., 2013). Cufflinks (Trapnell et al., 2012) was used for identifying differentially expressed genes between wild-type and knock out group. Genes with q-value < 0.05 and fold change greater than 1.5 were considered as significantly differential expressed genes. In addition, Pathway and GO enrichment analysis was performed using ChIPpeakAnno package (Zhu, 2013; Zhu et al., 2010). The RNaseq data were deposited into the GEO repository and the accession number is GSE115181.
Chromatin Immunoprecipitation-qPCR Analysis
ChIP assays were performed using ChIP-IT® Express Chromatin Immunoprecipitation Kit according to the manufacturer’s instructions (Active Motif, Cat # 53008). Briefly, freshly dissected tissues were fixed with 1% formaldehyde, washed with cold PBS and lysed in lysis buffer. After sonication, protein-DNA complexes were incubated with Tead4 (SCBT) or c-Jun (Cell Signaling) antibodies-coupled protein G beads at 4°C overnight. After elution and reverse cross-link, DNA was purified for subsequent PCR analysis. The antibodies used for ChIP were described in the KEY RESOURCES TABLE. The primers used for real-time PCR of the promoter regions were described in Table S2.
Quantitative Real-Time PCR
Total RNA of animal tissues and human UM cells was isolated using Trizol reagent (Thermo Fisher). cDNA was prepared using Superscript II Reverse Transcriptase (Thermo Fisher), and the amount of transcripts were quantified using Sybr Mastermix (Kapa Bioscience), with the respective oligonucleotides (Table S2) in Applied Biosystems 7300 RT-PCR systems. The number of copies of each gene was normalized to the housekeeping gene GAPDH. All qPCR experiments were conducted in biological triplicates, error bars represent mean ± standard deviation, and Student’s t test was used to generate p values (* = p value ≤ 0.05; ** = p value ≤ 0.01).
Cell Culture, Treatment and Lentiviral Infection
92–1 cells (human uveal melanoma cells, female) were cultured in RPMI supplemented with 10% FBS and treated with MEK inhibitor PD0325901 (Selleckchem) at 10nM for various analyses. To generate DN-TEAD4 lentiviral expression vector, the cDNA fragments encoding human TEAD4 were PCR cloned into a pGIPZ-based lentiviral vector. For lentiviral infection, pGIPZ constructs were transfected along with the packing plasmids into growing HEK293T cells (human embryonic kidney cells, female). Viral supernatants were collected 48 hours after transfection, and target cells were infected in the presence of polybrene and underwent selection with puromycin for 3–4 days before subsequent analyses.
Luciferase Reporter Analysis
Tead-Luc (8xGTIIC-luciferase) was a gift from Stefano Piccolo (Addgene # 34615), and AP1-Luc (3xAP-1 in pGL3-basic) was a gift from Alexander Dent (Addgene # 40342). For 8xGTIIC-Luc reporter assay, HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen) with 8xGTIIC-Luc reporter construct together with the expression plasmids of Renila-luciferase, DN-TEAD4, YAP-5SA or TAZ-4SA (a gift from Dr. Kun-liang Guan, UCSD) for 48 hours. For AP1-Luc reporter assay, 92–1 cells were transfected using Lipofectamine 2000 (Invitrogen) with AP1-Luc reporter construct together with the expression plasmids of Renila-luciferase and DN-TEAD4 with or without 10nM PD0325901 treatment for 48 hours. Luciferase activity was measured as using the Dual-Luciferase reporter assay system (Promega, Cat # E1910) according to the manufacturer’s protocol. All luciferase reporter experiments were conducted in biological triplicates, error bars represent mean ± standard deviation, and Student’s t test was used to generate p values. (* = p value ≤ 0.05; ** = p value ≤ 0.01).
MTT, Soft Agar Colony Formation, and Cell Migration Assays
MTT assay was performed using CellTiter 96® Non-Radioactive Cell Proliferation Assay (Promega, Cat # G4000). For anchorage-independent soft-agar colony formation assay, 92–1 cells were seeded at a density of 6000 cells/well in a 6-well plate of 0.3% agarose in RPMI media containing 10% FBS. Colonies from 12 fields of view were counted 14 days later. Assays were conducted in triplicates, and standard deviation was used to calculate error bars. For transwell migration assays, 5×104 cells were plated in the top chamber with the non-coated membrane (24-well insert; pore size, 8 mm; BD Biosciences) and in medium without serum, and medium supplemented with serum was used as a chemoattractant in the lower chamber. The cells were incubated for 24h and cells that did not migrate or invade through the pores were removed by a cotton swab. Cells on the lower surface of the membrane were stained with the Diff-Quick Staining Set (Dade) and counted.
QUANTIFICATION AND STATISTICAL ANALYSIS
No statistical method was used to predetermine sample size. The experiments were not randomized. For biochemical experiments we performed the experiments at least three independent times. Experiments for which we showed representative images were performed successfully at least 3 independent times. No samples or animal were excluded from the analysis. The investigators were not blinded to allocation during experiments and outcome assessment. Student’s t test was used to generate p values (* = p value ≤ 0.05; ** = p value ≤ 0.01). The variance was similar between groups that we compared.
DATA AND CODE AVAILABILITY
The accession number for the RNA-Seq data of Lats1/2 deleted mouse uveal melanoma is GEO: GSE115181.
Supplementary Material
In Brief.
Li et al. utilize an AAV-based ocular injection method to specifically manipulate Hippo/YAP and Ras signaling in mouse uveal melanocytes. They reveal the role of YAP/TAZ in uveal melanoma formation and suggest that the Hippo/ YAP-Ras/MAPK interaction during tumor growth can be exploited to develop a therapeutic strategy for uveal melanoma.
Highlights.
Modeling uveal melanoma (UM) via AAV-based Cre delivery into uveal tract
Lats1/2 kinases suppress UM formation in uveal melanocytes
Lats1/2 deletion cooperates with Kras activation to promote UM progression
Dual inhibition of YAP/TAZ and Ras/MAPK synergizes to suppress UM cell growth
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (R01DK099510 and R01CA238270 to J.M.; EY023570 to C.P.). X.W. is supported by the Samuel M. Fisher Memorial-MRA (Melanoma Research Alliance) Established Investigator Award, the Department of Defense (grant W81XWH-17-1-0361), and the NIH (grants R01CA181537 and R01DK107651). Y.T.I. is supported by NIH grants (R01DK083450 and GM107457). The authors thank Annie Pang for technical assistance and members of the Punzo and Mao labs for helpful discussions.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.03.021.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Carvajal RD, Sosman JA, Quevedo JF, Milhem MM, Joshua AM, Kudchadkar RR, Linette GP, Gajewski TF, Lutzky J, Lawson DH, et al. (2014). Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. JAMA 311, 2397–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wu Q, Tan L, Porter D, Jager MJ, Emery C, and Bastian BC (2014). Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene 33, 4724–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wu Q, Depeille P, Chen P, Thornton S, Kalirai H, Coupland SE, Roose JP, and Bastian BC (2017). RasGRP3 mediates MAPK pathway activation in GNAQ mutant uveal melanoma. Cancer Cell 31, 685–696.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L, Berube J, Fromont A, and Bell JB (2004). Ability of scalloped deletion constructs to rescue sd mutant wing phenotypes in Drosophila melanogaster. Genome 47, 849–859. [DOI] [PubMed] [Google Scholar]
- Cotton JL, Li Q, Ma L, Park JS, Wang J, Ou J, Zhu LJ, Ip YT, Johnson RL, and Mao J (2017). YAP/TAZ and hedgehog coordinate growth and patterning in gastrointestinal mesenchyme. Dev. Cell 43, 35–47.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, and Der CJ (2010). Ras history: the saga continues. Small GTPases 1, 2–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, Zaidi MR, Ksander BR, Merlino G, Sodhi A, et al. (2014). Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell 25, 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rendueles ME, Ricarte-Filho JC, Untch BR, Landa I, Knauf JA, Voza F, Smith VE, Ganly I, Taylor BS, Persaud Y, et al. (2015). NF2 Loss Promotes Oncogenic RAS-Induced Thyroid Cancers via YAP-Dependent Transactivation of RAS proteins and Sensitizes Them to MEK Inhibition. Cancer Discov 5, 1178–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golec DP, Henao Caviedes LM, and Baldwin TA (2016). RasGRP1 and RasGRP3 are required for efficient generation of early thymic progenitors. J. Immunol 197, 1743–1753. [DOI] [PubMed] [Google Scholar]
- Graham SM, Cox AD, Drivas G, Rush MG, D’Eustachio P, and Der CJ (1994). Aberrant function of the Ras-related protein TC21/R-Ras2 triggers malignant transformation. Mol. Cell. Biol 14, 4108–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, and Camargo FD (2013). The hippo tumor suppressor network: from organ size control to stem cells and cancer. Cancer Res 73, 6389–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JL, Urtatiz O, and Van Raamsdonk CD (2015). Oncogenic G Protein GNAQ Induces Uveal Melanoma and Intravasation in Mice. Cancer Res 75, 3384–3397. [DOI] [PubMed] [Google Scholar]
- Hubbard KB, and Hepler JR (2006). Cell signalling diversity of the Gqalpha family of heterotrimeric G proteins. Cell. Signal 18, 135–150. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, and Tuveson DA (2001). Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15, 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y, Wu CJ, Sadanandam A, Hu B, et al. (2014). Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsubara KM, Manson DK, and Carvajal RD (2016). Selumetinib for the treatment of metastatic uveal melanoma: past and future perspectives. Future Oncol 12, 1331–1344. [DOI] [PubMed] [Google Scholar]
- Larive RM, Abad A, Cardaba CM, Hernández T, Cañamero M, de Álava E, Santos E, Alarcón B, and Bustelo XR (2012). The Ras-like protein R-Ras2/TC21 is important for proper mammary gland development. Mol. Biol. Cell 23, 2373–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hu DN, Zhao H, McCormick SA, Nordlund JJ, and Boissy RE (2006). Uveal melanocytes do not respond to or express receptors for alpha-melanocyte-stimulating hormone. Invest. Ophthalmol. Vis. Sci 47, 4507–4512. [DOI] [PubMed] [Google Scholar]
- Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, Zhu LJ, Goel HL, Mercurio AM, Park JS, et al. (2016). Tead and AP1 Coordinate Transcription and Motility. Cell Rep 14, 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, and Pan D (2012). Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev 26, 1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AR, Ran L, Guan Y, Sher JJ, Hitchman TD, Zhang JQ, Hwang C, Walzak EG, Shoushtari AN, Monette S, et al. (2018). GNA11 Q209L Mouse Model Reveals RasGRP3 as an Essential Signaling Node in Uveal Melanoma. Cell Rep 22, 2455–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Pan D (2010). The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, et al. (2014). KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158, 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AD, Bergman L, and Seregard S (2005). Uveal melanoma: epidemiologic aspects. Ophthalmol. Clin. North Am 18, 75–84, viii. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, Simpson EM, Barsh GS, and Bastian BC (2009). Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457, 599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, et al. (2010). Mutations in GNA11 in uveal melanoma. N. Engl. J. Med 363, 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaqué JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, Debant A, Seeger MA, Ksander BR, Teramoto H, and Gutkind JS (2013). A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol. Cell 49, 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh A, Ma S, Langellotto F, Gao G, and Punzo C (2013). Retinal gene delivery by rAAV and DNA electroporation. Curr. Protoc. Microbiol 28, 14D.4.1–14D.4.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, and Olson EN (2011). Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal 4, ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. (2013). Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA 110, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Lu L, Yanger K, Wang W, Sohn BH, Stanger BZ, Zhang M, Martin JF, Ajani JA, Chen J, et al. (2016). Large tumor suppressor homologs 1 and 2 regulate mouse liver progenitor cell proliferation and maturation through antagonism of the coactivators YAP and TAZ. Hepatology 64, 1757–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. (2012). Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, Zhao L, Peyman G, Ouyang H, Jiang W, et al. (2014). Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell 25, 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, and Guan KL (2015). Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 163, 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, and Piccolo S (2015). Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17, 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Cordenonsi M, and Piccolo S (2016). YAP/TAZ at the Roots of Cancer. Cancer Cell 29, 783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, and Overbeek PA (1999). Tyrosinase-related protein 2 promoter targets transgene expression to ocular and neural crest-derived tissues. Dev. Biol 216, 154–163. [DOI] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21, 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ (2013). Integrative analysis of ChIP-chip and ChIP-seq dataset. Methods Mol. Biol 1067, 105–124. [DOI] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, and Green MR (2010). ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-Seq data of Lats1/2 deleted mouse uveal melanoma is GEO: GSE115181.