Abstract
We report the complete chloroplast genome sequence of the important nutritious millet crop Indian barnyard millet, Echinochloa frumentacea Link (Poaceae). The size of the circular chloroplast genome is 139 593 bp in length with a typical quadripartite structure, containing pair of inverted repeats of 22 618 bp, flanked by large single copy and small single copy regions of 81 839 bp, 12 518 bp, respectively. Overall GC content of the genome was 38.6% and consists of 112 individual genes, including 77 protein coding genes, 30 tRNA genes, four rRNA genes and one conserved open reading frame. In addition, phylogenetic analysis with grass species has revealed that E. frumentacea is diverged around 1.9–2.7 million years with its close relatives, E. oryzicola and E. crus-galli.
Keywords: Chloroplast genome, Echinochloa, E. frumentacea, Indian barnyard millet
The chloroplast is a vital organelle genome responsible for the photosynthesis. Chloroplast genomes have non-meiotic and uni-parental inheritance mostly by maternal inheritance (Reboud & Zeyl 1994) and 120–217 kb in size with about 130 conserved genes and relatively diverse intergenic spaces (IGS) (Rivarola et al. 2011, Wang & Messing 2011, Nah et al. 2015). The chloroplast genomes have highly conserved genome structure, organization, gene order which render them as main targets to understand plant genetic diversity and evolution (Moore et al. 2010, Ye et al. 2014).
The genus Echinochloa belongs to the family Poaceae which consists of 50 species including two domesticated species, Indian barnyard millet or Indian sawa millet (E. frumentacea) and Japanese barnyard millet (E. utilis or E. esculanta) (Aoki & Yamaguchi 2008). E. frumentacea (2n=6× =54) is one of the oldest domesticated millet in the semi-arid tropics of Asia and Africa (Hilu 1994) and expected to be domesticated from the hexaploid progenitor E. colona. It is nutritionally a rich cereal crop which serves as a food and fodder, contains vitamins, minerals, sulphur-containing amino acids and phyto chemicals even superior to the staple cereals such as rice and wheat (Yabuno 1983). Despite the above facts, the barnyard millet genome has been largely unexplored.
Here we report complete chloroplast genome of the E. frumentacea Link (Poaceae). Genomic DNA sample of Indian barnyard millet popular variety CO(KV)2 was obtained from Agricultural College and research Institute, Madurai, TNAU, India. Genomic DNA of the four-week old plant was used to decode the genome by Illumina MiSeq sequencing platform with 300 × 2 pair-end library. Whole genome sequences of E. frumentacea were assembled under CLC genome assembler to achieve the complete and error-free chloroplast genome (cpDNA) based on the guidance of our previous paper (Kim et al. 2015). The cpDNA was annotated by the Dual Organellar Genome Annotator (DOGMA) (Wyman et al. 2004) and tRNAscan-SE (Schattner et al. 2005). The cpDNA sequence with complete annotation information was deposited at GenBank database under the accession number KU242342.
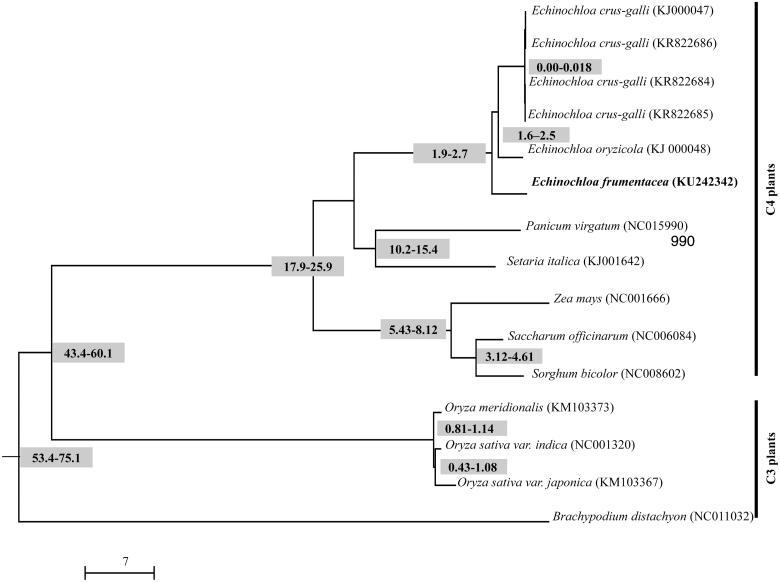
The chloroplast genome has a total length of 139 593 bp with a characteristic quadripartite structure, consisting of LSC region of 81 839 bp, two IR regions of 22 618 bp and SSC region of 12 518 bp. The chloroplast genome revealed the GC content of 38.6%, was predicted to encode 112 genes, including 77 protein coding genes, 30 tRNA genes, four rRNA genes and one conserved open reading frame (ORF). Phylogenetic analysis was performed using MEGA6 based on the complete cpDNA of E. frumentacea with reported close relatives of grass or poaceae family which not only differentiated the C3 and C4 plants but also revealed the inter-and intra-species diversity in the Echinochloa genus (Figure 1). The molecular divergence clock analysis using the BEAST has revealed that E. frumentacea diverged with its close relatives, E. oryzicola and E. crus-galli around 1.9–2.7 million years (Drummond et al. 2012; Ye et al. 2014). This newly characterized chloroplast genome of E. frumentacea will provide a valuable resource for population and evolutionary studies of the genus Echinochloa.
Figure 1.
Phylogenetic tree based on the complete chloroplast genome sequences of E. frumentacea and 14 other grass species. Neighbor-joining tree was generated by MEGA6 with 1000 bootstrap replications. The molecular divergence time denoted on the node was calculated using Yule process with the reference divergence time of rice and sorghum (50 mya). The corresponding GenBank number of the cpDNA including the outgroup Brachypodium distachyon was denoted in the parenthesis.
Disclosure statement
None of the authors report any conflict of interest. The authors alone are responsible for the content and writing of the paper.
This research was supported by the Golden Seed Project (Center for Horticultural Seed Development, No. 213003-04-3-SB430), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS).
References
- Aoki D, Yamaguchi H.. 2008. Genetic relationship between Echinochloa crus-galli and Echinochloa oryzicola accessions inferred from internal transcribed spacer and chloroplast DNA sequences. Weed Biol Manage. 8:233–242. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilu K. 1994. Evidence from RAPD markers in the evolution of Echinochloa millets (Poaceae). Plant Systemat Evol. 189:247–257. [Google Scholar]
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H.. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE.. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci. 107:4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nah G, Im J-H, Kim J-W, Kim K, Lim J, Choi AY, Choi I-Y, Yang T-J, Park T-S, Lee D, et al. 2015. The complete chloroplast genomes of three Korean Echinochloa crus-galli accessions. Mitochondrial DNA 1–2. 10.3109/19401736.2015.1089499. [DOI] [PubMed] [Google Scholar]
- Reboud X, Zeyl C.. 1994. Organelle inheritance in plants. Heredity 72:132–140. [Google Scholar]
- Rivarola M, Foster JT, Chan AP, Williams AL, Rice DW, Liu X, Melake-Berhan A, Creasy HH, Puiu D, Rosovitz M.. 2011. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS One 6:e21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Messing J.. 2011. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS One 6:e24670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yabuno T. 1983. Biology of Echinochloa species. Proceeding of the Conference on Weed Control in Rice. 307–318. [Google Scholar]
- Ye C-Y, Lin Z, Li G, Wang Y-Y, Qiu J, Fu F, Zhang H, Chen L, Ye S, Song W, et al. 2014. Echinochloa chloroplast genomes: insights into the evolution and taxonomic identification of two weedy species. PLoS One. 9:e113657. [DOI] [PMC free article] [PubMed] [Google Scholar]