Abstract
Common walnut (Juglans regia L.) is cultivated in temperate regions worldwide for its wood and nuts. The complete chloroplast genome of J. regia was sequenced using the Illumina MiSeq platform. This is the first complete chloroplast sequence for the Juglandaceae, a family that includes numerous species of economic importance. The chloroplast genome of J. regia was 160 367 bp in length, with 36.11% GC content. It contains a pair of inverted repeats (26 035 bp) which were separated by a large single copy (89 872 bp) and a small single copy region (18 425 bp). A total of 137 genes were annotated, which included 86 protein-coding genes, three pseudogenes (two ycf15 and one infA), 40 tRNA genes and eight rRNA genes. The neighbour-joining phylogenetic analysis with the reported chloroplast genomes showed that common walnut chloroplasts are most closely related to those of the Fagaceae family.
Keywords: Complete chloroplast genome, economic tree, Juglans regia
Juglans L. is one of the eight living genera in the family Juglandaceae consisting of 21 extant taxa (Manning 1978; Aradhya et al. 2007). The common walnut (Juglans regia L.) is the most economically important member of the genus Juglans because of its high-quality timber and nutritious nuts (Pollegioni et al. 2015). It is native to the mountainous regions of central Asia, but it has become the most widespread tree nut cultivated in the world (Chen et al. 2014).
Chloroplasts play an important role in photosynthesis in green plants and participate in the biosynthesis of starch, fatty acids and amino acids (Neuhaus & Emes 2000). The DNA sequence of the chloroplast genome can be used as a super barcode or a resource for research in phylogeography, genetic diversity and evolution. Although many members of the Juglandaceae are economically important, no member of the family has had its complete chloroplast sequence published.
For this study, we collected the fresh, healthy leaves from J. regia growing in the orchard of Northwest University, Shaanxi, China. DNA was extracted using methods described by Zhao and Woeste (2011). We sequenced the complete chloroplast genome of J. regia with the Illumina MiSeq sequencing platform (Sangon Biotech, Shanghai, China). We assembled the chloroplast genome using SPAdes (http://bioinf.spbau.ru/spades) and annotated with software CpGAVAS (http://www.biomedcentral.com/1471-2164/13/715) (Liu et al. 2012). We determined the complete chloroplast genome sequence of J. regia was 160 367 bp in length (GenBank accession number KT963008). The GC content was 36.11%. Chloroplasts are typically AT-rich, and the GC content of the J. regia chloroplast was similar to values previously reported for most other vascular species (e.g., 36.9% in Quercus aliena Blume; Lu et al. 2015). The large single copy (LSC) and small single copy (SSC) contained 89 872 and 18 425 bp, respectively, while the inverted repeat (IR) was 26 035 bp in length. This chloroplast genome contained 137 functional genes, including 86 protein-coding genes, three pseudogenes (two ycf15 and one infA), 40 tRNA genes and eight rRNA genes. There were 12 protein-coding genes, 14 tRNA and all eight rRNA genes duplicated in the IR regions. The LSC region contained 62 protein-coding and 25 tRNA genes, whereas the SSC region contained 12 protein-coding and one tRNA gene. Fourteen genes contained one or two introns, including the protein-coding genes, rps16, atpF, rpoC1, ycf3 (three introns), clpP, petB, petD, rpl16, rpl2, ndhB and rps12.
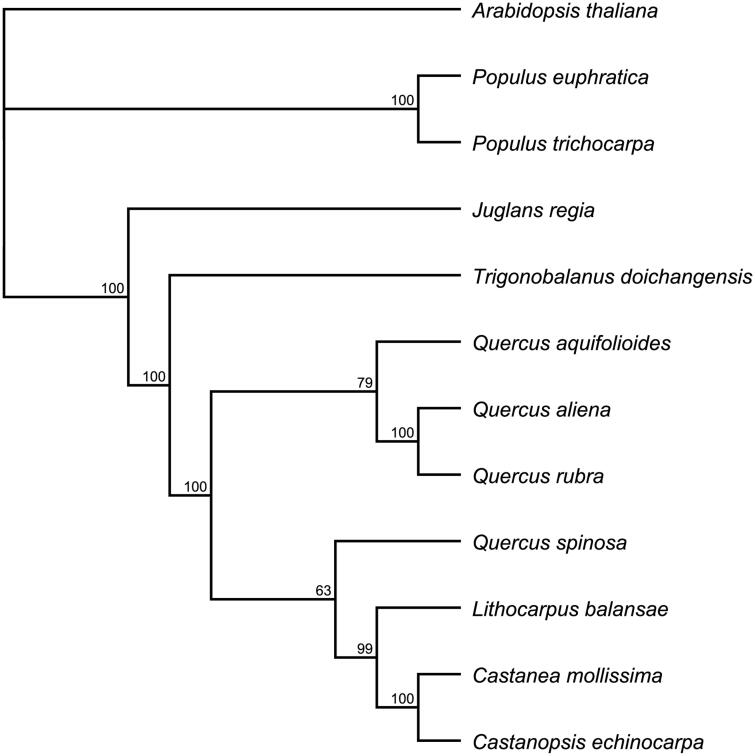
The phylogenetic relationship of J. regia to other plant families was evaluated by comparing the J. regia chloroplast to chloroplasts from 11 other species (eight species in the Fagales), downloaded from NCBI). A neighbour-joining analysis was performed using the software Geneious version 8.0.2 (http://www.geneious.com/) (Figure 1). The phylogenetic tree showed that J. regia (Juglandaceae) was most closely related to members of Fagaceae family and the genus Populus (Figure 1). The newly characterized J. regia complete chloroplast genome will provide essential data for further study on the phylogeny and evolution of the genus Juglans and of the Juglandaceae, for molecular breeding, and potentially for genetic engineering.
Figure 1.
Phylogenetic relationships among 12 chloroplast genomes. The 12 species can be divided into three independent clades: Fagales, Salicales and Brassicales. Arabidopsis thaliana (Brassicales) was used as an outgroup. Bootstrap support values are given at the nodes. Chloroplast genome accession number used in this phylogeny analysis: Arabidopsis thaliana: NC000932; Populus euphratica: NC024747; Populus trichocarpa: NC009143; Trigonobalanus doichangensis: KF990556; Quercus aquifolioides: NC026913; Quercus aliena: NC026790; Quercus rubra: JX970937; Quercus spinosa: NC026907; Lithocarpus balansae: KP299291; Castanea mollissima: NC014674; Castanopsise chinocarpa: NC023801.
Declaration of interest
This work was supported by the National Natural Science Foundation of China (No. 41471038, No. 31200500 and No. J1210063), the Program for Excellent Young Academic Backbones funding by Northwest University. Mention of a trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture and does not imply its approval to the exclusion of other products or vendors that also may be suitable.
References
- Aradhya MK, Potter D, Gao F, Simon CJ.. 2007. Molecular phylogeny of Juglans (Juglandaceae): a biogeographic perspective. Tree Genet Genom. 3:363–378. [Google Scholar]
- Chen LN, Ma QG, Chen YK, Wang BQ, Pei D.. 2014. Identification of major walnut cultivars grown in China based on nut phenotypes and SSR markers. Sci Hortic. 168:240–248. [Google Scholar]
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X.. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics 13:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Hou M, Du FK, Li J, Yin K.. 2015. Complete chloroplast genome of the Oriental white oak: Quercus aliena Blume. Mitochondrial DNA 1–3. DOI: 10.3109/19401736.2015.1053074. [DOI] [PubMed] [Google Scholar]
- Manning WE. 1978. The classification within the Juglandaceae. Ann Mo Bot Gard. 65:1058–1087. [Google Scholar]
- Neuhaus HE, Emes MJ.. 2000. Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol. 51:111–140. [DOI] [PubMed] [Google Scholar]
- Pollegioni P, Woeste KE, Chiocchini F, Lungo SD, Olimpier I, Virginia T, Clark J, Hemery GE, Mapelli S, Malvolti ME. 2015. Ancient humans influenced the current spatial genetic structure of common walnut populations in Asia. PLoS One 10:e0135980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Woeste KE.. 2011. DNA markers identify hybrids between butternut (Juglans cinerea L.) and Japanese awlnut (Juglans ailantifolia Carr.). Tree Genet Genom. 7:511–533. [Google Scholar]