Abstract
Neural stem cells (NSCs) in the developing and postnatal brain have distinct positional identities that dictate the types of neurons they generate. Although morphogens initially establish NSC positional identity in the neural tube, it is unclear how such regional differences are maintained as the forebrain grows much larger and more anatomically complex. We found that the maintenance of NSC positional identity in the murine brain requires a mixed-lineage leukemia 1 (Mll1)–dependent epigenetic memory system. After establishment by sonic hedgehog, ventral NSC identity became independent of this morphogen. Even transient MLL1 inhibition caused a durable loss of ventral identity, resulting in the generation of neurons with the characteristics of dorsal NSCs in vivo. Thus, spatial information provided by morphogens can be transitioned to epigenetic mechanisms that maintain regionally distinct developmental programs in the forebrain.
One Sentence Summary:
Epigenetic memory of positional identity in NSCs.
Introduction
Early in embryonic development, morphogens provide positional information to NSCs, inducing the expression of region-specific transcription factors (1–3). The morphogen sonic hedgehog (SHH) drives ventral NSC identity across the neural tube, including the telencephalon, where it induces the expression of ventral transcription factors such as NK homeobox 2 (Nkx2–1) (4). NSCs of the postnatal ventricular-subventricular zone (V-SVZ) also have distinct positional identities along the dorsal-ventral axis (5) and arise from embryonic NSCs before embryonic day 13.5 (E13.5) (6, 7). V-SVZ NSCs maintain the positional information of their embryonic precursors, including the expression of region-specific transcription factors, throughout adulthood (6), making them a well-suited model system in which to study the mechanisms underlying long-term maintenance of positional identity. In particular, embryonic NSCs in the ventrally located medial ganglionic eminence (MGE) express Nkx2–1 and give rise to the population of Nkx2–1+ NSCs in the postnatal ventral V-SVZ (8, 9) (Fig. 1A).
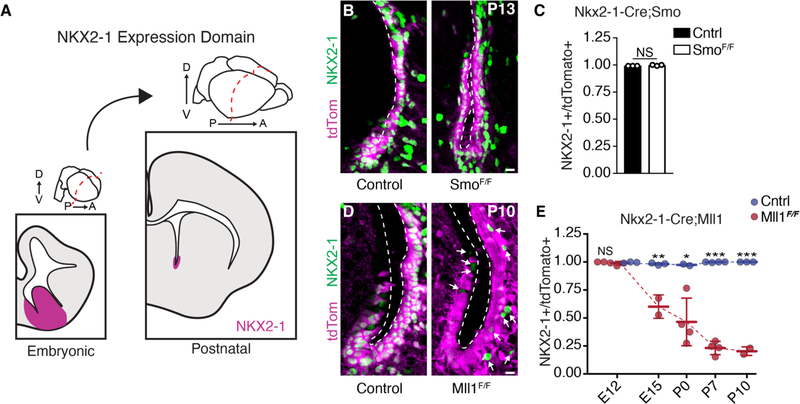
Fig. 1. Mll1, but not SHH signaling, is required to maintain NKX2–1 expression in postnatal V-SVZ.
(A) NKX2–1 expression in the germinal zones of embryonic forebrain and postnatal V-SVZ. NKX2–1 domain shown in magenta. D, dorsal; V, ventral; P, posterior; A, anterior. (B) Representative images of NKX2–1 (green) and tdTomato (magenta) expression in P13 coronal sections from ventral V-SVZ of Nkx2–1-Cre;SmoF/+ and Nkx2–1-Cre;SmoF/F mice. Ventricular walls are demarcated by a dashed line.
(C) Quantification of NKX2–1+;tdTomato+ double-positive V-SVZ cells in Nkx2–1-Cre:SmoF/+ (control) and Nkx2–1-Cre:SmoF/F animals at P13 (mean ± SD; NS, not significant; two-tailed student’s t test).
(D) Representative images of NKX2–1 (green) and tdTomato (magenta) expression in ventral V-SVZ of P10 Nkx2–1-Cre;Mll1F/+ and Nkx2–1-Cre;Mll1F/F mice. Ventricular walls are demarcated by a dashed line. Arrows indicate tdTomato−;NKX2–1+ cells. (E) Quantification of NKX2–1+;tdTomato+ double-positive V-SVZ cells from Nkx2–1-Cre:Mll1F/+ (control) and Nkx2–1-Cre:Mll1F/F mice over the course of development (mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed student’s t test). Scale bars, 10 mm [(B) and (D)].
MLL1, not SHH, maintains NKX2–1 expression in NSCs in vivo
Although it has been hypothesized that ongoing morphogen signaling is required to maintain the positional identity of NSCs (2, 10), other studies have suggested that NSC positional identity is maintained in a cell-autonomous fashion (5). Around E8.0, SHH signaling induces the expression of Nkx2–1 in the ventral neural tube (11), and deletion of Shh at ~E10 reduces NKX2–1 expression in the dorsal aspect of the E12.5 MGE (12). To determine whether ongoing SHH signaling was required to maintain Nkx2–1 expression in V-SVZ NSCs, we used the Nkx2–1-Cre driver to delete conditional alleles of the SHH receptor Smoothened (SmoF) and followed the fate of recombined cells with a tdTomato transgenic Cre reporter (Ai14). Although deletion of Smo at ~E8.25 has been shown to abolish the initiation of Nkx2–1 expression in ventral NSCs (13), the majority (99.7 ± 0.45%, n = 3 animals) of tdTomato+ cells in the postnatal V-SVZ of Nkx2–1-Cre; SmoF/F;Ai14 mutant mice remained NKX2–1+ (Fig. 1, B and C), as did Nkx2–1-Cre:SmoF/+ controls (99.2 ± 0.01%, n = 3 animals, P = 0.10). These data indicate that once Nkx2–1 is expressed, canonical SHH signaling is no longer required to maintain the Nkx2–1+ population of ventral V-SVZ precursors, and instead they are maintained by another mechanism.
Mixed-lineage leukemia 1 (Mll1) is the mammalian homolog of the Drosophila gene trithorax, and these genes are required to maintain—but not induce—the proper regional expression of Hox cluster genes that determine the anterior-posterior body plan during early embryogenesis (14–16). The vertebrate telencephalon develops from the most anterior aspect of the neural tube, and telencephalic NSCs remain regionally distinct throughout development and into postnatal life (9, 17, 18). To investigate whether Mll1 is required to maintain Nkx2–1 in ventral NSCs through embryonic and postnatal development, we used the Nkx2–1-Cre driver to delete conditional alleles of Mll1 (Mll1F) (19). Nkx2–1-Cre;Mll1F/F;Ai14 animals were born at expected Mendelian frequencies but died by postnatal day 13 (P13). Nkx2–1-Cre;Mll1F/F; Ai14 mutants exhibited a progressive loss of NKX2–1 in tdTomato+ cells as compared with Nkx2–1-Cre;Mll1F/+ controls beginning at E15.5 and reached an 80% reduction by P10 (Fig. 1, D and E, and fig. S1A)—the decrease occurring in a posterior-to-anterior gradient over this developmental period (fig. S1, B and C). In Nkx2–1-Cre;Mll1F/F;Ai14 mutants, the expression of the cell cycle marker Ki67 in tdTomato+ cells was not different from that in controls (fig. S1, D and E), indicating that changes in NSC proliferation do not account for the decrease in NKX2–1+/tdTomato+ cells. Mice with germline null mutation of Mll1 at E10 had normal expression of NKX2–1 in ventral NSCs (fig. S1F), indicating that Mll1 is not required for the initial expression of Nkx2–1. Thus, Mll1 is required for the maintenance, but not the initial induction, of Nkx2–1 in vivo.
Mll1−/− NSCs adopt a dorsal-like transcriptional profile
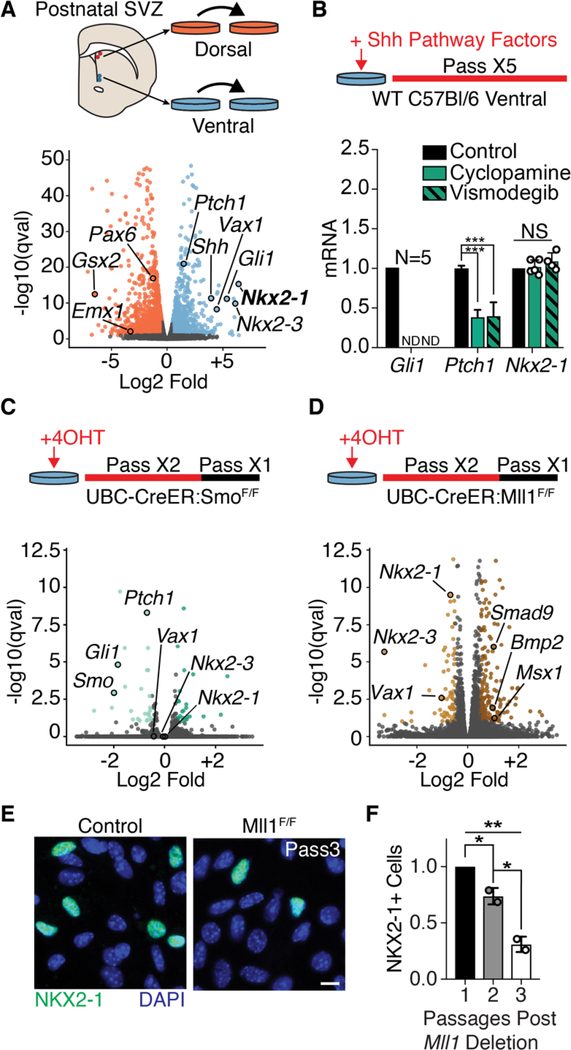
To further investigate the mechanisms that maintain NSC positional identity, we used postnatal V-SVZ NSC cultures that maintain their positional identity through serial passage in vitro (5, 20) (Fig. 2A). Dorsally derived NSC cultures had higher expression of transcription factors found in the dorsal V-SVZ in vivo—e.g., paired box 6 (Pax6), empty spiracles homeobox 1 (Emx1), and GS homeobox 2 (Gsx2)—and ventral NSC cultures were enriched for factors such as Nkx2–1, ventral anterior homeobox 1 (Vax1), and NK2 homeobox 3 (Nkx2–3) (Fig. 2A and data S1). Ventral NSCs expressed Shh as well as patched 1 (Ptch1) and GLI-kruppel family member GLI1 (Gli1) (Fig. 2A), two genes that are downstream transcriptional readouts of SHH signaling. While the addition of Smo pharmacological inhibitors cyclopamine or vismodegib reduced Gli1 and Ptch1 mRNA levels, the expression of Nkx2–1 remained unchanged, as assessed by quantitative polymerase chain reaction (qPCR) (Fig. 2B) and immunocytochemistry (fig. S2, A and B). Sequestration of SHH ligand by the 5E1 antibody resulted in decreased expression of Ptch1 and Gli1 but did not affect Nkx2–1 expression (fig. S2, C to E). We also genetically disrupted SHH signaling by deriving ventral V-SVZ cultures from SmoF/F mice carrying the UBC-Cre-ERT2 transgene (21) and adding 4-hydroxytamoxifen (4-OHT) to induce recombination. RNA sequencing (RNA-seq) analysis of Smo-deleted cultures identified 59 differentially expressed genes (27 down-regulated, 32 up-regulated) as com- pared with UBC-CreERT2;SmoF/+ controls (Fig. 2C, fig. S2F, and data S2). With Smo deletion, Smo, Gli1, and Ptch1 were all significantly down-regulated, but the expression of Nkx2–1, Vax1, and Nkx2–3 remained unchanged. Thus, neither pharmacological nor genetic disruption of SHH signaling reduces the expression of transcriptional markers of ventral V-SVZ NSCs.
Fig. 2. Mll1-dependent NSC transcriptional identity does not require SHH signaling.
(A) Derivation of region-specific NSC cultures from the dorsal and ventral P7 mouse V-SVZ (top panel). Volcano plot of differential gene expression by RNA-seq in dorsal and ventral NSC cultures. Dorsal enriched genes, orange; ventral-enriched genes, light blue. (B) Pharmacological SHH pathway inhibition assay timeline (top panel). qPCR from ventral V-SVZ NSC cultures exposed to SHH pathway inhibitors (mean ± SD, ***P < 0.001, two-tailed student’s t test; ND, not detected). Gli1 and Nkx2–1 analysis from 4 to 5 biological replicates, Ptch1 in technical triplicate. (C) Schematic of ubiquitous Smo deletion from ventral NSC cultures by 4-OHT administration (top). Volcano plot of differential gene expression by RNA-seq in ventral UBC-CreER; SmoF/+ and UBC-CreER; SmoF/F NSC cultures. Genes up-regulated, dark green; down-regulated, light green; not significantly changed, gray. (D) Schematic of ubiquitous Mll1 deletion from ventral NSC cultures by 4-OHT administration (top). Volcano plot of differential gene expression by RNA-seq in ventral UBC-CreER;Mll1F/+ and UBC-CreER; Mll1F/F NSC cultures. Genes up-regulated, dark brown; down-regulated, light brown; not significantly changed, gray. (E) Representative image of NKX2–1+ cells in ventral UBC-CreER;Mll1F/+ and UBC-CreER;Mll1F/F NSC cultures. Scale bar, 10 mm. (F) Quantification of NKX2–1+ cells after in vitro deletion of Mll1 by UBC-CreER relative to baseline at first passage (mean ± SD, *P < 0.05, **P < 0.01, one-way analysis of variance with Tukey’s multiple comparison test).
To study Mll1 deletion in ventral V-SVZ NSCs, we derived cultures from UBC-CreERT2;Mll1F/F mice. After 4-OHT–induced deletion of Mll1 (fig. S2G), we performed RNA-seq, identifying 272 differentially expressed genes (81 down- regulated, 191 up-regulated) as compared with UBC-CreERT2;Mll1F/+ controls (Fig. 2D and data S3). Genes down-regulated in Mll1-deleted cultures were almost entirely distinct from those down-regulated in Smo-deleted cultures (fig. S2H). Whereas neither Gli1 nor Ptch1 was decreased by Mll1-deletion (fig. S2I), the expression of ventral genes including Nkx2–1, Vax1, and Nkx2–3 was down-regulated (Fig. 2D and fig. S2J). Similar to the progressive loss of NKX2–1 observed in vivo (Fig. 1E), the proportion of NKX2–1+ cells in Mll1-deleted cultures decreased with serial passage (Fig. 2, E and F) without differences in cell proliferation, as measured by incorporation of the thymidine analog 5-ethynyl-2’-deoxyuridine (fig. S2K) or cell death (fig. S2L). SHH pathway inhibition did not increase the rate or magnitude of Nkx2–1 loss after Mll1-deletion (fig. S3, A to D), indicating that the gradual nature of Nkx2–1 loss was not due to persistent SHH signaling. Instead, inhibition of cell proliferation with either mitomycin C or colchicine (fig. S3, E and F) attenuated Nkx2–1 loss in Mll1-deleted cells over time (fig. S3, G to I), suggesting that the progressive loss of Nkx2–1 relates to serial cell division. In Mll1-deleted ventral NSCs, we also observed increased levels of dorsally enriched genes including limb and CNS expressed 1 (Lix1) as well as bone morphogenetic protein (BMP) pathway genes such as bone morphogenic protein 2 (Bmp2), SMAD family member 9 (Smad9), and inhibitor of DNA binding 3 (Id3) (Fig. 2D and data S3). BMP signaling has multiple developmental functions including the dorsalization of the telencephalon (22), leading to the expression of the dorsal marker Msx1 (23), which was also up-regulated in Mll1-deleted NSCs (Fig. 2D). However, inhibition of BMP signaling with LDN-193189 (24, 25) in Mll1-deleted cells did not prevent the loss of Nkx2–1 expression (fig. S3, J to M), suggesting that the loss of this aspect of ventral identity is not secondary to increased BMP signaling. Together, these results indicate that, in addition to losing the expression of ventral markers, ventral NSCs also gain some transcriptional characteristics of dorsal NSCs after the deletion of Mll1.
Ventral NSC transcriptional identity requires continuous MLL1 function
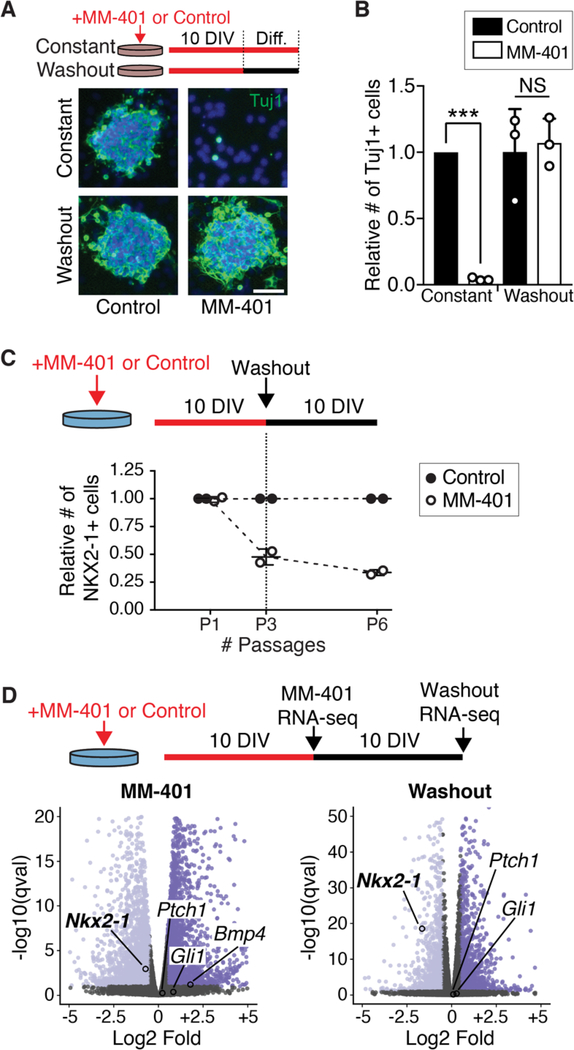
MM-401 is a small-molecule drug that specifically blocks MLL1 function and is reversible upon drug removal (26) (hereafter referred to as washout). Deletion of Mll1 inhibits neurogenesis from V-SVZ NSCs (27), and consistent with these prior results, the presence of MM-401 in ventral V-SVZ cultures abolished the production of new neurons in culture (Fig. 3A). After MM-401 washout, NSCs produce the same number of new neurons as control-treated cells (Fig. 3B and fig. S4A) with no difference in cell death or proliferation (fig. S4, B and C).
Fig. 3. Ventral NSC transcriptional identity requires continuous MLL1 function.
(A) Treatment timeline of V-SVZ NSCs with the MLL1-specific inhibitor MM-401 (or vehicle control) under proliferative conditions for 10 days in vitro (DIV), followed by differentiation (Diff.) with or without drug washout (top). Representative images of Tuj1 (green) and DAPI (4′,6-diamidino-2- phenylindole, blue) after differentiation. Scale bar, 50 mm. (B) Quantification of Tuj1+ cells after differentiation of MM-401– treated NSCs (mean ± SD, ***P < 0.001, two-tailed student’s t test). (C) Treatment timeline of ventral V-SVZ NSCs with MM-401 and subsequent washout under proliferative conditions (top). Quantification of NKX2–1+ cells normalized to control-treated group at each time point. (D) Volcano plot of differential gene expression by RNA-seq in ventral NSCs treated with MM-401 (left) and then after washout (right) under proliferative conditions. Genes upregulated, dark purple; down-regulated, light purple; not significantly changed, gray.
Although transient MLL1 inhibition did not impair the ability of V-SVZ NSCs to differentiate into neurons in vitro, we considered the possibility that NSC positional identity might have been durably affected. Consistent with Mll1 deletion in ventral NSCs (Figs. 1, D and E, and 2, D to F), including those from adult mice (fig. S4, D and E), the presence of MM-401 reduced the proportion of NKX2–1+ cells as compared with control-treated cultures (Fig. 3C), without changes to the expression levels of Gli1 and Ptch1 (fig. S4C). Even after 10 days of MM-401 washout (five serial passages), the expression of Nkx2–1 did not recover (Fig. 3D and fig. S4, F and G). Similarly, transient inhibition of MLL1 in ventral NSCs from adult (P60) mice also resulted in stable loss of Nkx2–1 expression (fig. S4, D and H), suggesting that MLL1 activity is required for NSC positional identity throughout life. In addition to Nkx2–1, ~28% of the genes down-regulated in the presence of MM-401 remained down-regulated after drug washout, as revealed by RNA-seq (data S4 and S5). Among the genes that were down-regulated in the presence of MM-401, Nkx2–1 was the most down-regulated after washout of MM-401 (fig. S4G). Although it has been suggested that NKX2–1 protein can autoregulate its own expression (28, 29) (fig. S5A), in ventral NSCs in vivo, active transcription of the Nkx2–1 locus did not require NKX2–1 protein (fig. S5, B to D). Thus, continuous MLL1 function is required for the transcriptional memory of markers of ventral identity in proliferating NSC cultures.
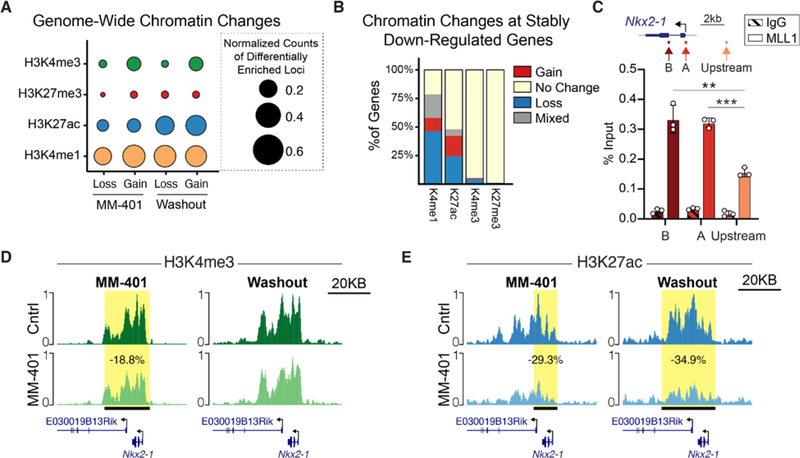
MLL1 can regulate chromatin via its methyltransferase domain, recruitment of acetyltransferases, and antagonism of polycomb repressive factors (30–36). We therefore performed genome-wide analysis of modifications related to histone methylation (H3K4me3 and H3K4me1), acetylation (H3K27ac), and poly- comb factor repression (H3K27me3) using Cleavage Under Targets and Release Using Nuclease (CUT&RUN) (37). In ventral V-SVZ cultures, H3K4me1, a mark associated with transcriptional enhancers, and H3K27ac, a mark associated with active enhancers and promoters, were the most commonly decreased chromatin marks during MM-401 treatment (Fig. 4A). These changes corresponded to the differential gene expression observed with this ongoing inhibition of MLL1 activity (fig. S6A). Ten days after MM-401 washout, the genome-wide landscape of NSCs remained enriched for changes of H3K4me1 and H3K27ac levels (Fig. 4A). Fifty-three percent of loci with decreased H3K4me1 and 51% of loci with decreased H3K27ac under MLL1 inhibition remained decreased after MM-401 washout (fig. S6B). This “stable” loss of H3K4me1 at putative enhancer sequences correlated significantly with stably down-regulated gene expression (fig. S6C). Within gene bodies, the stable loss of H3K27ac also corresponded to stably down-regulated gene expression (fig. S6D). Focusing on genes that remained down-regulated after MM-401 washout, we found that decreases in H3K4me1 and/or H3K27ac enrichment were the most common chromatin changes (Fig. 4B).
Fig. 4. Chromatin state changes to ventral NSCs after transient MLL1 inhibition.
(A) Genome-wide quantification of chromatin changes in MM-401–treated and MM-401–washout ventral NSC cultures. Circle size represents the number of sites that have either a gain or loss of a chromatin mark in the treatment group compared with control, normalized to the size of genome covered by statistically called peaks for that chromatin mark (X10,000). (B) Quantification of chromatin changes within stably down-regulated genes after MM-401 washout. (C) Locations of qPCR primers at the Nkx2–1 locus (top). ChIP-qPCR analysis of MLL1 enrichment at Nkx2–1 (mean ± SD, **P < 0.01, ***P < 0.001, two-tailed student’s t test, in technical triplicate). IgG, immunoglobulin G; kb, kilobases. (D and E) CUT&RUN traces of (D) H3K4me3 and (E) H3K27ac enrichment over the Nkx2–1 locus in control- and MM-401–treated cultures (left) and after 10 days of washout (right). Horizontal black bar with yellow highlight depicts genomic region with statistically significant differential chromatin enrichment.
At the Nkx2–1 locus, MLL1 protein was enriched near the transcriptional start site, as indicated by chromatin immunoprecipitation (ChIP) analysis (Fig. 4C). Treatment with MM-401 resulted in a small but statistically significant decrease (18.8%, Padj = 0.009) in H3K4me3 enrichment across the Nkx2–1 gene body (Fig. 4D, left, and fig. S6D). However, this decrease in H3K4me3 was transient as there was no difference in H3K4me3 enrichment detected after the washout of MM-401 (Fig. 4D, right, and fig. S6D). In contrast, we observed a durable loss of H3K27ac enrichment across the Nkx2–1 gene body both during MM-401 treatment (Fig. 4E, left, and fig. S6E) and after washout of the inhibitor (Fig. 4E, right, and fig. S6E). Together, these results indicate that transient MLL1 inhibition leads to persistent changes to the chromatin landscape that relate to stable differential gene expression.
Transient MLL1 inhibition alters developmental fate of ventral NSCs
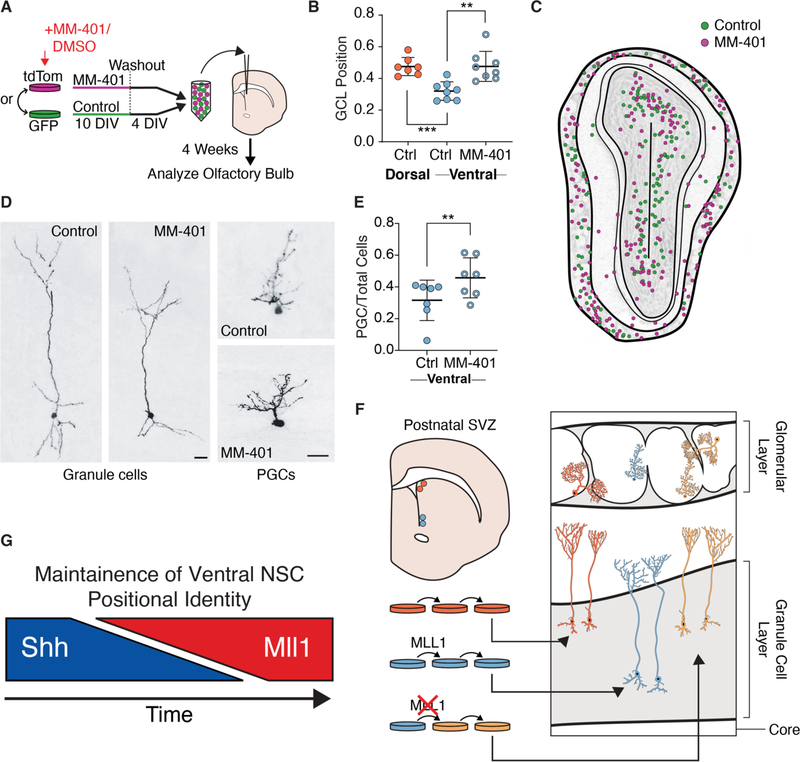
To investigate whether transient inhibition of MLL1 activity affected the developmental potential of ventral NSCs, we studied the fate of their daughter cells after transplantation back to the V-SVZ. Throughout life, the V-SVZ produces new neurons for the olfactory bulb, and NSC positional identity underlies which olfactory bulb neuronal subtypes are generated (5). In vivo, dorsal NSCs generate superficial granule cells, whereas ventral NSCs generate deep granule cells (5, 9), and such differences in developmental potential are maintained in vitro even after transplantation to ectopic V-SVZ locations (5) (e.g., ventral NSCs still generate deep granule cells when grafted to the dorsal V-SVZ). Thus, deep and superficial granule cells are distinct neuronal subtypes that arise from different populations of NSCs with distinct positional identities related to their location in the walls of the V-SVZ. We treated ventral NSC cultures with MM-401 or vehicle control for 10 days, after which the cells were cultured under washout conditions for 4 days before transplantation to the V-SVZ of adult mice (Fig. 5A). MM-401-washout and control NSCs were labeled with either tdTomato or green fluorescent protein (GFP) and mixed together in equal numbers before being cotransplanted into the V-SVZ. In experiments performed in parallel, consistent with previous findings (5), control NSCs from the dorsal V-SVZ generated granule cell neurons more superficial than those generated from ventral V-SVZ NSCs (Fig. 5B). Four weeks post-transplantation, the number of olfactory bulb neurons derived from MM-401-washout and control NSCs was similar (fig. S7A), again indicating that transient MLL1 inhibition does not impair the ability of NSCs to differentiate into neurons (Fig. 3, A and B). Furthermore, graft-derived neurons had normal morphologies and were dispersed throughout the olfactory bulb (Fig. 5, C and D, and fig. S7, B and C). However, instead of producing deep granule cells, MM-401-washout ventral NSCs produced granule cells whose location was similar to those born from dorsal NSC controls (Fig. 5B). MM-401-washout cells also generated a greater number of periglomerular cells (Fig. 5E), an olfactory bulb neuron subtype preferentially born from the anterior V-SVZ (5). Thus, after transient MLL1 inhibition, the normal developmental potential of ventral V-SVZ NSCs is altered, producing neuronal subtypes more typical of an anterior-dorsal NSC identity (Fig. 5F).
Fig. 5. Transient MLL1 inhibition alters NSC positional identity.
(A) Derivation and treatment of NSC cultures labeled with GFP or tdTomato for transplantation into adult V-SVZ. (B) Granule cell layer (GCL) position of transplant-derived olfactory bulb (OB) GC interneurons per animal (mean ± SD, **P < 0.01, ***P < 0.001, two-tailed student’s t test, three separate rounds of transplantation). Dorsal-derived cells, orange; ventral-derived cells, light blue; control-treated cells, solid circles; MM-401–treated cells, rings. (C) Map of ventral NSC transplant-derived neurons within OB, pooled from two animals. MM-401–treated cells (magenta) and control cells (green). (D) Representative GC and periglomerular (PGC) OB interneurons generated by transplanted ventral NSCs. Scale bars, 20 mm. (E) Relative proportion of transplant-derived PGCs produced by control and MM-401–treated ventral NSCs (mean ± SD, **P < 0.01, two-tailed student’s t test). (F) Schematized results of MM-401–mediated MLL1 inhibition followed by washout and transplantation. GC interneurons derived from transplanted NSCs are color coded to indicate positional identity: dorsal, orange; ventral, light blue; “dorsal-like,” yellow. (G) Generalized model of the regulation of ventral NSC positional identity over the course of development, where SHH signaling is initially required but later shifts to a state of MLL1-dependence.
Outlook
Our results support a model in which MLL1 functions as part of a transcriptional memory system in telencephalic NSCs, retaining key aspects of NSC positional identity that underlie the generation of proper neuronal diversity. After ventral NSC identity is established by SHH in the early forebrain, MLL1 is required to maintain this positional information (Fig. 5G). As key aspects of ventral NSC identity are not lost by inhibition of SHH signaling, we propose that the maintenance of regional differences in NSCs during telencephalic development can become independent of the morphogen gradients that initially pattern the neural tube. MLL1 can remain localized to target genes through mitosis, allowing for the rapid reestablishment of transcription after cell cycle exit (38, 39). In ventral NSCs, MLL1 may be targeted to loci by SHH-induced transcription factors, with MLL1 remaining localized over serial cell divisions, resulting in persistent chromatin-state changes even after SHH withdrawal.
The mechanism(s) by which chromatin regulators maintain specific patterns of gene expression over serial cell division remain poorly understood. We found persistent, genome-wide changes in H3K4me1 and H3K27ac after transient inhibition of MLL1 (Fig. 4A). MM-401 blocks the interaction between MLL1 and WD repeat-containing protein 5 (WDR5), which disrupts the localization of MLL1 to chromatin (27). A lack of proper MLL1 localization may lead to both the local loss of H3K4me1, which is one of the histone modifications catalyzed by MLL1 (34), and loss of H3K27ac, which is catalyzed by CREB (cAMP response element–binding protein) binding protein (CBP), a histone acetyltransferase that interacts directly with MLL1 (35, 36). Whereas many loci exhibited persistent changes of H3K4me1 and H3K27ac after MM-401 washout, losses of H3K4me3 enrichment were relatively few and mostly transient, suggesting that other histone methyltransferases can compensate for loss of MLL1 activity. Rather than acting solely through its methyltransferase function, localized MLL1 protein may serve as a genomic “bookmark” for the reestablishment of gene expression after mitosis (38, 39).
This model provides an alternative understanding of how the boundaries for populations of regionally distinct NSCs are maintained as the cells continue to proliferate—expanding the germinal zones by many orders of magnitude— and while the brain undergoes complex morphological changes. Our finding that MLL1 plays a role in the positional identity of NSCs may inform future methods for producing specific neuronal subtypes from NSCs in vitro for clinical or research purposes or provide additional insights into the neurodevelopmental defects of Wiedemann-Steiner syndrome, which is caused by mutations in MLL1 (40).
Supplementary Material
Acknowledgements:
We would like to acknowledge Cristina Guinto for her help and expertise in transplantation experiments.
Funding: Supported by NIH grant 1R01NS091544, VA grant 5I01 BX000252, the Shurl and Kay Curci Foundation, the LoGlio Foundation, and the Hana Jabsheh Initiative to (D.A.L.). Work in the Alvarez-Buylla laboratory is supported by NIH grants R01 NS028478 and a generous gift from the John G. Bowes Research Fund. A.A.-B. is the Heather and Melanie Muss Endowed Chair and Professor of Neurological Surgery at UCSF.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: All datasets generated from this study are available on the GEO repository under the accession number GSE145871.
References and Notes:
- 1.Ericson J, Briscoe J, Rashbass P, van Heyningen V, Jessell TM, Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. Cold Spring Harbor symposia on quantitative biology 62, 451–466 (1997). [PubMed] [Google Scholar]
- 2.Fuccillo M, Joyner AL, Fishell G, Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nature reviews. Neuroscience 7, 772–783 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Sur M, Rubenstein JL, Patterning and plasticity of the cerebral cortex. Science 310, 805–810 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Ericson J et al. , Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell 81, 747–756 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Merkle FT, Mirzadeh Z, Alvarez-Buylla A, Mosaic organization of neural stem cells in the adult brain. Science 317, 381–384 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Fuentealba LC et al. , Embryonic Origin of Postnatal Neural Stem Cells. Cell 161, 1644–1655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merkle FT, Tramontin AD, Garcia-Verdugo JM, Alvarez-Buylla A, Radial glia give rise to adult neural stem cells in the subventricular zone. Proceedings of the National Academy of Sciences of the United States of America 101, 17528–17532 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delgado RN, Lim DA, Embryonic Nkx2.1-expressing neural precursor cells contribute to the regional heterogeneity of adult V-SVZ neural stem cells. Developmental biology 407, 265–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young KM, Fogarty M, Kessaris N, Richardson WD, Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 8286–8296 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ihrie RA et al. , Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron 71, 250–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL, Longitudinal organization of the anterior neural plate and neural tube. Development 121, 3923–3933 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Xu Q, Wonders CP, Anderson SA, Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development 132, 4987–4998 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Fuccillo M, Rallu M, McMahon AP, Fishell G, Temporal requirement for hedgehog signaling in ventral telencephalic patterning. Development 131, 5031–5040 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Geisler SJ, Paro R, Trithorax and Polycomb group-dependent regulation: a tale of opposing activities. Development 142, 2876–2887 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G, Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ, Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Alvarez-Buylla A, Kohwi M, Nguyen TM, Merkle FT, The heterogeneity of adult neural stem cells and the emerging complexity of their niche. Cold Spring Harbor symposia on quantitative biology 73, 357–365 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Delgado RN, Lim DA, Maintenance of Positional Identity of Neural Progenitors in the Embryonic and Postnatal Telencephalon. Front Mol Neurosci 10, 373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jude CD et al. , Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell stem cell 1, 324–337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delgado RN, Lu C, Lim DA, Maintenance of neural stem cell regional identity in culture. Neurogenesis 3, e1187321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruzankina Y et al. , Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell stem cell 1, 113–126 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bond AM, Bhalala OG, Kessler JA, The dynamic role of bone morphogenetic proteins in neural stem cell fate and maturation. Developmental neurobiology 72, 1068–1084 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuta Y, Piston DW, Hogan BL, Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 124, 2203–2212 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Cuny GD et al. , Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett 18, 4388–4392 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers SM et al. , Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol 30, 715–720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao F et al. , Targeting MLL1 H3K4 Methyltransferase Activity in Mixed-Lineage Leukemia. Molecular cell 53, 247–261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim DA et al. , Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das A et al. , Thyroid transcription factor-1 (TTF-1) gene: identification of ZBP-89, Sp1, and TTF-1 sites in the promoter and regulation by TNF-alpha in lung epithelial cells. American journal of physiology. Lung cellular and molecular physiology 301, L427–440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oguchi H, Kimura S, Multiple transcripts encoded by the thyroid-specific enhancer-binding protein (T/EBP)/thyroid-specific transcription factor-1 (TTF-1) gene: evidence of autoregulation. Endocrinology 139, 1999–2006 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G, Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171, 34–57 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ, MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol 21, 2249–2258 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milne TA et al. , MLL targets SET domain methyltransferase activity to Hox gene promoters. Molecular cell 10, 1107–1117 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Dou Y et al. , Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13, 713–719 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Patel A, Dharmarajan V, Vought VE, Cosgrove MS, On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. The Journal of biological chemistry 284, 24242–24256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Guzman RN, Goto NK, Dyson HJ, Wright PE, Structural basis for cooperative transcription factor binding to the CBP coactivator. J Mol Biol 355, 1005–1013 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Cosgrove MS, Patel A, Mixed lineage leukemia: a structure-function perspective of the MLL1 protein. FEBS J 277, 1832–1842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skene PJ, Henikoff S, An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blobel GA et al. , A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Molecular cell 36, 970–983 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bina M et al. , Discovery of MLL1 binding units, their localization to CpG Islands, and their potential function in mitotic chromatin. BMC Genomics 14, 927 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones WD et al. , De novo mutations in MLL cause Wiedemann-Steiner syndrome. American journal of human genetics 91, 358–364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.