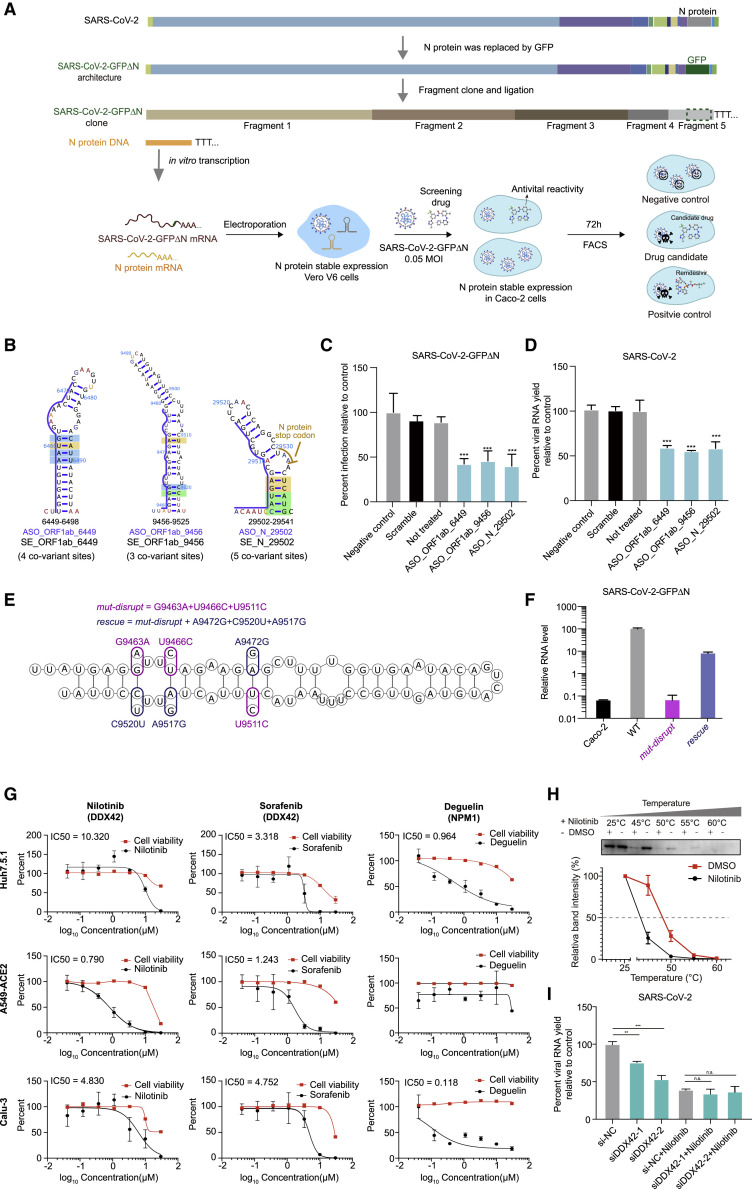
Figure 7.
Validation of ASO and compounds with antiviral activity targeting conserved RNA structural elements and predicted RBPs using a SARS-CoV-2 N trans-complementation system and the bona fide virus
(A) Schematic for a SARS-CoV-2 N trans-complementation system that uses a pseudo SARS-CoV-2 virus in which the sequence encoding viral protein N is replaced with GFP (SARS-CoV-2-GFPΔN). SARS-CoV-2-GFPΔN only amplifies in Caco-2 cells that are actively expressing (complementing) the required viral N protein. We infected Caco-2 cells with MOI of 0.05. Drugs/compounds were added at the same time, at a concentration of 10 μM. SARS-CoV-2-GFPΔN accumulation was detected by FACS after 3 days.
(B) Three conserved structural elements within the ORF1ab and N coding regions. Blue lines indicate the binding sites of ASOs. Nucleotides are colored by icSHAPE reactivity scores, with red and yellow colors indicating reactive nucleotides. Nucleotides with a color background were predicted as co-variant base pairs.
(C and D) The infection ratios of SARS-CoV-2-GFPΔN in Caco-2 cells (C) and the viral yield for bona fide SARS-CoV-2 in Huh7.5.1 cells (D) both decreased upon treatment with an ASO targeting conserved structures compared to controls, including a “Negative control” control treated with an ASO targeting the ORF1ab protein coding region (16,114––16,168 nt) which has a long stem but no conserved covariation, a “Scramble” control treated with a non-targeting ASO, and a “Not treated” control with no ASO treatment. Data represent the mean ± SEM; n = 3 biological replicates. n.s., not significant. ∗∗∗p < 0.005, ∗∗p < 0.01, and ∗p < 0.05 using one-way ANOVA and post hoc Student’s t test.
(E) Secondary structure model of SE_ORF1ab_6449 with designed mutations. Purple circles represent designed mutations for mut-disrupt; the rescue includes both the mut-disrupt mutations and the mutations indicated with pink circles.
(F) qPCR quantitation of relative viral RNA level from pellets of Caco-2 cells infected for 48 h. The Caco-2 cells without infection were used as the negative control. Data represent the mean ± SEM; n = 3 biological replicates.
(G) qPCR quantification of viral titers for equal volume supernatant from Huh7.5.1 cells (top), A549 cell with ACE2 protein stable expression (middle), and Calu3 cells (bottom), infected with the bona fide SARS-CoV-2 virus (MOI = 0.05), 48 h post infection. Drug concentrations ranged from 0.04 μM to 30 μM. Dose-response curves for infectivity (black) and cell viability (red) are shown. Data are normalized to the average of DMSO-treated samples (0.1%) and represent mean ± SEM for n = 3 independent experiments.
(H) CETSA curves for DDX42, with or without Nilotinib (100 μM), measured in cell lysates at the indicated temperatures. The black curve is the Nilotinib treatment, and the red curve is the negative control. Data represent the mean ± SEM; n = 3 biological repeats.
(I) The yield of SARS-CoV-2 in Huh7.5.1 cells after viral infection for 48 h (MOI = 0.05), with different siRNA transfection and with/without Nilotinib treatment as indicated. si-NC, non-targeting scramble siRNA; siDDX42-1 and siDDX42-2, two siRNAs targeting DDX42. Data are normalized to the si-NC control and represent the mean ± SEM; n = 3 biological replicates.
n.s., not significant. ∗∗∗p < 0.005, ∗∗p < 0.01, and ∗p < 0.05 using one-way ANOVA and post hoc Student’s t test.