Abstract
The worldwide demand for SARS-CoV-2 RT-PCR testing resulted in a shortage of diagnostic kits. RNA extraction step constitutes a major bottleneck to perform diagnostic. The aim of this study was to assess performances of different extraction-free SARS-CoV-2 RT-PCR assays compared to a reference RT-PCR assay.
The panel of evaluation consisted of 94 samples: 69 positive and 25 negative for SARS-CoV-2 by reference RT-PCR. Three extraction-free RT-PCR assays were assessed: (i) PrimeDirect® Probe RT-qPCR Mix (Takara), (ii) PrimeScript®RT-PCR (Takara), and (iii) SARS-CoV-2 SANSURE®BIOTECH Novel Coronavirus (Sansure).
The overall sensitivity of PrimeDirect, PrimeScript and Sansure assays was 55.1 %, 69.6 % and 69.6 %, respectively. The sensitivity increased among samples with Ct<30: 91.9 % (n = 34/37), 89.2 % (n = 33/37) and 94.6 % (n = 35/37) for PrimeDirect, PrimeScript and Sansure assays, respectively. The specificity was 88 %, 100 % and 100 % for PrimeDirect, PrimeScript and Sansure assays, respectively.
In the present study, we showed a good sensitivity of extraction-free PCR assays, especially for high viral loads (Ct<30), except PrimeDirect that displayed imperfect sensitivity and specificity. Despite a lower sensitivity for low viral loads, extraction-free reagents can provide a valuable option, cheaper, easier and less reagent consuming for SARS-CoV-2 diagnostic, especially in laboratory with lower experience and equipment for molecular assays.
Keywords: SARS-CoV-2, RT-PCR, Extraction-Free
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is commonly diagnosed by reverse transcription polymerase chain reaction (RT-PCR) to detect viral RNA in patient samples, but RNA extraction constitutes a major bottleneck in current testing. The massive demand for SARS-CoV-2 RT-PCR testing resulted in a worldwide shortage of diagnostic reagents including RNA extraction kits which is still a major challenge. To overcome the lack of reagents and to increase the capacities for SARS-CoV-2 testing various approaches like pooling strategies, direct RT-PCR without extraction step using primary material or isothermal methods were developed and numerous assays are currently available (Ganguli et al., 2020; Jalandra et al., 2020). One of the approach is to perform RT-PCR but skipping the extraction step. This approach confers several advantages like extraction step and reagents-sparing, analysis time-reducing and expanding the number of non-specialized laboratories able to perform COVID-19 diagnosis.
The aim of this study was to assess analytical performances of three different extraction-free SARS-CoV-2 RT-PCR assays compared to reference RT-PCR, including extraction step, on the same samples panel.
2. Methods
2.1. Naso-pharyngeal specimens
This evaluation was performed on nasopharyngeal swabs collected in Viral Transport Medium (VTM) sent to the Virology Laboratory of Bichat-Claude Bernard University Hospital, Paris, France, for testing SARS-CoV-2 RT-PCR as part of routine clinical care. The panel, constituted between March and July 2020, comprised 94 samples: 69 positive for SARS-CoV-2 and 25 negative for SARS-CoV-2. All specimens were aliquoted and stored at −20 °C.
2.2. SARS-Co-V-2 RT-PCR assays
The routine SARS-CoV-2 diagnostic was performed using two assays: (i) RealStar® SARS-CoV-2 RT-PCR Kit 1.0 (Altona diagnostics, France) (Visseaux et al., 2020), quoted after “Altona”, which can be associated to different extraction and amplification devices, and (ii) the Cobas® SARS-CoV-2 kit used on the Cobas® 6800 system (Cobas 6800; Roche Diagnostics, Mannheim, Germany) (Wirden et al., 2020) quoted after “Cobas”.
Three extraction-free RT-PCR assays were assessed. The PrimeDirect® Probe RT-qPCR Mix (TaKaRa, Kusatsu, Shiga, Japan), quoted after “PrimeDirect”, which is designed to skip the extraction step. The One-step PrimeScript® RT-PCR (TaKaRa), quoted after “PrimeScript”, initially designed after an extraction step. For both “PrimeDirect” and “PrimeScript” RT-PCR assays, we used primers and probes targeting E and RdRp SARS-CoV-2 genes, previously described by Paris Pasteur Institute. Primers and probe used were as follows for E gene: E-SARBECO-F1: ACAGGTACGTTAATAGTTAATAGCGT, E-SARBECO-R2: ATATTGCAGCAGTACGCACACA, and E-SARBECO P1 (FAM/BHQ1): ACACTAGCCATCCTTACTGCGCTTCG and for RdRp gene (IP2-12669-Fw: ATGAGCTTAGTCCTGTTG, IP2-12759-Rv: CTCCCTTTGTTGTGTTGT, IP2-12696-Probe (HEX/BHQ1) AGATGTCTTGTGCTGCCGGTA; IP4-14059-Fw: GGTAACTGGTATGATTTCG, IP4-14146-Rv: CTGGTCAAGGTTAATATAGG, IP4-14084-Probe (FAM/BHQ1) TCATACAAACCACGCCAGG).
We have also evaluated the SARS-CoV-2 SANSURE® BIOTECH Novel Coronavirus (Sansure Biotech, Changsha, China), quoted after “Sansure”, a ready-to-use commercial kit designed for extraction-free step, targeting N and ORF1ab SARS-CoV-2 genes (Table 1 ).
Table 1.
Sample pre-treatment and PCR cycling parameters with the different extraction-free RT-PCR assays for detection of SARS-CoV-2 virus in clinical specimens.
| 2019 nCoV kit SANSURE | PrimeDirect Probe RT-qPCR Mix TAKARA | One Step PrimeScript ™ III RT-qPCR Mix TAKARA | ||
|---|---|---|---|---|
| Pretreatment | Specimen volume | 10 μL | NA | 10 μL |
| Sample Release Reagent | 10 μL | NA | 40 μL NaCl | |
| Inactivation | 10 mn at room temperature | NA | NA | |
| PCR | Volume of sample or pretreated sample | 20 μL | 5 μL | 5 μL |
| Final master mix volume | 50 μL | 25 μL | 25 μL |
| Temperature | Time | Cycles | Temperature | Time | Cycles | Temperature | Time | Cycles | ||
|---|---|---|---|---|---|---|---|---|---|---|
| PCR conditions | Reverse transcription | 50 °C | 30 mn | 1 | 52°C | 5 mn | 1 | |||
| Initial activation | 95 °C | 1 mn | 1 | 90°C | 3 mn | 1 | 95 °C | 10 s | 1 | |
| Reverse transcription | 60 °C | 5 mn | 1 | |||||||
| Denaturation | 95 °C | 15 s | 45 | 95 °C | 5 s | 45 | 95 °C | 5 s | 45 | |
| Annealing/Extension | 60 °C | 30 sec | 45 | 58°C | 30 sec | 45 | 58°C | 30 sec | 45 | |
| Additional step | 25 °C | 10 s | 1 | |||||||
| Real-time PCR platforms | QuantStudio 5 | QuantStudio 5 and ABI 7500 | QuantStudio 5 | |||||||
| NA: Not Applicable | ||||||||||
For all extraction-free RT-PCR assays, nasopharyngeal swabs and PCR mix were directly deposited in the PCR tube, these manipulations and tubes lock were performed in a microbiological safety class II cabinet.
Samples pre-treatments steps were performed according to the manufacturer's recommendations.
2.3. Statistical analyses
Statistical analyses were performed using Microsoft Excel and the figure was realized with GraphPad Prism software.
2.4. Ethics
All samples were collected as part of routine clinical care and participants were not opposed to the collection of their data.
3. Results
3.1. Sensitivity and specificity of three extraction-free RT-PCR assays
Sensitivity and specificity of the three extraction-free RT-PCR assays were calculated taking as a reference the gold standard RT-PCR complete assay Altona or Roche (Table 2 ).
Table 2.
Performance characteristics of three extraction-free SARS-CoV-2 RT-PCR assays compared to a standard RT-PCR assay.
| Test | Sensitivity |
Specificity | ||
|---|---|---|---|---|
| Ct <30 | Ct >30 | Overall | ||
| PrimeDirect | 91.9 % (34/37) | 12.5 % (4/32) | 55.1 % (38/69) | 88 % (22/25) |
| PrimeScript | 89.2 % (33/37) | 46.9 % (15/32) | 69.6 % (48/69) | 100 % (25/25) |
| Sansure | 94.6 % (35/37) | 40.6 % (13/32) | 69.6 % (48/69) | 100 % (25/25) |
The overall sensitivity of the PrimeDirect assay was 55.1 % (n = 38/69, 95 %CI = 43.3–66.9). Stratifying according to the Cycle threshold (Ct), the sensitivity of the PrimeDirect assay was 91.9 % (95 %CI = 83.0–100) for samples with Ct <30 cycles (n = 34/37) and 12.5 % (95 %CI 0.9–24.1) for samples with Ct >30 cycles (n = 4/32). The specificity of the PrimeDirect assay was 88.0 % (n = 22/25, 95 %CI = 75–100). Regarding the 3 negative samples detected by this assay, it was always with high Ct (31.1 for E gene, 38.2 and 40.6 for RdRp gene). With a Ct threshold at 35, sensitivity was 64.4 % (95 %CI = 52.2–76.6) for samples with Ct <35 cycles (n = 38/59) and none of the samples with Ct <35 has been detected.
The overall sensitivity of the PrimeScript assay was 69.6 % (n = 48/69, 95 %CI = 58.6–80.6). Stratifying according to the Ct, the sensitivity of the PrimeScript assay was 89.2 % (95 %CI = 79.1–99.3) for samples with Ct <30 cycles (n = 33/37) and 46.9 % (95 %CI = 29.3–64.5) for samples with Ct >30 cycles (n = 15/32). The specificity of the PrimeScript assay was 100 % (n = 25/25). With a Ct threshold at 35, sensitivity was 79.7 % (95 %CI = 69.2–90.2) for samples with Ct <35 cycles (n = 47/59) and only 2 of the 10 samples with Ct <35 have been detected.
The overall sensitivity of the Sansure assay was 69.6 % (n = 48/69, 95 %CI = 58.6–80.6). Stratifying according to the Ct, the sensitivity of the Sansure assay was 94.6 % (95 %CI = 87.2–100) for samples with Ct <30 cycles (n = 35/37) and 40.6 % (95 %CI = 23.3–57.9) for samples with Ct >30 cycles (n = 13/32). The specificity of the Sansure assay was 100 % (n = 25/25). With a Ct threshold at 35, sensitivity was 74.6 % (95 %CI = 63.4–85.8) for samples with Ct <35 cycles (n = 44/59) and half of the 10 samples with Ct <35 have been detected.
3.2. Correlations of Ct between reference and extraction-free RT-PCR assays
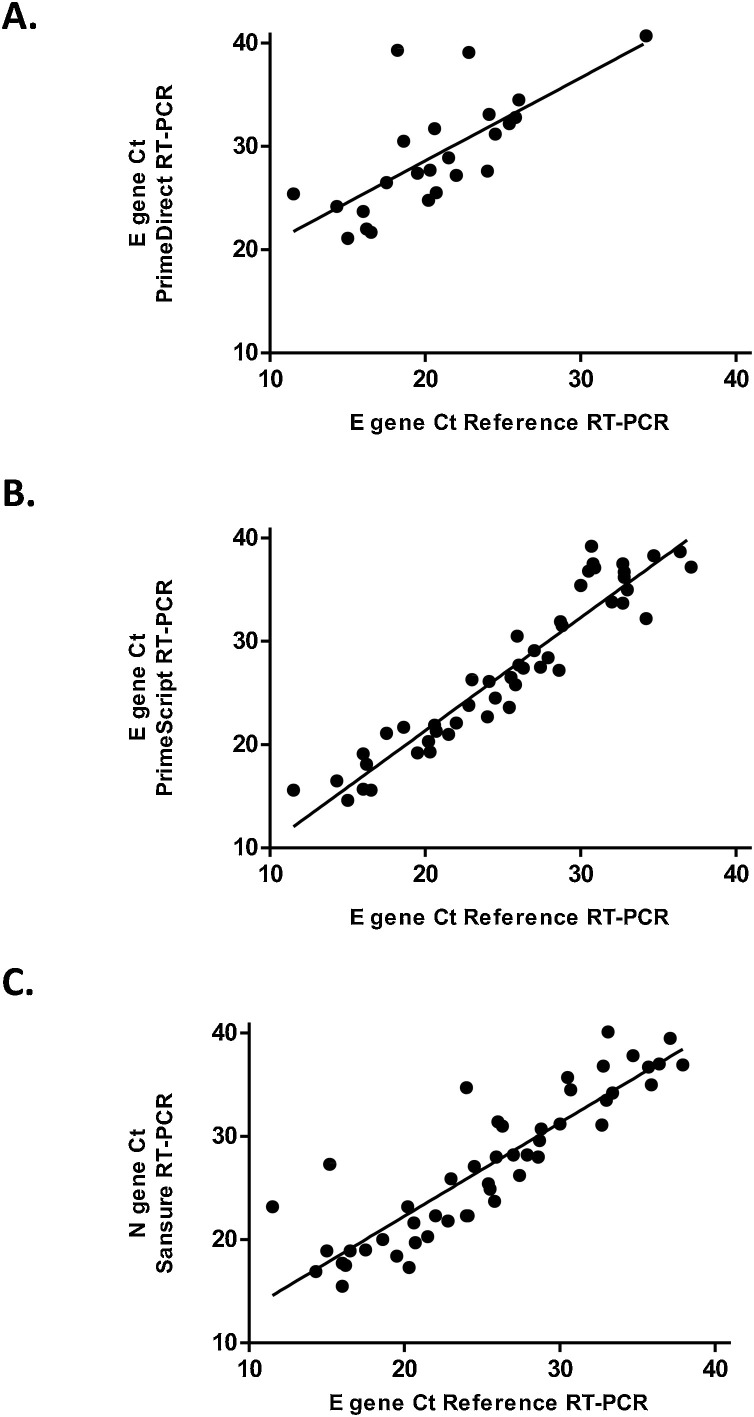
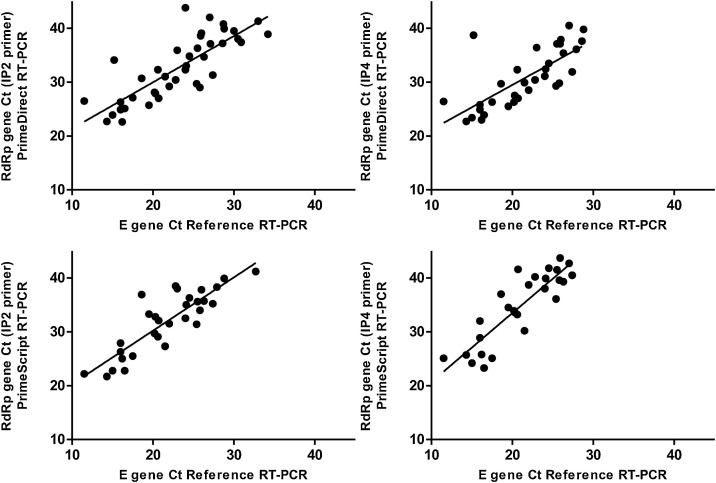
The correlation between E gene Ct issued from the reference RT-PCR assay and E gene Ct with RdRp gene Ct issued from PrimeDirect or PrimeScript assays was calculated for samples positive in both reference and evaluated assays (Figs. 1 & 2 ). Similarly, the correlation between E gene Ct issued from the reference RT-PCR assay and or N gene Ct for Sansure assay was also calculated. The R2 correlation coefficient was 0.501 (n = 24, p = 0.001), 0.904 (n = 48, p < 0.0001), and 0.787 (n = 49, p < 0.0001), for PrimeDirect, PrimeScript and Sansure assays, respectively (Fig. 1).
Fig. 1.
Correlations between Cycle Threshold (Ct) of SARS-CoV-2 between reference RT-PCR assays (E gene) and three extraction-free SARS-CoV-2 RT-PCR assays for E and N genes.
A: PrimeDirect, B: PrimeScript and C: Sansure).
Fig. 2.
Correlations between Cycle Threshold (Ct) of SARS-CoV-2 between reference RT-PCR assays (E gene) and three extraction-free SARS-CoV-2 RT-PCR assays for RdRP gene (amplicons IP2 and IP4).
4. Discussion
In the present study, we report a good sensitivity of three extraction-free SARS-CoV-2 RT-PCR assays compared to a standard reference RT-PCR assay for samples with Ct above 30.
Among the three extraction-free SARS-CoV-2 RT-PCR assays evaluated, two showed a similar overall sensitivity of 69.6 %, PrimeScript and Sansure. The remaining one, PrimeDirect, showed a lower overall sensitivity of 55.1 %, despite being initially designed to be used as an extraction-free assay.
As expected, we observed an increased sensitivity of all three assays for samples with high viral load, with a cut-off at 30 Ct. Thus, there is clear drop in sensitivity among samples with Ct >30, showing a decrease from 91.9%–12.5% with PrimeDirect, from 89.2%–46.9% with PrimeScript, and from 94.6%–40.6% with Sansure. For samples with Ct <30, observed sensitivities were similar in all three assays. A previous study assessing PrimeDirect among 91 samples showed higher sensitivity rate than in our study (Lübke et al., 2020). This study showed an overall sensitivity of 84.9 % rising to 95.8 % among samples with Ct <35 while it is 55.1 % and 91.9 % among samples with Ct <30 in our study (Lübke et al., 2020). One explanation to this difference could be that they used fresh specimen in the study of Lübke et al. (Lübke et al., 2020), while samples tested in our study undergoes a single frozen cycle. Another previous study has reported the reliability of extraction-free SARS-CoV-2 RT-PCR among a large panel of clinical samples with a sensitivity of 96 % when compared to Cobas assay (Smyrlaki et al., 2020). It is important to note that samples with high cycle threshold are unlikely to have infectious potential, since most of them displayed negative culture. In a meta-analysis, a cut-off RT-PCR Ct > 30 was associated with non-infectious samples (Jefferson et al., 2020).
A caveat in specificity was observed with the PrimeDirect kit, showing a specificity of 88 % despite the two others assays PrimeScript and Sansure showing a specificity of 100 %.
Following this evaluation, PrimeScript and Sansure kits appear satisfactory and useful in diagnostic despite a lower sensitivity for low viral loads, a result that is expected when using an extraction-free assay, generally using smaller sample volumes and more sensitive to PCR inhibition.
PrimeScript kit is an open kit requiring the use of primers ordered separately and validated before use. This assay can also be adapted to other viral targets or other pathogens as needed. Its cost is also significantly lower than existing commercial kits, with or without extraction. On the other hand, Sansure kit is a complete ready-to-use kit also including all the material necessary for the nasopharyngeal sample collection and testing. This latter is at higher price compared to the other extraction-free assays, but is still less expensive than a RT-PCR assay with extraction.
One limitation of this study is that we did not exhaustively assess the limit of detection (LOD) of the different assays. The LOD of the main reference assays used in this work, the Altona assay, with extraction, has been estimated in our laboratory at approximately 625 copies/mL (Visseaux et al., 2020), while the WHO reference assay presented LOD between 1000–1250 copies/mL (Visseaux et al., 2020; Wirden et al., 2020). Another limitation of this study is to have used frozen samples, a conservation process that may have an impact on the Ct of the RT-PCR assay.
The present study showed that two extraction-free RT-PCR assays (PrimeScript and Sansure) are a suitable alternative to classical RT-PCR assays for detecting SARS-CoV-2 in nasopharyngeal samples despite a lack of sensitivity for low viral loads. This testing strategy of extraction-free RT-PCR will help to expand SARS-CoV-2 testing by enlarging the number of laboratories able to perform it and by bypassing extraction reagents and equipment needs.
Author statement
BV, NHF, CC and DD were involved in the conception, design, data collection, analysis and report writing. GC, QLH, VMF, FD and HI performed analysis. CC, GC, NHF and DD participated in manuscript preparation. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc Natl Acad Sci. 2020;117(37):22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalandra R., Yadav A.K., Verma D., Dalal N., Sharma M., et al. Strategies and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed. Pharmacother. 2020;129 doi: 10.1016/j.biopha.2020.110446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson T., Spencer E.A., Brassey J., Heneghan C. Viral cultures for COVID-19 infectious potential assessment - a systematic review. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1764. Dec 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübke N., Senff T., Scherger S., Hauka S., Andrée M., et al. Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials. J. Clin. Virol. 2020;130 doi: 10.1016/j.jcv.2020.104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrlaki I., Ekman M., Lentini A., Rufino de Sousa N., Papanicolaou N., et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat. Commun. 2020;11(1):4812. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visseaux B., Le Hingrat Q., Collin G., Ferré V.M., Storto A., et al. Evaluation of the RealStar® SARS-CoV-2 RT-PCR kit RUO performances and limit of detection. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirden M., Feghoul L., Bertine M., Nere M.L., Le Hingrat Q., et al. Multicenter comparison of the Cobas 6800 system with the RealStar RT-PCR kit for the detection of SARS-CoV-2. J. Clin. Virol. 2020;130 doi: 10.1016/j.jcv.2020.104573. [DOI] [PMC free article] [PubMed] [Google Scholar]