Abstract
The emergence of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) from China has become a global threat due to the continuous rise in cases of Coronavirus disease 2019 (COVID-19). The problem with COVID-19 therapeutics is due to complexity of the mechanism of the pathogenesis of this virus. In this review, an extensive analysis of genome architecture and mode of pathogenesis of SARS-CoV-2 with an emphasis on therapeutic approaches is performed. SARS-CoV-2 genome consists of a single, ~29.9 kb long RNA having significant sequence similarity to BAT-CoV, SARS-CoV and MERS-CoV genome. Two-third part of SARS-Cov-2 genome comprises of ORF (ORF1ab) resulting in the formation of 2 polyproteins, pp1a and pp1ab, later processed into 16 smaller non-structural proteins (NSPs). The four major structural proteins of SARS-CoV-2 are the spike surface glycoprotein (S), a small envelope (E), membrane (M), and nucleocapsid (N) proteins. S protein helps in receptor binding and membrane fusion and hence plays the most important role in the transmission of CoVs. Priming of S protein is done by serine 2 transmembrane protease and thus plays a key role in virus and host cell fusion. This review highlights the possible mechanism of action of SARS-CoV-2 to search for possible therapeutic options.
Keywords: Severe acute respiratory syndrome coronavirus-2, Coronavirus disease 19, Drug targets
1. Introduction
Coronaviruses (CoVs) are exceptionally diverse, positive-sense, enveloped, and single-stranded RNA viruses from Nidovirales (order) of the Coronaviridae family [1]. These cause a wide array of diseases affecting respiratory, hepatic, neurological, and enteric systems with varied intensity amid animals and humans [2]. Based on serotype and genotype, CoVs are sorted into four genera viz. α, β, γ, and δ [3]. The first two CoVs i.e. α- and β-affect mammals while γ-CoVs infect avian species and δ-CoVs mammals as well as aves. In the past, six CoVs species had the potential to cause human diseases however, in immune-competent individuals, four of these six can only cause common cold [4,5]. The other two species were severe acute respiratory syndrome CoV (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). These two were much more morbific than others and can ensure deadly disease. The newly identified CoV referred to as SARS-CoV-2 is far more contagious as compared to other earlier recognized human CoVs SARS-CoV (2002) and MERS-CoV (2013). Additionally, SARS-CoV-2 is also initiated by zoonotic transmission as were SARS-CoV and MERS-CoV. SARS-CoV-2 is very fatal as it is transferred among humans through sneezing and making contacts [5].
SARS-CoV and MERS-CoV are very similar in sequence and encode crucial enzymes and proteins like spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. This similarity provides a clue that both these share the same pathogenesis mechanism which can be further used in common therapeutic targeting.
On 11th February 2020, the World Health Organization (WHO) has given the title of ‘Coronavirus disease (COVID-19)’ to the infection caused by this species of CoV. According to WHO, this disease is now pandemic worldwide and is a health emergency of international concern as it has taken more than two million lives around the globe. Due to the lack of understanding of the mechanism of pathogenesis, an effective therapeutic option is unavailable. Only timely diagnosis of this disease will help to control its spread by social distancing and quarantine [6,7].
At present, the entire world is dealing with this global pandemic condition and hence, there is an immediate need to develop strategies that can be implicated in therapeutics of COVID-19 [7,8]. Thus, the need of the hour is to perform an extensive analysis of its genomics and its comparison with other pathogenic human CoVs as this detailed insight will provide a platform to figure out the origin of SARS-CoV-2 pathogenesis. This will further help us in designing therapeutics strategies to combat COVID-19.
2. Genome structure of SARS-CoV-2
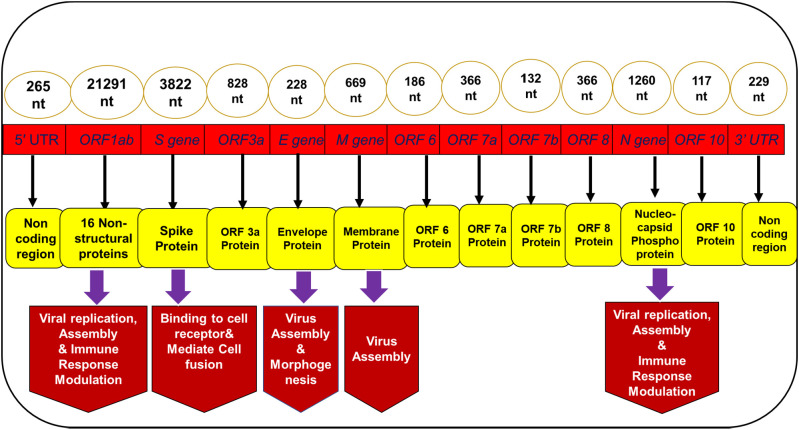
The genome size of the SARS-CoV-2 is ~29.9 kb [9]. It consists of approximately 13 to 15 ORFs which are flanked by 5′ and 3′ UTRs [10] (Fig. 1 ). These ORFs are ordered in such a way that they constitute a replicase assembly which has the potential of encoding around 27 distinct structural and non-structural proteins (NSPs) [11]. First ORF (ORF 1ab) covers two-third part of the genome and translates into 7096 residues long polyprotein (PP) [9]. PP pp1a and pp1b are encoded by ORF1a and ORF1b respectively, which further produces 16 NSPs after processing through virally encoded proteases. All these 16 proteins, mainly involved in viral replication, assembly, and immune response modulation are conserved in all SARS viruses that belong to the same family [9]. The four major structural proteins of SARS-CoV-2 are spike surface glycoprotein (S) a small envelope protein (E), membrane (M), and nucleocapsid (N) proteins. There is a difference in epidemiological dynamics of SARS-CoV-2 and SARS-CoV and MERS-CoV and hence former is comparatively more infectious and lethal.
Fig. 1.
Schematic representation of the genomic organization of SARS-CoV-2 depicting the architecture.
3. Insight into Pathogenesis of SARS-CoV-2
Various organs like lungs, heart, arteries, kidney, intestine, etc. possess Angiotensin-converting enzyme 2 (ACE2) attached to their cell membrane. ACE2 is a monocarboxypeptidase that plays an essential role in maintaining the balance of the renin-angiotensin system [12]. This enzyme acts as a cell receptor for SARS-CoV [13]. It is through this enzyme that SARS-CoV virion particles enter into the cell and its genome is then translated into PPs, via the host ribosome, which further gets processed by proteolysis. PP degradation is mediated by main protease (Mpro) and papain-like protease (PLpro) which chop it into smaller fragments to support replication and helps in forming new virions [14]. The resulting PP1a and PP1ab are processed into the individual NSPs that form the viral replication and transcription complex [15]. The PPs move into endoplasmic reticulum (ER) membranes and transfer through the ER-to-Golgi intermediate compartment, where contact with N-encapsidated, newly formed genomic RNA fallouts in budding into the lumen of secretory vesicular compartments, and then virions are secreted from the infected cell by the process of exocytosis [15]. Similarly, SARS-CoV-2 also uses ACE2 as a way in the ACE2-expressing cells, thus it can be assumed that both targets a similar spectrum of cells. According to literature, macrophages and pneumocytes are the main targets of SARS-CoV [16]. The extra-pulmonary spread of SARS-CoV was also reported as ACE2 expression is not restricted to lungs only. A high transmission rate of SARS-CoV-2 in contrast to SARS-CoV can be attributed to more efficient exploitation of cellular attachment factors that mediates vigorous infections of ACE2 expressing cells in the upper respiratory tract. ACE2 occurs mainly in type II alveolar epithelial cells (pneumocytes). It is with these ACE2 molecules, the S-protein of SARS-CoV-2 interact Expression of ACE2 is slightly elevated in males in comparison to females which can be attributed to higher incidences of COVID-19 in men. CoVs have been known to modulate the affecting cells by their cytocidal activity and immune-mediated mechanisms [17]. CoVs infection results in cytopathic effects, as well as apoptosis and cell degradation.
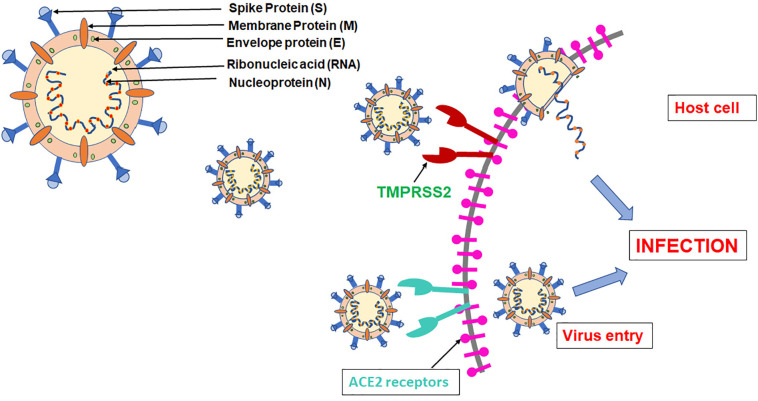
The binding of ACE to SARS-CoV-2, as was in SARS-CoV, may lead to its increased expression, resulting in alveolar damage. Based on biophysical and structural analysis, in contrast to SARS-CoV, SARS-CoV-2 showed approximately 10–20 times higher affinity for ACE2 [18]. Finally, it can be said that SARS-CoV-2 entry in the host cell is initiated by binding to the ACE2 receptor. Apart from ACE2, there is another host cell factor that plays a key role in virion entry is the serine protease, Transmembrane protease serine 2 (TMPRSS2) [19]. TMPRSS2 plays a key role in pathogenesis by cleaving S protein at S1/S2 and the S2′ site and helps in S protein priming that allows virion entry into the host cell and ultimately viral and cellular membrane fusion. Thus, TMPRSS2 can also serve as a potential drug target. Post binding, ss RNA of SARS-CoV-2 gets attached to the host cell ribosomes which lead to the translation of two co terminals large PPs. Fig. 2 shows a brief outline of various factors performing different responsibilities in the life cycle of SARS-CoV-2. The PPs thus formed are further processed into 16 distinct NSPs by the action of two main proteolytic enzymes namely SARS-CoV-2 Mpro and PLpro [14]. These smaller components further play different roles such as viral assembly, immune response modulation, and others. Mpro can be considered as an important drug target among CoVs owing to its function in the processing of PPs that result from the translation of viral RNA. CoVs Like all pathogens, the mechanism of SARS-CoV-2 pathogenesis follows innate and the adaptive immune system as well [20].
Fig. 2.
Graphical representation of important components of SARS-CoV-2 and showing SARS-CoV-2 entry in the host cells.
4. Potential therapeutic drug targets
4.1. Spike (S) protein
The envelope spike (S) protein plays a crucial role in CoVs infection and pathogenesis [21,22]. SARS-CoV-2 has a highly glycosylated S protein that belongs to trimeric class I viral fusion glycoprotein. S protein has 1273 amino acid residues forming three subunits namely, S1, S2, and S2′. They undergo structural changes during the process of viral and host membrane fusion (Li, 2016). S1 and S2 domains are responsible for receptor binding and membrane fusion respectively, while S2′, a cleaved subunit of S protein, acts as a fusion peptide [23]. The process of viral infection is initiated when the virus binds to the human ACE2 cell surface receptor with its S1 subunit [19].
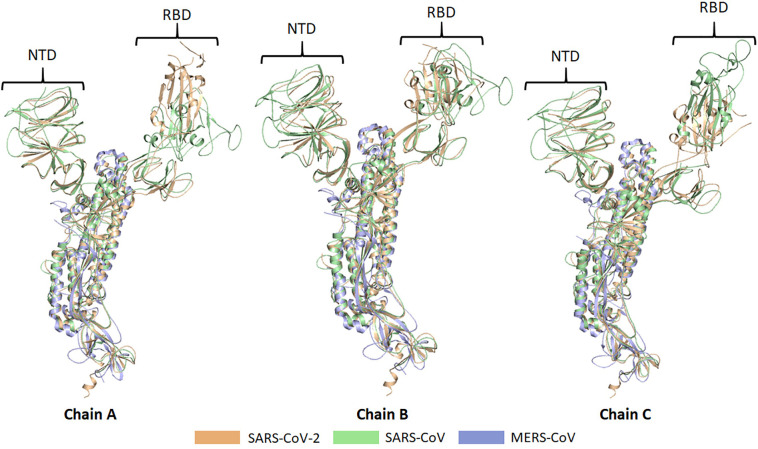
The head region of S1 is known as receptor binding domain (RBD) which recognizes ACE2, with Glu394 of RBD and Lys31 of ACE2 playing a crucial role in receptor and S protein interaction [24]. This binding destabilizes the pre-fusion trimer that leads to the release of the S1 subunit and ultimately the transition of the S2 subunit to a more stable post-fusion conformation [25]. For attachment to host cell receptor, RBD of S1 endures hinge-like conformational changes that can conceal or reveal determinants of receptor binding transiently. These two states are known as “down” and “up” conformation with down conformation being referred to as stable and receptor unapproachable while up conformation as less stable receptor approachable state [26]. The function of the S2 subunit is to cause membrane fusion between the virions and the host cell. For this, S2 exists in three different conformations namely; pre-fusion native state, hairpin intermediate state, and post-fusion hairpin state viz. RBD is the most unpredictable feature of SARS-CoV-2 with maximum variation in the receptor-binding motif. This variation can be directly linked to variation in the mechanism of pathogenesis among different CoVs. An understanding of changing conformations of S protein that results in the entry of the virion into the mammalian cell can provide a breakthrough in COVID-19 therapeutics. Our recent study suggested that out of six residues of RBD of S1 subunit of SARS-CoV-2 (Leu455, Phe486, Gln493, Ser494, Asn501, and Tyr505) that are crucial for binding to ACE2, five differ from SARS-CoV [27]. The similarity between S protein of SARS-CoV-2, SARS CoV, and MERS CoV is that in all these it exists in homologous trimeric conformation having three chains viz. A, B and C [28,29]. Chain A and C show a high degree of a structural anomaly in the N terminal domain (NTDs) and RBDs as compared to chain B when they were aligned and visualized in PyMOL (Fig. 3 ). These deviations can provide a platform to develop different approaches that can be implicated in COVID-19 therapeutics. The trimeric spike glycoprotein (S) of SARS-CoV-2 is a key target for virus neutralizing antibodies6 and the prime candidate for vaccine development [30]. Recent research reported the preclinical development of two BNT162b vaccine candidates, which contain lipid-nanoparticle (LNP) formulated nucleoside-modified mRNA encoding SARS-CoV-2 spike glycoprotein-derived immunogens [30].
Fig. 3.
Structural comparison of spike (S) proteins trimeric conformation. The S protein of the SARS-CoV-2, SARS-CoV, and MERS-CoV exists in homologous trimeric conformation consisting of three chains named chain A, B, and C. These chains are aligned and visualized in PyMOL which revealed a high degree of structural deviations in the N-terminal domains (NTDs) and receptor-binding domains (RBDs) of the chain A and C compared to that of chain B. The structural coordinates of the S protein of the SARS-CoV-2, SARS-CoV, and MERS-CoV were taken from the RCSB Protein Data Bank with PDB IDs: 6VSB, 6ACD, and 5W9H, respectively.
4.2. Envelope (E) protein
The E protein plays a key part in the virion life cycle, particularly during viral morphogenesis and assembly [31,32]. It is another structural protein that can serve as a potential drug target in COVID-19 therapeutics because it can form a pentamer and function as an ion channel, thus named as E channel or viroporin. Moreover, there are regions of high similarity in E proteins of BAT-CoV, SARS-CoV, and SARS-CoV-2. Interestingly, there is a subtle disparity among SARS-CoV-2 and MERS-CoV E proteins. These proteins can form ion channels, an important function in virus-host interaction [33]. Ion conductivity is considered a beneficial feature for viruses and thus in its pathogenesis [34]. E protein ion channel activity is also required for inflammasome activation [35]. E protein also plays an important role in intracellular protein trafficking as well as regulation and hence this is another aspect of E protein that can be used in COVID-19 therapeutics.
4.3. Membrane (M) protein
Another example of transmembrane glycoprotein is M protein which has three transmembrane domains, a characteristic trait of the membrane proteins. These are the structural blocks made up of 222 amino acids that provide a framework to the virion particle and aids in the structural assembly of the virus. This structural protein functions in harmony with E, N, and S proteins and aids in RNA packaging [36]. Another role played by M protein is intracellular homeostasis. M protein also helps in viral-specific humoral response and can elicit efficient neutralizing antibodies in SARS patients [37]. Like E proteins, there is a similarity among M proteins of BAT-CoV, SARS-CoV, and SARS-CoV-2 and subtle differences in M proteins of SARS-CoV-2 and MERS-CoV.
4.4. Nucleoprotein (N)
Structurally, N proteins comprise three domains arranged as an N-terminal RNA-binding domain (NTD), followed by a Ser/Arg (SR)-rich central linker region and a C-terminal dimerization domain (CTD). These N proteins help in packing viral RNA into viral nucleocapsid. The viral genomes are released into the network of the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) with the help of these N proteins. The central core of N protein has ~140 amino acid residues long RNA binding domain which is arranged in a ‘bead on string manner’ and helps in its binding with viral RNA. Targeting this RNA-binding domain of N protein can be a possible therapeutic option in COVID-19 treatment. In addition to this, N proteins also assist viral RNA transcription and replication. It also plays an important role in cellular processes like cell cycle, cytoskeleton reorganization, and host cell apoptosis. All these important functional aspects of N proteins enable them as potential drug targets that can be implicated in COVID-19 therapeutics. There is a high sequence similarity among N protein of BAT-CoV, SARS-CoV, and SARS-CoV-2 which implicates that Abs recognizing N proteins of SARS-CoV will be successful against SARS-CoV-2 also and thus, N proteins can be used as a diagnostic tool.
4.5. Replicase polyprotein
Apart from the above discussed structural proteins, Replicase proteins also play a key part in SARS-CoV-2 pathogenesis. The major part of the replicase genome is covered by the ORF1ab gene which encodes two major PPs i.e., pp1a and pp1ab. The replicase PP is made up of mainly three domains; the macro domain, the papain-like domain, and the main protease. These multifunctional proteins guide the degradation of host RNA and replication of viral RNA [38]. These PPs are subsequently processed into 16 smaller NSPs through proteolytic enzymes, mainly the Mpro and PLpro [14], which cleaves the C and N terminal ends of these PPs respectively [39]. The function of viral RNA transcription and replication is performed by a specific RNA dependent RNA polymerase (RdRp) region present in ORF 1ab. As per the literature, similar to structural proteins, high sequence similarity is observed in 3CLpro of BAT-CoV, SARS-CoV, and SARS-CoV-2, unlike MERS-CoV which doesn't show such similarity.
4.6. Main protease (Mpro or 3CLpro) and papain-like protease (PLPRO)
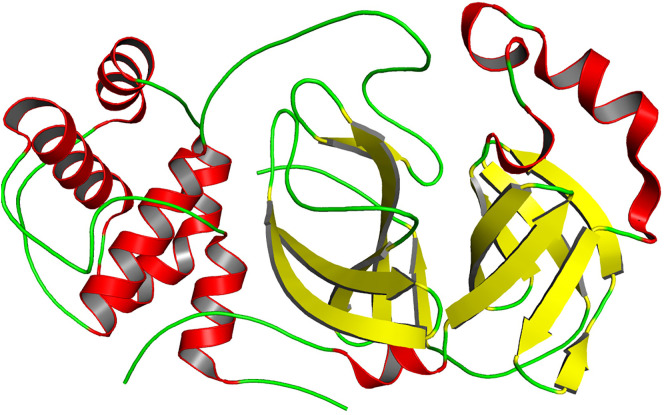
Mpro is a vital cog of SARS-CoV-2 that can be an attractive drug target in COVID-19 therapeutics [14]. It is a crucial enzyme that works in cooperation with other components and helps in the replication and transcription of viral RNA Mpro is also the most studied target for CoVs drugs as it is responsible for the processing of polyproteins required for the assembly of virus drugs. Fig. 4 depicts the structure of this main protein depicting the secondary structural content. Mpro of SARS-CoV-2 is a 306 amino acid residues long protein, having three domains in its structure viz. domain I (residues 8–101), domain II (residues 102–184), and domain III (residues 201–303). Domain III and II are connected by a 15 residues long loop (residues 185–200) having Cys-His catalytic dyad. SARS-CoV and SARS-CoV-2 share ~96% similarity in their Mpro sequence. Just like its counterparts in other CoVs, the substrate-binding site of Mpro in SARS-CoV-2 is located in the cleft between domain I and II [40,41]. It cleaves PPs to generate NSPs that form a replicase-transcriptase complex (RTC). The ability of Mpro to control this vital step of SARS-CoV-2 highlights its importance that can be used in drug discovery. SARS-CoV-2 Mpro recognizes and acts remarkably at fewer than 11 cleavage sites of Leu-Gln↓ (Ser, Ala, Gly) of the polyprotein replicase 1ab [42]. A recent study reported the crystal structure of SARS-CoV-2 providing a platform to develop COVID-19 therapeutics targeting Mpro [14]. Design and development of safe and potential for SARS-CoV-2 can be achieved via targeting conserved enzymes residues of Mpro [[43], [44], [45]].
Fig. 4.
Structural representation of the SARS-CoV-2 main protease (3CLpro or Mpro) (PDB ID: 6YB7).
5. RNA-dependent RNA polymerase
A multisubunit replication and transcription complex of NSPs arbitrate the replication of SARS-CoV-2 The catalytic subunit (Nsp12) of the RNA-dependent RNA polymerase (RdRp) enzyme is the core component of this complex.Nsp12 has little activity by itself and requires additional factors (Nsp7 and Nsp8) for proper functioning [46]. RdRp is treated as a potential drug spot for developing antivirals. It is a well-defined target for Remdesivir, an antiviral drug. The active site region of SARS-CoV-2 RdRp is consists of many critical residues including Lys545, Arg555, Asp623, Ser682, Thr687, Asn691, Ser759, Asp760, and Asp761. These residues where Remdesivir is bound can be served as a platform for the development of powerful, effective, and selective inhibitors of SARS-CoV-2 RdRp [47].
6. Non-structural protein
These refer to proteins that are not included in viral particles but function inside the infected cell playing several important roles viz. viral replication, controlling early transcription and modulates immune response [9]. NSPs are also found to be associated with helicase activity. Table 1 lists all the NSPs along with their proposed functions.
Table 1.
Functional importance of non-structural proteins (NSPs) of SARS-CoV-2.
| S. No | Protein | Function |
|---|---|---|
| 1 | Nsp1 | The exact function is biologically unique and unknown. Forms a previously unknown complex β-barrel fold with several unique structural features and contributes to the degradation of mRNA [51]. It is also involved in innate immune response antagonism [52]. |
| 2 | Nsp2 | A replicase product, has no special known function but fund to involved in modulation of host cell survival signaling pathway by interacting with host PHB and PHB2 [53] |
| 3 | Nsp3 | Binds to viral RNA, nucleocapsid protein, as well as other viral proteins, and contributes in polyprotein processing [54]. It has also an important role in innate immune response antagonism. |
| 4 | Nsp4 | Plays a role in membrane rearrangement in association with Nsp3 thereby affecting viral replication. |
| 5 | Nsp5 | 3C-like proteinase and main proteinase involved in viral polyprotein processing during replication [55]. |
| 6 | Nsp6 | Transmembrane domain, plays a role in the initial induction of autophagosomes from the host endoplasmic reticulum. |
| 7 | Nsp7 | An RNA-dependent RNA polymerase works in association with Nsp8 [55]. Also stimulates the polymerase activity of Nsp12. The stoichiometric ratio of nsp7 and nsp8 is found to less than nsp12 [56]. Nsp7 and nsp8 increase nsp12 binding to the template-primer RNA [56]. |
| 8 | Nsp8 | Replicase capable of de novo initiation and has been proposed to operate as a primase in complex with nsp7. Crystallized together with the 10-kDa nsp7, forming a hexadecameric, dsRNA-encircling ring structure [i.e. Nsp (7 + 8), consisting of 8 copies of both Nsps] [57]. The nsp12-nsp7-nsp8 complex also showed RNA polymerization activity on a poly-U template upon addition of ATP [56]. |
| 9 | Nsp9 | Single-stranded RNA-binding protein mediate both viral replication and virulence [58]. |
| 10 | Nsp10 | Acts as a stimulatory factor along with Nsp16 to execute its MTase activity, therefore plays an essential role in viral mRNAs cap methylation [59]. It also forms complex with Nsp14 and stimulates MTase activity. |
| 11 | Nsp11 | Unknown |
| 12 | Nsp12 | RNA-dependent RNA polymerase and also has nucleotidyltransferase activity [46] |
| 13 | Nsp13 | The helicase unwinds the double-stranded RNA segment into single strands by hydrolyzing NTPs, involved in replication and transcription. |
| 14 | Nsp14 | Nsp14 has two enzymatic activities, an N7 methyltransferase activity and an exonuclease activity, involved in the unique proofreading system of CoVs [60] |
| 15 | Nsp15 | Mn(2+)-dependent Endoribonuclease activity [61]. A highly conserved nidovirus component with endoribonuclease activity acts in conjunction with the viral replication complex to limit the exposure of viral dsRNA to host dsRNA sensors [62]. |
| 16 | Nsp16 | 2′-O-ribose methyltransferase involved in MTase activity [63]. |
7. Host cell proteases
SARS CoV-2 enters into the host cell with the help of its S protein whose S1 subunit binds to the receptor on the cell surface. In the cell line, other key players involved in this entry were found to be endosomal cysteine proteases cathepsin B and L (CatB/L) [48] and the serine protease TMPRSS2 [49] which are involved in the priming of S protein. The important fact is that only the activity of TMPRSS2 is crucial for viral spread and pathogenesis unlike the dispensable activity of CatB/L activity [50] (Fig. 2). Another study suggested that TMPRSS2 is a critical host cell SARS-CoV-2 also playing a role in its spread and pathogenesis [50]. A TMPRSS2 inhibitor approved for clinical use, camostat mesylate, blocked the entry and can provide a new avenue in COVID-19 therapeutics.
8. Conclusion and future directions
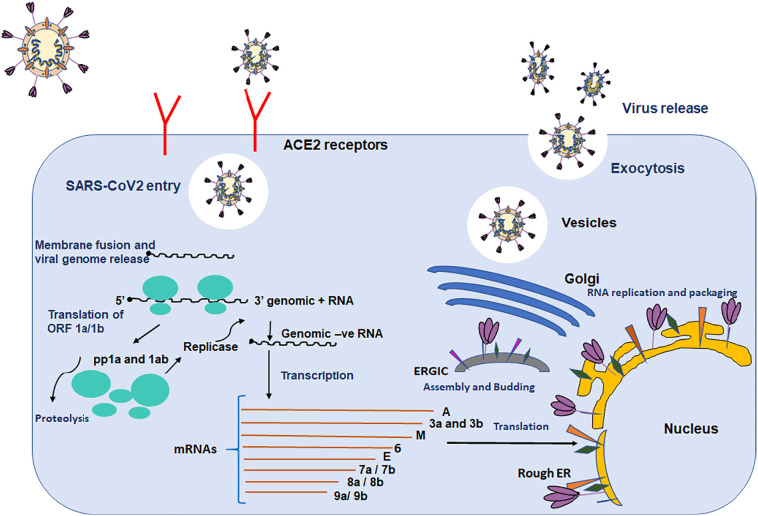
At present, SARS-CoV-2 is offering a major risk across the globe and there is no specific drug available to treat COVID-19. In the past, antiviral drugs have been used to treat other human CoVs, but these have been rendered ineffective due to structural differences in SARS-CoV-2 as compared to other human CoVs. SARS-CoV-2 is far more pathogenic in contrast to other human CoVs. The SARS-CoV-2 vaccine is still under development, and there is no specific drug at present and all other trial drugs have also been unsuccessful. A comparative genomics-based approach with earlier known human CoVs can provide a breakthrough in COVID-19 therapeutics. Fig. 5 shows the SARS-CoV-2 life cycle with the depiction of different target sites that can be retorted in COVID-19 therapeutics. Thus, the need of the hour is to perform an extensive analysis of its genomics and its comparison with other pathogenic human CoVs as this detailed insight will provide a platform to understand the molecular basis of SARS-COV-2 pathogenesis. This review article provides an extensive investigation of the genome of SARS-COV-2 and its comparison with other human CoVs that enable us to identify the molecular way of pathogenesis. It also provides a brief insight into important proteins of SARS-CoV-2 that can act as possible drug targets and enlightened the differences in the structure of these proteins in comparison to other human CoVs. Table 2 lists various drugs against different targets of SARS-CoV-2 having clinical effectiveness and can be implicated in COVID 19 therapeutics. That have been found to have clinical effectiveness in COVID-19 therapy targeting various targets of SARS-CoV-2. This review provides deeper insights into the SARS-CoV-2 genome that delineates the mechanism of action of SARS-CoV-2 which can be implicated in COVID-19 therapeutics.
Fig. 5.
Diagrammatic representation of the SARS-CoV-2 life cycle depicting different target sites that can be implicated in COVID-19 therapeutics.
Table 2.
List of drugs having clinical effectiveness in COVID-19 therapy targeting various targets of SARS-CoV-2.
| S. No. | Drug | Use | Target | Mechanism of action | Clinical trial | Ref. |
|---|---|---|---|---|---|---|
| 1 | Atazanavir | HIV | SARS-CoV-2 Mpro | Atazanavir could adjust in Mpro active site and can I inhibit its activity resulting in a disruption in viral replication. | Phase 2: NCT04459286 | [64] |
| 2 | Baricitinib | Rheumatoid arthritis | Human AP2-associated protein kinase 1 (AAK1); Janus kinase (JAK) 1 and 2 | Can block the entry and infectivity of SARS CoV-2 in pneumocytes by impairing AAK1 that are involved in virus endocytosis; also inhibit the intracellular signaling pathway of cytokines IL-2, 6, 10 and INF-γ, a granulocyte-macrophage colony-stimulating factor that is enhanced in severe SARS CoV-2 infection | Phase 2: NCT04321993; Phases 2 and 3: NCT04320277; Phase 3 NCT04421027 |
[[65], [66], [67]] |
| 3 | Mefuparib hydrochloride (CVL218) | Cancer | N protein; poly-ADP-ribose polymerase 1(PARP1) | Can target N protein to reduce its RNA binding and thus impede viral replication; inhibit the production of IL-6 by CpG oligodeoxynucleotide 1826 in peripheral blood mononuclear cells | Phase 1 | [68] |
| 4 | Pemirolast, nitrofurantoin isoniazid pyruvate, eriodictyol | Numerous | ACE2 receptor | Can interact with ACE2 receptor more efficiently and inhibit undesirable S protein to ACE2 interaction. | – | [69] |
| 5 | Cepharanthine, ergoloid, hypericin | Numerous | S protein | Can cause favorable ring-protein interaction which blocks host recognition | – | [69] |
| 6 | Remdesivir | Ebola | RdRP | Nucleoside (adenosine) analogue RdRP inhibitor which inhibits RNA synthesis and can result in premature termination | Phase 3: NCT04292899 | [70,71] |
| 7 | Chloroquine/hydroxychloroquine | Malaria, lupus and rheumatoid arthritis | Affect both early and late stage of viral replication | keep the virus out of host cells by disturbing ACE2 glycosylation and breaking down the production of viral proteins by inhibiting endosomal acidification. | Phase 2 and 3: NCT04353336 | [70,72,[74], [75], [76]] |
| 8 | Lopinavir/ritonavir combination | HIV | 3CLpro | Disrupt the process of viral replication and release from the cell. | Phase 2: NCT0427668 | [77,78] |
| 9 | Nafamostat or camostat | Pancreatitis | Serine protease TMPRSS2 | Acts as an antagonist to the serine protease TMPRSS2; Prevents membrane fusion by reducing the release of cathepsin B. | Phase 2 and 3: NCT04418128 Phase 2: NCT04625114 | [79] |
| 10 | Famotidine | Heartburn | PLpro | Possibly bind PLpro which is known to be essential to the entry of SARS-CoV-2 | Phase 3: NCT04504240 | [80] |
| 11 | Umifenovir | Influenza | Viral lipid membrane | Can bind viral lipid membrane and affect cellular trafficking of the virus | Phase 4: NCT04350684 | [81] |
| 12 | Nitazoxanide | Influenza; diarrhoea | Not known | Can suppress maturation of the viral hemagglutinin and the viral transcription factor immediate-early 2 (IE2) as well as by activating the translation INF2α. | Phase 2: NCT04552483 | [82] |
| 13 | Ivermectin | Influenza; dengue; broad-spectrum antiparasitic | Not known | Can inhibit expression of the viral N protein and IL-6; Inhibit viral IMPα/β1-mediated nuclear import, causing a reduction in viral replication; Can also work by binding and destabilizing cell-transport proteins used to enter the nucleus. | Phase 1: NCT04343092 | [83] |
| 14 | Teicoplanin | Gram-positive bacterial infection | Not known | Inhibit the activity of cathepsin L which potentially plays an important role in blocking viral entry in the cells | – | [84] |
| 15 | Tocilizumab/sarilumab (mAb) | Rheumatoid arthritis | IL-6 receptor antagonists | Inhibition of IL-6 may attenuate pulmonary inflammation and fibrosis | Phase 3 and Phase 2/3: NCT04315298 | [84] |
| 16 | Anti TNF-α agents | Rheumatoid arthritis | TNF-α | TNF-α blockage leads to down-regulation of pro-inflammatory mediators, including IL-1, IL-6, and granulocyte-macrophage colony-stimulating factor as well as cytokines and acute-phase proteins | – | [84] |
Declaration of competing interest
None.
Acknowledgments
Acknowledgements
A.S. is thankful to Dr. D.S. Kothari Postdoctoral Fellowship ((BSR)/BL/19-20/0119). M.S.K., F.M.H., and M.T.R. acknowledge the generous support from the Deanship of Scientific Research at King Saud University, Riyadh, Kingdom of Saudi Arabia (Grant No. RGP-215).
CRediT authorship contribution statement
Anas Shamsi: Conceptualization; Data curation; Formal analysis Validation; Visualization; Roles/Writing – original draft; Writing – review & editing, Taj Mohammad: Resources; Software; Validation; Visualization; Roles/Writing – original draft; Writing – review & editing, Saleha Anwar: Methodology; Writing – original draft; Investigation, Samreen Amani: Writing – review & editing, Mohd Shahnawaz Khan: Funding acquisition; Resources, Fohad Mabood Husain: Funding acquisition; Validation, Md. Tabish Rehman: Funding acquisition, Visualization, Asimul Islam: Investigation; Methodology; Project administration; Writing – review & editing; Software; Supervision, Md Imtaiyaz Hassan: Investigation; Methodology; Writing – review & editing.
References
- 1.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.-Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F.-W., Lau S.K.-P., Woo P.C.-Y. The emerging novel Middle East respiratory syndrome coronavirus: the “knowns” and “unknowns”. J. Formos. Med. Assoc. 2013;112(7):372–381. doi: 10.1016/j.jfma.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong A.C., Li X., Lau S.K., Woo P.C. Global epidemiology of bat coronaviruses. Viruses. 2019;11(2):174. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asrani P., Hasan G.M., Sohal S.S., Hassan M.I. Molecular basis of pathogenesis of coronaviruses: a comparative genomics approach to planetary health to prevent zoonotic outbreaks in the 21st century. OMICS: A Journal of Integrative Biology. 2020;11:634–644. doi: 10.1089/omi.2020.0131. [DOI] [PubMed] [Google Scholar]
- 6.Asrani P., Eapen M.S., Chia C., Haug G., Weber H.C., Hassan I., Sohal S.S. 2020. Diagnostic Approaches in COVID-19: Clinical Updates, Expert Review of Respiratory Medicine. (null-null) [DOI] [PubMed] [Google Scholar]
- 7.Kumari P., Singh A., Ngasainao M.R., Shakeel I., Kumar S., Lal S., Singhal A., Sohal S.S., Singh I.K., Hassan M.I. Potential diagnostics and therapeutic approaches in COVID-19. Clin. Chim. Acta. 2020;510:488–497. doi: 10.1016/j.cca.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatima U., Rizvi S.S.A., Fatima S., Hassan M.I. Impact of hydroxychloroquine/chloroquine in COVID-19 therapy: two sides of the coin. J. Interf. Cytokine Res. 2020;10:469–471. doi: 10.1089/jir.2020.0105. [DOI] [PubMed] [Google Scholar]
- 9.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92(6):595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabish M. Alternative splicing of ACE2 possibly generates variants that may limit the entry of SARS-CoV-2: a potential therapeutic approach using SSOs. Clin. Sci. 2020;134(10):1143–1150. doi: 10.1042/CS20200419. [DOI] [PubMed] [Google Scholar]
- 13.Oudit G., Kassiri Z., Jiang C., Liu P., Poutanen S., Penninger J., Butany J. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur. J. Clin. Investig. 2009;39(7):618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020:1–16. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shieh W.-J., Hsiao C.-H., Paddock C.D., Guarner J., Goldsmith C.S., Tatti K., Packard M., Mueller L., Wu M.-Z., Rollin P. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum. Pathol. 2005;36(3):303–309. doi: 10.1016/j.humpath.2004.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki T., Otake Y., Uchimoto S., Hasebe A., Goto Y. Genomic characterization and phylogenetic classification of bovine coronaviruses through whole genome sequence analysis. Viruses. 2020;12(2):183. doi: 10.3390/v12020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M., Kuate S., Überla K., Niedrig M., Pöhlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78(12):6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qing E., Gallagher T. SARS coronavirus redux. Trends Immunol. 2020;41(4):271–273. doi: 10.1016/j.it.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020:1–5. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.-J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. 2017;114(42):11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8 doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta Mol. basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walls A.C., Xiong X., Park Y.-J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176(5):1026–1039. doi: 10.1016/j.cell.2018.12.028. (e15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A. Immunogenic BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021:1–10. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 31.Li S., Yuan L., Dai G., Chen R.A., Liu D.X., Fung T.S. Regulation of the ER stress response by the ion channel activity of the infectious bronchitis coronavirus envelope protein modulates virion release, apoptosis, viral fitness, and pathogenesis. Front. Microbiol. 2020;10:3022. doi: 10.3389/fmicb.2019.03022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Liu L. The membrane protein of severe acute respiratory syndrome coronavirus functions as a novel cytosolic pathogen-associated molecular pattern to promote beta interferon induction via a Toll-like-receptor-related TRAF3-independent mechanism. MBio. 2016;7(1) doi: 10.1128/mBio.01872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Torres J., Aguilella V.M., Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432(2):485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieto-Torres J.L., DeDiego M.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Castaño-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10(5) doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Enjuanes L., Aguilella V.M. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2013;1828(9):2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers as a potential target for antiviral development. Antivir. Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pang H., Liu Y., Han X., Xu Y., Jiang F., Wu D., Kong X., Bartlam M., Rao Z. Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J. Gen. Virol. 2004;85(10):3109–3113. doi: 10.1099/vir.0.80111-0. [DOI] [PubMed] [Google Scholar]
- 38.Graham R.L., Sparks J.S., Eckerle L.D., Sims A.C., Denison M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133(1):88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-H. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59(14):6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020:1–5. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 41.Mengist H.M., Fan X., Jin T. Designing of improved drugs for COVID-19: crystal structure of SARS-CoV-2 main protease M pro. Signal Transduction and Targeted Therapy. 2020;5(1):1–2. doi: 10.1038/s41392-020-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raj V., Park J.G., Cho K.-H., Choi P., Kim T., Ham J., Lee J. Assessment of antiviral potencies of cannabinoids against SARS-CoV-2 using computational and in vitro approaches. Int. J. Biol. Macromol. 2021;168:474–485. doi: 10.1016/j.ijbiomac.2020.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shamsi A., Mohammad T., Anwar S., AlAjmi M.F., Hussain A., Rehman M., Islam A., Hassan M. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020;40(6) doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohammad T., Shamsi A., Anwar S., Umair M., Hussain A., Rehman M.T., AlAjmi M.F., Islam A., Hassan M.I. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: towards the development of effective COVID-19 therapy. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Molecular Informatics. 2020;39 doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;369:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019;93(6):e01815–e01818. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almeida M.S., Johnson M.A., Herrmann T., Geralt M., Wüthrich K. Novel β-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81(7):3151–3161. doi: 10.1128/JVI.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., Guo F., Zhao Z., Zhou Z., Xiang Z., Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11(1):020–17665. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stobart C.C., Sexton N.R., Munjal H., Lu X., Molland K.L., Tomar S., Mesecar A.D., Denison M.R. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J. Virol. 2013;87(23):12611–12618. doi: 10.1128/JVI.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Te Velthuis A.J., van den Worm S.H., Snijder E.J. The SARS-coronavirus nsp7+ nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Littler D., Gully B., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. BioRxiv. 2020;23(7) doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Brooun A., Griffith M., Moy K., Yadav M.K., Velasquez J., Buchmeier M.J. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J. Virol. 2006;80(16):7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113(6):701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 2005;353(5):1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. 2017;114(21):E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7(10) doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Computational and Structural Biotechnology Journal. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cantini F., Niccoli L., Matarrese D., Nicastri E., Stobbione P., Goletti D. Baricitinib therapy in COVID-19: a pilot study on safety and clinical impact. The Journal of Infection. 2020;2:318–356. doi: 10.1016/j.jinf.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalil A.C., Patterson T.F., Mehta A.K., Tomashek K.M., Wolfe C.R., Ghazaryan V., Marconi V.C., Ruiz-Palacios G.M., Hsieh L., Kline S. Baricitinib plus remdesivir for hospitalized adults with Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (London, England) 2020;395(10223):e30. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ge Y., Tian T., Huang S., Wan F., Li J., Li S., Yang H., Hong L., Wu N., Yuan E. bioRxiv; 2020. A Data-driven Drug Repositioning Framework Discovered a Potential Therapeutic Agent Targeting COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spinner C.D., Gottlieb R.L., Criner G.J., López J.R.A., Cattelan A.M., Viladomiu A.S., Ogbuagu O., Malhotra P., Mullane K.M., Castagna A. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. Jama. 2020;324(11):1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hung I.F.-N., Lung K.-C., Tso E.Y.-K., Liu R., Chung T.W.-H., Chu M.-Y., Ng Y.-Y., Lo J., Chan J., Tam A.R. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Borba M.G.S., Val F.F.A., Sampaio V.S., Alexandre M.A.A., Melo G.C., Brito M., Mourão M.P.G., Brito-Sousa J.D., Baía-da-Silva D., Guerra M.V.F. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Netw. Open. 2020;3(4):e208857. doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PubMed] [Google Scholar]
- 75.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modeling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;(5) doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ko M., Jeon S., Ryu W.-S., Kim S. bioRxiv; 2020. Comparative Analysis of Antiviral Efficacy of FDA-approved Drugs Against SARS-CoV-2 in Human Lung Cells: Nafamostat is the Most Potent Antiviral Drug Candidate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoffmann M., Schroeder S., Kleine-Weber H., Müller M.A., Drosten C., Pöhlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020;64(6) doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.A., Urquiza J., Ramírez D., Alonso C., Campillo N.E. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020;63(21):12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 80.Pepperrell T., Pilkington V., Owen A., Wang J., Hill A.M. Review of safety and minimum pricing of nitazoxanide for potential treatment of COVID-19. J. Virus Erad. 2020;6(2):52. doi: 10.1016/S2055-6640(20)30017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ceccarelli G., Alessandri F., d’Ettorre G., Borrazzo C., Spagnolello O., Oliva A., Ruberto F., Mastroianni C.M., Pugliese F., Venditti M. Is teicoplanin a complementary treatment option for COVID-19? The question remains. Int. J. Antimicrob. Agents. 2020;56(2) doi: 10.1016/j.ijantimicag.2020.106029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chakraborty C., Sharma A.R., Bhattacharya M., Sharma G., Lee S.S., Agoramoorthy G. COVID-19: consider IL6 receptor antagonist for the therapy of cytokine storm syndrome in SARS-CoV-2 infected patients. J. Med. Virol. 2020;92(11):2260–2262. doi: 10.1002/jmv.26078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson P.C., Richards D., Tanner H.L., Feldmann M. Accumulating evidence suggests anti-TNF therapy needs to be given trial priority in COVID-19 treatment. The Lancet Rheumatology. 2020;2(11):e653–e655. doi: 10.1016/S2665-9913(20)30309-X. [DOI] [PMC free article] [PubMed] [Google Scholar]