Abstract
In this study, we first determined the complete mitochondrial genome of the Triplophysa (Hedinichthys) yarkandensis (Day). The genome was 16,566 bp in length, containing 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and a non-coding region (D-loop region). The overall base composition of T. (H.) yarkandensis (Day) in descending order was T 28.20%, A 27.12%, C 25.61% and G 19.07%, with a slight A + T bias. The complete mitochondrial genome sequence could provide useful information for phylogenetic analysis and studies of population genetics of T. (H.) yarkandensis (Day).
Keywords: Genome, mitochondrial, Triplophysa (Hedinichthys) yarkandensis (Day)
Triplophysa (Hedinichthys) yarkandensis (Day 1876), also called Goutou fish (Dog-head), widespread and endemic to Tarim River with the world’s highest altitude, belongs to the genus of Cyprinidformes, Cobitidae, Nemachilinae, Triplophysa, Hedinichthys (Day 1876; Zhu 1989; Wang 1995). In order to provide useful information for the future research of genetic diversity and phylogenetics, this study first reported complete mitochondrial DNA sequence of T.(H.) yarkandensis (Day) (GenBank accession no. KT192439).
Triplophysa (H.) yarkandensis (Day) (voucher no. GYQ201408018) were obtained from the Tarim River, China (40°54′N; 81°31′E) using otter trawl, and conserved laboratory of wild fish research at Tarim University. Initially, the sample fish samples were identified based on both the morphologic features and the mitochondrial gene, COI. Tissue samples for molecular analysis were preserved in 95% ethanol. Whole genomic DNA was extracted from muscle tissue of individual specimens using the phenol–chloroform extraction method. Polymerase chain reaction (PCR) was performed. The PCR products were sequenced by Shanghai Personal Biotechnology Co, Ltd, Shanghai, China.
Sequences were assembled using Generious 4.5.3 (http://www.geneious.com). BioEdit 7.0 was used for sequence alignment. The neighbour-joining (NJ) methods were used to construct the phylogenetic trees. The NJ trees were obtained with 10,000 bootstrap replications using MEGA 5.0.
The total length of the complete mitogenome was 16,566 bp, with the base composition of 27.12% for A, 28.20% for T, 25.61% for C, 19.07% for G, in the order T > A > C > G feature occurring in the T. (H.) yarkandensis (Day). It contained the typical structure, including two ribosomal RNA subunits (12S rRNA and 16S rRNA), 13 protein-coding genes, 22 transfer RNA genes (tRNA) and a non-coding control region (D-loop region). Twelve protein-coding genes were encoded on the heavy strand, whereas ND6 was encoded on the light strand. Most protein-coding genes initiated with ATG except for CO1 which began with GTG. Five protein-coding genes (ND2, CO2, CO3, ND3 and CYTB) terminated with T–; four protein-coding genes (ATP6, ATP8, ND4L and ND6) terminated with TAA. Three protein-coding genes (ND1, ND5 and COX1) terminated with TAG whereas ND4 gene terminated with TA–. The lengths of 12 SrRNA and 16S rRNA were 953 bp and 1672 bp, respectively. They were located between the tRNA-phe and tRNA-Leu genes and separated by the tRNA-Val gene. The 22 tRNA genes ranged from 66 bp to 76 bp in size, which were distributed in rRNA and protein-coding genes. The complete mitochondrial genome D-loop region of T. (H.) yarkandensis (Day) was located between tRNA-Pro and tRNA-Phe with 918 bp in length (Figure 1).
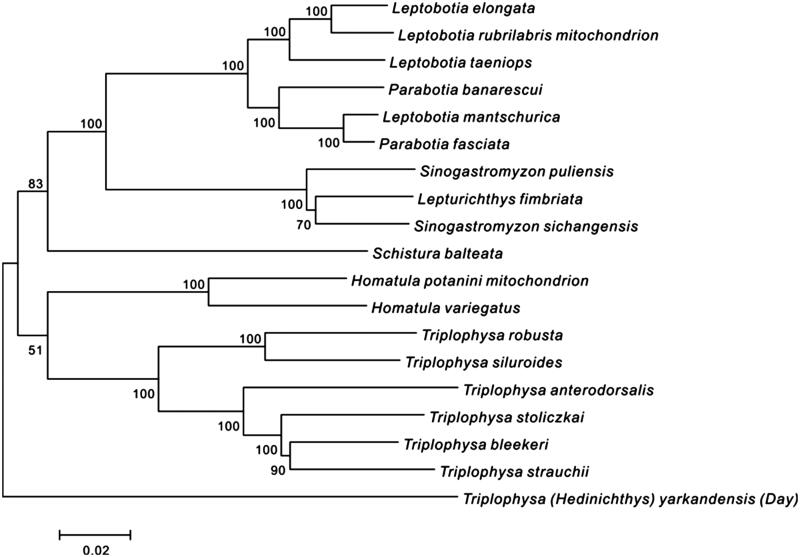
Figure 1.
Phylogenetic tree based on mitochondrial genome nucleotide sequences of T. (H.) yarkandensis (Day) (KT192439) and the other 18 kinds of fish set using the ML method. Genetic distances are listed above the branches. GenBank accession numbers of the sequences were used for the tree as follows: Leptobotia mantschurica (AB242170.1); Schistura balteata (AB242172.1); Sino gastromyzon puliensis (FJ605359.1); Leptobotia elongate (JQ230103.1); Triplophysa stoliczkai (JQ663847.1); Triplophysa bleekeri (JQ686729.1); Homatula variegatus (JX144893.1); Leptobotia rubrilabris (KF534784.1); Sinogastromyzonsichangensis (KF711948.1); Triplophysa anterodorsalis (KJ739868.1); Triplophysa siluroides (KJ781206.1); Lepturichthys fimbriata (KJ830772.1); Homatula potanini (KM017732.1); Leptobotia taeniops (KM386686.1); Parabotia banarescui (KM393222.1); Parabotia fasciata (KM393223.1); Triplophysa strauchii (KP297875.1).
As can be seen clearly from the graph above, Triplophysa (Hedinichthys) yarkandensis (Day) stands at the bottom of the phylogenetic tree, suggesting that compared with the other Cobitidae fishes, Triplophysa (Hedinichthys) yarkandensis (Day) is an outgroup. This is because of the whole classification of Triplophysa, Triplophysa (Hedinichthys) yarkandensis (Day) is a subfamily, while the other Triplophysa fishes, such as Triplophysamicrop and Triplophysarosa, are subgenuses of Triplophysa. The data of the phylogenetic tree support the classification of Triplophysa by Zhu (1989), Wu (1992) and others on the basis of morphology and geographical distribution of Triplophysa.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This research was supported by the National Natural Sciences Foundation of China (No. 31360635).
References
- Day F. 1876. On the fishes of York and proceedings. Zool Soc Lond. 53:781–807. [Google Scholar]
- Wang DZ. 1995. The changes of fishes fauna and protections of aboriginal fishes in the Tarim River. Arid Zone Res. 12:54–59. [Google Scholar]
- Wu YF, Wu CZ.. 1992. The fishes of the Qinghai-Xizang Plateau [M]. Chengdu: Sichuan Science and Technology Press; p. 256–259. [Google Scholar]
- Zhu SQ. 1989. The Loaches of the subfamily Nemacheilinae in China (Cypriniformes: Cobitidae) [M]. Nanjing: Jiangsu Science and Technology Press; p. 68–132. [Google Scholar]