Abstract
Cellular and humoral immunity to SARS-CoV-2 is critical to control primary infection and correlates with severity of disease. The role of SARS-CoV-2-specific T cell immunity, its relationship to antibodies, and pre-existing immunity against endemic coronaviruses (huCoV), which has been hypothesized to be protective, were investigated in 82 healthy donors (HDs), 204 recovered (RCs), and 92 active COVID-19 patients (ACs). ACs had high amounts of anti-SARS-CoV-2 nucleocapsid and spike IgG but lymphopenia and overall reduced antiviral T cell responses due to the inflammatory milieu, expression of inhibitory molecules (PD-1, Tim-3) as well as effector caspase-3, -7, and -8 activity in T cells. SARS-CoV-2-specific T cell immunity conferred by polyfunctional, mainly interferon-γ-secreting CD4+ T cells remained stable throughout convalescence, whereas humoral responses declined. Immune responses toward huCoV in RCs with mild disease and strong cellular SARS-CoV-2 T cell reactivity imply a protective role of pre-existing immunity against huCoV.
Keywords: COVID-19, SARS-CoV-2, antiviral T cell immunity, humoral immunity, endemic human coronavirus, antibody, convalescence, caspases, chemokine receptors, cell death
Graphical Abstract

COVID-19 varies from asymptomatic infection to multiorgan failure, but data on cellular immunity against SARS-CoV-2 during disease and beyond are lacking. Bonifacius et al. show a beneficial effect of preexisting immunity to endemic coronaviruses during disease and stable cellular immunity with concomitant decrease of humoral responses early during convalescence.
Introduction
SARS-CoV-2 infection was declared a pandemic by the World Health Organization (WHO) on March 11, 2020. Clinical manifestations of the resulting COVID-19 disease range from asymptomatic infection to acute respiratory failure and death. Seven to ten days after initial symptoms, COVID-19 progresses in a minority of patients to a severe illness requiring hospitalization, intensive care treatment, and mechanical ventilation (Huang et al., 2020; Zhou et al., 2020). Most patients have a mild course of COVID-19. The reasons for this as well as the question of whether and how long acquired COVID-19 immunity might protect against rechallenge, as described in rhesus macaques (Chandrashekar et al., 2020), are still unknown. However, cases of reinfection have been reported (AlFehaidi et al., 2020; Iwasaki, 2021).
In this context, reliable high-level evidence regarding adaptive antiviral immunity, its correlation with humoral responses, the temporal course of disease, and the potentially protective role of immunity against endemic coronaviruses (huCoV) is still lacking. The earliest detectable antibody (Ab) responses occur 3 days after the onset of symptoms, but seroconversion occurs within 7–14 days in the majority of patients (Huang et al., 2020; Krammer and Simon, 2020; Zhou et al., 2020). Abs against the nucleocapsid (N) and the spike (S) protein are commonly detected (Amanat et al., 2020; Ni et al., 2020; Sekine, 2020) and Abs against the immunogenic receptor-binding domain (RBD) of the S protein act as potently neutralizing Abs by binding to human angiotensin-converting enzyme 2 (Ju et al., 2020; Vabret et al., 2020; Zhou et al., 2020). It has been reported that COVID-19 is accompanied by changes in the immune cell compartment including increased numbers of Ab-producing plasmablasts, and this correlated with disease severity (Kuri-Cervantes et al., 2020; Mathew et al., 2020). One group found a correlation between neutralizing Ab titers and the number of antiviral T cells (Grifoni et al., 2020; Ni et al., 2020). SARS-CoV-2-specific CD4+ and CD8+ T cells recognizing peptides derived from N, S, and membrane (M) protein are generally detected in 70%–100% of active and recovered patients (Baruah and Bose, 2020; Grifoni et al., 2020; Huang et al., 2020; Leung et al., 2020; Ni et al., 2020; Peng et al., 2020a; Vabret et al., 2020; Weiskopf et al., 2020). Specific T cells are also detected in Ab-seronegative family members with asymptomatic or mild disease (Sekine, 2020) and in a small proportion of non-exposed individuals, indicating cross-reactivity in individuals infected with huCoV (Braun et al., 2020; Kissler et al., 2020; Weiskopf et al., 2020).
In this study, samples collected from 82 non-infected controls (healthy donors, HDs), 204 recovered COVID-19 patients (RCs), and 92 hospitalized, active COVID-19 patients (ACs) were analyzed at different time points. The study focused on key issues regarding the relationship, magnitude, and composition of humoral and cellular SARS-CoV-2 immunity as well as the question of whether Abs, functional, and phenotypic characteristics of antiviral T cells, plasma cytokines and chemokines, and pre-existing huCoV-specific T cells are associated with the outcome of COVID-19. ACs exhibited generally low T cell responses, which largely were not dependent on lymphopenia, but rather on the role of programmed cell death protein 1 (PD-1) and T cell immunoglobulin and mucin domain-3 (Tim-3) as well as caspase-mediated apoptosis in T cells. High IgG ratios of SARS-CoV-2 Abs were observed in ACs, suggesting an early role of the humoral immune response. The fact that RCs exhibited broad and strong T cell responses and HDs had pre-existing T cells against M and S protein yet no SARS-CoV-2 Abs suggests a potentially protective role of pre-existing T cell immunity. In follow-up samples from RCs, Ab levels decreased over time while T cell frequencies remained stable.
Our results suggest that T cell immunity is important for lasting protection against SARS-CoV-2. Monitoring of COVID-19 patients and SARS-CoV-2-naive donors and the resulting knowledge about antiviral T cell and other immune cell function in combination with the occurrence of specific Abs will lead to a better understanding of the pathology, protective mechanisms, disease outcome predictors, and the success of vaccination strategies.
Results
SARS-CoV-2 IgG ratios correlate with disease severity and decrease over time
Three cohorts were investigated in this study: HDs (n = 82), RCs (n = 204), and ACs (n = 92). None of the subjects had a history of transplantation or malignant disease. 90 ACs were hospitalized for COVID-19 and 2 for COVID-19-unrelated reasons (WHO score 2). 27 ACs were treated with remdesivir, 3 with dexamethasone, and 17 with both. None of the subjects received convalescent plasma. The average cohort age was 44 (range 18–68) years for HDs, 43 (19–68) years for RCs, and 60 (21–92) years for ACs (Figure S1A). SARS-CoV-2 ELISA for detection of Abs against N and S1 in plasma revealed that three HDs had IgG ratios defined as intermediate or positive results for one of the tested Abs, but negative results for the other (Figure 1 A). Assessing anti-SARS-CoV-2 Abs in HDs with low pre-test probability requires confirmation of positive results from single measurements by alternative serology tests or functional assays (Behrens et al., 2020). Additional western blot-based testing of these samples using SARS-CoV-2 recomLine was negative. Thus, these results were considered false positive. In the RC cohort, n = 151 (75.9%) and n = 169 (84.9%) subjects had detectable loads of Abs against N and S1, respectively (double-positive: n = 137 [68.8%]), while n = 10 (5.0%) did not. Overall, SARS-CoV-2 IgG ratios in ACs were slightly higher than in RCs and significantly higher than in HDs. S1- and N-specific SARS-CoV-2 Abs were detected in 62.0% and 72.8% of ACs, respectively. All ACs except one seroconverted for at least one of the tested Abs during weekly follow-up monitoring (time post positive PCR: mean 13 days; range 0–49 days) (Figure 1B). Significant correlations between anti-N and -S1 IgG ratios were observed in both RCs and ACs (Figure 1C).
Figure 1.
SARS-CoV-2 IgG ratios decline and COVID-19-associated humoral and cellular immune profile regresses throughout recovery
Shown are the SARS-CoV-2 humoral and cellular immune profiles of healthy donors (HD, green), recovered COVID-19 patients (RC, red), and patients with active COVID-19 (AC, blue).
(A) SARS-CoV-2 N and S1 IgG ratios in HDs, RCs, and ACs (first sample).
(B) Seroconversion during COVID-19, expressed as anti-N and -S1 IgG ratios during weekly follow-up of ACs. Dashed lines indicate cutoff values for negative (< 0.8) and intermediate IgG ratios (0.8–1.1).
(C) Left: Correlation between anti-N and -S1 IgG ratios in RCs and ACs (first sample). Right: Association between WHO score and SARS-CoV-2 antibodies. Dashed lines indicate cutoff values for negative and intermediate IgG ratios.
(D) Left: Correlation between disease severity score (DSS) or time of convalescence and SARS-CoV-2 IgG ratios. Right: SARS-CoV-2 IgG ratios over time in individual RCs. Each symbol and color represent data from one RC; numbers behind the lines indicate p values for the corresponding individual.
(E and F) Cellular profile was determined by flow cytometric analysis of whole blood.
(A–C) n = 76–82 (HD), n = 199–204 (RC), and n = 92 (AC; first sample).
(D) Left: n = 199; right: n = 16.
(E–F) n = 63–79 (HD), n = 171 (RC), and n = 91–92 (AC).
(A, E, and F) Kruskal-Wallis test and Dunn’s test for multiple comparisons or (C and D) linear regression analysis was used to calculate statistical significance: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figures S1–S3.
According to the Ordinal Scale for Clinical Improvement (WHO, 2020), all HDs had a WHO score of 0 (uninfected), 2.0% of RCs had a WHO score of 1 (ambulatory, no limitation of activities), 94.4% of 2 (ambulatory, limitation of activities), 3.1% of 3 (hospitalized, mild disease, no oxygen therapy), and 0.5% of 6 (hospitalized, severe disease, intubation, and mechanical ventilation). 60.9% of ACs had a WHO score of 1–4 (ambulatory to hospitalized, mild disease), and 39.1% of 5–7 (hospitalized, severe disease) (Figure 1C). The WHO scores for all cohorts significantly correlated with anti-N and -S1 IgG ratios. Since most RCs had mild disease with heterogeneous symptoms for a mean duration of 12 days (range 1–73 days), we used an internal Disease Severity Score (DSS) to further classify their symptoms. The results were as follows: no symptoms (n = 4, 2.0%), flu-like symptoms (n = 159, 81.1%), diarrhea or nausea (n = 20, 10.2%), fever (n = 90, 45.9%), respiratory symptoms (n = 138, 70.4%), neurological symptoms (n = 88, 44.9%), hospitalization (n = 7, 3.6%), and ventilation (n = 1, 0.5%). DSS significantly correlated with IgG ratios (Figure 1D). No correlation between DSS and age or sex was found (Figure S1B). Symptom duration in RCs correlated with age in males but not females (Figure S1B). Within RCs, no correlation between time of convalescence and IgG ratios was found (Figure 1D). The majority of ACs presented with high IgG ratios early during disease, and these remained stable or even increased during active disease (Figure 1B). However, analysis of individual RCs revealed a significant decrease in anti-N and -S1 IgG ratios over time (Figure 1D).
In summary, RCs and ACs were heterogeneous with respect to disease severity and IgG ratios and a correlation between disease severity and anti-N and -S1 IgG ratio during active disease and early recovery was found. High IgG ratios developed throughout the course of active disease indicating a functional humoral immune response. Overall, it appears that Ab responses peak at around day 50 followed by a decline thereafter.
Cellular immune profile is altered during COVID-19 and regresses throughout recovery
The observed decrease in IgG ratios shortly after recovery suggested that humoral immune responses against SARS-CoV-2 alone did not provide long-term immunity. Immune phenotyping of whole blood samples was performed to characterize the cellular profile during and after SARS-CoV-2 infection (Figures 1E and 1F; Figures S1–S3). Compared to RCs and HDs, ACs had a distinct infection-associated B cell phenotype as indicated by significantly decreased numbers of total CD19+ B cells along with decreased numbers of transitional (IgD+CD27-CD38hi), naive (CD27-CD38+CD24-), marginal zone (IgD+CD27+), non-switched (IgD+CD27+CD38lo), and switched memory (IgD-CD27+CD38lo) B cells (Figure 1E). ACs had significantly increased numbers of Ab-producing plasmablasts (CD27hiCD38hi) along with high anti-N and -S1 IgG ratios (Figure 1A). The regress of humoral immunity during convalescence was clearly indicated by a decrease of plasmablasts and an increase of transitional, naïve, and marginal zone B cells, coming closer to steady-state conditions found in the healthy population (Rudolf-Oliveira et al., 2015). The development of long-term humoral immunity elicited by SARS-CoV-2 infection was indicated by higher numbers of memory B cells in RCs when compared to ACs.
Consistent with previous reports (Huang and Pranata, 2020), ACs presented with overall reduction of lymphocyte and T cell counts (Figure 1F). All three cohorts had comparable CD4+ and CD8+ T cell ratios (Figure S1C), but RCs with intermediate DSS (3–7) had a slightly higher CD4/CD8 ratio than those with more severe disease (data not shown). In ACs, the T cell phenotype was characterized by decreased naive T cells (Tn) and a concomitant increase of CD4+ central memory (CD4+ Tcm) and CD8+ effector memory T cells re-expressing CD45RA (CD8+ Temra) (Figures S1C and S1D). The fraction of CD4+ Tcm cells remained elevated during recovery (Figures S1D). ACs had reduced numbers of γδ T cells and NK cells compared to HDs (Figure 1F). These cell subsets increased in RCs compared to HDs during convalescence. Compared to RCs and HDs, ACs had increased granulocyte and monocyte counts, which positively correlated with disease severity (Figure 1F; Figure S3A). Follow-up samples from individual RCs indicated that cellular immunity, determined mostly by the number of CD45+ lymphocytes and granulocytes, regressed to a steady state (Figure S3B).
Overall, ACs exhibited the characteristic immunological cellular profile associated with the development of lymphopenia throughout the course of disease, distinguishing them from RCs and HDs. These results suggest a strong pro-inflammatory response during active disease and the restoration of cellular immunity during recovery.
Recovery from mild COVID-19 is associated with a broad anti-SARS-CoV-2 T cell repertoire
Frequencies of antiviral T cells against peptide pools derived from SARS-CoV-2, huCoV strains OC43 and 229E, Respiratory Syncytial Virus (RSV), Influenza A Virus (IAV), and Cytomegalovirus (CMV) were enumerated by interferon-gamma (IFN-γ)-enzyme-linked immunospot assay (ELISPOT). Each subject was classified as a non-responder (NR), low (LR), intermediate (IR), or high responder (HR) based on the results (Figure 2 ). 75.6% of HDs did not have SARS-CoV-2-specific T cells against any of the tested pools, whereas 15.5%, 11.9%, and 8.5% had M-, S-, and S2-specific T cells, respectively (Figure 2A). Most RCs had T cells against a broad variety of SARS-CoV-2-derived antigens (Figure 2B). 64.2%–72.1% exhibited T cells against M, N, S1, and S2, whereas only 45.6% had detectable T cells against S. Six of the ten RCs without SARS-CoV-2-specific Abs had specific T cells. In RCs, highest T cell frequencies were observed for M, S1, and S2; nevertheless, most RCs were classified as LR or IR. 85.9% of ACs exhibited SARS-CoV-2-specific T cells covering a broad spectrum of antigens throughout the course of disease (Figure S4A), but T cell frequencies were generally lower and very heterogeneous. Most responders were found for S1 (40%) and S2 (32%) (Figure 2C), and T cells against M, S1, and S2 were most frequent (mean number of spots per well [spw] 8.2, 10.2, and 11.0, respectively).
Figure 2.
Recovery from mild COVID-19 is associated with a broad anti-SARS-CoV-2 T cell repertoire
(A–C) IFN-γ ELISPOT data from (A) healthy donors (HD, green, n = 48–82), (B) recovered COVID-19 patients (RC, red, n = 110–204), and (C) patients with active COVID-19 (AC, blue, n = 86–92; first sample) are depicted as the number of spots per well (spw)/2.5x105 PBMCs on the left side. For CMV_pp65 only values for seropositive individuals are depicted. The corresponding frequencies of non- (NR), low (LR), intermediate (IR), and high responders (HR) are shown on the right side.
(D) Inter-cohort comparison of antiviral T cell frequencies determined by ELISPOT assay normalized to T cell frequencies within PBMCs, depicted as spw/1.0x104 CD3+ T cells. Statistical significance was calculated by Kruskal-Wallis test and Dunn’s test for multiple comparisons.
(A–C) Statistically significant differences to negative control (NC) are depicted. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S4.
Overall T cell immunity, measured as the frequency of T cells specific for CMV phosphoprotein 65 (CMV_pp65) in CMV-seropositive subjects, was markedly lower in ACs than in RCs and HDs (mean spw: 34.2, 185.0, and 191.3, respectively). All CMV-seropositive RCs and HDs had detectable CMV_pp65-specific T cells, whereas 40 of 78 (51.3%) CMV-seropositive ACs did not. While RCs and HDs had comparable T cell frequencies specific for OC43_S and 229E_S, ACs had markedly lower frequencies against these antigens and against those from RSV and IAV. RCs had lower frequencies of RSV- and IAV-specific T cells than HDs even though they had recovered from lymphopenia. Moreover, RCs with no detectable SARS-CoV-2-specific T cells also had lower fractions of responders for RSV and IAV (data not shown), indicating an overall reduction of T cell immunity in these subjects.
To adjust for differences due to the observed reduction of T cell frequencies in ACs, ELISPOT data were normalized to the number of CD3+ T cells within peripheral blood mononuclear cells (PBMCs) (Figure 2D; Figure S4B). The comparison showed that the differences in antiviral T cell frequencies observed in the three cohorts were independent of CD3+ T cell numbers.
In summary, SARS-CoV-2-specific T cells were detected in all three cohorts. Post-recovery T cell responses were heterogeneous, and RCs exhibited the highest frequencies of antiviral T cells for M-, N-, and S-derived peptide pools. Both RCs and ACs had reduced RSV- and IAV-specific T cell frequencies compared to controls. In general, ACs had a reduced antiviral T cell repertoire, which did not seem to be caused by a reduction of T cell numbers alone.
Antiviral T cell repertoire remains stable during recovery from mild COVID-19
The finding that SARS-CoV-2 IgG ratios and infection-associated B cell subsets decreased during recovery from COVID-19 suggests an important role of T cell immunity (Figures 1D and 1E; Figure S3B). Analysis of the RC cohort showed that increasing DSS scores were generally associated with increasing M- and S-specific T cell frequencies but, paradoxically, subjects with a DSS > 6 had lower SARS-CoV-2-specific T cell frequencies (Figure 3 A). Frequencies of T cells against M-, N-, and S-derived peptide pools not only remained stable but even became higher over the course of the convalescence period (Figure 3B). Analysis of follow-up samples from individual RC subjects collected up to 102 days after symptom onset revealed that T cell frequencies for SARS-CoV-2, huCoV, RSV, and IAV remained mostly stable (Figure 3C) and suggest the development of potentially protective T cell immunity during moderate COVID-19.
Figure 3.
Antiviral T cell repertoire remains stable during recovery from mild COVID-19
(A) Antiviral T cell frequencies in relation to disease severity in recovered COVID-19 patients (RCs; red, n = 136–204).
(B) SARS-CoV-2_M-, N-, S-, S1-, and S2-specific T cell frequencies during convalescence in RCs (n = 110–178). Statistical significance was calculated by linear regression analysis.
(C) Frequencies of T cells specific for SARS-CoV-2 (upper panel), OC43, 229E, respiratory syncytial virus (RSV) and influenza A virus (IAV) (lower panel) in follow-up samples from RCs (n = 4–15).
See also Figure S4.
Together with the finding that active COVID-19 is associated with high SARS-CoV-2 IgG ratios but overall low T cell functionality, these results indicate that humoral immune responses plays an early role, whereas resolved mild COVID-19 appears to be associated with stable T cell immunity rather than long-lasting humoral immunity.
SARS-CoV-2 T cell frequencies partially correlate with SARS-CoV-2 IgG ratios
The diverse range of SARS-CoV-2 IgG ratios and T cell repertoires partly correlated with disease severity. To gain insight into whether there is an association between the establishment of humoral and cellular immunity against SARS-CoV-2, we investigated the relationship between SARS-CoV-2 IgG ratios and SARS-CoV-2-specific T cells in RCs and ACs. The results, shown as IgG ratios in the T cell responder groups, suggest that IgG ratios increased with increasing antiviral T cells (Figure 4 ). In the RC cohort, HR (mean IgG ratio 2.88) and IR (ratio 2.62) for SARS-CoV-2_M had significantly higher SARS-CoV-2_N IgG ratios than NR (ratio 1.90; Figure 4A). Within the AC cohort, SARS-CoV-2_N IR had significantly higher anti-N IgG ratios (ratio 5.31) than NR (ratio 3.10). Similar tendencies were found for M- and S-specific T cells. In the case of S1-specific T cells, both RCs and ACs with intermediate or high frequencies of antiviral T cells had higher amounts of Abs against N (RCs HR ratio 3.62, ACs IR ratio 4.52) and S1 protein (RCs HR 5.59, ACs IR 5.83) than NR or LR (N: RCs NR 1.93, RCs LR 2.09, ACs NR 2.55; S1: RCs LR 2.78, ACs NR 3.17; Figures 4A and 4B).
Figure 4.
SARS-CoV-2 T cell frequencies partially correlate with SARS-CoV-2 IgG ratios
Recovered (RC, red, n = 131–199) and active COVID-19 patients (AC, blue, n = 90; first sample) were classified as non- (NR), low (LR), intermediate (IR), and high responders (HR) for the indicated antigens based on ELISPOT results. Dashed lines indicate cutoff values for negative and intermediate IgG ratios.
(A and B) Association between SARS-CoV-2-specific T cells and (A) anti-N or (B) anti-S1 IgG ratios. Statistical significance was calculated by Kruskal-Wallis test and Dunn’s test for multiple comparisons. ∗p < 0.05.
Overall, higher frequencies of SARS-CoV-2-specific T cells were found with higher SARS-CoV-2 IgG ratios, indicating some connection between humoral and cellular immunity in moderate cases of COVID-19.
COVID-19 patients with pre-existing T cell immunity against endemic coronaviruses have higher SARS-CoV-2_S-specific T cell frequencies
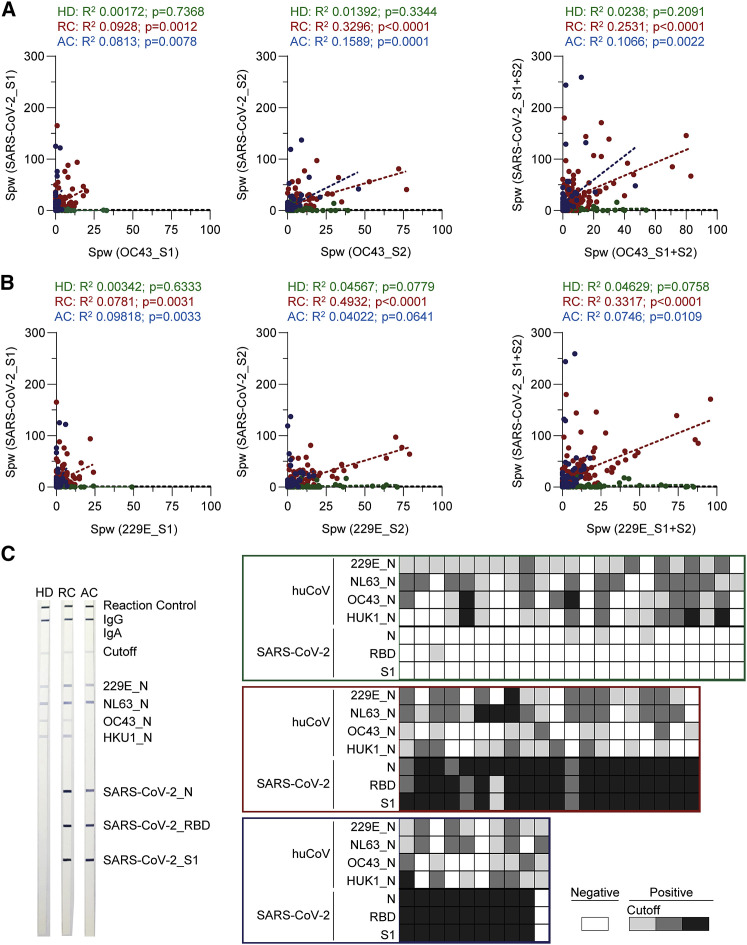
To study whether pre-existing T cell immunity against endemic huCoV strains OC43 and 229E was connected to SARS-CoV-2 T cell immunity, we correlated T cell frequencies for S-derived peptide pools S1 and S2 of SARS-CoV-2 with those for huCoV strains OC43 and 229E (Figure 5 ). As mentioned, HDs were heterogeneous with respect to OC43- and 229E-specific T cell frequencies, and some had high frequencies of SARS-CoV-2-specific T cells (Figure 2B). Although no significant correlation between frequencies of antiviral T cells against OC43 and 229E with SARS-CoV-2_S1- or S2-specific T cells was observed (Figures 5A and 5B), this did not exclude the possible presence of cross-reactive T cells. In RCs and in some ACs, subjects with higher OC43 or 229E_S-specific T cell frequencies exhibited higher numbers of SARS-CoV-2_S1 and SARS_CoV-2_S2-specific T cells. When testing plasma from 23 HDs, 20 RCs, and 10 ACs for the presence of huCoV-specific Abs, 61%, 45%, and 60% had OC43_N- and 87%, 75%, and 80% had 229E_N-specific Abs, respectively (Figure 5C). Some of those RCs lacked detectable huCoV-specific Abs but had huCoV-specific T cells. The latter finding is in line with reports showing waning huCoV-specific Abs over time but rapid functional immune responses upon second infection (Sariol and Perlman, 2020). Although ACs were heterogeneous with respect to SARS-CoV-2 T cells and their T cell responses against the common cold viruses were generally reduced, their SARS-CoV-2_S-specific T cells significantly correlated with OC43- and 229E-specific T cell frequencies, hinting toward cross-reactivity.
Figure 5.
COVID-19 patients with pre-existing T cell immunity against endemic coronaviruses have higher SARS-CoV-2_S-specific T cell frequencies
(A) Correlation between OC43_S- and SARS-CoV-2_S-specific T cells in healthy donors (HD, green, n = 69), recovered (RC, red, n = 136) and active COVID-19 patients (AC, blue, n = 92; first sample).
(B) Correlation between 229E_S- and SARS-CoV-2_S-specific T cells in HDs (green, n = 69), RCs (red, n = 136) and ACs (blue, n = 92; first sample).
(A and B) Star-shaped symbols indicate RCs tested for huCoV-specific antibodies. Statistical significance was calculated by linear regression analysis.
(C) Exemplary western blots for SARS-CoV-2 and huCoV and summarized results from selected HDs (green), RCs (red) and ACs (blue, first sample). Each column represents data from one subject; the legend indicates signal strength.
In summary, ACs and RCs with T cell immunity against huCoV strains OC43 and 229E exhibited higher SARS-CoV-2_S-specific T cell numbers. These results suggest that pre-existing cellular immunity against endemic huCoV might be beneficial for the development of SARS-CoV-2 T cell immunity and for disease outcome.
Overall impairment of T cell function contributes to reduced antiviral T cell immunity during COVID-19
There is evidence that a subset of COVID-19 cases is characterized by the development of a cytokine storm (Huang et al., 2020) as well as lymphopenia (Huang and Pranata, 2020). To gain insight into the underlying mechanism, cytokine and chemokine concentrations in plasma and expression of the respective receptor molecules were analyzed. As reported before for acute respiratory infection (Schögler et al., 2016), plasma IFN-γ-induced protein (IP-10) concentrations were significantly increased in ACs (mean 528.2 pg/mL) compared to HDs (mean 116.1 pg/mL) and RCs (mean 113.4 pg/mL) (Figure 6 A). Frequencies of T cells expressing the corresponding cellular receptor, CXCR3, were significantly lower in ACs (mean 29.91%) than in HDs (mean 45.10%) and RCs (mean 48.87%). Moreover, ACs had significantly increased amounts of T cell polarization-associated cytokines IL-6 and IL-12p70, a sign of enhanced IP-10-induced T cell polarization (Figure 6B). While the frequencies of CXCR3+ T cells and IP-10 concentrations in plasma were independent of disease severity, IL-6 concentrations in ACs were positively correlated with disease severity (Figures S5A and S5B). Plasma IL-2 concentrations in ACs were slightly lower than those in HDs and RCs, and those in RCs were higher than those in HDs (Figure 6B). In addition to CXCR3, independently of disease severity, frequencies of T cells expressing CCR2 were significantly reduced in ACs (mean 16.33%) compared to HDs (mean 27.92%) and RCs (mean 29.82%), whereas concentrations of its ligand, CCL2 (MCP-1), were highest in RCs, followed by ACs and HDs (Figure 6C; Figure S5C). Significantly higher amounts of soluble CD25 (sCD25) were detected in ACs compared to HDs and RCs but did not correlate with T cell counts in blood (Figure 6D; Figure S5D). ACs had high numbers of plasmablasts in conjunction with increased levels of sCD27, and this positively correlated with disease severity.
Figure 6.
Overall impairment of T cell function contributes to reduced antiviral T cell immunity during COVID-19
Chemokine and immune checkpoint receptor expression on T cells as well as caspase-3 and/or -7 and caspase-8 activity in T cells were determined by flow cytometry, and plasma of healthy donors (HD, green, n = 24–43), recovered COVID-19 patients (RC, red, n = 47–66) and active COVID-19 patients (AC, blue, n = 65–92; first sample) was analyzed by LEGENDPlex assay. Statistical significance was calculated by Kruskal-Wallis test and Dunn’s test for multiple comparisons: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S5.
The reduced antiviral T cell functionality observed by ELISPOT is not necessarily caused by reduced T cell numbers but rather influenced by inhibitory molecules and immune checkpoint receptors. ACs had elevated frequencies of T cells expressing PD-1 (mean 28.79%) compared to HDs (mean 24.49%) and RCs (mean 21.31%), whereas soluble PD-1 concentration was comparable between the three cohorts (Figure 6E). Compared to RCs and HDs, ACs had increased plasma programmed death ligand 1 (PD-L1) and decreased PD-L2 concentrations, independent of disease severity (Figure 6E; Figure S5E). Consistent with this high PD-L1 and PD-1 expression, ACs also had increased plasma IL-10 concentrations compared to RCs and HDs. In contrast to PD-1, the frequency of Tim-3+ T cells was reduced in ACs (mean 30.18%) compared to HDs (mean 41.80%) and RCs (mean 38.59%) (Figure 6F). Likewise, soluble Tim-3 (sTim-3) in ACs was higher than in RCs and HDs and was positively correlated with disease severity (Figure 6F; Figure S5F). In addition to increased PD-1 and PD-L1 expression, ACs also had significantly higher amounts of the Tim-3 ligand galectin-9 and, consequently, higher fractions of potentially apoptotic T cells, as indicated by frequencies of cells with active caspase-3 and/or -7 and caspase-8 (mean 11.13% and 8.26%), compared to HDs (mean 2.31% and 1.76%) and RCs (mean 3.09% and 3.63%), respectively (Figure 6G). Similar to PD-L1 and PD-L2, the amount of galectin-9 and frequencies of cells with active effector caspases did not correlate with disease severity (Figure S5G).
The signature of SARS-CoV-2-specific T cell recall responses is diverse and CD4-mediated
Since T cell immunity appears to be maintained after the resolution of COVID-19, we analyzed the SARS-CoV-2-specific inflammatory signature in more detail. In vitro peptide pool-stimulated PBMCs from selected RCs were tested for cytokine and chemokine release and evaluated by ELISPOT assay to determine antiviral T cell frequencies (Figures 7A and 7B). SARS-CoV-2_M-, N-, and S-derived peptide pools induced high secretion of essential immune mediators, with concentrations close to those reached in response to CMV_pp65 in CMV-seropositive donors (Figure 7A). Upregulation of T helper 1 (Th1) and Th2 cytokines IL-2, IL-4, IL-10, and tumor necrosis factor-α (TNF-α) as well as of pro-inflammatory chemokines IP-10, MCP-1, and IL-8 occurred after stimulation with SARS-CoV-2_M, N, S, S1, and S2, which was higher than that obtained after stimulation with S-derived peptide pools from OC43 and 229E strains. This finding might be connected to the lower T cell frequencies against these antigens compared to SARS-CoV-2 (Figure 7B). As expected, none of the antigens resulted in an increase in transforming growth factor-β (TGF-β) secretion. The highest degree of upregulation was observed for IFN-γ, which was comparable for all peptide pools. Granzyme B (Gzmb) concentrations in the same culture supernatants were measured by ELISA (Figure 7C). None of the antigens induced Gzmb concentrations as high as those found in CMV-seropositive subjects stimulated with CMV_pp65. However, marked increases in Gzmb were observed after stimulation with SARS-CoV-2_M, N, and S as well as after stimulation with OC43_S1 and S2.
Figure 7.
The signature of SARS-CoV-2-specific T cell recall responses is diverse and CD4-mediated
(A–C) Cell culture supernatants of PBMCs of recovered COVID-19 patients (RCs) stimulated with the specified peptide pools for 20 h were analyzed by (A and B) LEGENDPlex assay (n = 8; CMV_pp65: seropositive subjects only [n = 6]) or (C) ELISA (n = 7; CMV_pp65: seropositive subjects only [n = 5]). Error bars indicate standard deviation.
(B) Data are shown as fold change to unstimulated controls. For key to color code, see legend (n = 8).
(C) Statistical significance was calculated by Friedman’s test and Dunn’s test for multiple comparisons; CMV_pp65 was excluded from the analysis. ∗p < 0.05.
(D) Frequencies of IFN-γ and granzyme B (Gzmb)-producing cells in response to the specified peptide pools were determined by FluoroSpot. Exemplary and summarized results for n = 5 cases are shown.
(E and F) PBMCs from RC were stimulated with the specified peptide pools for 5 h, followed by flow cytometric analysis. Graphs show frequencies of IFN-γ+ TNF-α- (yellow), IFN-γ+ TNF-α+ (green), and IFN-γ- TNF-α+ (blue) cells within CD4+ (E) and CD8+ (F) T cell subsets (n = 8; CMV_pp65: seropositive subjects only [n = 6]).
See also Figure S6.
The stimulating potential of the highly immunodominant peptide pools was further affirmed by IFN-γ and Gzmb FluoroSpot assay (Figure 7D). Despite appreciable frequencies of IFN-γ and Gzmb single-producing cells, the fraction of IFN-γ and Gzmb double-producing cells was comparably low for all tested antigens, indicating the engagement of different T cell subsets. To identify the main involved T cell populations, intracellular IFN-γ and TNF-α expression was measured after antigenic stimulation in PBMCs from RCs (Figures 7E and 7F). Among CD4+ T cells, high frequencies of IFN-γ+ TNF-α+ double- and TNF-α+ single-positive cells were detected in response to SARS-CoV-2_M-, N-, and S-derived peptide pools (Figure 7E). The highest frequencies were observed within CD4+ Tem cells and in response to the SARS-CoV-2_S2 peptide pool. HuCoV-derived peptide pools induced mainly TNF-α. CD8+ T cell responses to SARS-CoV-2 and common cold peptide pools were considerably lower than those to CMV_pp65 (CMV-seropositive donors only) (Figure 7F). In summary, these findings show that the SARS-CoV-2 immune response mainly exerts a Th1 and Th2 signature and that CD4+ Tem cells produce the most cytokines in response to SARS-CoV-2 antigens.
SARS-CoV-2-specific T cells for adoptive immunotherapy can be enriched from RCs
Since ACs with severe disease presented with high SARS-CoV-2-specific Abs yet low SARS-CoV-2-specific T cell frequencies, adoptive T cell transfer might be a promising treatment strategy. Therefore, an IFN-γ cytokine secretion assay (CSA) was performed to evaluate the suitability of SARS-CoV-2 overlapping peptide pools M, N, and/or S as target antigens for the generation of clinical-grade SARS-CoV-2-specific T cells (Figures S6A–S6D) (Priesner et al., 2016). The highest IFN-γ+ T cell frequencies were detected after stimulation with all three peptide pools combined (mean 0.3%). However, the enrichment efficiency was higher for the respective peptide pools alone, while N resulted in the highest purity (mean 48.0%). In line with our finding that the majority of SARS-CoV-2-specific T cells were CD4+, we observed a slightly enhanced proportion of CD4+ T cells in the enriched cell fractions (data not shown). The cytotoxic potential of T cells specific for SARS-CoV-2 and CMV_pp65 was examined in CD4+ and CD8+ T cells sorted according to their IFN-γ production after stimulation and CSA (Figures S6E–S6G). Messenger RNA (mRNA) expression of IFN-γ was generally higher in stimulated CD4+ T cells than CD8+ T cells and was highest in response to S, but not as high as for CMV_pp65 (Figure S6E). Regarding GZMB and perforin (PRF1) mRNA, the highest expression was observed within positive fractions of CD8+ T cells after stimulation with N (Figures S6F and S6G).
Overall, SARS-CoV-2-specific T cells against M, N and S can be efficiently generated with acceptable purity and with CD8+ T cells that are more cytotoxic than CD4+ T cells.
Discussion
A better understanding of the humoral and cellular players involved in COVID-19 disease progression and recovery is urgently needed to guide treatment strategies, predict disease outcome, and determine whether patients develop long-lasting immunity. In this study, we determined SARS-CoV-2 IgG ratios and T cell frequencies against a variety of SARS-CoV-2 proteins in HDs, RCs, and ACs in a cross-sectional manner. The analysis was extended to antigens derived from huCoV strains 229E and OC43 and antigens from RSV, IAV, and CMV. Longitudinal analysis revealed a quantitative reduction of the humoral SARS-CoV-2-specific immune response, together with constant T cell immunity up to 102 days after symptom onset. Pre-existing antiviral T cells against huCoV had beneficial effects on the development of effective T cell immunity, although during active disease a markedly reduced antiviral T cell immunity was observed.
Humoral immunity is defined by varying titers of neutralizing Abs (Amanat et al., 2020; Ni et al., 2020; Wu et al., 2020). Recent reports that COVID-19 patients become seronegative early during convalescence (Long et al., 2020; Seow et al., 2020; Ward et al., 2020) raise the question of whether recovery from COVID-19 confers immunity to reinfection, as was shown in a rhesus macaque model (Chandrashekar et al., 2020). On the other hand, it was found that IgG and neutralizing Abs remained stable for up to four months, whereas IgA and IgM decayed rapidly (Isho et al., 2020; Iyer et al., 2020). To date, a few confirmed cases of SARS-CoV-2 reinfection have been reported, and reinfection resulted in a worse course of disease in some cases (AlFehaidi et al., 2020; Iwasaki, 2021). Although it was recently reported that the number of N-specific T cells correlates with neutralizing Ab titers in convalescent subjects (Ni et al., 2020; Swadling and Maini, 2020), our study revealed no correlation in RCs but in ACs. Moreover, individuals from both cohorts with high SARS-CoV-2_S1-specific T cell frequencies had significantly higher S1 Ab values than those with low T cell frequencies, indicating that S-specific T cell responses correlated with S-specific Ab responses (Peng et al., 2020b).
It has been assumed that a certain degree of cross-reactive coronavirus immunity might influence susceptibility to COVID-19 (Braun et al., 2020; Le Bert et al., 2020; Mateus et al., 2020). Braun and colleagues suggest that pre-existing S-reactive SARS-CoV-2 CD4+ T cells might be protective (Braun et al., 2020). In line with that, SARS-CoV-2-specific T cells mainly directed against M and S were detected in a fraction of SARS-CoV-2-seronegative controls in this study. Moreover, we found evidence of a correlation between huCoV_S-specific and SARS-CoV-2_S-specific T cells in RCs and ACs, implying a possibly protective role of pre-existing, potentially cross-reactive T cells (Selin et al., 2006).
The immune cell composition has been described as a partial predictor for discriminating between mild and severe COVID-19 (Odak et al., 2020; Velavan and Meyer, 2020) and extended lymphopenia with reduced T cell numbers correlates with disease severity (Chen et al., 2020; Ni et al., 2020; Wang et al., 2020). In this study, lymphocyte counts did not correlate with disease severity in hospitalized COVID-19 patients. We found that the immune cell profile was restored in patients with moderate symptoms after a relatively short period of convalescence. Moreover, M- and S-specific T cell frequencies were higher in RCs with an intermediate DSS than in those with a low DSS, whereas subjects with a high DSS had less SARS-CoV-2-specific T cells, indicating a connection between disease severity and the establishment of T cell immunity.
ACs exhibited a highly inflammatory milieu, which might be one reason for the reduced T cell immunity. Various mechanisms contributing to T cell loss are suspected, but it is assumed that the inflammatory milieu characterized by high IL-6, IL-10 and TNF-α amounts is responsible (Diao et al., 2020; Wang et al., 2020). ACs indeed, exhibited high amounts of IL-6 and IL-10, possibly driven by IP-10-induced T cell polarization. We found that the reduced number of functional antiviral T cells detected was not solely caused by a general reduction of T cell numbers, since T cell responses to CMV_pp65 were also reduced during COVID-19 and this was independent of circulating T cell numbers in the blood. Reduced frequencies of CXCR3+ and CCR2+ T cells might be indicative of enhanced migration, leading to a reduction of memory T cells. On the other hand, high amounts of plasma IP-10 were detected in ACs and continuously high IP-10 concentrations result in CXCR3 internalization (Metzemaekers et al., 2018), thereby possibly reducing cell migration to the place of infection and inhibiting overshooting inflammation. Moreover, it has been shown that the CCR2 and CCR5 inhibitor cenicriviroc, also known as an HIV-1 inhibitor via blockade of the HIV-1 co-receptor CCR5, inhibits SARS-CoV-2 replication in vitro (Okamoto et al., 2020).
Zhang et al. recently reported that sCD25 concentrations were increased in COVID-19 patients and negatively correlated with T cell numbers (Zhang et al., 2020). We also observed a significant increase in sCD25 in plasma of ACs compared to RCs and HDs, but the increase did not correlate with blood T cell count. In addition to a general T cell loss during SARS-CoV infection, T cell priming was shown to be negatively affected by elevated plasma IL-6 and reduced DC migration (Sariol and Perlman, 2020; Yoshikawa et al., 2009; Zhao et al., 2009). PD-L2, mainly produced by activated DCs, was significantly reduced, indicating impaired T cell priming during COVID-19. In contrast, ACs exhibited high concentrations of PD-L1 and high frequencies of PD-1+ T cells, suggesting T cell exhaustion or anergy, which is in line with the observed high amounts of IL-10 (Zou and Chen, 2008).
PD-1 expression can be a sign of T cell activation during acute viral infection (Ahn et al., 2018). However, the PD-L1-PD-1 axis may also lead to enhanced T cell apoptosis, further supported by a significant increase in frequencies of T cells with active caspases. In addition, there is in vitro evidence that caspase-8-dependent cell death may be induced by a murine coronavirus as well as SARS-CoV-2 (Li et al., 2020; Zheng et al., 2020). In contrast to PD-1, Tim-3+ T cell frequencies were reduced in ACs, while we observed increased amounts of sTim-3 in plasma of ACs. sTim-3 has been reported to reduce T cell activation by reducing IL-2 production (Yasinska et al., 2019); however, we did not find any decrease in IL-2. In addition, mechanisms including cell migration (IP-10-CXCR3 axis) and inhibitory molecules (PD-L1-PD-1 axis) as well as cell death (caspases-3, -7 and -8) contributed to the overall reduction of functionality in ACs. Apparently, the plasma cytokine milieu may have the potential to discriminate between mild and severe disease, suggesting their utility as prognostic markers.
In the majority of COVID-19 cases, the adaptive immune system is trained to control the virus within one to two weeks, but a minority progress to severe disease and require ICU admission (Huang et al., 2020; Rees et al., 2020). Impaired T cell function and immunosuppression may contribute to viral persistence. On the other hand, exhaustion of T cells may also prevent immunopathology (Cornberg et al., 2013). We found that ACs have generally reduced T cell immunity, as evidenced by low frequencies of antiviral T cells and the absence of CMV_pp65-reactive T cells in some CMV-seropositive ACs. The fact that ACs also had high anti-N and -S1 IgG ratios constitutes further evidence that neutralization of SARS-CoV-2 infection occurs via S1-specific Abs covering the RBD of S protein (Zhou et al., 2020). However, Abs alone appear to be insufficient for viral clearance. Adoptive transfer of SARS-CoV-2-enriched T cells from COVID-19 convalescent donors might be a therapeutic option for patients with severe disease (Ferreras et al., 2020; Leung et al., 2020), but is restricted by the availability of HLA-matched donors with sufficient SARS-CoV-2-specific T cell frequencies. The risk of graft-versus-host disease (GvHD) due to alloreactivity and the risk of induction of cytokine storm syndrome and lung damage are major concerns (Mehta et al., 2020; Ong et al., 2020). Ferreras et al. postulated that in vitro depletion of Tn cells via CD45RA would result in immediately available, ‘off-the-shelf’ SARS-CoV-2-specific memory T cell products (Ferreras et al., 2020).
Throughout this study, one HD became infected with SARS-CoV-2 and was diagnosed with WHO 3 and DSS 8 COVID-19. The T cell repertoire analysis revealed a low frequency of SARS-CoV-2-specific T cells prior to infection and an LR for M-, N-, and S-specific T cells. The subject had decreased lymphocyte numbers, diminished overall T cell responses, and increased amounts of plasma IP-10 and IL-8 early during recovery. The impact of pre-existing SARS-CoV-2-reactive T cells in naive individuals is a subject of high interest. Prospective studies in larger cohorts are needed to determine whether their presence correlates with pathology or protection from COVID-19.
A better understanding of the role of the adaptive immune system during the course of SARS-CoV-2 infection and beyond will improve COVID-19 diagnosis and prognosis and is crucial for the successful development of vaccines and cell-based therapies. Active COVID-19 patients included in this study had high SARS-CoV-2 IgG ratios yet generally reduced antiviral T cell frequencies compared to recovered COVID-19 patients, suggesting that T cells are important for the successful clearance of infection. This study provides evidence that the generally low T cell responses observed during COVID-19 are not exclusively dependent on lymphopenia, but rather on the role of PD-1 and Tim-3 as well as caspase-mediated cell death. Our study indicates that SARS-CoV-2-specific cellular immunity may be more stable and longer lasting than humoral immunity. In addition, our findings suggest that the presence of pre-existing SARS-CoV-2-specific T cells and T cells against epitopes from endemic huCoV might be protective.
Limitations of study
Our study provides evidence for a contribution of pre-existing memory CD4+ and CD8+ T cells against huCoV strains 229E and OC43, but pre-existing humoral responses against these strains and further huCoV strains were not considered. Moreover, we reported one case with informative immune responses before and after SARS-CoV-2 exposure. Collecting data on pre-existing cellular and humoral immunity prior to infection and during the course of the disease is very important to elucidate the role of pre-existing immunity in more detail. The results of our study are further limited to T cell responses against the huCoV S proteins and do not take into account other structural and non-structural proteins, nor HLA diversity and the resulting peptide presentation. Finally, our study relies on detection of IFN-γ secreting T cells after short-term stimulation, wheras other effector molecules such as TNF-α or Gzmb as well as the detection of lower frequencies of antiviral T cells after prolonged re-stimulation may provide further insight into the importance of these effector cells for viral clearance.
STAR★Methods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-Cy7 anti-CD45 | BD Biosciences | Cat#641417, Clone 2D1; RRID: 2800453 |
| AlexaFluor 700 anti-CD45 | BioLegend | Cat#368514, Clone 2D1; RRID: 2566374 |
| Pacific Blue anti-CD45 | BioLegend | Cat#304022, Clone HI30; RRID: 493655 |
| FITC anti-CD3 | BD Biosciences | Cat#345764, Clone SK7 |
| PE anti-CD3ÜBackspace | BioLegend | Cat#300407, Clone UCHT1; RRID: AB_314061 |
| APC anti-CD8 | BD Biosciences | Cat#345775, Clone SK1; RRID: 2868803 |
| PE-Cy7 anti-CD8 | BioLegend | Cat#344712, Clone SK1; RRID: 2044008 |
| PerCP anti-CD4 | BD Biosciences | Cat#345770, Clone SK3; RRID: 2868798 |
| AlexaFluor 700 anti-CD19 | BD Biosciences | Cat#557921, Clone HIB19; RRID: 396942 |
| BV510 anti-CD19 | BioLegend | Cat#302242, Clone HIB19; RRID: 2568668 |
| PE anti-CD56 | BD Biosciences | Cat#345812, Clone NCAM16.2; RRID: 2629216 |
| BV510 anti-CD14 | BD Biosciences | Cat#563079, Clone MϕP9; RRID: 2737993 |
| BV605 anti-CD45RA | BioLegend | Cat#304134, Clone HI100; RRID: 2563814 |
| BV421 anti-CD62L | BioLegend | Cat#304828, Clone DREG-56; RRID: 2562914 |
| BV510 anti-CD62L | BioLegend | Cat#304844, Clone DREG-56; RRID: 2617003 |
| FITC anti-CD62L | BioLegend | Cat#304804, Clone DREG-56; RRID: 314464 |
| APC-Cy7 anti-CD20 | BioLegend | Cat#302314, Clone 2H7; RRID: 314262 |
| BV421 anti-CD27 | BioLegend | Cat#356418, Clone M-T271; RRID: 2562559 |
| APC anti-CD38 | BioLegend | Cat#356606, Clone HB-7; RRID: 2561902 |
| PerCP anti-CD24 | BioLegend | Cat#311114, Clone ML5; RRID: 2561284 |
| PE anti-IgD | BioLegend | Cat#348204, Clone IA6-2; RRID: 10553900 |
| FITC anti-IgM | BioLegend | Cat#314506, Clone MHM-88; RRID: 493009 |
| PE anti PD-1 | BioLegend | Cat#329906, Clone EH12.2H7; RRID: 940483 |
| AlexaFluor 700 anti-CD4 | BD Biosciences | Cat#557922, Clone RPA-T4; RRID: 396943 |
| PE anti-IFN-γ | BioLegend | Cat#502509, Clone 4S.B3; RRID: 315234 |
| APC anti-TNF-α | BioLegend | Cat#502912, Clone Mab11; RRID: 315264 |
| PE-Cy7 anti-TCRγ/δ | BD Biosciences | Cat#655410, Clone 11F2; RRID: 2870377 |
| BV510 anti-CXCR3 | BioLegend | Cat#353726, Clone G025H7; RRID: 2563642 |
| BV605 anti-CCR2 | BioLegend | Cat#357214, Clone K036C2; RRID: 2563876 |
| BV421 anti-Tim-3 | BioLegend | Cat#345008, Clone F38-2E2; RRID: 11218598 |
| mAb-AP IFNγ T-Track | Lophius | Cat#123600002 |
| Biological Samples | ||
| Blood samples HD (peripheral blood of healthy adult donors from alloCELL registry) | Hannover Medical School | http:/allocell.org |
| Plasma samples HD (peripheral blood of healthy adult donors from alloCELL registry) | Hannover Medical School | http:/allocell.org |
| Blood samples RC (recovered COVID-19 patients) | Hannover Medical School | N/A |
| Plasma samples RC (recovered COVID-19 patients) | Hannover Medical School | N/A |
| Blood samples AC (patients with active COVID-19) | Hannover Medical School, University Hospital Essen | N/A |
| Plasma samples AC (patients with active COVID-19) | Hannover Medical School, University Hospital Essen | N/A |
| AB-Serum | CCPro | Cat#S-41-M |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 7AAD | BD Biosciences | Cat#559925; RRID: 2869266 |
| CellEvent Caspase-3/7 Green Detection Reagent | Invitrogen | Cat#C10423 |
| CaspGLOW Fluorescein Active Caspase-8 Staining Kit | Invitrogen | Cat#88-7005-42; RRID: 2574940 |
| TruCount Tubes | BD Biosciences | Cat#340334 |
| Lysing Solution | BD Biosciences | Cat#349202; RRID: 2868862 |
| Lysing Solution | Beckman Coulter | Cat#A07799 |
| RPMI1640 | LONZA | Cat#BE12-702F |
| Lymphosep | CCPro | Cat#PL-15-L |
| PBS | LONZA | Cat#BE17-512F |
| WFI | Lonza | Cat#BE17-724F |
| TexMACS Medium | Miltenyi Biotec | Cat#170-076-307 |
| AutoMACS Rising Solution | Miltenyi Biotec | Cat#130-091-222 |
| BCIP/NBT | Serva | Cat#15246.01 |
| Streptavidin-Alkaline Phosphatase | Mabtech | Cat#12360002-S |
| PMA | Sigma | Cat#P1585 |
| Ionomycin | Sigma | Cat#I9657 |
| Brefeldin A | BioLegend | Cat#420601 |
| anti-CD3/anti-CD28 Dynabeads | Thermo Fisher Scientific | Cat#111.31D |
| Staphylococcus Enterotoxin B (SEB) | Sigma | Cat#S4881 |
| CMVpp65 peptide pool | Miltenyi Biotec | Cat#130-093-435 |
| SARS-CoV-2 M peptide pool | Miltenyi Biotec | Cat#130-126-703 |
| SARS-CoV-2 N peptide pool | Miltenyi Biotec | Cat#130-126-699 |
| SARS-CoV-2 S peptide pool | Miltenyi Biotec | Cat#130-126-701 |
| SARS-CoV-2 S peptide pool vial 1 and vial 2 | JPT | Cat#PM-WCPV-S |
| SARS-CoV-2 VEMP | JPT | Cat#PM-WCPV-VEMP |
| SARS-CoV-2 NS6 peptide pool | JPT | Cat#PM-WCPV-NS6 |
| SARS-CoV-2 AP3A peptide pool | JPT | Cat#PM-WCPV-AP3A |
| SARS-CoV-2 NS7A peptide pool | JPT | Cat#PM-WCPV-NS7A |
| SARS-CoV-2 NS7B peptide pool | JPT | Cat#PM-WCPV-NS7B |
| SARS-CoV-2 NS8 peptide pool | JPT | Cat#PM-WCPV-NS8 |
| SARS-CoV-2 ORF10 peptide pool | JPT | Cat#PM-WCPV-ORF10 |
| SARS-CoV-2 ORF9B peptide pool | JPT | Cat# PM-WCPV-ORF9B |
| SARS-CoV-2 Y14 peptide pool | JPT | Cat#PM-WCPV-Y14 |
| huCoV 229E | JPT | Cat#PM-229E-S-1 |
| huCoV OC43 | JPT | Cat#PM-OC43-S-1 |
| human Respiratory Syncytial Virus RSV (HRSVA nucleoprotein, NP) | JPT | Cat#PM-HRSVB-NCPN |
| human Influenza virus (matrix protein 1, MP1) | JPT | Cat#PM-INFA-MP1-H1N1 |
| Critical Commercial Assays | ||
| recomLine CMV IgG | Mikrogen | Cat#5572 |
| recomLine SARS-CoV2 IgG | Mikrogen | Cat#7374 |
| anti-SARS-COV-2 S1 spike protein domain/receptor binding domain IgG ELISA | Euroimmun | Cat#EI 2606-9601 G, A |
| anti-SARS-COV-2 NCP IgG ELISA | Euroimmun | Cat#EI 2606-9601-2 G, M |
| IFN-γ Secretion Assay | Miltenyi Biotec | Cat#130-054-201 |
| MS Column | Miltenyi Biotec | Cat#130-042-201 |
| Ready-MTP basic IFNγ T-Track | Lophius | Cat#12100010 |
| FluorSpot assay | Mabtech | Cat#FSP-0110-10 |
| IntraPrep Permeabilizaton Reagent | Beckman Coulter | Cat#A07803 |
| Human Essential Immune Response Panel LegendPlex | BioLegend | Cat#740930 |
| HU Immune Checkpoint Panel LegendPlex | BioLegend | Cat#740866 |
| Granzyme B ELISA | Invitrogen | Cat#BMS2027; RRID: 2575322 |
| TaqMan Gene Expression Master Mix | Applied Biosystems | Cat#4369016 |
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| Deposited Data | ||
| analyzed data | This paper | N/A |
| Oligonucleotides | ||
| GAPDH TaqMan® Gene Expression Assays | Applied Biosystems | Hs_027588991_g1 |
| IFNγ TaqMan® Gene Expression Assays | Applied Biosystems | Hs_00989291_m1 |
| Granzyme B TaqMan® Gene Expression Assays | Applied Biosystems | Hs_01554355_m1 |
| Perforin TaqMan® Gene Expression Assays | Applied Biosystems | Hs_00169473_m1 |
| Software and Algorithms | ||
| BD FACSDiva Software version 8.0.1 | BD Biosciences | N/A |
| FlowJo™ v10.6.2 | FlowJoTM LLC, BD Biosciences | https://www.flowjo.com |
| Prism Version 8.2.0 | GraphPad Software | https://www.graphpad.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be direct to and will be fulfilled by the Lead Contact, Britta Eiz-Vesper (eiz-vesper.britta@mh-hannover.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The published article includes all datasets generated or analyzed during this study. This study did not generate codes.
Experimental model AND subject details
Study population
The study was approved by the Internal Review Board of Hannover Medical School (MHH, approval number 3639_2017, 9001_BO-K, 9255_BO_K_2020), and all patients and donors were recruited from MHH. Following written informed consent, peripheral blood samples from 82 adult healthy donors (HDs) from the alloCELL registry (controls), 204 recovered COVID-19 patients (RCs) and 92 patients with active COVID-19 (ACs) were tested between April and October 2020. PBMCs were isolated from either whole blood samples (RCs and ACs) or from residual blood samples from platelet and plasma apheresis disposables used for routine collection (RCs and controls). SARS-CoV-2-negative samples collected from healthy adult donors in 2020 were used for the analysis. Age and sex of all patients and donors are displayed in Figure 1. Samples from RCs and ACs were collected 29-175 days and 0-49 days from the time of the first positive SARS-CoV-2 reverse transcriptase polymerase chain reaction (RT-PCR) test or start of their symptoms, respectively. Up to five follow-up samples from ACs were obtained within five weeks of first sampling. Follow-up samples from RCs were obtained between 5 and 154 days after first sampling.
Clinical definition of cohort
All recovered and active COVID-19 cases were confirmed by SARS-CoV-2 RT-PCR testing of upper respiratory tract (nose or throat) swabs in accredited laboratories using Real-Time-PCR-Tests SARS-CoV-2 RUO (R-biopharm), Allplex 2019-nCOV Assay (Seegene), RealStar SARS-CoV-2 RT-PCR Kit (Altona Diagnostics), Xpert Xpress SARS-CoV-2 (Cepheid) or Abbott RealTime SARS-CoV-2 assay (Abbott). According to WHO recommendations, severity was graded as “uninfected” (score 0), “ambulatory” (score 1-2), “hospitalized, mild or severe disease” (score 3-7), or “death” (score 8). RCs were additionally scored based on the following symptoms: flu-like symptoms (dizziness, headache, chills, runny nose; 1 point), gastrointestinal complaints (diarrhea, vomiting, excessive; 1 point), fever (3 points), cough and respiratory problems (sore throat, lung pain, shortness of breath; 2 points) neurological symptoms (impaired sense of taste or smell; 3 points), hospitalization (5 points), and hospitalization with ventilation (5 points); the score range was 0-20.
Method details
Serological testing by ELISA and western blot
All patients and donors were pretested for CMV as described previously using commercially available IgG ELISA and IgG western blot kits (recomLine CMV IgG, Mikrogen, Neuried, Germany) (Tischer et al., 2014). SARS-CoV-2 serology was performed by ELISA (anti-SARS-COV-2 S1 spike protein domain IgG and anti-SARS-COV-2 NCP, referred to as N; IgG; Euroimmun, Lübeck, Germany) according to the manufacturer’s instructions. Antibody amounts are expressed as IgG ratio (optical density divided by calibrator); values below 0.8 were defined as negative, those between 0.8 and 1.1 were classified as intermediate, and values above 1.1 were defined as positive. Western blot tests (recomLine SARS-CoV2 IgG, Mikrogen, Neuried, Germany) were performed in selected individuals from all three cohorts to detect IgG directed against N protein of huCoV strains and against N, RBD and S1 proteins of SARS-CoV-2.
Cellular immune profiling by flow cytometry
All flow cytometric analyses were performed using the FACSCanto 10c cytometer (BD Biosciences, Heidelberg, Germany) and BD FACSDiva Software version 8.0.1. Cellular immune status (Panel 1), T cell activation and exhaustion (Panel 2) and B cell phenotype (Panel 3) were determined using specific markers for innate leukocytes, T cells and B cells. Briefly, cell frequencies and total cell numbers in whole blood samples were analyzed on a single-cell platform using TruCount™ tubes (BD Biosciences). Cells were stained for 30 min at room temperature using anti-CD45 allophycocyanin-H7 (APC-H7) or AlexaFluor 700 (AF-700), anti-CD3 fluorescein isothiocyanate (FITC), anti-CD8 allophycocyanin (APC), anti-CD4 peridinin chlorophyll protein complex (PerCP), anti-CD19 AF-700 or Brilliant Violet (BV510), anti-CD56 phycoerythrin (PE), anti-CD14 Brilliant Violet (BV510), anti-CD45RA BV605 and anti-CD62L BV421 or BV510, anti-CD20 APC-cyanine 7 (APC-Cy7), anti-CD27 BV421, anti-CD38 APC, anti-CD24 PerCP, anti-IgD PE, anti-IgM FITC and anti-PD-1 PE monoclonal antibodies (BioLegend and BD Biosciences) either before (Panels 1 and 2) or after (Panel 3) lysis of erythrocytes using 1x Lysing Solution according to the manufacturer’s instructions (Panels 1 and 2: BD Biosciences, Panel 3: Beckman Coulter, Brea, CA, USA).
Detection of antiviral T cells by IFN-γ ELISPOT
SARS-CoV-2-specific T lymphocytes were detected by IFN-γ ELISPOT assay as previously described (Bieling et al., 2017). Briefly, PBMCs were isolated from blood samples by discontinuous density gradient centrifugation, resuspended in culture medium (CM) consisting of RPMI-1640 (Lonza, Vervies, Belgium) supplemented with 10% human AB serum (C.C.pro, Oberdorla, Germany) at a concentration of 1x107 cells/mL, seeded in 24-well plates and rested overnight. Rested PBMCs were co-cultured in anti-IFN-γ pre-coated ELISPOT plates (Lophius Biosciences, Regensburg, Germany) for 16-18 h at a density of 2.5x105 cells/well with specific antigens of interest. Overlapping peptide pools against SARS-CoV-2 M, N and S proteins (Miltenyi Biotec, Bergisch Gladbach, Germany) were used to stimulate each sample; they were extended to the following SARS-CoV-2 proteins if PBMC counts allowed: S1 and S2, envelope small membrane protein VEMP, accessory protein 3A (AP3A), nonstructural proteins NS6, NS7A, NS7B and NS8, open reading frame proteins ORF10 and ORF9B, and Y14 (JPT, Berlin, Germany). The S peptide pool covered mainly the C-terminal domain of SARS-CoV-2 S protein supplemented with selected N-terminal epitopes, whereas S1 and S2 together covered the whole protein sequence. Pools were used at a final concentration of 1 μg of each peptide/mL peptide pool. Antigens of the S1 and S2 epitopes of huCoV strains 229E and OC43, antigens derived from human RSV (nucleoprotein, NP), IAV (matrix protein 1, MP1) (all supplied by JPT), and CMV phosphoprotein 65 (pp65; Miltenyi Biotec) were also analyzed.
Cells stimulated with staphylococcal enterotoxin B (1 μg/mL, SEB, Merck, Taufkirchen, Germany) served as positive controls, and PBMCs incubated in media alone as negative controls (NC). IFN-γ secretion was detected using streptavidin-alkaline phosphatase (Mabtech Stockholm, Sweden) and revealed by 5-13 bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium (BCIP/NBT Liquid Substrate, Merck, Darmstadt, Germany). Spots were counted using AID ELISPOT 8.0 on an AID iSpot spectrum reader system (both from AID, Strassberg, Germany). Means of duplicate well readings were calculated and expressed as the number of spots per well (spw). The positive test threshold was set at ≥ 3 spw or > 2xNC + 1. Donors were divided into four groups based on spw values as follows: high responders (HR): ≥ 50 spw or 47 spw + 2xNC), intermediate responders (IR): ≥ 10 spw or 7 spw + 2xNC), low responders (LR): ≥ 3 spw or 2xNC), and non-responders (NR): < 3 spw or 2xNC. PBMCs were stained with anti-CD45 APC-H7, anti-CD3 FITC, anti-CD4 PerCP and anti-CD8 APC (BioLegend and BD Biosciences) for calculation of spots/10,000 CD3+ T cells.
Detection of IFN-γ and granzyme B by FluoroSpot
SARS-CoV-2-specific T cell function was determined by IFN-γ and Gzmb effector molecule detection by FluoroSpot assay (Mabtech). Briefly, PBMCs were seeded into pre-coated FluoroSpot plates in CM at a density of 2.5x105 cells/well and stimulated with indicated peptide pools for a period of 45 h. Negative and positive controls were carried out as described for IFN-γ ELISPOT assay. Following detection according to the manufacturer’s instruction, spots were identified using the filter for FITC to identify IFN-γ-producing cells and using the filter for Cy3 to identify Gzmb-producing cells. Results were expressed as the number of spots per well (spw).
IFN-γ CSA for enrichment of antiviral T cells
The IFN-γ CSA (Miltenyi Biotec) was performed as previously described (Tischer et al., 2014). After overnight resting in TexMACS™ Medium (Miltenyi Biotec), 1x107 isolated PBMCs were stimulated with peptide pools of SARS-CoV-2 M, N and/or S protein (Miltenyi Biotec) alone or in combination at a final concentration of 1 μg of each peptide/mL peptide pool. Unstimulated PBMCs served as negative controls. Activated IFN-γ-secreting T cells were specifically captured during the magnetic cell sorting enrichment process using anti-IFN-γ PE antibodies and paramagnetic anti-PE microbeads. Aliquots of the respective cell fractions collected before and after enrichment were used for analysis of IFN-γ+ T cell subsets by multicolor flow cytometry. The distribution of viable and dead cells in these fractions was analyzed by 7-amino-actinomycin D (7-AAD) staining (BD Biosciences). The percentage of viable IFN-γ+ cells was determined by staining the cells with anti-CD45 APC-H7, anti-CD3 FITC, anti-CD8 APC and anti-CD4 AF-700 mAbs (all from BD Biosciences). At least 10,000 events were acquired in the viable CD45+ leukocyte gate in each test (system: FACSCanto10c, BD Biosciences). CD3+ IFN-γ+ CD8+ IFN-γ+ and CD4+ IFN-γ+ T cell populations were gated based on the scatter properties of viable 7AAD- CD45+ CD3+ T cells.
Intracellular cytokine staining
After overnight resting, 1x106 isolated PBMCs were stimulated with peptide pools derived from SARS-CoV-2 M, N, S, S1 and S2 proteins (Miltenyi Biotec and JPT) at a final concentration of 1 μg of each peptide per ml peptide pool. Unstimulated PBMCs served as negative controls, and cells stimulated with phorbol 12-myristate 13-acetate (PMA; 10 ng/mL) and ionomycin (500 ng/mL, Sigma Aldrich) served as positive controls. Antigens from epitopes of huCoV strains OC43 and 229E as well as RSV_NP, IAV_MP1 (all JPT) and CMV_pp65 (Miltenyi Biotec) were also analyzed. After 1 h of incubation, 5 μg/mL Brefeldin A was added to each well (BioLegend). Following a total stimulation time of 5 h, cells were harvested and extracellularly stained with anti-CD45 Pacific Blue, anti-CD4 PerCP, anti-CD8 PE-Cy7, anti-CD45RA BV605 and anti-CD62L FITC. Subsequently, cells were fixed and permeabilized using the IntraPrep Kit according to the manufacturer’s instructions (Beckman Coulter). Cells were intracellularly stained with anti-IFN-γ PE and anti-TNF-α APC. Samples were acquired on a FACSCanto 10c system (BD Biosciences), and at least 50,000 events in the CD45+ lymphocyte gate were analyzed in each test.
Cytokine profiling in cell culture supernatants
After overnight resting, 1x106 PBMCs isolated from RCs were stimulated with peptide pools derived from SARS-CoV-2 M, N, S, S1 and S2 proteins (Miltenyi Biotec. and JPT) at a final concentration of 1 μg/mL. Unstimulated PBMCs served as negative controls, and cells stimulated with anti-CD3/anti-CD28 Dynabeads (Thermo Fisher Scientific) served as positive controls. S1 and S2 antigens of huCoV strains 229E and OC43 as well as RSV_NP, IAV_MP1 and CMV_pp65 antigens were also analyzed. After 20 h of stimulation, cell culture supernatants were collected and stored at -20°C until further processing. Cytokine and chemokine secretion was determined using the Human Essential Immune Response Panel (BioLegend).
Cytokine and chemokine profiling by LEGENDPlex
Plasma samples and antigen-stimulated cell culture supernatants were collected from all three cohorts as described above and analyzed by LEGENDPlex™ using the Human Essential Immune Response Panel and the HU Immune Checkpoint Panel 1 (BioLegend). Gzmb concentrations in cell culture supernatants were determined by ELISA according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA).
Chemokine receptor expression on T cells
PBMCs from all three cohorts were stained with anti-CD45 APC-H7 and anti-CD3 FITC (BD Biosciences) at room temperature and washed once, followed by staining with anti-CCR2 BV605 and anti-CXCR3 BV510 (both Biolegend) at 37°C for 30 min. After final washing, samples were acquired on a FACSCanto 10c system (BD Biosciences), and at least 50,000 events in the CD45+ lymphocyte gate were analyzed in each test.
Caspase-3 and/or -7 and -8 activity in T cells
PBMCs from each cohort were stained with anti-CD3 PE (BioLegend) at room temperature and washed once, followed by staining with CellEvent™ Caspase-3/7 Green Detection Reagent or eBioscience CaspGLOW™ Fluorescein Active Caspase-8 Kit (both Invitrogen) according to the manufacturer’s instructions. Finally, cells were washed and dead cells stained with 7-AAD. Subsequently, samples were acquired on a FACSCanto 10c system (BD Biosciences), and at least 50,000 events in the CD45+ lymphocyte gate were analyzed in each test.
RT-PCR analysis of SARS-CoV-2-specific T cells
Messenger RNA amounts of IFN-γ (IFNG), perforin (PRF1) and granzyme B (GZMB) were analyzed as previously described (Bunse et al., 2015). Briefly, total cellular RNA (RNeasy Mini Kit; QIAGEN, Hilden, Germany) was isolated from CD4+ IFN-γ+ and CD8+ IFN-γ+ T cells and their IFN-γ-negative counterparts after stimulation with overlapping peptide pools covering the M, N and S protein of SARS-CoV-2 or CMV_pp65, using 1 μg of each peptide per ml peptide pool. Subsequent cytokine detection (Cytokine Detection Kit, Miltenyi Biotec) and FACS sorting was performed using anti-CD3 FITC, anti-CD8 APC and anti-CD4 APC-Cy7 (BD Biosciences and BioLegend). The cDNA was amplified using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Darmstadt, Germany). Messenger RNA amounts were quantified using inventoried mixes and TaqMan Gene Expression Master Mix (both from Applied Biosystems). Constitutively expressed glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the reference gene. All tests were performed in duplicate, and all samples from each donor were measured on a single plate to reduce inter-assay variability.
Quantification and statistical analysis
Data were analyzed using Microsoft Excel 2010 (Microsoft Corporation, Redmond), FlowJo™ v10 (FlowJo™ LLC, BD Biosciences) and BD FACSDiva v8.0.1 (BD Biosciences). Graphs for statistical analysis were generated using Prism Version 8.2.0 (GraphPad Software, San Diego, California, USA). Statistical analysis was performed using a linear regression model and the Kruskal-Wallis or Friedman test followed by multiple comparison correction. Significant differences were calculated and expressed as p values (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Statistical details of all experiments are described in each figure legend.
Acknowledgments
This research was funded in part by grants from the state of Lower Saxony (14 - 76103-184 CORONA-12/20), the Federal Ministry of Health (ZMVI1-2520COR804), and the German Research Foundation (DFG; Research Unit 2830 and SFB 900/B11 project ID 158989968). We are grateful to all study participants for providing samples and to the MVZ Laboratory Limbach Hannover GbR for RT-PCR analysis. Furthermore, we would like to thank Dörthe Rokitta, Nicole Neumann, Sarina Lukis, Elvira Schulde, Milena Stietzel, Juliane Ebersold, Sophie Meyer, Bach Uy Vu, Leona Schmaltz, and Marion Hitzigrath for technical support, as well as the team at the Cell Sorting Core Facility of Hannover Medical School, which was supported in part by the Braukmann-Wittenberg Heart Foundation and the DFG.
Author contributions
Conceptualization: A.B., B.M.K., G.M.N.B., M.C., O.W., R.B., and B.E.V. Methodology: A.B., S.T., D.G., A.V., A.C.D., M.V.S., and B.E.V. Validation: A.B., S.T., A.V., A.C.D., A.R.M.K., J.M., M.V.S., and G.M.N.B. Formal analysis: A.B., S.T., D.G., A.V., A.C.D., N.G., and M.V.S. Investigation: A.B., S.T., A.V., A.C.D., D.G., N.G., C.L., M.B., I.P., M.V.S., and S.D. Resources: U.K., M.Y., M.M.H., Y.L., T.W., J.M., G.M.N.B., M.C., S.D., J.J.S., M.B., C.L., O.W., R.B., and B.E.V. Writing the original draft: A.B., S.T., and B.E.V. Visualization: A.B. and S.T. Supervision: B.E.V. Project administration: B.E.V. Grant acquisition: R.B. and B.E.V.
Declaration of interests
The authors declare that they have no competing interests. The funders played no role in designing the study, in collecting, analyzing, or interpreting the data, in writing the manuscript, or in the decision to publish the results.
Published: February 9, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2021.01.008.
Supplemental information
References
- Ahn E., Araki K., Hashimoto M., Li W., Riley J.L., Cheung J., Sharpe A.H., Freeman G.J., Irving B.A., Ahmed R. Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. USA. 2018;115:4749–4754. doi: 10.1073/pnas.1718217115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlFehaidi A., Ahmed S.A., Hamed E. A case of SARS-CoV-2 re-infection. J. Infect. 2020;25 doi: 10.1016/j.jinf.2020.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020;92:495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens G.M.N., Cossmann A., Stankov M.V., Schulte B., Streeck H., Förster R., Bosnjak B., Willenzon S., Boeck A.L., Thu Tran A., et al. Strategic Anti-SARS-CoV-2 Serology Testing in a Low Prevalence Setting: The COVID-19 Contact (CoCo) Study in Healthcare Professionals. Infect. Dis. Ther. 2020;9:837–849. doi: 10.1007/s40121-020-00334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling M., Tischer S., Kalinke U., Blasczyk R., Buus S., Maecker-Kolhoff B., Eiz-Vesper B. Personalized adoptive immunotherapy for patients with EBV-associated tumors and complications: Evaluation of novel naturally processed and presented EBV-derived T-cell epitopes. Oncotarget. 2017;9:4737–4757. doi: 10.18632/oncotarget.23531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Bunse C.E., Fortmeier V., Tischer S., Zilian E., Figueiredo C., Witte T., Blasczyk R., Immenschuh S., Eiz-Vesper B. Modulation of heme oxygenase-1 by metalloporphyrins increases anti-viral T cell responses. Clin. Exp. Immunol. 2015;179:265–276. doi: 10.1111/cei.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., et al. Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated With Drastically Elevated Interleukin 6 Level in Critically Ill Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornberg M., Kenney L.L., Chen A.T., Waggoner S.N., Kim S.K., Dienes H.P., Welsh R.M., Selin L.K. Clonal exhaustion as a mechanism to protect against severe immunopathology and death from an overwhelming CD8 T cell response. Front. Immunol. 2013;4:475. doi: 10.3389/fimmu.2013.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras C., Pascual-Miguel B., Mestre-Durán C., Navarro-Zapata A., Clares-Villa L., Martín-Cortázar C., De Paz R., Marcos A., Vicario J.L., Balas A., et al. SARS-CoV-2 specific memory T lymphocytes from COVID-19 convalescent donors: identification, biobanking and large-scale production for Adoptive Cell Therapy. bioRxiv. 2020 doi: 10.3389/fcell.2021.620730. 2020.2010.2023.352294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I., Pranata R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): systematic review and meta-analysis. J. Intensive Care. 2020;8:36. doi: 10.1186/s40560-020-00453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A. What reinfections mean for COVID-19. Lancet Infect. Dis. 2021;21:3–5. doi: 10.1016/S1473-3099(20)30783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng E., Kamruzzaman M., Garcia-Beltran W.F., et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Simon V. Serology assays to manage COVID-19. Science. 2020;368:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Leung W., Soh T.G., Linn Y.C., Low J.G.-H., Loh J., Chan M., Chng W.J., Koh L.P., Poon M.L.-M., Ng K.P., et al. SUCCESSFUL MANUFACTURING OF CLINICAL-GRADE SARS-CoV-2 SPECIFIC T CELLS FOR ADOPTIVE CELL THERAPY. medRxiv. 2020 doi: 10.1101/2020.04.24.20077487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang Y., Guan Z., Li H., Ye M., Chen X., Shen J., Zhou Y., Shi Z.L., Zhou P., Peng K. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 2020;5:235. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369 doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., HLH Across Speciality Collaboration, UK COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzemaekers M., Vanheule V., Janssens R., Struyf S., Proost P. Overview of the Mechanisms that May Contribute to the Non-Redundant Activities of Interferon-Inducible CXC Chemokine Receptor 3 Ligands. Front. Immunol. 2018;8:1970. doi: 10.3389/fimmu.2017.01970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X., et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odak I., Barros-Martins J., Bošnjak B., Stahl K., David S., Wiesner O., Busch M., Hoeper M.M., Pink I., Welte T., et al. Reappearance of effector T cells is associated with recovery from COVID-19. EBioMedicine. 2020;57:102885. doi: 10.1016/j.ebiom.2020.102885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Toyama M., Baba M. The chemokine receptor antagonist cenicriviroc inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;182:104902. doi: 10.1016/j.antiviral.2020.104902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E.Z., Chan Y.F.Z., Leong W.Y., Lee N.M.Y., Kalimuddin S., Haja Mohideen S.M., Chan K.S., Tan A.T., Bertoletti A., Ooi E.E., Low J.G.H. A Dynamic Immune Response Shapes COVID-19 Progression. Cell Host Microbe. 2020;27:879–882.e2. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Broad and strong memory CD4 + and CD8 + T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients. bioRxiv. 2020;8 doi: 10.1101/2020.06.05.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Oxford Immunology Network Covid-19 Response T cell Consortium. ISARIC4C Investigators Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priesner C., Esser R., Tischer S., Marburger M., Aleksandrova K., Maecker-Kolhoff B., Heuft H.G., Goudeva L., Blasczyk R., Arseniev L., et al. Comparative Analysis of Clinical-Scale IFN-γ-Positive T-Cell Enrichment Using Partially and Fully Integrated Platforms. Front. Immunol. 2016;7:393. doi: 10.3389/fimmu.2016.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E.M., Nightingale E.S., Jafari Y., Waterlow N.R., Clifford S., B Pearson C.A., Group C.W., Jombart T., Procter S.R., Knight G.M. COVID-19 length of hospital stay: a systematic review and data synthesis. BMC Med. 2020;18 doi: 10.1186/s12916-020-01726-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf-Oliveira R.C., Gonçalves K.T., Martignago M.L., Mengatto V., Gaspar P.C., de Moraes A.C., da Silva R.M., Bazzo M.L., Santos-Silva M.C. Determination of lymphocyte subset reference ranges in peripheral blood of healthy adults by a dual-platform flow cytometry method. Immunol. Lett. 2015;163:96–101. doi: 10.1016/j.imlet.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Sariol A., Perlman S. Lessons for COVID-19 Immunity from Other Coronavirus Infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schögler A., Stokes A.B., Casaulta C., Regamey N., Edwards M.R., Johnston S.L., Jung A., Moeller A., Geiser T., Alves M.P. Interferon response of the cystic fibrosis bronchial epithelium to major and minor group rhinovirus infection. J. Cyst. Fibros. 2016;15:332–339. doi: 10.1016/j.jcf.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T. Critical Review of the Literature on Chest CT and Coronavirus Disease (COVID-19): Data Adjustment. AJR Am. J. Roentgenol. 2020;215:W27. doi: 10.2214/AJR.20.23435. [DOI] [PubMed] [Google Scholar]
- Selin L.K., Brehm M.A., Naumov Y.N., Cornberg M., Kim S.K., Clute S.C., Welsh R.M. Memory of mice and men: CD8+ T-cell cross-reactivity and heterologous immunity. Immunol. Rev. 2006;211:164–181. doi: 10.1111/j.0105-2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O’Byrne A., Kouphou N., Galao R.P., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swadling L., Maini M.K. T cells in COVID-19 - united in diversity. Nat. Immunol. 2020;21:1307–1308. doi: 10.1038/s41590-020-0798-y. [DOI] [PubMed] [Google Scholar]
- Tischer S., Dieks D., Sukdolak C., Bunse C., Figueiredo C., Immenschuh S., Borchers S., Stripecke R., Maecker-Kolhoff B., Blasczyk R., Eiz-Vesper B. Evaluation of suitable target antigens and immunoassays for high-accuracy immune monitoring of cytomegalovirus and Epstein-Barr virus-specific T cells as targets of interest in immunotherapeutic approaches. J. Immunol. Methods. 2014;408:101–113. doi: 10.1016/j.jim.2014.05.011. [DOI] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Sinai Immunology Review Project Immunology of COVID-19: Current State of the Science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan T.P., Meyer C.G. Mild versus severe COVID-19: Laboratory markers. Int. J. Infect. Dis. 2020;95:304–307. doi: 10.1016/j.ijid.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H., Cooke G., Atchison C., Whitaker M., Elliott J., Moshe M., Brown J.C., Flower B., Daunt A., Ainslie K., et al. Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. medRxiv. 2020 doi: 10.1016/j.lanepe.2021.100098. 2020.2010.2026.20219725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. COVID-19 Therapeutic Trial Synopsis. [Google Scholar]
- Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., Gu C., Zhang R., Tu C., Xie Y., et al. Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898 e5. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasinska I.M., Sakhnevych S.S., Pavlova L., Teo Hansen Selnø A., Teuscher Abeleira A.M., Benlaouer O., Gonçalves Silva I., Mosimann M., Varani L., Bardelli M., et al. The Tim-3-Galectin-9 Pathway and Its Regulatory Mechanisms in Human Breast Cancer. Front. Immunol. 2019;10:1594. doi: 10.3389/fimmu.2019.01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa N., Yoshikawa T., Hill T., Huang C., Watts D.M., Makino S., Milligan G., Chan T., Peters C.J., Tseng C.T. Differential virological and immunological outcome of severe acute respiratory syndrome coronavirus infection in susceptible and resistant transgenic mice expressing human angiotensin-converting enzyme 2. J. Virol. 2009;83:5451–5465. doi: 10.1128/JVI.02272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang X., Li X., Xi D., Mao R., Wu X., Cheng S., Sun X., Yi C., Ling Z., et al. Potential contribution of increased soluble IL-2R to lymphopenia in COVID-19 patients. Cell. Mol. Immunol. 2020;17:878–880. doi: 10.1038/s41423-020-0484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Van Rooijen N., Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Williams E.P., Malireddi R.K.S., Karki R., Banoth B., Burton A., Webby R., Channappanavar R., Jonsson C.B., Kanneganti T.D. Impaired NLRP3 inflammasome activation/pyroptosis leads to robust inflammatory cell death via caspase-8/RIPK3 during coronavirus infection. J. Biol. Chem. 2020;295:14040–14052. doi: 10.1074/jbc.RA120.015036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W., Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. This study did not generate codes.